Abstract
The specific functions of the prodomains of TGFβ superfamily members are largely unknown. Interactions are known between prodomains of TGFβ-1–3 and latent TGFβ-binding proteins and between prodomains of BMP-2, -4, -7, and -10 and GDF-5 and fibrillins, raising the possibility that latent TGFβ-binding proteins and fibrillins may mediate interactions with all other prodomains of this superfamily. This possibility is tested in this study. Results show that the prodomain of BMP-5 interacts with the N-terminal regions of fibrillin-1 and -2 in a site similar to the binding sites for other bone morphogenetic proteins. However, in contrast, the prodomain of GDF-8 (myostatin) interacts with the glycosaminoglycan side chains of perlecan. The binding site for the GDF-8 prodomain is likely the heparan sulfate chain present on perlecan domain V. These results support and extend the emerging concept that TGFβ superfamily prodomains target their growth factor dimers to extracellular matrix macromolecules. In addition, biochemical studies of prodomain·growth factor complexes were performed to identify inactive complexes. For some members of the superfamily, the prodomain is noncovalently associated with its growth factor dimer in an inactive complex; for others, the prodomain·growth factor complex is active, even though the prodomain is noncovalently associated with its growth factor dimer. Results show that the BMP-10 prodomain, in contrast to BMP-4, -5, and -7 prodomains, can inhibit the bioactivity of the BMP-10 growth factor and suggest that the BMP-10 complex is like TGFβ and GDF-8 complexes, which can be activated by cleavage of the associated prodomain.
Keywords: Bone Morphogenetic Protein (BMP), Fibrillin, Glycosaminoglycan, Heparan Sulfate, Transforming Growth Factor beta (TGFbeta), Myostatin, Perlecan
Introduction
Members of the transforming growth factor β (TGFβ) superfamily of growth factors are translated as precursor molecules that are proteolytically processed by the furin family of proprotein convertases before their secretion. This proteolytic event yields two products, an N-terminal prodomain (pd)3 and a C-terminal mature growth factor dimer (gfd), which is disulfide cross-linked. TGFβ and other growth factors of this family are secreted as stable pd·gfd complexes consisting of a gfd noncovalently associated with two pds (1–5). Although much effort has been spent on investigating how TGFβ superfamily gfds act as powerful signaling molecules that trigger important cellular and developmental events through receptor interactions followed by intracellular signal transduction (6), little is known about the function of pds. Most insight in this regard comes from biochemical studies of TGFβ-1.
The TGFβ-1 pd performs at least two functions as follows. It confers latency to its gfd, and it interacts with extracellular matrix (ECM) molecules, the latent TGFβ-binding proteins (LTBPs). The TGFβ-1 pd, also called latency-associated peptide (LAP), remains associated with TGFβ-1 gfd, forming the small latent complex (SLC). In this complexed form, the TGFβ-1 gfd is rendered inactive, because LAP blocks the interaction of TGFβ with its receptors or because LAP induces a conformational change in TGFβ such that it cannot interact with its receptors (7). LAP also targets the SLC to the ECM through interactions with LTBPs (8).
Recently, we demonstrated that the pds of bone morphogenetic proteins (BMP) -4, -7, and -10 and growth and differentiation factor-5 form stable complexes with their gfds and mediate high affinity interactions with fibrillin-1 and -2 proteins (9). Because fibrillin microfibrils are important architectural scaffolds in the connective tissue space, these findings extended the concept that TGFβ superfamily pds function as facilitators of interactions with the ECM. In addition, these findings expanded the repertoire of interactions between TGFβ superfamily members and the fibrillin family of proteins, which includes the LTBPs.
Like LAP and TGFβ-1, the noncovalent interaction of the pd/gfd of GDF-8 (also called myostatin) results in an inactive complex (10). However, our study of the BMP-7 pd·gfd complex demonstrated that, unlike the TGFβ-1 SLC and GDF-8 complex, BMP-7 pd can be competitively displaced by type II BMP receptors (11). Our data showed that TGFβ superfamily pds differ in their ability to render the growth factor complex inactive and that a variety of modes of activation is possible. Furthermore, the implications of these data were that the required mechanisms of activation depend on the specific interactions of the pd with its gfd and possibly also with its ECM binding partners. Therefore, it is now important to define the specific functions of each of the TGFβ superfamily pds, rather than to assume that all members of this family share similar functions.
In this study, we identify two new interactions between TGFβ superfamily pds and ECM molecules. We found that the BMP-5 pd binds to fibrillin-1 and -2. In addition, we investigated whether other TGFβ superfamily pds might interact with ECM molecules other than the fibrillins and LTBPs. Using blot overlay assays to screen cell culture medium proteins for interactions with pds, we identified a novel interaction between the pd of GDF-8 (myostatin) and the ECM molecule, perlecan. We also tested additional pds for their ability to complex with their gfds and to form inactive complexes. All together, our findings now clearly show that TGFβ superfamily pds use a variety of mechanisms to target growth factors to the ECM and to control growth factor signaling and activation. Therefore, the specific functions of the remaining pds should be defined experimentally.
EXPERIMENTAL PROCEDURES
Recombinant Proteins
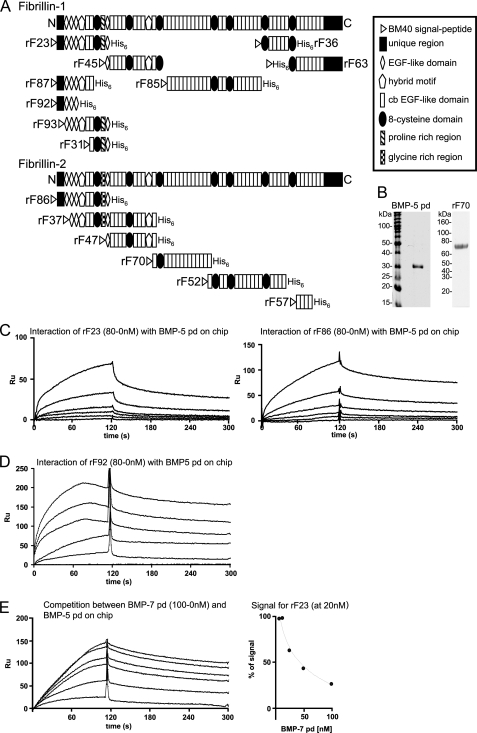
Construction, transfection, purification, and characterization of recombinant human fibrillin-1 and -2 polypeptides rF23 (12), rF31 and rF37 (13), rF45 (14), rF52 (15), rF36, rF63, rF85, rF86, rF87, F92, and rF93 and pds of BMP-4, -7, and -10 (9) were as described. Full-length mouse perlecan was extracted and purified from mouse Engelbreth-Holm-Swarm sarcoma (16), and recombinant mouse perlecan domains (I, II, III-1, III-2, III-3, IV-1, IV-2, and V with and without GAG side chains) were produced as described previously (17–21). Human gfds BMP-4, -5, -7, and -10 and mouse GDF-8 gfd and pd containing 0.1% BSA as stabilizing protein and human BMP-1 reconstituted in 25 mm HEPES, 0.01% Brij-35 at pH 7.5 were all purchased from R&D Systems (Minneapolis, MN). The expression vector for the fibrillin-2 polypeptide rF70 (mRNA coding region, 2862–4696 coding for cbEGF10-cbEGF22) was constructed by generating an NheI- and XhoI-flanked PCR product using a human FBN2 cDNA clone (22) as template and the following primers: rF70-S (5′-cgtagctagcagatgttaatgatggtgaggtgttcc-3′) and rF70-AS (5′-ctagctcgagtcagtgatggtgatggtgatgaacacaacccacaccagttgg-3′). rF70-AS also introduced the coding region for a His6 tag and a stop codon at the 3′-end. The NheI-XhoI-digested PCR fragment was ligated with an NheI-XhoI-restricted pCEP-SP vector. The rF70 polypeptide was harvested and purified from the media of stably transfected 293/EBNA cells, followed by molecular sieve chromatography, as described previously (12). A Coomassie Blue-stained quality control gel of rF70 is shown in Fig. 1B. Domain structures of fibrillin polypeptides used in this study are shown in Fig. 1A; perlecan domains are shown in Fig. 7A.
FIGURE 1.
BMP-5 pd interacts with N-terminal polypeptides of fibrillin-1 and -2. A, schematic representation of the recombinant fibrillin polypeptides used in this study. B, Coomassie Blue-stained gel of the bacterially expressed, purified, and alkylated BMP-5 pd, after SDS-PAGE in a nonreducing 12.5% gel, demonstrates purity of the sample. In addition, a Coomassie Blue-stained gel of the newly described fibrillin-2 polypeptide rF70 is shown. C, selected sensorgrams from SPR experiments with immobilized BMP-5 pd and soluble ligands rF23 or rF86 show interactions. The fibrillin polypeptides where diluted from 80 to 0 nm in HBS-EP buffer. D, SPR sensorgram with immobilized BMP-5 pd and titrated concentrations of rF92. E, BMP-5 and BMP-7 pds compete for the same binding site in fibrillin-1. Left, sensorgram shows concentration-dependent decrease of the BMP-5 pd-rF23 interaction signal, when BMP-7 pd is preincubated with rF23. BMP-5 pd was immobilized, and the fibrillin-1 N-terminal polypeptide rF23 was injected at a constant concentration of 20 nm in the presence of increasing concentrations (0–100 nm) of BMP-7 pd. Right, inhibition curve shows competition of the BMP-7 pd for the BMP-5 pd-binding site in fibrillin-1. The signal in RU obtained for rF23 at 20 nm without competitor was set as 100%. The decrease of the 100% rF23 signal was graphed against the inhibitor concentration to determine the inhibition constant (I50) for the competition reaction. To account for variations of the rF23 signal because of the buffer change caused by the addition of different amounts of competitor, a buffer matched control sensorgram without competitor was generated for each BMP-7 concentration. This control signal served as the 100% reference signal for each competitor concentration.
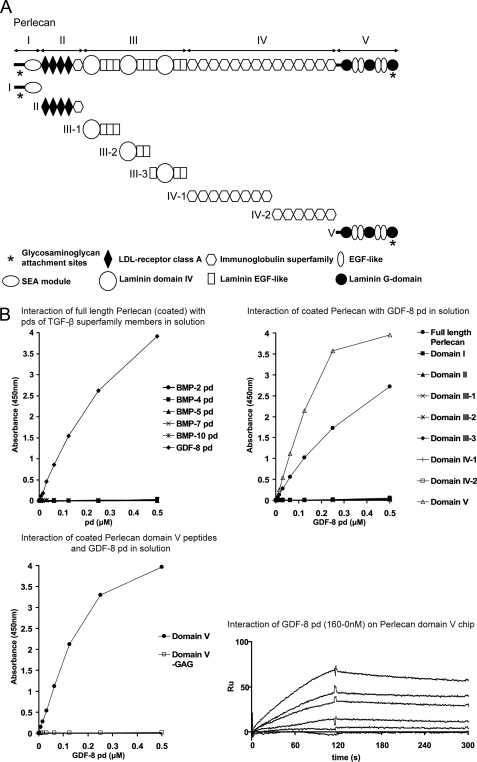
FIGURE 7.
GDF-8 pd binds specifically to the GAG chains of domain V of perlecan. A, schematic diagram of the perlecan recombinant polypeptides used in this study. B, upper left graph, ELISA interaction assay with full-length perlecan (purified from mouse Engelbreth-Holm-Swarm sarcoma) coated on the wells showed specific interaction only with the GDF-8 pd. Pds of TGF-β superfamily members were titrated at different concentrations (0.5–0 μm) in solution, and anti-His6 antibody was used for detection of bound pds. Upper right graph, ELISA interaction study with coated recombinant polypeptides spanning full-length perlecan revealed that GDF-8 pd binds exclusively to domain V of perlecan (open triangles). Lower left graph, ELISA binding assay with coated domain V and nonmodified domain V (without GAG chains) confirmed that GDF-8 pd binds specifically to the GAG side chain and not to the protein core of domain V. Lower right sensorgrams, SPR with immobilized perlecan domain V and GDF-8 pd flowing in solution at different concentrations allowed the determination of a KD of 11 nm.
To obtain the cDNA for the human BMP-5 pd (NM_021073), a total RNA preparation of MG63 cells was generated by using TRIzolTM reagent (Invitrogen) and subsequently reverse-transcribed using the Bio-Rad iScriptTM cDNA synthesis kit (Bio-Rad). The coding region for the BMP-5 pd was amplified from this cDNA source by PCR using the PlatinumTM Pfx DNA polymerase system (Invitrogen) and the following primers: forward, 5′-ggaattccatatggacaatcatgttcactccagt-3′, and reverse, 5′-catgggatcctcagtgatggtgatggtgatgtctcacggatcgaagaagtac-3′. The 5′-primer introduced an NdeI restriction site, whereas a BamHI site and six histidine residues in tandem followed by a termination signal were added to the downstream primer. The product was cloned into an NdeI/BamHI-digested pET11a vector so that the final construct contained the entire pd coding sequence starting from the predicted endogenous signal peptide cleavage site, including the predicted furin cleavage site followed by a C-terminal His6 tag and a stop codon. The vector construct was transformed into competent cells of Escherichia coli DH5α, and the insert structure was verified by restriction analysis and DNA sequencing. The BMP-5 pd was overexpressed in E. coli BL21 (DE3) cells and purified using chelating chromatography under the same conditions as described previously for the BMP-7 pd (23). The BMP-5 pd showed a tendency to form dimers because of a free cysteine and was therefore alkylated with 4-vinylpyridine after affinity chromatography, as described before (Fig. 1B) (24).
Antibodies
The following antibodies were used: polyclonal antibody (pAb) 9543 (anti-fibrillin-1) (25), pAb 0868 (anti-fibrillin-2) (26), polyclonal anti-perlecan domain II (18), monoclonal anti-BMP-7 pd antibody (mab2) (3), monoclonal anti-BMP-5 gfd (mab715), and anti His6-tag (both from R&D Systems).
Other Reagents
Other reagents included the ECL® chemiluminescence kit (Pierce), E. coli BL21 (DE3) and the pET11a vector (Stratagene, La Jolla, CA), and DH5-α cells (Invitrogen). The following established cell cultures were obtained from American Type Culture Collection (ATCC, Manassas, VA): A204 (human rhabdomyosarcoma, HTB-82); SW1353 (human chondrosarcoma, HTB-94); MG63 (human osteosarcoma, CRL-1427); WISH (human transformed amniotic epithelial cells, CCL-25); and mouse C2C12 myoblasts (CRL-1772). Human embryonic kidney cells (293/EBNA) were purchased from Invitrogen. Neonatal skin fibroblasts were derived from explant cultures of human foreskin obtained at circumcision.
Velocity Sedimentation
For reference analyses, 10 μg of BMP-5 pd and 5 μg of BMP-5 gfd were each dialyzed separately into TBS containing 1 m urea. Aliquots (200 μl) were then pipetted onto the top of a 5–20% (w/v) sucrose gradient (3.6-ml total volume), buffered with TBS containing 1 m urea, formed in Polyallomer tubes (11 × 3 × 60 mm; Beckman, Brea, CA). Ultracentrifugation experiments were performed for 22 h, 15 min at 42,000 rpm (ω2t, 1.55·1012) at 4 °C in a Beckman L8-M ultracentrifuge using a Beckman SW 60Ti rotor. After a small hole was pricked with a pin in the bottom of the tubes, 8-drop fractions were collected. Fractions were TCA-precipitated, separated by nonreducing SDS-PAGE on 12.5% (w/v) acrylamide gels, and analyzed by Western blot analysis. Protein loading was checked by Ponceau S stain. Nitrocellulose membranes were developed with the SuperSignalTM kit (Pierce) according to the manufacturer's instructions.
Reconstitution of the BMP-5 complex was achieved by dialysis of the BMP-5 pd (1.32 μm) and BMP-5 gfd (0.66 μm) in 200 μl of TBS, 1 m urea. After dialysis, the sample was analyzed by velocity sedimentation as described above.
Surface Plasmon Resonance (SPR)
Binding analyses were performed using a BIAcoreX (Biacore Life Sciences, GE Healthcare). The pds of tested TGF-β superfamily members (500 resonance units (RU)) or perlecan domain V carrying GAG chains (1700 RU) were covalently coupled to a CM5 sensor chip (research grade) using the amine coupling kit following the manufacturer's instructions (Biacore Life Sciences). The choice of which proteins to couple to the chip and which to use as soluble ligands was made based on ELISA binding assays (data not shown). Although both orientations were positive in ELISA, the orientation that yielded the best results was selected for optimization using SPR. According to the manufacturer's instructions, the amounts of immobilized ligand RUs were chosen based on the following formula: Rmax = RL × (MWA/MWL) × Sm, where Rmax is the maximum RUs to be expected as a read-out response (given 100% of the ligand is active and 100% of the ligand-binding sites are filled); RL is the amount of immobilized RUs of ligand, Mw,A/Mw,L is the ratio of molecular weights between analyte and ligand, and Sm is the number of binding sites. Binding responses due to analyte interaction with the surface-coupled ligand were normalized by subtraction of background binding to plain control flow cells.
Binding assays were performed at 25 °C in 10 mm HEPES buffer, pH 7.4, containing 0.15 m NaCl, 3 mm EDTA, and 0.005% (v/v) P20 surfactant (HBS-EP buffer, BIAcore Life Sciences). Fibrillin polypeptides, gfds, or the GDF-8 pd were diluted in HBS-EP buffer and then injected at several concentrations and different flow rates over the immobilized ligands. Based on the formula recommended for immobilized RUs, all observed response RUs upon analyte injection (0–160 nm) were in the expected range. For competition assays, rF23 was preincubated at a constant concentration of 20 nm with the competitor BMP pd at concentrations of 400 to 5 nm prior to injection. To account for variations of the rF23 signal because of buffer changes caused by the addition of different amounts of competitor, we generated for each competition sensorgram a buffer-matched control without competitor, which was set in each case as the 100% reference signal. The surface was regenerated with a pulse of 10 mm glycine, pH 1.7. Kinetic constants were calculated by nonlinear fitting (1:1 interaction model with mass transfer) to the association and dissociation curves according to the manufacturer's instructions (BIAevaluation 3.0 software). Apparent equilibrium dissociation constants (KD) were then calculated as the ratio of kd/ka.
Blot Overlay Assay
Equal numbers of cells (1 × 106) were plated in a 6-well plate in serum-containing medium. The next day, serum-containing medium was replaced by serum-free medium. Serum-free medium was collected after 1 day in culture. 1 ml of medium from various cell lines was trichloroacetic acid (TCA)-precipitated, separated by nonreducing SDS-PAGE on a 5% (w/v) polyacrylamide gel, and electrotransferred onto a nitrocellulose membrane. Transferred proteins were blocked by incubation in 5% nonfat dry milk in TBS at room temperature for 1 h. Pds diluted in 2% milk in TBS (0.5 μm) were incubated with the membrane for 3 h, and bound pd was detected using an anti-His6 tag antibody (R&D Systems), followed by incubation of enzyme-coupled secondary antibody and substrate development with the SuperSignalTM kit (Pierce).
ELISA Binding Assays
Multiwell plates were coated with purified full-length mouse perlecan (0.01 μm) or recombinant mouse perlecan polypeptides (0.1 μm) in 100 μl/well of coating buffer (15 mm Na2CO3 and 35 mm NaHCO3, pH 9.2) at 4 °C overnight. Coated wells were blocked with 5% nonfat dry milk in TBS at room temperature for 1 h. Polyhistidine-tagged pds of TGF-β superfamily members (100 μl/well) were serially diluted 1:2 in 2% milk in TBS and incubated for 3 h. For detection of the bound ligands, a monoclonal anti-His6 antibody (R&D Systems) diluted in 2% milk in TBS was used, followed by a final incubation with enzyme-conjugated secondary antibody. Color reaction was achieved using 3,3′,5,5′-tetramethylbenzidine (Sigma) and was stopped with 0.1 n HCl. Absorbance was read at 450 nm using a Molecular Devices Emax plate spectrophotometer.
Enzymatic Digests
For digests with heparinase III (Sigma), 1 ml of cell culture medium from A204 cells was dialyzed against 20 mm Tris-HCl, pH 7.5, containing 4 mm calcium acetate and 100 μg/ml bovine serum albumin (BSA). For chondroitinase ABC (Sigma) digests, 1 ml of cell culture medium was dialyzed against 50 mm Tris, pH 8.0, 60 mm sodium acetate, containing 0.02% BSA. Each solution was subsequently digested at 37 °C with 2 units of heparinase III or chondroitinase ABC enzyme for 2 h. After TCA precipitation, the samples were separated by nonreducing SDS-PAGE and subjected to blot overlay analysis.
BMP Reporter Assays
Subconfluent C2C12 cells were washed, trypsinized, and seeded into 96-well plates (Costar, Corning, Lowell, MA) at a density of 30,000 cells/well in Dulbecco's modified Eagle's medium (DMEM) (MediaTech, Hendon, VA) without serum with or without 50 ng/ml BMP-4, -5, and -10 gfds, 100 ng/ml BMP-7 gfd, or 300 ng/ml BMP-7 complex for 6 h. To test for inhibition of gfd activity by pds, BMP gfds and pds were preincubated at the indicated molar ratios in PBS for at least 2 h prior to addition to cells. To determine whether BMP-1 activates the BMP-10 pd·gfd complex, 100 ng of BMP-10 gfd and 2.12 μg of BMP-10 pd (molar ratio 1:16) in 100 μl of PBS were dialyzed against 50 mm Tris-HCl, 150 mm NaCl, pH 7.5, and subsequently incubated with 200–800 ng of BMP-1 for 12 h at 37 °C. This solution was diluted 1:10 with serum-free DMEM, and 100 μl of it was added to C2C12 cells seeded in 100 μl of serum-free DMEM, resulting in a final concentration of 50 ng/ml BMP-10 gfd, 1.06 μg/ml BMP-10 pd, and 100–400 ng/ml BMP-1.
After incubation with gfds and complexes, total RNA from cells was harvested using TRIzol® reagent (Invitrogen), and the RNA from eight equally treated wells was combined. Total RNA preparations were quantified by photospectrometry. 0.1 μg of RNA per sample was reverse-transcribed using the Bio-Rad iScriptTM cDNA synthesis kit. Samples in triplicate were amplified with primers for Id3, a BMP-responsive element, using the iTaqTM SYBR Green Supermix (Bio-Rad) in an iQTM5 multicolor real time PCR detection system (Bio-Rad). Analysis of data was performed using the 2−ΔΔCt method (27) and quantitated relative to the ARBO P0 gene. Gene expression was normalized to samples where cells were incubated with corresponding amounts of BSA, which provided an arbitrary constant for comparative fold expression.
RESULTS
BMP-5 pd Interacts with Fibrillin
Because BMP-5 and BMP-7 belong to a subfamily within the BMPs (28), we tested whether the BMP-5 pd binds to fibrillin polypeptides in a similar manner as BMP-7 pd (3, 11). BMP-5 pd (Fig. 1B) was coupled to a BIAcore sensor chip, and recombinant fibrillin polypeptides (Fig. 1A) were tested, using SPR technology. The BMP-5 pd interacted with high affinities (KD = 6 and 28 nm) with rF23, a fibrillin-1 polypeptide composed of the N terminus followed by the subsequent nine domains, and its homologous fibrillin-2 polypeptide, rF86 (Fig. 1C and Table 1). In contrast, a fibrillin-2 polypeptide, rF37, which lacks the N terminus and begins with the second epidermal growth factor (EGF)-like domain and ends with the 10th calcium-binding EGF-like domain, failed to bind to the BMP-5 pd (Table 1), indicating that the major BMP-5 pd-binding site in fibrillin-2 is located within the first two N-terminal domains.
TABLE 1.
Dissociation constants (KD in nm) of interactions between the BMP-5 pd and various fibrillin polypeptides
NB means no binding.
| rF23 | 6 |
| rF87 | 14 |
| rF92 | 8 |
| rF93 | NB |
| rF31 | NB |
| rF45 | 33·103 |
| rF85 | NB |
| rF36 | NB |
| rF63 | 380 |
| rF86 | 28 |
| rF37 | NB |
| rF47 | NB |
| rF70 | NB |
| rF52 | NB |
| rF57 | 22·103 |
The BMP-5 pd also interacted with a moderate to weak affinity (KD = 380 nm to 22 μm) with polypeptides rF63 (fibrillin-1) and rF57 (fibrillin-2), both representing domains in the C-terminal regions of fibrillin-1 and -2. In addition, a weak (33 μm) binding site was identified in the fibrillin-1 polypeptide rF45, which begins with EGF4 and ends with the third 8-cysteine domain. rF47, a fibrillin-2 polypeptide identical in domain structure to rF45, except that it lacks the third 8-cysteine domain, showed no binding to BMP-5 pd. The fibrillin-2 polypeptide rF70, which begins with cbEGF10 and contains the third 8-cysteine domain, also showed no binding. No binding to other polypeptides, which together span all of the remaining domains in fibrillin-1 and almost all of fibrillin-2, was detected (Fig. 1A and Table 1).
To further define the binding site of the BMP-5 pd in fibrillin-1, immobilized BMP-5 pd was tested with recombinant fibrillin-1 polypeptides that represent subregions of rF23 (Fig. 1A). Recombinant fibrillin-1 polypeptides rF31 and rF93 failed to bind to the BMP-5 pd, whereas rF87 and rF92 interacted with dissociation constants of 8–14 nm (Table 1 and Fig. 1D), comparable with the KD values obtained with rF23 (6 nm). These results showed that the high affinity BMP-5 pd-binding site resides in the N terminus of fibrillin-1.
Competition of the BMP-5 pd with pds of BMP-4, -7, and -10
Previously, we defined a high affinity binding site for other pds of the BMP/growth and differentiation factor family of growth factors (9). To determine whether the BMP-5 pd shares the same binding site in the N terminus of fibrillin-1 as other family members, competition studies were performed. rF23 (at a constant concentration of 20 nm) was incubated with increasing amounts of competitor pds (0–100 or 0–400 nm), followed by injection of the mixture onto a BIAcore CM5 chip immobilized with the BMP-5 pd. The pds of BMP-4, -7, and -10 competed with inhibition constants (I50) ranging from 30 nm to 28 μm with the BMP-5 pd for rF23 binding (Table 2 and Fig. 1E). These results suggest that all the tested pds bind to a common site in rF23, the N terminus of fibrillin-1.
TABLE 2.
Inhibition constants (I50 in nm) of competitor BMP pds with BMP-5 pd
| BMP-4 | 28·103 |
| BMP-7 | 30 |
| BMP-10 | 64 |
Comparison of BMP-5 pd·gfd Complex Formation to BMP-4, -7, and -10 and GDF-8
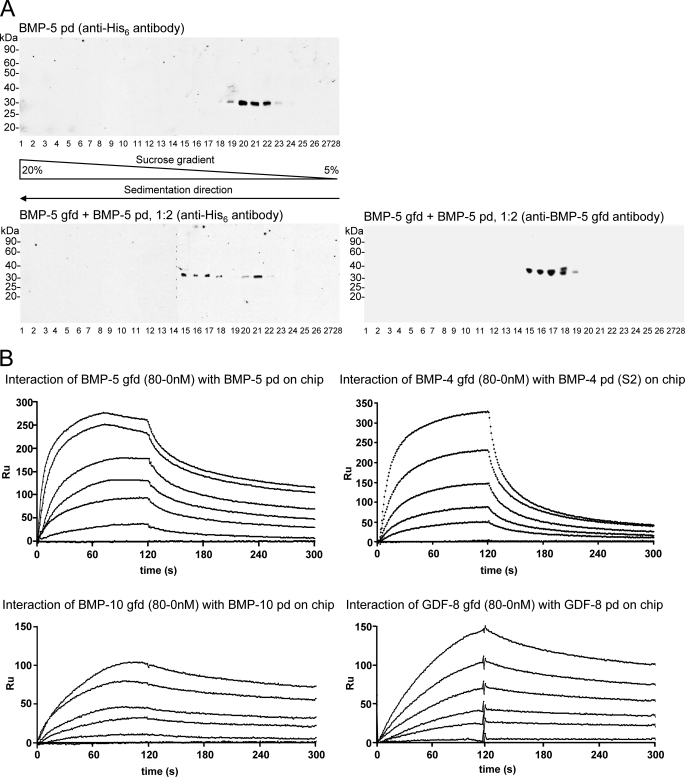
We have shown that pds of TGF-β superfamily members form complexes with their cognate gfds, using a velocity sedimentation approach (9). Recombinantly expressed pds were dialyzed together with cognate commercially available gfds and were then subjected to velocity sedimentation. Using this method, reconstituted pd·gfd complexes were shown to be similar in stoichiometry, molecular mass, and hydrodynamic shape to native, recombinantly expressed BMP-7 complexes (9). Therefore, we first tested whether the BMP-5 gfd can form a complex with its pd, using the velocity sedimentation approach.
Commercially available BMP-5 gfd was incubated with BMP-5 pd and expressed in E. coli, in TBS containing 1 m urea overnight, and the migration of the mixture through a 5–20% sucrose gradient was analyzed after velocity sedimentation. The addition of 1 m urea was necessary because of the low solubility of the pd. The entire gradient was collected in fractions beginning from the bottom to the top of the tube. Each fraction was TCA-precipitated and analyzed by nonreducing SDS-PAGE, followed by immunoblotting using monoclonal antibodies specific for the BMP-5 gfd and the polyhistidine-tagged pd. A reference run with BMP-5 pd alone showed peak signals in fractions 20–22 (Fig. 2A, upper panel). When incubated together with the BMP-5 gfd at a molar ratio of 1:2 (BMP-5 gfd/BMP-5 pd), the mixture revealed a shift of the BMP-5 pd to the middle of the gradient (fractions 15–18, Fig. 2A, lower left panel). Residual signals in fractions 20–22 indicated the presence of some uncomplexed pd. The same stripped blot, reprobed with anti-BMP-5 gfd, demonstrated that BMP-5 gfd was present in the same fractions with the BMP-5 pd (fractions 15–19, Fig. 2A, lower right panel), indicating that complex formation with the pd had occurred. These results for BMP-5 pd·gfd complex formation were similar to our previous findings for BMP-4 and -7 and GDF-5 and -8 (9).
FIGURE 2.
BMP-5 pd forms a growth factor complex with its pd. A, BMP-5 gfd forms a complex with its pd. Top panel, reference signal for the bacterially expressed BMP-5 pd peaks in fraction 20–21 (immunoblotted with anti-His6 antibody) after ultracentrifugation through a 5–20% sucrose gradient containing TBS, 1 m urea. The direction of sedimentation through the sucrose gradient is indicated below the fraction numbers. Lower panel, reconstitution of the BMP-5 complex using commercially available BMP-5 gfd and bacterially expressed BMP-5 pd was monitored by velocity sedimentation in sucrose gradients. Both the gfd and the pd signals peak in fractions 15–18, indicating that BMP-5 gfd and pd migrated together as a complex through the gradient. This represents a shift of four fractions further down in the gradient when compared with the BMP-5 pd reference signal (top panel). A minor amount of pd remains uncomplexed in fractions 20–21 (lower panel). B, gfds and pds of TGFβ superfamily members interact with high molecular affinities. SPR sensorgrams show interaction between the BMP-5 gfd (in solution) and the BMP-5 pd immobilized on a sensorchip. Gfds of BMP-4, BMP-10, and GDF-8 (all in solution) also bind their corresponding pds. High affinity KD values were determined from these sensorgrams (Table 4).
To further compare complex formation between BMP-4, -5, and -10 and GDF-8, we quantitated the molecular dissociation constants (KD) for each pd-gfd pair, using SPR. Pds were immobilized on biosensor chips, and gfds were used as soluble ligands at different concentrations. KD values for the interactions between the pds and gfds of BMP-4, -5, and -10 and GDF-8 were obtained from fitted sensorgrams (Fig. 2B). The KD values ranged between 4 and 13 nm (Table 3), indicating a high affinity interaction between all tested pds and gfds.
TABLE 3.
Dissociation constants (KD in nm) of interactions between pds and their cognate gfds
| pd on chip | gfd in solution |
|---|---|
| BMP-4 | 13 |
| BMP-5 | 4 |
| BMP-10 | 7 |
| GDF-8 | 8.3 |
BMP-4, -5, and -7 pd·gfd Complexes Are Not Latent
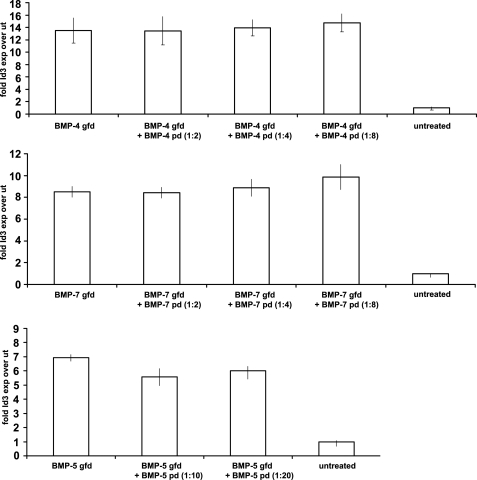
Some pd·gfd complexes of TGF-β family members, such as TGF-β and GDF-8, are latent (1, 2, 29), although others, like BMP-7 and BMP-9, are active (4, 11). Therefore, we tested whether binding of the BMP-4 or -5 pd to its gfd inhibits gfd bioactivity. As a bioactivity read-out, we used the sensitive and rapid response of Id3, a BMP-responsive element, upon BMP stimulation in C2C12 cells (11). Subconfluent C2C12 cells were seeded in serum-free cell culture media in the presence of BMP components for 6 h. Afterward, total RNA was extracted, and real time quantitative PCR for Id3 was performed.
BMP-4, -5, and -7 gfds induced 7–14-fold increases in Id3 expression, compared with expression by untreated cells (Fig. 3). At 50 ng/ml, BMP-5 was able to induce a 7-fold increase of Id3 expression after 6 h (Fig. 3, bottom graph). However, when BMP-5 gfd was first dialyzed against a 10- or 20-fold molar excess of BMP-5 pd and then added to the cells, there was no significant reduction in bioactivity (Fig. 3, bottom graph). Because BMP-5 forms a pd·gfd complex using only a 2-fold molar excess of pd (Fig. 2A), this result suggests that the BMP-5 pd does not have the ability to confer latency to its gfd. The same result was obtained when pds of BMP-4 or BMP-7 were added in increasing molar excesses to their gfds (Fig. 3, upper and middle graphs). Because previous results showed successful complex formation using these molar ratios of BMP-4 and -7 pds to gfds (9), we conclude that binding of BMP-4 and -7 pds to their gfds does not render these complexes inactive.
FIGURE 3.
Complex formation between the pds and gfds of BMP-4, -5, and -7 does not inhibit gfd bioactivity. BMP bioactivity assays were performed in which C2C12 cells were stimulated with gfds of BMP-4, -5, and -7 in the absence or presence of increasing amounts of pds. BMP gfds and pds were used in the indicated molar ratios to form pd·gfd complexes. The expression of the BMP-responsive element Id3 was measured and graphed relative to cells incubated with BSA (untreated, ut). Error bars indicate standard deviations from median values obtained from triplicate samples. None of the tested pds was able to inhibit the bioactivity of their cognate gfds.
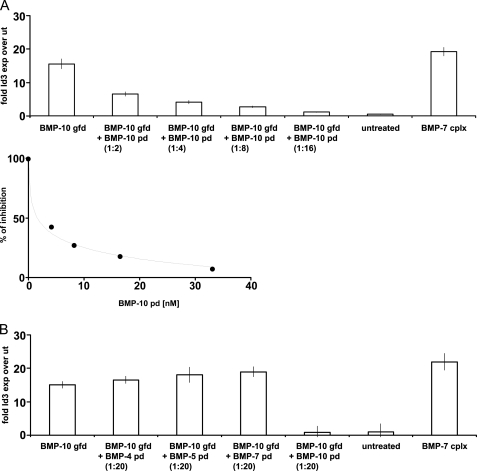
BMP-10 pd Confers Latency to Its gfd
The BMP-9 pd·gfd complex was reported to be bioactive (4). Because BMP-9 and BMP-10 belong to the same subgroup of the TGF-β superfamily based on amino acid sequence similarities (26), we tested whether BMP-10 pd can inhibit BMP-10 gfd bioactivity, using C2C12 cells and Id3 expression as the read-out for BMP bioactivity. Native recombinant BMP-7 complex was used as a positive control for BMP bioactivity (Fig. 4) and also to show that BMP-7 pd·gfd complex is active in this assay.
FIGURE 4.
pd of BMP-10 confers latency to the BMP-10 gfd. A, top graph, titration of increasing amounts of BMP-10 pd with BMP-10 gfd suppressed BMP-10 bioactivity. Lower graph, suppression of BMP-10 bioactivity was obtained with 2.07 nm (50 ng/ml) BMP-10 pd. The inhibition constant for the BMP-10 pd in this bioactivity assay was 1.6 nm. B, BMP-10 pd inhibits BMP-10 gfd bioactivity specifically, because the pds of BMP-4, -5, and -7 failed to inhibit BMP-10 gfd bioactivity, even at a 20-fold molar excess. Cells treated with BMP-7 complex served as a positive control and BSA-treated cells (untreated) as negative control. Error bars represent standard deviations around median values obtained from triplicate samples.
Addition of BMP-10 gfd resulted in a 15-fold induction of Id3 expression, compared with untreated cells. However, addition of BMP-10 after prior complex formation with increasing molar excesses of BMP-10 pd showed dose-dependent inhibition of BMP activity (Fig. 4, upper graph). The inhibition constant (I50) for this reaction was 1.6 nm pd at a concentration of 2.07 nm (50 ng/ml) for the BMP-10 gfd (Fig. 4, middle graph). At a molar ratio of 1:16 (gfd/pd), 100% of BMP-10 gfd bioactivity was suppressed. To test how specific this effect was, we added pds of BMP-4, -5, and -7 in 20-fold molar excess to the BMP-10 gfd. Although BMP-10 pd reduced BMP bioactivity to the level of untreated cells, incubation of BMP-10 with the other pds resulted in no reduction of BMP-10 gfd bioactivity (Fig. 4, bottom graph).
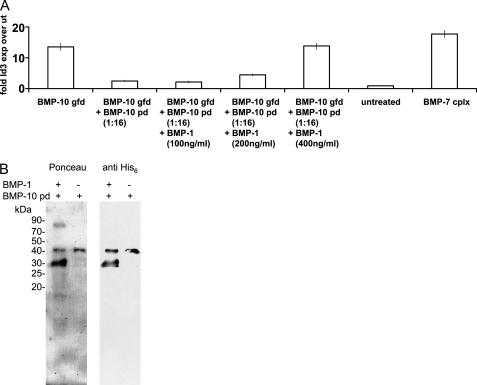
To further demonstrate that the BMP-10 pd confers latency to its gfd, we first incubated BMP-10 gfd with 16-fold molar excess of BMP-10 pd and subsequently added increasing amounts of BMP-1 to this mixture. Bioactivity assays showed that addition of BMP-1 could restore BMP-10 gfd bioactivity in a dose-dependent manner up to 100% (Fig. 5A). Incubation of BMP-1 with C-terminally His6-tagged BMP-10 pd revealed specific cleavage of the BMP-10 pd, resulting in a detectable C-terminal His6-tagged fragment of ∼30 kDa (Fig. 5B). BMP-1 failed to cleave pds of other BMPs (BMP-4, -5, and -7) (data not shown). These results indicate that BMP-10 pd, when cleaved, can no longer confer latency to BMP-10 gfd.
FIGURE 5.
BMP-1 activates the BMP-10 complex by cleaving the BMP-10 pd. A, suppressed BMP-10 gfd bioactivity caused by addition of BMP-10 pd was restored by increasing amounts of BMP-1. Error bars represent standard deviations around median values obtained from triplicate samples. B, Ponceau stain and anti-His6 tag Western blot detected a 30-kDa C-terminally His6-tagged fragment of BMP-10 pd, after cleavage by BMP-1.
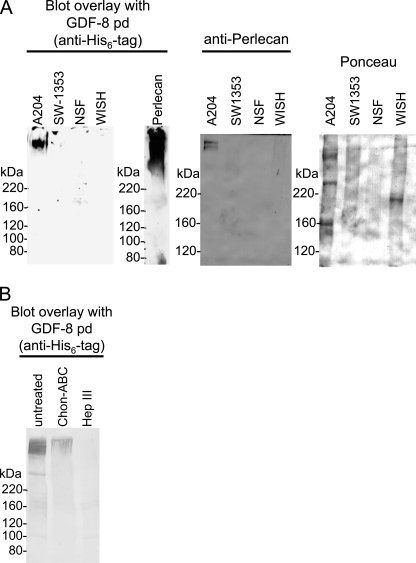
Pd of GDF-8 Binds to GAG Side Chains of Perlecan
To test whether growth factor pds interact with other ECM proteins, we performed screens using a blot overlay approach with total medium proteins from various cell lines and the pd of GDF-8. The GDF-8 pd showed no binding to fibrillin-1 in our previous studies (9) and was therefore used in this screen for other interactors. For this experiment, total medium proteins from various human cell lines (A204 (rhabdomyosarcoma), SW1353 (chondrosarcoma), neonatal skin fibroblasts, and WISH (transformed amniotic epithelial cells)) were TCA-precipitated from serum-free media collected after 2 days of incubation. Medium proteins were applied to nonreducing 5% SDS-PAGE, transferred to nitrocellulose, and incubated with polyhistidine-tagged GDF-8 pd, followed by blotting with an anti-His6 tag antibody. Blot overlay screens revealed that the GDF-8 pd interacted with a diffuse high molecular weight band at the top of the lane containing proteins secreted by human A204 rhabdomyosarcoma cells (Fig. 6A, left blot).
FIGURE 6.
GDF-8 pd binds to GAG chains of perlecan. A, left, a blot overlay assay showed that GDF-8 pd binds to a high molecular weight species secreted into the medium of A204 rhabdomyosarcoma cells. In this assay, medium proteins from SW1353 chondrosarcoma, neonatal skin fibroblasts (NSF), and WISH-transformed amniotic epithelial cells were negative. Middle, a blot overlay assay showed that GDF-8 pd binds to mouse perlecan from Engelbreth-Holm-Swarm sarcoma. Right, total medium proteins from A204, SW1353, neonatal skin fibroblasts, and WISH cells probed with anti-perlecan confirmed the presence of perlecan at the top of the gel in the A204 lane. Ponceau-stained lanes are also shown. B, enzymatic digestion of A204 medium with heparinase III abolished GDF-8 pd binding to perlecan. Digestion with chondroitinase ABC appeared to reduce the GDF-8 pd blot.
Based on the high molecular mass of 500 kDa or more and the nonhomogeneous appearance of the band, we speculated that the interaction partner could be the proteoglycan perlecan. Western blotting of A204 medium proteins with an anti-perlecan antibody revealed perlecan at the top of the gel (Fig. 6A, right blot). The anti-perlecan immunoblot also showed that perlecan is particularly enriched in the medium of A204 cells compared with the other cells tested. Blot overlay using purified full-length perlecan and GDF-8 pd demonstrated a diffuse positive reaction (Fig. 6A, middle blot). To further identify the A204-interacting protein as perlecan, A204 media were digested with enzymes specific for cleaving off GAG side chains consisting of chondroitin sulfate or heparan sulfate. Treatment with heparinase III completely abolished the GDF-8 pd interaction signal (Fig. 6B), whereas chondroitinase ABC was less effective. These results suggested that the GDF-8 pd interacts with the heparan sulfate side chains of perlecan and not with the perlecan core protein.
To check the specificity of this novel interaction with perlecan, ELISA-based solid phase interaction assays were performed with purified full-length perlecan adsorbed to the wells and polyhistidine-tagged pds of BMP-2, -4, -5, -7, and -10 and GDF-8 in solution. Incubation with anti-His6 resulted in a strong signal only for the GDF-8 pd, whereas all other tested pds were negative (Fig. 7B, top left). To define more precisely the binding site in perlecan, recombinant polypeptides spanning full-length perlecan (Fig. 7A) were adsorbed to wells, and GDF-8 pd was incubated in solution. GDF-8 pd interacted specifically with domain V of perlecan. Binding to all other tested perlecan polypeptides, including domain I which also carries multiple GAG side chains, was negative (Fig. 7B, top right). We also reconfirmed that the GDF-8 pd interacts exclusively with the GAG side chain and not with the core protein of domain V by testing a nonmodified form of domain V. Domain V without GAG side chains failed to bind GDF-8 pd (Fig. 7B, bottom left).
SPR technology was used to determine the molecular affinity between perlecan domain V and GDF-8 pd. Perlecan domain V was covalently coupled to a chip, and different concentrations (160 to 0 nm) of GDF-8 pd were tested as solution analytes. Calculated from the fitted sensorgrams (Fig. 7B, bottom right), the KD value (11 nm) revealed a high affinity interaction between perlecan and GDF-8 pd.
DISCUSSION
Results presented here contribute at least two novel insights into the function of pds of the TGFβ superfamily. First, although no binding was found between the GDF-8 pd and fibrillin (9), we demonstrate here that the GDF-8 pd interacts well with another ECM protein, perlecan. This novel finding reinforces the general concept that TGFβ-like growth factors are targeted to the ECM through specific interactions between pds and ECM structural macromolecules. Moreover, because previous examples of pd interactions with ECM molecules have been limited to fibrillin family members (3, 9), including the LTBPs (30), our new data demonstrate that TGFβ superfamily pds can interact with a larger repertoire of ECM structural macromolecules than previously suspected.
The interaction between GDF-8 and perlecan was shown using authentic proteins secreted by cells in culture and recombinant perlecan. After initial identification in a screen for ligands, perlecan was confirmed as a specific GDF-8 interactor using both ELISA and SPR approaches. Furthermore, experiments showed that GAG side chains present on perlecan domain V are responsible for the interaction with GDF-8 pd, because nonmodified domain V without GAG side chains failed to interact with GDF-8 pd. This conclusion was also supported by the failure of native heparinase-treated proteins to interact with GDF-8 pd in blot overlay assays. Therefore, in contrast to previously documented protein/protein interactions, we now show that TGFβ superfamily pds can also interact with GAG side chains.
Interestingly, binding of GDF-8 pd to perlecan domain I, which contains multiple heparan sulfate side chains, was negative. This finding implies that domain I and domain V carry heparan sulfate side chains of different composition tailored specifically to target different types of growth factors. For example, heparan sulfate chains of domain I are known to concentrate heparin-binding growth factors such as FGF-2 or PDGF (17, 31). In contrast, our data demonstrate interactions between TGFβ superfamily pds and domain V GAG chains. It may be worthwhile to speculate that, in concert, perlecan GAG chains may physically integrate signaling by growth factors targeted separately to domain I and domain V.
The main biological function of GDF-8 is to negatively regulate muscle mass (2, 32, 33). Recently, perlecan deficiency in mice was shown to result in skeletal muscle hypertrophy and reduced expression of GDF-8 (34), suggesting that perlecan and GDF-8 may together negatively regulate muscle mass. More in-depth studies are required to determine the molecular mechanisms by which perlecan and GDF-8 work together. However, our studies now establish a direct interaction between the GDF-8 pd and perlecan and are a first step toward unraveling the detailed molecular mechanisms underlying their concerted function.
In addition to identifying a second type of ECM interaction with TGFβ superfamily pds, results presented in this study indicate that the BMP-10 pd·gfd complex is inactive. Using a C2C12 cell-based assay and Id3 as a BMP signaling readout, we showed that the BMP-10 pd specifically inhibited BMP-10 gfd bioactivity in a dose-dependent manner. This was surprising, because BMP-9, the BMP family member with the highest homology to BMP-10, was reported to form bioactive complexes with its pd (4). Because large molar excesses of other BMP pds did not inhibit BMP-10 gfd bioactivity, inhibition appeared to be a specific property of the BMP-10 pd.
Our findings raise the possibility that BMP-10 exists in the form of a latent pd·gfd complex in vivo, similar to TGF-β or GDF-8. Latent pd·gfd complexes require activation. Proteolytic cleavage of the propeptide is believed to be one mechanism to activate latent TGF-β or GDF-8 (10, 35). We found that addition of increasing amounts of BMP-1 reversed the inhibition of BMP-10 gfd bioactivity by its pd. In addition, we showed that BMP-1 cleaves the BMP-10 pd, suggesting that BMP-10 pd cleavage by BMP-1 could be a specific activation mechanism for latent BMP-10 in vivo. Thus, although most homologous to BMP-9, BMP-10 is similar to GDF-8 in that the BMP-10 pd can confer latency to its gfd and can be activated by BMP-1. However, BMP-10 pd, unlike GDF-8 pd, interacts with fibrillins (9).
Recently, inhibition of BMP-10-driven vascularization was suggested as a potential antitumor therapy (36). The BMP-10 inhibitor used in this study was a soluble chimeric protein consisting of the gfd binding domain of activin receptor-like kinase-1 (ALK1) fused to the Fc domain of IgG1. Human ALK1-Fc binds BMP-10 gfd with a molecular affinity of 3.2 nm, which is similar to the KD value determined by us for the BMP-10 pd/gfd interaction (7 nm, Table 3). A 20-fold molar excess of ALK1-Fc suppressed BMP-10 bioactivity by 60%, presumably through competition with the native cellular receptor. However, a 2-fold molar excess of BMP-10 pd led to similar inhibition of BMP activity in our assay, suggesting that the BMP-10 pd might have the potential to be developed as a treatment option for conditions of pathologic vascularization driven by BMP-10.
Our previous studies showed that pds of BMP-2, -4, -7, and -10 and GDF-5 interact with fibrillin-1 and -2 (3, 9). We can now add the pd of BMP-5 to this list. BMP-5 pd binds with high affinity to the N-terminal domains of fibrillin-1 and -2. Competition studies with the pds of BMP-4, BMP-7, and BMP-10 showed that the BMP-5 pd binds to the same high affinity site present in the very N-terminal “unique” region of fibrillin-1 (9). In contrast to the BMP-7 pd (9), the BMP-5 pd also interacted with additional sites in fibrillin-1 and -2. The BMP-5 pd interacted with moderate affinity (380 nm) to a polypeptide representing the C-terminal end of fibrillin-1 (rF63) and with weak affinity to a polypeptide representing the region from the start of the 4th EGF-like domain, including the third 8-Cys domain (rF45). In addition, the BMP-5 pd showed a weak interaction site with a fibrillin-2 polypeptide near the C terminus (rF57). These results suggest that, although BMP-5 and BMP-7 are homologous members of the same BMP subfamily, the pds of BMP-5 and -7 might each have an individual set of exquisite molecular affinities for different sites on fibrillin microfibrils. These differences in molecular interactions may be used to specify differences in biological functions during organ development, growth, and homeostasis.
There are currently three major paradigms for the function of pds in pd·gfd complexes. TGFβ and GDF-8 pd·gfd complexes are stable, inactive complexes that are targeted to the ECM through specific molecular interactions mediated by the pds. In this paradigm, activation of the growth factor signal is facilitated by proteolytic cleavage of the pd and/or the binding protein. Our new studies add BMP-10 to this paradigm. The second paradigm is illustrated by BMP-2, which does not form a stable pd·gfd complex and is active. The third paradigm is exemplified by BMP-7, which forms a stable pd·gfd complex (3) that is “active” in solution because type II BMP receptors can compete with the pd for the gfd (11). Our new studies show that BMP-5 pd and gfd form a stable complex, like BMP-4, -7, and -10, and that the pds of BMP-4, -5, and -10 and GDF-8 bind to their respective gfds with similar high affinities. However, despite high affinity binding between the pd and gfd, the pds of BMP-4 and -5 did not confer latency to their gfds when tested in our C2C12 bioactivity assay (Fig. 3), whereas the BMP-10 pd did inhibit the activity of its gfd. Additional indirect support for placing BMP-4 and -5 in the third paradigm along with BMP-7 came from experiments showing that BMP-1 cleaves the BMP-10 pd but not the pds of BMP-4, -5, and -7.
In this study, we have provided additional evidence supporting the emerging concept that ECM structural macromolecules operate as platforms to which growth factors of the TGF-β superfamily are targeted and concentrated (37). In this regard, the pds perform the important role of molecular mediators that position the gfds onto the ECM scaffolds. In vivo evidence that fibrillin-1 and -2 regulate TGFβ and BMP signaling in osteoblasts has been recently published (38), demonstrating that these interactions are likely to be significant.
By showing that perlecan interacts with the GDF-8 pd, we have extended our original concept of “connective tissue pathways” that regulate growth factor signaling (39). Perlecan interacts with fibrillin-1 (40) and is also thought to act as a co-receptor mediating heparin-binding growth factor delivery and receptor signaling (41). Studies establishing the different interactions between pds of TGFβ superfamily members and ECM structural macromolecules, as well as between the structural macromolecules and between macromolecules and cells, will provide the necessary information for understanding the physical basis by which the connective tissue integrates growth factor signaling. Because the connective tissue is designed to integrate organ shape as well as development and function, connective tissue pathways must work in concert with the potent activities of growth factors. Future studies are needed to understand how these multiple interactions work in concert.
Acknowledgments
We thank Dr. Kerry Maddox and the Analytical Core Facility of the Portland Shriners Hospital for DNA sequencing and amino acid analysis. We also thank Glen M. Corson for generating some of the fibrillin-1 and -2 expression constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 AR049698 (to L. Y. S.). This work was also supported by Shriners Hospitals for Children (to L. Y. S.).
- pd
- prodomain
- BMP
- bone morphogenetic protein
- ECM
- extracellular matrix
- GAG
- glycosaminoglycan
- gfd
- growth factor dimer
- LAP
- latency-associated peptide
- LTBP
- latent TGFβ binding protein
- rF
- recombinant fibrillin
- RU
- resonance units
- SLC
- small latent complex
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Wakefield L. M., Smith D. M., Broz S., Jackson M., Levinson A. D., Sporn M. B. (1989) Growth Factors 1, 203–218 [DOI] [PubMed] [Google Scholar]
- 2. Lee S. J., McPherron A. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9306–9311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gregory K. E., Ono R. N., Charbonneau N. L., Kuo C. L., Keene D. R., Bächinger H. P., Sakai L. Y. (2005) J. Biol. Chem. 280, 27970–27980 [DOI] [PubMed] [Google Scholar]
- 4. Brown M. A., Zhao Q., Baker K. A., Naik C., Chen C., Pukac L., Singh M., Tsareva T., Parice Y., Mahoney A., Roschke V., Sanyal I., Choe S. (2005) J. Biol. Chem. 280, 25111–25118 [DOI] [PubMed] [Google Scholar]
- 5. Ge G., Hopkins D. R., Ho W. B., Greenspan D. S. (2005) Mol. Cell. Biol. 14, 5846–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 7. De Crescenzo G., Grothe S., Zwaagstra J., Tsang M., O'Connor-McCourt M. D. (2001) J. Biol. Chem. 276, 29632–29643 [DOI] [PubMed] [Google Scholar]
- 8. Nunes I., Gleizes P. E., Metz C. N., Rifkin D. B. (1997) J. Cell Biol. 136, 1151–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sengle G., Charbonneau N. L., Ono R. N., Sasaki T., Alvarez J., Keene D. R., Bächinger H. P., Sakai L. Y. (2008) J. Biol. Chem. 283, 13874–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolfman N. M., McPherron A. C., Pappano W. N., Davies M. V., Song K., Tomkinson K. N., Wright J. F., Zhao L., Sebald S. M., Greenspan D. S., Lee S. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15842–15846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sengle G., Ono R. N., Lyons K. M., Bächinger H. P., Sakai L. Y. (2008) J. Mol. Biol. 381, 1025–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reinhardt D. P., Sasaki T., Dzamba B. J., Keene D. R., Chu M. L., Göhring W., Timpl R., Sakai L. Y. (1996) J. Biol. Chem. 271, 19489–19496 [DOI] [PubMed] [Google Scholar]
- 13. Keene D. R., Jordan C. D., Reinhardt D. P., Ridgway C. C., Ono R. N., Corson G. M., Fairhurst M., Sussman M. D., Memoli V. A., Sakai L. Y. (1997) J. Histochem. Cytochem. 45, 1069–1082 [DOI] [PubMed] [Google Scholar]
- 14. Reinhardt D. P., Ono R. N., Notbohm H., Müller P. K., Bächinger H. P., Sakai L. Y. (2000) J. Biol. Chem. 275, 12339–12345 [DOI] [PubMed] [Google Scholar]
- 15. Corson G. M., Charbonneau N. L., Keene D. R., Sakai L. Y. (2004) Genomics 83, 461–472 [DOI] [PubMed] [Google Scholar]
- 16. Paulsson M., Yurchenco P. D., Ruben G. C., Engel J., Timpl R. (1987) J. Mol. Biol. 197, 297–313 [DOI] [PubMed] [Google Scholar]
- 17. Costell M., Mann K., Yamada Y., Timpl R. (1997) Eur. J. Biochem. 243, 115–121 [DOI] [PubMed] [Google Scholar]
- 18. Costell M., Sasaki T., Mann K., Yamada Y., Timpl R. (1996) FEBS Lett. 396, 127–131 [DOI] [PubMed] [Google Scholar]
- 19. Schulze B., Sasaki T., Costell M., Mann K., Timpl R. (1996) Matrix Biol. 15, 349–357 [DOI] [PubMed] [Google Scholar]
- 20. Hopf M., Göhring W., Kohfeldt E., Yamada Y., Timpl R. (1999) Eur. J. Biochem. 259, 917–925 [DOI] [PubMed] [Google Scholar]
- 21. Brown J. C., Sasaki T., Göhring W., Yamada Y., Timpl R. (1997) Eur. J. Biochem. 250, 39–46 [DOI] [PubMed] [Google Scholar]
- 22. Corson G. M., Chalberg S. C., Dietz H. C., Charbonneau N. L., Sakai L. Y. (1993) Genomics 17, 476–484 [DOI] [PubMed] [Google Scholar]
- 23. Kuo C. L., Isogai Z., Keene D. R., Hazeki N., Ono R. N., Sengle G., Bächinger H. P., Sakai L. Y. (2007) J. Biol. Chem. 282, 4007–4020 [DOI] [PubMed] [Google Scholar]
- 24. Hawke D. H., Yuan P. M. (1987) Applied Biosystems User Bulletin 28, Foster City, CA [Google Scholar]
- 25. Pereira L., Andrikopoulos K., Tian J., Lee S. Y., Keene D. R., Ono R., Reinhardt D. P., Sakai L. Y., Biery N. J., Bunton T., Dietz H. C., Ramirez F. (1997) Nat. Genet. 17, 218–222 [DOI] [PubMed] [Google Scholar]
- 26. Charbonneau N. L., Dzamba B. J., Ono R. N., Keene D. R., Corson G. M., Reinhardt D. P., Sakai L. Y. (2003) J. Biol. Chem. 278, 2740–2749 [DOI] [PubMed] [Google Scholar]
- 27. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 28. Newfeld S. J., Wisotzkey R. G., Kumar S. (1999) Genetics 152, 783–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gentry L. E., Nash B. W. (1990) Biochemistry 29, 6851–6857 [DOI] [PubMed] [Google Scholar]
- 30. Rifkin D. B. (2005) J. Biol. Chem. 280, 7409–7412 [DOI] [PubMed] [Google Scholar]
- 31. Whitelock J. M., Murdoch A. D., Iozzo R. V., Underwood P. A. (1996) J. Biol. Chem. 271, 10079–10086 [DOI] [PubMed] [Google Scholar]
- 32. McPherron A. C., Lawler A. M., Lee S. J. (1997) Nature 387, 83–90 [DOI] [PubMed] [Google Scholar]
- 33. Bogdanovich S., Krag T. O., Barton E. R., Morris L. D., Whittemore L. A., Ahima R. S., Khurana T. S. (2002) Nature 420, 418–421 [DOI] [PubMed] [Google Scholar]
- 34. Xu Z., Ichikawa N., Kosaki K., Yamada Y., Sasaki T., Sakai L. Y., Kurosawa H., Hattori N., Arikawa-Hirasawa E. (2010) Matrix Biol. 29, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lyons R. M., Keski-Oja J., Moses H. L. (1988) J. Cell Biol. 106, 1659–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mitchell D., Pobre E. G., Mulivor A. W., Grinberg A. V., Castonguay R., Monnell T. E., Solban N., Ucran J. A., Pearsall R. S., Underwood K. W., Seehra J., Kumar R. (2010) Mol. Cancer Ther. 9, 379–388 [DOI] [PubMed] [Google Scholar]
- 37. Sengle G., Charbonneau N. L., Ono R. N., Sakai L. Y. (2008) Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism, 7th Ed., Chapter 5, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 38. Nistala H., Lee-Arteaga S., Smaldone S., Siciliano G., Carta L., Ono R. N., Sengle G., Arteaga-Solis E., Levasseur R., Ducy P., Sakai L. Y., Karsenty G., Ramirez F. (2010) J. Cell Biol. 190, 1107–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Charbonneau N. L., Ono R. N., Corson G. M., Keene D. R., Sakai L. Y. (2004) Birth Defects Res. C Embryo Today 72, 37–50 [DOI] [PubMed] [Google Scholar]
- 40. Tiedemann K., Sasaki T., Gustafsson E., Göhring W., Bätge B., Notbohm H., Timpl R., Wedel T., Schlötzer-Schrehardt U., Reinhardt D. P. (2005) J. Biol. Chem. 280, 11404–11412 [DOI] [PubMed] [Google Scholar]
- 41. Jiang X., Couchman J. R. (2003) J. Histochem. Cytochem. 51, 1393–13410 [DOI] [PMC free article] [PubMed] [Google Scholar]