Abstract
The NFκB transcription factor is a key component of immune and inflammatory signaling as its activation induces the expression of antimicrobial reagents, chemokines, cytokines, and anti-apoptotic factors. Many pathogens encode effector proteins that target factors regulating NFκB activity and can provide novel insights on regulatory mechanisms. Given the link of NFκB dysfunction with inflammatory diseases and some cancers, these effectors have therapeutic potential. Here, screening enteropathogenic Escherichia coli proteins for those implicated in suppressing NFκB function revealed that eGFP-NleC, unlike eGFP, strongly inhibited basal and TNFα-induced NFκB reporter activity to prevent secretion of the chemokine, IL-8. Work involving NleC variants, chemical inhibitors, and immunoprecipitation studies support NleC being a zinc metalloprotease that degrades NFκB-IκBα complexes. The findings are consistent with features between residues 33–65 recruiting NFκB for proteasomal-independent degradation by a mechanism inhibited by metalloprotease inhibitors or disruption of a consensus zinc metalloprotease motif spanning NleC residues 183–187. This raises the prospect that mammalian cells, or other pathogens, employ a similar mechanism to modulate NFκB activity. Moreover, NleC represents a novel tool for validating NFκB as a therapeutic target and, indeed, as a possible therapeutic reagent.
Keywords: Cancer Therapy, Immunosuppressor, Inflammation, NF-kB Transcription Factor, NF-Kappa B, Protease, Proteasome
Introduction
Interaction of inflammatory stimuli, such as cytokines and microbial products, with their cognate receptors induces ubiquitination and phosphorylation cascades that alter the transcriptional profile of the cell to produce factors including antimicrobials, chemokines (e.g. IL-8), and cytokines (e.g. TNFα; see Refs. 1–4). A critical player in this process is the transcription factor nuclear factor κB (NFκB),3 which forms homo- or heterodimers composed of p65 (RelA), RelB, c-Rel, p50, and p52 proteins, with the p65/p50 dimer being the most abundant and associated with the canonical NFκB pathway. The p65/p50 dimer is retained within the cytoplasm bound to the inhibitor of κB (IκB) until receptor-mediated activation of the inhibitor of κB kinase (IKK) phosphorylates IκB, thereby triggering its proteasomal degradation to release NFκB for import into the nucleus. Signaling by many receptors converges at the IKK complex, composed of IKKα, IKKβ, and the NFκB essential modulator (IKKγ), but often involves distinct or overlapping upstream adaptors including TRAF and MyD88 and serine/threonine kinases such as IRAK, TGF-β-activated kinase 1, and RIP1 (1–4). The classic mechanism to terminate NFκB transcriptional activity involves the NFκB-dependent transactivation of IκB, which shuttles the transcription factor back into the cytoplasm (5, 6). However, additional regulatory mechanisms have been described including proteasome-dependent degradation (7) or processing of p65 by caspase and serine proteases to generate forms with inhibitory functions (8, 9). The importance of NFκB in immune and inflammatory signaling is reflected by the fact that its dysregulation is linked to many diseases including cancer, diarrhea, arthritis, inflammatory bowel disease, and neurodegenerative diseases (10, 11).
Given the co-evolution of micro-organisms with mammals, it is not surprising that many, mostly pathogens, inhibit NFκB signaling as part of their strategy to colonize normally privileged niches. Collectively, bacteria and viruses possess “effector” proteins that target most of the proteins known to play roles in transmitting signals that report the presence of foreign antigens (12–14). Examples of how pathogens inhibit NFκB signaling at diverse levels include the A52R protein of the vaccinia virus that acts as a dominant-negative homologue of MyD88 (15), the Yersinia YopP/J protein whose acetylation of IKKβ prevents activation (16, 17) and the Shigella OspG protein, which prevents IκB ubiquitination by targeting an E2 ubiquitin-conjugating enzyme (18). Another bacterium, enteropathogenic Escherichia coli (EPEC), has recently been reported to deliver at least three effectors into host cells to inhibit NFκB function. Although two of these effectors, NleB and NleE, are speculated to block signaling at the level of TGF-β-activated kinase 1 or IKKβ kinases (19, 20), the NleH protein binds the NFκB cofactor RPS3 (ribosomal protein S3) to inhibit the transcription of a subset of genes (21). In this study, we describe that the EPEC gene nleC encodes a protein which targets p65, p50, and IκBα proteins for degradation by a proteasome-independent mechanism. The findings suggest that NleC is a zinc metalloprotease that recruits NFκB complexes for degradation.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa cells (ATCC CCL-2) were maintained at 37 °C in DMEM (Invitrogen) supplemented with 10% fetal calf serum in a 5% CO2 environment. Hela cells seeded in 12- or 24-well plates (Nunc) were grown for 24 h (to ∼80% confluence) prior to introducing mammalian expression vectors by transfection using jetPRIME (PEQLAB Ltd.) following the manufacturer's recommendations. Transfection efficiency was routinely ∼60–80% (data not shown). When required, the proteasomal inhibitor MG132 (Calbiochem; final concentration, 25 μm), NFκB activation inhibitor (Calbiochem product no. 481406; final concentration, 0.28 μm), protease inhibitor mixture (Sigma-Aldrich, P8340 1/1000), GM6001 (final concentration, 25 μm), or EDTA (final concentration, 2.5 mm) were added to the medium just prior to transfection.
Plasmids
nleC gene constructs, including substitution and truncation variants thereof, were all generated by PCR and cloned into pEGFP-C1 (Clontech) using EcoRI/PstI restriction sites, with desired gene product confirmed by DNA sequencing (GATC Biotech). Studies included plasmids encoding the NFκB luciferase reporter protein (22) and IKK pathway components TRAF2, IKKα, IKKβ (23, 24), and p65 (kindly provided by professor Neil D. Perkins, Newcastle University).
Luciferase and IL-8 Assays
Approximately 24 h post-transfection, HeLa cells were either left untreated or treated with TNFα (final concentration, 50 ng/ml) for 30 min prior to addition of luciferase cell culture lysis buffer (Stratagene) following the manufacturer's protocol. Luciferase levels were measured using a FLUOstar Optima 413-3266 (BMG Labtech) plate reader with 96-well plates containing 100 μl luciferase assay solution (Promega) and 25 μl lysate per well. By contrast, IL-8 levels in HeLa supernatants were determined using the OptEIATM Human IL-8 ELISA (BD Biosciences) kit following the manufacturer's recommendations. These assays were routinely carried out ∼48 h post-transfection with or without an additional 3-h incubation with TNFα (final concentration, 50 ng/ml).
Co-immunoprecipitation
For co-immunoprecipitation, cells were transfected with plasmids encoding eGFP or eGFP-fusion proteins as described above. The cells were lysed under native conditions, and proteins were immunoprecipitated using GFP-Trap® beads (Chromotek) according to the manufacturer's recommendations.
Western Blot Analyses
Cells were either lysed using 1× SDS sample buffer or in PBS with 1% Triton and protease inhibitors for subsequent fractionation as described previously (25). Samples were equalized for protein content, separated via SDS-PAGE (10–12%), and transferred onto nitrocellulose membrane. Membranes were blocked in 5% skimmed milk powder in Tris-buffered saline, pH 7.5, containing 0.1% Tween (TBST) for 1 h followed by an overnight incubation at 4 °C in 5% BSA/TBST solution containing the appropriate antibody. Membranes were washed and incubated in 5% skim milk/TBST containing the respective HRP-conjugated secondary antibody (Jackson ImmunoResearch Laboratories). Blots were developed in SuperSignal West Pico chemiluminescent substrate (Pierce) according to the manufacturer's recommendations, and the signals were detected on Hyperfilm ECL (Amersham Biosciences). Primary antibodies used were NFκB p65, NFκB p50, IκBα, IKKα/β (Santa Cruz Biotechnology), p65, phospho-p65 (Cell Signaling Technologies), actin (Sigma), and GFP (Zymed Laboratories Inc.).
RESULTS
NleC Effector Inhibits NFκB Reporter Activity and IL-8 Secretion
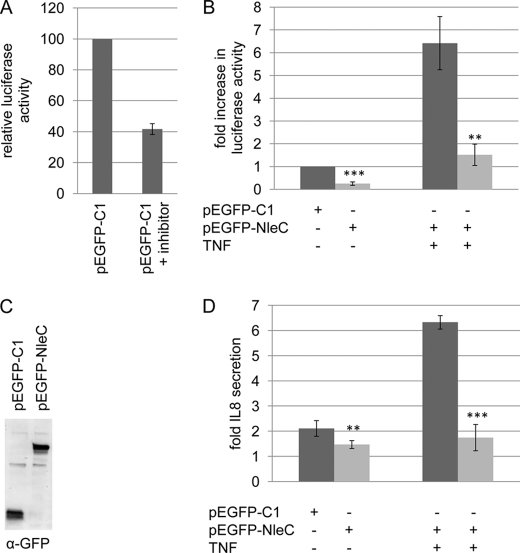
Bioinformatic analyses of the EPEC genome identified 21 putative effector genes (26, 27), of which one or more are predicted to inhibit NFκB function (25). Our previous work argued against critical roles for the Tir, Map, EspF-H, EspZ, EspG2, NleA, NleF, and NleH effectors (25), so other effector-encoding genes were cloned into mammalian expression vectors for screening. NFκB assays involved co-transfection studies using empty control plasmid (pEGFP-C1) and a vector encoding the luciferase protein under the transcriptional control of NFκB (via a 5× κΒ promoter). This analysis revealed basal levels of NFκB activity in HeLa cells mediated by multiple signaling pathways, as the “NFκB activation inhibitor” (Calbiochem; product no. 481406) reduced luciferase levels by only ∼60% (Fig. 1A). Co-transfection studies confirmed basal NFκB activity in pEGFP-transfected cells, with a dramatic reduction (∼75%) in cells transfected with the eGFP-NleC-expressing plasmid (Fig. 1B). Western blot analysis revealed similar eGFP and eGFP-NleC expression levels (Fig. 1C). TNFα induction of NFκB activity increased luciferase levels in control cells (∼6 fold) but not eGFP-NleC expressing cells (Fig. 1B). To probe the relationship of this inhibition to NFκB function, cells were assayed for IL-8 secretion levels, as the IL-8 gene is under the transcriptional control of NFκB (28). Consistent with the luciferase assay data, eGFP-expressing cells exhibited low basal levels of IL-8 secretion with reduced levels from eGFP-NleC expressing cells (Fig. 1D). Furthermore, TNFα treatment increased IL-8 secretion levels in control cells (∼3-fold) but not from eGFP-NleC-expressing cells (Fig. 1D). Thus, the nleC gene product of EPEC has a potent capacity to inhibit NFκB activity when expressed within host cells as an N-terminally tagged eGFP fusion protein.
FIGURE 1.
NleC inhibits NFκB-dependent protein expression and IL-8 secretion. A, relative change in NFκB-dependent luciferase activity in pEGFP (empty vector control)-transfected HeLa cells following treatment with NFκB activation inhibitor (Calbiochem product no. 481406). B, fold increase in NFκB-dependent luciferase activity. C, representative immunoblot showing similar expression levels of eGFP and eGFP-NleC in transfected cells. D, IL-8 secretion levels in untreated or TNFα-treated HeLa cells transfected with pEGFP-NleC or pEGFP vectors. Transfection with pEGFP-NleC inhibits luciferase activity and IL-8 secretion. Data shown are mean (±S.D.) of three experiments done in triplicate with level of significance (Student's t test) indicated. **, p ≤ 0.01; ***, p ≤ 0.005 as compared with empty vector control.
NleC Inhibits NFκB Function at Level of Its Constituent Proteins
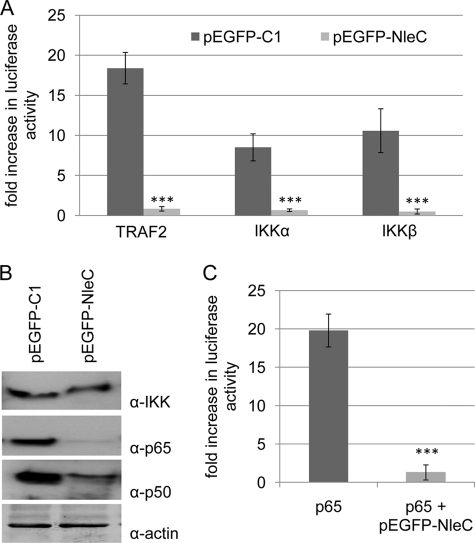
Ectopic expression of NleC inhibits basal NFκB luciferase activity by 75–80% compared with ∼60% for the NFκB activation inhibitor (Fig. 1B versus 1A), thereby suggesting that NleC inhibits signaling through multiple pathways. As signaling to NFκB converges at the level of IKK complex activation, it was predicted that NleC targets proteins at or below the IKK complex. To examine this, HeLa cells were co-transfected with vectors encoding (i) IKK complex components, (ii) the luciferase reporter protein, and (iii) eGFP or eGFP-NleC. Examination of reporter-gene expression (Fig. 2A) revealed that luciferase activity driven by plasmid expression of IKKα or IKKβ was dramatically reduced (∼95%) in pEGFP-NleC, compared with pEGFP transfected cells (Fig. 2A). Consistent with this finding was a similar NleC-specific decrease in luciferase activity driven by plasmid expression of an upstream NFκB pathway component; the TNF receptor-associated factor 2 (TRAF2; Fig. 2A). Thus, the data support the contention that NleC interferes with NFκB function by targeting the IKKα/β complex, or downstream components such as IκB or NFκB itself. Western blot analysis failed to detect NleC-mediated changes in the cellular level or phosphorylation status of IKK (Fig. 2B and data not shown). By contrast, the analysis revealed a dramatic decrease in the cellular levels of the NFκB components, p65 and p50, relative to the actin loading control (Fig. 2B), thereby suggesting that NleC targets the transcription factor for degradation. In support of this premise, luciferase activity induced by plasmid expression of p65 was, in essence, abolished in pEGFP-NleC-transfected cells (Fig. 2C).
FIGURE 2.
NleC inhibits NFκB function by targeting is constituents. Fold increase in NFκB-dependent luciferase activity of Hela cells co-transfected with vectors encoding pEGFP or pEGFP-NleC and NFκB pathway components TRAF2, IKKα, or IKKβ (A) and p65 (C). Transfection with pEGFP-NleC inhibits luciferase activity associated with plasmid expression of all interrogated IKK pathway components. Data shown are mean (±S.D.) of three experiments done in triplicate with level of significance (Student's t test) indicated. ***, p ≤ 0.005 as compared with empty vector control. B, representative immunoblot probing for actin, IKK (α and β), p65, and p50 demonstrates similar levels of IKK and loading control, actin, with decreased cellular levels of p65 and p50 in pEGFP-NleC-transfected cells.
Critical Role for NleC Residues 237–266 in Cellular Loss of p65 and p50
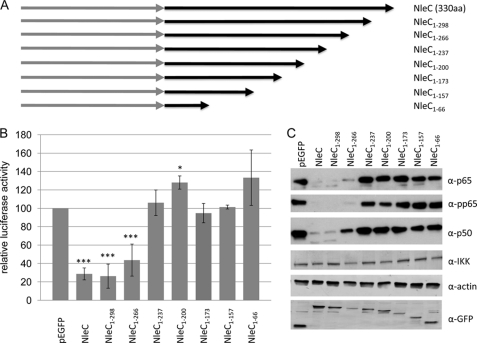
Bioinformatic analyses of the 330-amino acid residue NleC protein failed to provide clues on how it might induce the cellular loss of NFκB components. Hence, a series of C-terminal truncations were generated in an attempt to define regions critical for the process. Fig. 3A provides a schematic of the variants used in co-transfection studies with the NFκB luciferase reporter construct. These studies confirmed the potent inhibitory activity of full-length NleC, whereas similar findings with the NleC1–298 and NleC1–266 variants revealed that the C-terminal 64 residues are dispensable (Fig. 3B). However, removing another 29 (NleC1–237 variant) or more residues (Fig. 3A) abolished the inhibitory activity of NleC (Fig. 3B). Western blot analysis confirmed the expression of each variant (Fig. 3C) and also unlinked inhibitory defects from reduced protein levels, as one of the poorest expressed proteins (pEGFP-NleC1–266) inhibited luciferase activity nearly as effectively as NleC (Fig. 3, B and C). This suggests that feature(s) between residues 237 and 266 are required, directly or indirectly, for the inhibitory process. However, the C-terminal NleC domain is not sufficient for the process as an additional variant (eGFP-NleC200–330) did not reduce luciferase reporter activity (data not shown). Probing cellular extracts for p50, p65, and phosphorylated p65 correlated the failure to inhibit luciferase activity with a defect in inducing the cellular loss of these NFκB components, whereas the levels of IKK and actin control proteins remain unaffected (Fig. 3C). The partial inhibitory defect of the pEGFP-NleC1–266 variant may relate to reduced expression levels or a contributing role of amino acid residues 267 to 298 in the inhibitory process. This work implies that feature(s) located between residues 237 and 266 play a critical, direct or indirect, role in enabling NleC to induce the targeted loss of NFκB complex components from the host cell.
FIGURE 3.
Key role for NleC residues 237–266 in inducing cellular loss of p65 and p50. A, schematic of NleC C-terminal truncation variants constructed and screened in the NFκB luciferase reporter assay. B, relative luciferase activity of cells expressing NleC, and variants thereof, relative to pEGFP-transfected cells. NleC1–266, but not NleC1–237, inhibits NFκB luciferase reporter activity revealing a critical role for residues between 237–266. Data shown are mean (±S.D.) of three experiments done in triplicate with level of significance (Student's t test) indicated. *, p ≤ 0.05; ***, p ≤ 0.005 as compared with empty vector control. C, representative immunoblot probed for p65 (and phosphorylated activation-associated form; pp65), p50, IKK (α and β), actin, and GFP that links loss of NFκB luciferase reporter activity with an inability to deplete p65 and p50 from HeLa cells. aa, amino acids.
NleC Induces Proteasome-independent Degradation of p65 and p50
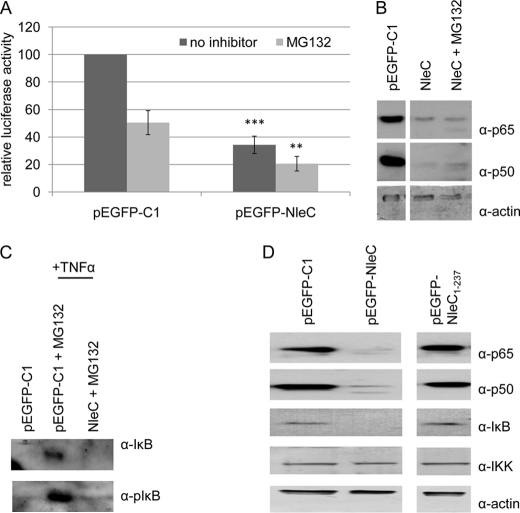
Several mechanisms have been described for regulating NFκB function, including proteasome-dependent degradation and p65 processing by caspase and serine proteases (7–9). To examine whether NleC is exploiting or mimicking these processes, Western blot analyses were carried out with antibodies against N- or C-terminal p65 domains using extracts from cells treated, or not, with the MG132 proteasome inhibitor. This drug reduced basal NFκB activity of control and pEGFP-NleC transfected cells by ∼50% (Fig. 4A) suggesting that the NleC inhibitory process occurs independently of the proteasomal degradation system. Moreover, the decrease in luciferase activity implies that proteasomal degradation antagonizes the NleC inhibitory process. Western blot analysis confirmed that MG132 treatment failed to prevent NleC-mediated cellular loss of p65 or p50 (Fig. 4B) and verified the ability of the drug to inhibit IκB degradation as it allowed phospho-IκB to accumulate within TNFα-treated control cells (Fig. 4C). Interestingly, probing for IκB in extracts from non-stimulated HeLa cells revealed its cellular loss, unlike upstream IKK, from cells expressing eGFP-NleC, but not eGFP or the NleC1–237 variant that does not induce cellular loss of p50 or p65 (Fig. 4D). Thus, NleC appears to induce the proteasome-independent loss of p50, p65, and IκBα. Furthermore, the inhibitory process appears to be independent of known p65 processing events, as probing cellular extracts with antibodies that specifically recognize the N- or C-terminal domains failed to identify p65 processed forms (Fig. 4B and data not shown). Collectively, these data suggest that NleC targets p65, p50, and IκBα for proteasome-independent degradation.
FIGURE 4.
NleC proteasomal-independent degradation of p65, p50, and IκBα. A, relative amount of NFκB luciferase reporter activity in pEGFP and pEGFP-NleC transfected cells treated, or not, with the proteasomal inhibitor MG132 revealing that the drug reduces basal NFκB activity and increases the inhibitory activity of NleC. Data shown are mean (±S.D.) of three experiments done in triplicate with level of significance (Student's t test) indicated. **, p ≤ 0.01; ***, p ≤ 0.005 as compared with pEGFP-transfected cells. B–D, representative immunoblots probing for p65, p50, IκB, pIκB (phosphorylated form targeted for proteasomal degradation), actin, and IKK(α and β) proteins in cellular extracts from cells transfected with pEGFP, pEGFP-NleC, or pEGFP-NleC1–237 incubated, or not, with MG132. Data verify the inhibitory activity of MG132 on TNFα-treated cells (prevents proteasomal-dependent degradation of pIκB; see C) and ability of eGFP-NleC, but not eGFP or eGFP-NleC1–237 proteins, to deplete p50, p65, and/or IκBα from untreated and MG132-treated cells.
N-terminal NleC Features Are Required to Recruit NFκB
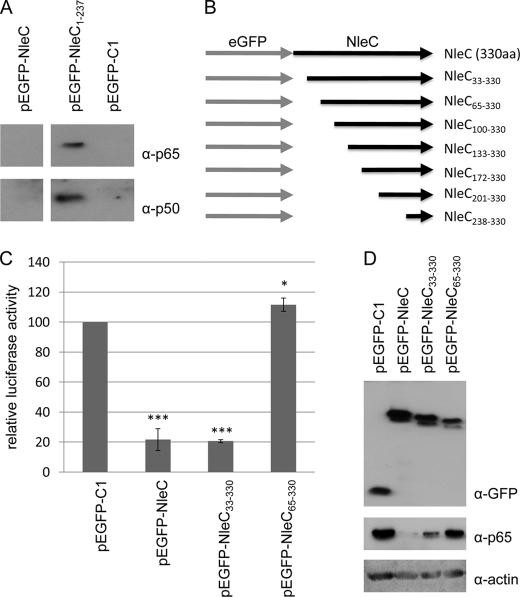
To test whether NleC interacts with NFκB, GFP-Trap beads were used to isolate eGFP, eGFP-NleC, and the eGFP-NleC1–237 fusion protein that does not induce NFκB degradation. Probing immunoprecipitates revealed that p65 and p50 interact, directly or indirectly, with NleC as they can be isolated with eGFP-NleC1–237 but not eGFP (Fig. 5A). The absence of p65 or p50 from eGFP-NleC co-immunoprecipitates (Fig. 5A) presumably reflects their rapid degradation. Thus, features located between NleC residues 1 and 237 are involved in recruiting NFκB components for degradation. To define domain(s) required in this interaction process, a series of N-terminal truncations was generated and screened in the luciferase reporter assay. Fig. 5B provides an illustration of the generated variants, with screening data revealing a dispensable role for the first 32 residues (Fig. 5C). By contrast, features located between residues 33 and 64 (NleC65–330) are critical for the inhibitory process (Fig. 5C) and, thus, all remaining variants failed to inhibit luciferase activity (data not shown). Western blot analyses confirmed variant expression, with the ability of eGFP-NleC33–330 but not eGFP-NleC65–330 to induce the cellular loss of a NFκB component illustrated (Fig. 5D and data not shown). Importantly, p65 and p50 could be co-immunoprecipitate with the eGFP-NleC1–66 but not eGFP-NleC65–330 variant (see Fig. 6C) revealing that N-terminal features (apparently located between residues 33–64) play a critical and sufficient role in recruiting NFκB.
FIGURE 5.
Interaction of p65 and p50 with eGFP-NleC1–237 and critical role for NleC residues 33–64. A, representative immunoblot of anti-GFP immunoprecipitate isolated from cells transfected with pEGFP, pEGFP-NleC, or pEGFP-NleC1–237 probed for p65 and p50. The data reveals that p65 and p50 can be isolated with eGFP-NleC1–237 but not eGFP or eGFP-NleC, the latter presumably due to rapid p65/p50 degradation. B, schematic of NleC N-terminal truncation variants constructed and screened in NFκB luciferase reporter assay. C, relative luciferase activity of cells expressing NleC and indicated variants, compared with control pEGFP-transfected cells. Data shown are mean (±S.D.) of three experiments done in triplicate with level of significance (Student's t test) indicated. *, p ≤ 0.05; ***, p ≤ 0.005 as compared with empty vector control. NleC33–330 but not NleC65–330 inhibits NFκB luciferase reporter activity revealing a critical role for residues between 33 and 64. D, representative immunoblot probed for GFP, p65, and actin that links loss of NFκB luciferase reporter activity of the NleC65–330 variants with a major defect in depleting p65 from the cell.
FIGURE 6.
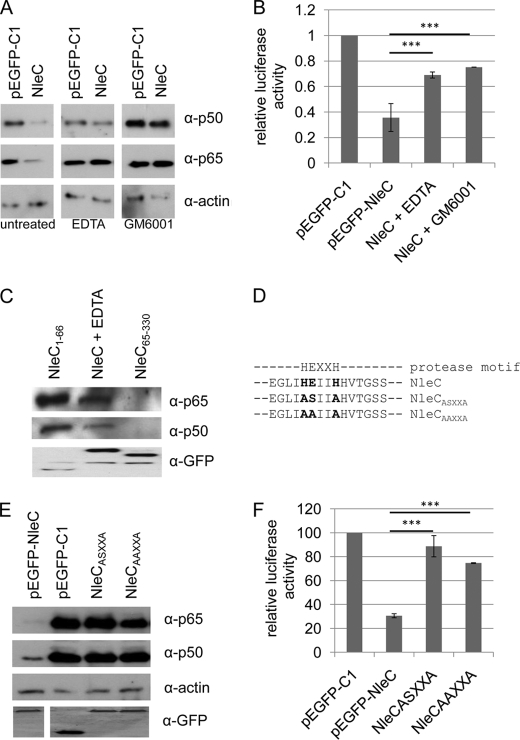
Metalloprotease inhibitors and disruption of the consensus zinc metalloprotease motif of NleC abolish p65 and p50 degradation. A, representative immunoblot of cellular extracts from pEGFP, pEGFP-NleC, and pEGFP-NleC transfected treated, or not, with metalloprotease inhibitors (EDTA or GM6001). B, relative amount of NFκB luciferase reporter activity for cells described in A. C, representative immunoblot demonstrating co-immunoprecipitating of p65 and p50 with eGFP-NleC1–66, but not eGFP-NleC65–330, with EDTA-mediated inhibition of protease activity revealing that full length NleC interacts with p65 and p50. The consensus zinc metalloprotease motif of NleC and destructive substitutions (D) with immunoblot (E) and NFκB luciferase reporter activity studies revealing the key role for this feature in the cellular loss of p50/p65 and NFκB activity (F), respectively, are shown. Luciferase data are mean (±S.D.) of three experiments done in triplicate with level of significance (Student's t test) indicated. ***, p ≤ 0.005 as compared with empty vector control.
NleC Acts as a Zinc Metalloprotease
To provide insight into the degradation process a variety of nonspecific and specific inhibitors (including those of calpain, caspase, and autophagy proteases) were screened for their ability to prevent NleC-mediated cellular loss of p65. Importantly, only one reagent, a commercial inhibitor mixture (Sigma; product no. P8340) led to the detection of NleC and p65 in the same cellular extract and hence prevented degradation (data not shown). Interestingly, this inhibition was replicated by EDTA as illustrated in NFκB reporter luciferase assay (Fig. 6B) and Western blot analyses (Fig. 6A), thereby indicating a key role for a metalloprotease. Indeed, p65 and p50 could be co-immunoprecipitated with NleC if the cells were pretreated with EDTA (Fig. 6C), indicating an importance for metal ions in the degradation process. The involvement of a metalloprotease was supported by obtaining similar results with a metalloprotease-specific inhibitor, GM6001 (Fig. 6, A and B). Although bioinformatics failed to reveal NleC homology to proteases, examination of the protein sequence identified a consensus zinc metalloprotease HEXXH motif (29, 30) spanning residues 183–187 (Fig. 6D). To directly address the idea that NleC acts as a metalloprotease that targets NFκB, the consensus motif was destroyed (Fig. 6D). NFκB luciferase reporter activity and p65/p50 Western detection assays revealed highly similar findings to those obtained with studies using protease inhibitors (Fig. 6E versus Fig. 6A and Fig. 6B versus Fig. 6F). Collectively, the work implies that NleC inhibits NFκB pathway signaling by specifically recruiting p65/p50 transcription factor complexes for degradation by its function as a metalloprotease.
DISCUSSION
Here, it is demonstrated that the nleC gene of enteropathogenic E. coli encodes a protein whose expression as an eGFP-fusion protein within HeLa cells potently inhibits the basal- and TNFα-stimulated activity of the NFκB transcription factor. This finding has applications for understanding and controlling the function of this critical component of mammalian inflammatory, immune modulation, and anti-apoptotic responses as its dysfunction is linked to inflammatory diseases and some cancers (10, 11). Although a variety of proteins delivered into host cells by bacterial pathogens have been described to inhibit NFκB activity (12, 13), this study reveals that NleC represents a novel mechanism. Thus, ectopic expression of NleC is shown to lead to a dramatic decrease in the cellular levels of both p65 (RelA) and p50 that comprise the most abundant form of the dimeric transcription factor in the canonical NFκB signaling pathway (3). Although most pathogen-encoded proteins target upstream kinases, ubiquitinases, and adaptor molecules (12, 13), some target NFκB components as illustrated by chlamydia infection leading to p65 processing, whereas another EPEC effector, NleH, binds RPS3 to inhibit transcription of genes under the control of a NFκB-RPS3 complex (21, 31). By contrast, ectopic NleC expression induced the rapid proteasomal-independent loss of p65 without evidence of processing intermediates. Interestingly, the proteasomal inhibitor augmented NleC-mediated inhibition of NFκB activity to suggest that the effector may be subjected to proteasomal degradation. Importantly, NleC expression also induced the cellular loss of p50 and IκBα, but not the upstream pathway components IKKα or IKKβ, thereby demonstrating degradation specificity. IκB acts to retain NFκB within the cytoplasm until IKKα/β-induced phosphorylation triggers its proteasomal-dependent degradation and release of NFκB for nuclear import (2). This suggests that NleC targets NFκB-IκBα complexes. Proteasome-independent, not dependent, degradation of NFκB components represents a novel inhibitory mechanism.
Fluorescent microscopy studies revealed a pool of eGFP-NleC within the nucleus, unlike the similarly sized eGFP-NleH protein that accumulates at the cell periphery (21) (data not shown). As NleC is significantly larger than the cut-off size for free nuclear entry (<50 kDa), this suggests that it may carry a novel nuclear translocation signal (not evident by bioinformatic analysis; data not shown) or enters by associating with nuclear-targeted protein(s). Of interest, the EPEC EspF effector protein enters the nucleus by a process dependent on a domain with no recognizable signal sequence (32). Given that NleC is normally delivered into the cytoplasm and degrades p65, p50, and IκBα, it is likely that its target relates to the cytoplasmic NFκB-IκBα complex. Given the multifunctional nature of EPEC effectors (26), it is possible that nuclear import represents a distinct function or perhaps relates to targeting NFκB-IκBα complexes being shuttled to the cytoplasm. Time course studies (epifluorescent microscopy and Western blot analysis) have failed to resolve whether NleC has a preference for cytoplasmic or nuclear pools of NFκB (data not shown). The mechanism and role of nuclear NleC import in the NFκB inhibitory process deserves further investigation.
Interestingly, ectopic NleC expression inhibited the basal NFκB activity in immortalized HeLa cells to a greater extent than the NFκB activation inhibitor from Calbiochem (product no. 481406). This suggests that this basal activity involves additional non-canonical pathways, of which some may be sensitive to NleC. It is likely that such pathways relate to other NFκB subunits composed of RelB, c-Rel, and/or p52 proteins. Moreover, as NleC expression depletes cellular p50, this implies that it would also alter the expression of genes regulated by p50 homodimers (33). Studies are underway to define whether NleC degrades specific isoforms of NFκB (p65, RelB, c-Rel, p105/p50, p100/p52), IκB (e.g. IκBβ, IκBγ, IκBϵ, Bcl3), NFκB-associated cofactors (e.g. RPS3, CBP/p300) and/or co-functional transcription factors (e.g. AP-1, STAT).
The idea, that NleC employs a novel mechanism for inhibiting NFκB function was evidenced by the finding that loss of transcription activity not only involves proteasome-independent degradation of p65, p50, and IkBα in the absence of p65 processing intermediates but was linked to NleC being a metalloprotease. The metalloprotease premise was supported by multiple lines of evidence. First, the NFκB inhibitory activity of NleC was dramatically reduced by pretreating cells with metalloprotease inhibitors (EDTA or GM6001) unlike chemicals that interfere with caspase, calpain, or autophagosome-related proteases. Moreover, NleC carries a consensus zinc metalloprotease motif spanning residues 183–187 whose disruption had a similar affect as adding metalloprotease inhibitors. A minor inhibitory activity for metalloprotease-treated NleC and the NleC metalloprotease motif-disrupted variant probably relates to the recruitment of NFκB, directly or indirectly, interfering with its transcription activity. Indeed, p65 and p50 could only be isolated with full-length NleC when it lacked its protease function. Importantly, residues within the first 66 residues of NleC were shown to be sufficient and essential to recruit p65 and p50. Interestingly, the first 32 residues were dispensable for the inhibitory process thereby suggesting that features residing between residues 33–66, directly or indirectly, recruit canonical NFκB components. Further studies are necessary to define the residues and mechanism (direct or indirect) by which p65, p50 and IκBα are recruited to NleC for degradation. The requirement for residues 237–266 (upstream of the zinc metalloprotease motif) but not residues 267–330 in the inhibitory process presumably reflects an indirect deleterious consequence on a NleC protein structure needed for its protease activity. Collectively, the data is consistent with a model (see Fig. 7) whereby NleC, directly or indirectly, interacts with NFκB-IκBα complexes via features residing between residues 33 and 64 for degradation though its function as a zinc metalloprotease.
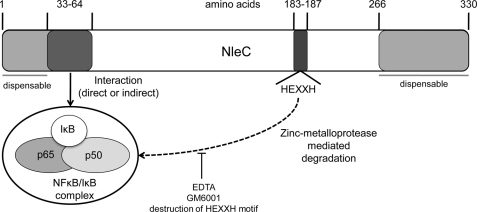
FIGURE 7.
Model for NleC inhibition of NFκB activity. NleC residues 1–66 are sufficient to, directly or indirectly, recruit p50 and p65 to degrade these proteins as well as upstream IκB, but not IKK or TRAF2, proteins by a process inhibited by adding metalloprotease inhibitors (EDTA and GM6001) or disrupting the consensus zinc metalloprotease motif (HEXXH) encompassing residues 183–187.
Little is known about the role of NleC in the context of pathogenesis, as deleting the nleC gene does not impact on virulence, at least in interrogated strains and models (34, 35). Studies using a disease-relevant small intestinal model suggest that NFκB cellular levels remain relatively unchanged following EPEC infections where NFκB function has been inhibited (25). Recent work suggests that such inhibition is due to the activity of the NleB and NleE effectors predicted to act at the level of IKK or the upstream kinase TGF-β-activated kinase 1 (19, 20). Nevertheless, these studies are consistent with contributions for other effectors, a premise illustrated by the documented role for NleH (21). Although EPEC can deliver plasmid-expressed NleC into host cells, an interrogated EPEC strain displayed little evidence of native nleC gene transcription or NleC production under the examined conditions (36). Preliminary studies support EPEC delivery of plasmid-expressed NleC in host cells where it decreases cellular levels of p65 and p50 (data not shown). Why EPEC have evolved mechanisms to limit the level of NleC within host cells and/or the ability of the effectors to degrade NFκB remains to be defined. Despite the apparent subtle role for NleC in inhibiting NFκB function within the context of an infection, the protein has a potent NFκB-specific degradation activity that may be useful for inhibiting the function of this transcriptional factor whose dysfunction is linked to inflammatory diseases and some cancers (10, 11). Thus, the novel NFκB-specific protease property of NleC has interesting applications. Firstly, as proteins (such as the >120-kDa functionally active β-galactosidase) can be delivered into cells in culture and whole animal models (37), NleC may be useful not only as a NFκB research tool but also in validating NFκB as a therapeutic target in disease samples or, indeed, as a therapeutic reagent.
Acknowledgments
We thank professors Luke O'Neill (Trinity College, Dublin, Ireland), Neil D. Perkins (Newcastle University, Newcastle, UK), Jurg Tschopp (University of Lausanne, Lausanne, Switzerland), and Martin E. Dorf (Harvard Medical School, Boston, MA) for kindly providing mammalian expression vectors encoding IKK pathway components and professor Drew Rowan for supplying GM6001. We also thank professors Neil D. Perkins and Derek Mann (Newcastle University) for advice, comments, and feedback on the manuscript.
This work was supported in part by Wellcome Trust Grant 083313 (to B. K.).
- NFκB
- nuclear factor κB
- IκB
- inhibitor of NFκB
- IKK
- IκB kinase
- eGFP
- enhanced GFP
- EPEC
- enteropathogenic E. coli.
REFERENCES
- 1. O'Dea E., Hoffmann A. (2009) Wiley Interdiscip Rev. Syst. Biol. Med. 1, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vallabhapurapu S., Karin M. (2009) Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 3. Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 4. Hiscott J., Nguyen T. L., Arguello M., Nakhaei P., Paz S. (2006) Oncogene 25, 6844–6867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chiao P. J., Miyamoto S., Verma I. M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Finco T. S., Baldwin A. S. (1995) Immunity 3, 263–272 [DOI] [PubMed] [Google Scholar]
- 7. Saccani S., Marazzi I., Beg A. A., Natoli G. (2004) J. Exp. Med. 200, 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ravi R., Bedi A., Fuchs E. J., Bedi A. (1998) Cancer Res. 58, 882–886 [PubMed] [Google Scholar]
- 9. Franzoso G., Biswas P., Poli G., Carlson L. M., Brown K. D., Tomita-Yamaguchi M., Fauci A. S., Siebenlist U. K. (1994) J. Exp. Med. 180, 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sethi G., Sung B., Aggarwal B. B. (2008) Exp. Biol. Med. 233, 21–31 [DOI] [PubMed] [Google Scholar]
- 11. Kumar A., Takada Y., Boriek A. M., Aggarwal B. B. (2004) J. Mol. Med. 82, 434–448 [DOI] [PubMed] [Google Scholar]
- 12. Sansonetti P. J., Di Santo J. P. (2007) Immunity 26, 149–161 [DOI] [PubMed] [Google Scholar]
- 13. Bhavsar A. P., Guttman J. A., Finlay B. B. (2007) Nature 449, 827–834 [DOI] [PubMed] [Google Scholar]
- 14. Schröder M., Bowie A. G. (2007) Biochem. Soc. Trans. 35, 1512–1514 [DOI] [PubMed] [Google Scholar]
- 15. Bowie A., Kiss-Toth E., Symons J. A., Smith G. L., Dower S. K., O'Neill L. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10162–10167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruckdeschel K., Mannel O., Richter K., Jacobi C. A., Trülzsch K., Rouot B., Heesemann J. (2001) J. Immunol. 166, 1823–1831 [DOI] [PubMed] [Google Scholar]
- 17. Mittal R., Peak-Chew S. Y., McMahon H. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18574–18579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim D. W., Lenzen G., Page A. L., Legrain P., Sansonetti P. J., Parsot C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newton H. J., Pearson J. S., Badea L., Kelly M., Lucas M., Holloway G., Wagstaff K. M., Dunstone M. A., Sloan J., Whisstock J. C., Kaper J. B., Robins-Browne R. M., Jans D. A., Frankel G., Phillips A. D., Coulson B. S., Hartland E. L. (2010) PLoS Pathog. 6, e1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nadler C., Baruch K., Kobi S., Mills E., Haviv G., Farago M., Alkalay I., Bartfeld S., Meyer T. F., Ben-Neriah Y., Rosenshine I. (2010) PLoS Pathog. 6, e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao X., Wan F., Mateo K., Callegari E., Wang D., Deng W., Puente J., Li F., Chaussee M. S., Finlay B. B., Lenardo M. J., Hardwidge P. R. (2009) PLoS Pathog. 5, e1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Radons J., Gabler S., Wesche H., Korherr C., Hofmeister R., Falk W. (2002) J. Biol. Chem. 277, 16456–16463 [DOI] [PubMed] [Google Scholar]
- 23. Rothe M., Sarma V., Dixit V. M., Goeddel D. V. (1995) Science 269, 1424–1427 [DOI] [PubMed] [Google Scholar]
- 24. Ling L., Cao Z., Goeddel D. V. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3792–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruchaud-Sparagano M. H., Maresca M., Kenny B. (2007) Cell Microbiol. 9, 1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dean P., Kenny B. (2009) Curr. Opin. Microbiol. 12, 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iguchi A., Thomson N. R., Ogura Y., Saunders D., Ooka T., Henderson I. R., Harris D., Asadulghani M., Kurokawa K., Dean P., Kenny B., Quail M. A., Thurston S., Dougan G., Hayashi T., Parkhill J., Frankel G. (2009) J. Bacteriol. 191, 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kunsch C., Lang R. K., Rosen C. A., Shannon M. F. (1994) J. Immunol. 153, 153–164 [PubMed] [Google Scholar]
- 29. Jongeneel C. V., Bouvier J., Bairoch A. (1989) FEBS Lett. 242, 211–214 [DOI] [PubMed] [Google Scholar]
- 30. Hooper N. M. (1994) FEBS Lett. 354, 1–6 [DOI] [PubMed] [Google Scholar]
- 31. Lad S. P., Li J., da Silva Correia J., Pan Q., Gadwal S., Ulevitch R. J., Li E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2933–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dean P., Scott J. A., Knox A. A., Quitard S., Watkins N. J., Kenny B. (2010) PLoS Pathog. 6, e1000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmitz M. L., Baeuerle P. A. (1991) EMBO J. 10, 3805–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marchés O., Wiles S., Dziva F., La Ragione R. M., Schüller S., Best A., Phillips A. D., Hartland E. L., Woodward M. J., Stevens M. P., Frankel G. (2005) Infect Immun. 73, 8411–8417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kelly M., Hart E., Mundy R., Marchès O., Wiles S., Badea L., Luck S., Tauschek M., Frankel G., Robins-Browne R. M., Hartland E. L. (2006) Infect Immun. 74, 2328–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roe A. J., Tysall L., Dransfield T., Wang D., Fraser-Pitt D., Mahajan A., Constandinou C., Inglis N., Downing A., Talbot R., Smith D. G., Gally D. L. (2007) Microbiology 153, 1350–1360 [DOI] [PubMed] [Google Scholar]
- 37. Orange J. S., May M. J. (2008) Cell Mol. Life Sci. 65, 3564–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]