Abstract
Eukaryotic cell cycle progression is mediated by phosphorylation of protein substrates by cyclin-dependent kinases (CDKs). A critical substrate of CDKs is the product of the retinoblastoma tumor suppressor gene, pRb, which inhibits G1-S phase cell cycle progression by binding and repressing E2F transcription factors. CDK-mediated phosphorylation of pRb alleviates this inhibitory effect to promote G1-S phase cell cycle progression. pRb represses transcription by binding to the E2F transactivation domain and recruiting the mSin3·histone deacetylase (HDAC) transcriptional repressor complex via the retinoblastoma-binding protein 1 (RBP1). RBP1 binds to the pocket region of pRb via an LXCXE motif and to the SAP30 subunit of the mSin3·HDAC complex and, thus, acts as a bridging protein in this multisubunit complex. In the present study we identified RBP1 as a novel CDK substrate. RBP1 is phosphorylated by CDK2 on serines 864 and 1007, which are N- and C-terminal to the LXCXE motif, respectively. CDK2-mediated phosphorylation of RBP1 or pRb destabilizes their interaction in vitro, with concurrent phosphorylation of both proteins leading to their dissociation. Consistent with these findings, RBP1 phosphorylation is increased during progression from G1 into S-phase, with a concurrent decrease in its association with pRb in MCF-7 breast cancer cells. These studies provide new mechanistic insights into CDK-mediated regulation of the pRb tumor suppressor during cell cycle progression, demonstrating that CDK-mediated phosphorylation of both RBP1 and pRb induces their dissociation to mediate release of the mSin3·HDAC transcriptional repressor complex from pRb to alleviate transcriptional repression of E2F.
Keywords: Cell Cycle, E2F Transcription Factor, Phosphorylation Enzymes, Retinoblastoma (Rb), Transcription Regulation, Retinoblastoma-binding Protein 1 (RBP1)
Introduction
The eukaryotic cell cycle is an evolutionarily conserved process that regulates cell division from unicellular organisms such as yeast through to humans. In response to developmental cues, appropriate growth conditions, and stimulation by mitogenic growth factors, cell division is promoted by activation of the key enzymes responsible for promoting cell cycle progression, the cyclin-dependent kinases (CDKs).4 These dimeric enzymes consist of a cyclin regulatory subunit, which binds and activates a CDK protein kinase subunit (1). In mammalian cells, progression through the different cell cycle phases is mediated by different CDKs. Therefore, cyclin D/CDK4/6 controls progression through G1 phase (2, 3) followed by cyclin E/CDK2 kinase activity promoting G1-S phase progression (4). Cyclin A/CDK2 is important during S phase and cyclin A/CDK1 activity peaks during G2 phase (5). Finally, cyclin B/CDK1 activity is required for mitosis (6, 7). CDKs mediate cell cycle progression through phosphorylation of protein substrates to alter their biological function(s). Different CDKs phosphorylate distinct substrates to promote progression through different cell cycle phases due to their differential temporal activities, substrate specificities, and subcellular localization, although some substrates are phosphorylated by different CDKs whose activities temporally overlap (8–10).
One important CDK substrate is the retinoblastoma gene product, pRb, a tumor suppressor protein that controls G1-S phase cell cycle progression in mammalian cells (11, 12). pRb inhibits cell cycle progression largely through binding and inhibition of the E2F family of transcription factors, whose activity is required for the transcription of genes necessary for S phase progression (13). pRb inhibits E2F transcriptional activity by two mechanisms, including binding and inhibition of the E2F transactivation domain (14), and by recruiting members of the histone deacetylase (HDAC) family to E2F promoters (15, 16). HDAC-mediated deacetylation of the core histones at E2F promoters leads to a tighter association between core histones and DNA, impairing access of transcriptional co-activators leading to transcriptional inhibition (17).
pRb contains several domains, including a so-called pocket region as well as N- and C-terminal regions (18–20). The pocket of pRb is ∼45 kDa, comprising of two domains, A and B, which are separated by a spacer region. The pRb pocket domain plays an important role in E2F transcriptional inhibition, as it is responsible for recruitment of the HDAC multisubunit transcriptional repressor complex (21, 22). Although initial studies suggested that the HDAC complex binds directly to pRb (15, 16), subsequent studies demonstrated that recruitment of this complex to pRb is mediated by the retinoblastoma-binding protein 1 (RBP1) (22–28). RBP1 binds to the pRb pocket region via an LXCXE motif (23), which is present in many pRb-binding proteins (29). RBP1 also binds the Sin3-associated protein of the 30-kDa (SAP30) subunit, which in turn binds to the Sin3 scaffolding protein (26). Sin3 binds several subunits (30), including SAP18 (31), SAP45 (32), retinoblastoma-associated proteins of 46 kDa (RbAP46) and 48 kDa (RbAP48) (21, 31) as well as HDACs. RBP1, thus, acts as a bridging protein to recruit the mSin3·HDAC complex to pRb to repress E2F-mediated transcription (22, 24, 27, 28).
During G1-S phase cell cycle progression the cumulative phosphorylation of pRb by cyclin D/CDK4/6 and cyclin E/A/CDK2 on different sites is necessary to achieve full inactivation of its repressor function and activation of E2F-mediated transcription (13, 18, 20, 33–35). Phosphorylation on several pRb sites leads to disruption of E2F and HDAC binding to abolish pRb-mediated inhibition of E2F transcription (13, 18, 20, 36).
Previous work in this laboratory has utilized an in vitro phosphorylation screen of recombinant proteins expressed from λ-phage cDNA expression library to identify the ubiquitin conjugating enzyme hHR6A as a novel CDK substrate (37). The utility of this method for identifying novel substrates of CDKs and other protein kinases has also been validated by other studies (37–39). In addition to hHR6A, we isolated a protein termed Sin3-associated protein of 180 kDa (SAP180) as a putative CDK substrate in this screen. SAP180 belongs to the AT-rich interaction domain 4 (ARID4) family of DNA-binding proteins (25, 40), which binds to the mSin3 transcriptional repressor complex (32) and is referred to as ARID4B. SAP180 (ARID4B) is closely related to ARID4A, which is also known as RBP1, sharing 34% identity and 50% similarity at the amino acid sequence level (27, 32). Due to the important role of RBP1 (ARID4A) in regulating E2F function through recruitment of the mSin3·HDAC complex to pRb (24, 41), we investigated if RBP1 is regulated by CDK-mediated phosphorylation.
RBP1 contains a consensus ARID DNA binding sequence of ∼100 amino acids (residues 314–409), common to other members of the ARID DNA binding family proteins (40). RBP1 also possesses a Tudor domain at the N-terminal region (residues 58–114) as well as a chromatin organization modifier (Chromo) domain (residues 593–633). Tudor domains facilitate interaction with the methylated lysine residue of histone H3 (42), whereas Chromo domains, which span between 30 and 70 amino acids, are found in proteins involved in the assembly of protein complexes on chromatin (43–45). RBP1 binds to SAP30 via repressor region 2 (amino acids 1167–1257) to recruit the mSin3·HDAC complex and induce transcriptional repression (22, 24). RBP1 also contains a transcriptional repressor region 1 (amino acids 241 and 542), which encompasses the ARID domain and induces repression independent of HDACs through an unknown mechanism (24).
At a whole animal level, RBP1-deficient mice (Arid4a−/−) are viable but display an increased mortality rate (46). RBP1-deficient mice display an age-related cytopenia and thrombocytopenia and develop myelofibrosis and hepatosplenomegaly (46). They develop a myelodysplastic/myeloproliferative disorder. Approximately 12% of RBP1-deficient mice with myelodysplastic/myeloproliferative disorder further developed acute myeloid leukemia, indicating the functional importance of RBP1 as murine leukemia suppressor (46).
In the present study we demonstrate that RBP1 is a novel CDK substrate, which is phosphorylated on serines 864 and 1007. These sites are located N and C terminus to the RBP1 LXCXE pRb binding motif (amino acids 957–961). CDK-mediated phosphorylation of RBP1 or pRb reduced their interaction, whereas phosphorylation of both proteins leads to maximal dissociation. These findings indicate that concurrent CDK-mediated phosphorylation of RBP1 and pRb underpins the mechanism for dissociation of the mSin3·HDAC transcriptional repressor complex from E2F during cell cycle progression.
EXPERIMENTAL PROCEDURES
Plasmids, Cell Lines, and Antibodies
Human RBP1 cDNA was cloned into mammalian pCMV Tag2A (Stratagene), Escherichia coli pGEX 4T-1 (GE Healthcare), and baculovirus pFastBAC Tri-EX expression vectors. The C-terminal region of RBP1 was cloned into pGEX4T-1 to generate GST-RBP1784–1257. Deletion constructs of RBP1 were cloned into the pET15b (Novagen) to generate His6-RBP1784–930 and His6-RBP1937–1073.
Human embryo kidney 293 (HEK293), HEK293 with T-large antigen (HEK293T), and human breast adenocarcinoma (MCF-7) cell lines were cultured in Dulbecco's modified eagle's medium (Sigma) supplemented with 10% fetal bovine serum (SAFC Bioscience) at 37 °C with 5% CO2. Spodoptera frugiperda (Sf9) cells were cultured in SF900 II SFM media (Invitrogen) at 27 °C.
Mouse monoclonal antibodies against FLAG epitope (M2, Sigma, F1804), Penta-His (Qiagen, 34660), and pRb (Calbiochem, OP-66) and rabbit polyclonal antibodies against human SAP30 (Upstate, 06-875), human mSin3A (AK-11) (Santa Cruz, sc-767), and goat polyclonal anti-GST antibody (Amersham Biosciences, 27-4577-01) were used according to the manufacturer's instructions.
Expression and Purification of Recombinant Proteins
Recombinant GST-RBP1, GST-RBP1784–1257, His6-RBP1784–930, His6-RBP1937–1073, His6-SAP30, and MBP-pRb279–928 were expressed in E. coli strain Rosetta BL21 (DE3) pLysS (Novagen). Cultures were grown in LB medium containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol at 37 °C to an A600 nm of ∼0.8. GST-RBP1 and GST-RBP1784–1257 protein expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at either 10 °C for 24 h or room temperature (RT) for 3 h, respectively, whereas MBP-pRb279–928 expression was induced at RT with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside and 2 g/liter (∼11 mm) glucose for 3 h. Cells were lysed in PBS supplemented with 50 μg/ml lysozyme, 1% Triton X-100, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mm PMSF, and proteins were bound to either glutathione-agarose (Sigma) or amylose resin (New England Biolabs). Purification of recombinant proteins was carried out at 4 °C. The resin was washed extensively with 5 repeats of 1 ml of buffer W (50 mm HEPES, pH 7.5, 150 mm NaCl, 1% Tween 20, 0.5 mm DTT, 2 mm EDTA, 30 mm NaF, 20 mm NaPPi, 10 mm β-glycerol phosphate, 1 mm Na3VO4, 1 mm PMSF). GST-RBP1 was eluted with 500 μl of glutathione elution buffer (100 mm HEPES, pH 7.9, 150 mm NaCl, 1 mm DTT, 10% glycerol, 20 mm l-glutathione, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm PMSF), whereas MBP-pRb279–928 was eluted with 500 μl of maltose elution buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 1 mm DTT, 100 mm maltose, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm PMSF). His6-SAP30 expression was induced at RT, whereas His6-RBP1784–930 and His6-RBP1937–1073 expression was induced at 37 °C with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h. Cells were lysed with 50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 1% Nonidet P-40, 10 mm β-mercaptoethanol, 10 mm imidazole, 50 μg/ml lysozyme, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mm PMSF. After centrifugation, the supernatant was incubated with Ni2+-nitrilotriacetic acid resin (Qiagen) at 4 °C overnight. The resin was extensively washed with lysis buffer, and His6-SAP30 was eluted with 500 μl of imidazole elution buffer (50 mm NaH2PO4 pH7.7, 300 mm NaCl, 250 mm imidazole, 10 mm β-mercaptoethanol, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm PMSF). GST-RBP1, MBP-pRb279–928, and His6-SAP30 were dialyzed for three successive rounds against 600 ml of dialysis buffer (50 mm HEPES, pH 7.5, 1 mm DTT, 0.01% Tween 20, 5% glycerol, 0.5 mm PMSF).
For expression of baculoviral GST-RBP1, Sf9 insect cells (0.5 × 106 cells/ml) were infected with recombinant baculoviruses coding for GST-RBP1 at 27 °C for 3 days and lysed with buffer W. GST-RBP1 was purified using glutathione agarose and then dialyzed as described above.
CDK-mediated Phosphorylation of RBP1 in Vitro and Phosphoamino Acid Analysis
Recombinant cyclin/CDKs were prepared as described previously (8, 37). 2 μg of GST-RBP1 purified from Sf-9 insect cells immobilized on 10 μl of glutathione-agarose was dephosphorylated by treatment with 140 units of λ-phosphatase (New England Biolabs) at 30 °C for 1 h. After 3 × 1-ml washes with buffer W, GST-RBP1 was phosphorylated by purified cyclin/CDKs in a 30-μl volume as described previously (37, 47). Alternatively, His6-RBP1784–930 and His6-RBP1937–1073 purified on Ni2+-nitrilotriacetic acid resin were phosphorylated with purified cyclin A/CDK2 as described previously (37, 47). Samples were separated by SDS-PAGE, stained with Coomassie Brilliant Blue (Bio-Rad), and visualized by autoradiography. Phosphoamino acid analysis was performed as described previously (48).
In Vivo [32P]Orthophosphate Labeling of RBP1
FLAG-RBP1 was expressed in HEK293 cells after transfection with pCMV Tag2A-RBP1 plasmid using FuGENE HD. 48 h post-transfection, cells were grown for 4 h in medium containing 0.5 mCi/ml [32P]orthophosphate (MP Biomedicals) in the absence or presence of the CDK1/CDK2 inhibitor Roscovitine (50 μm) (Calbiochem) (49). Cells were lysed with 600 μl of ice-cold buffer W, syringed with a 25-gauge needle to shear DNA, and centrifuged at 16,100 × g at 4 °C for 15 min, and FLAG-RBP1 was immunoprecipitated from the supernatant with 20 μl of packed anti-FLAG M2-agarose (Sigma). Phosphorylated FLAG-RBP1 was separated by 8% SDS-PAGE and visualized by autoradiography or transferred to nitrocellulose before detection by immunoblotting with anti-FLAG antibody.
Cell Cycle Studies in MCF-7 Breast Cancer Cells
MCF-7 cells were transfected with either pCMV Tag2A or pCMV Tag2A-RBP1 plasmid and 2 h post-transfection, and cells were incubated with 10 nm estrogen receptor antagonist ICI 182780 (ICI, Tocris Bioscience) for 24 h to induce G0/G1-phase cell cycle arrest. The MCF-7 cells were then stimulated to synchronously re-enter the cell cycle by adding 100 nm 17β-estradiol (E2, Sigma), as described previously (41, 50). To monitor cell cycle progression, cells were collected at 15, 21, 27, 33, and 39 h after treatment with 17β-estradiol, fixed with 70% (v/v) ethanol at 4 °C overnight, and stained with 5-bromo-2-deoxyuridine (BrdU) to determine S phase cell population. Briefly, 2 h before cell collection, cells were pulsed with 1 μg/ml BrdU (Sigma). Once fixed, cells were collected by centrifugation at 500 × g 4 °C for 5 min and resuspended in 200 μl of ice-cold 0.1% Triton-X-100, 0.1 n HCl, left on ice for 1 min, and centrifuged at 500 × g at 4 °C for 5 min. The cell pellet was washed once with 1 ml of DNA denaturation buffer (150 μm NaCl, 15 μm trisodium citrate dihydrate) at 500 × g at RT for 5 min, resuspended in 200 μl of DNA denaturation buffer, and incubated at 90 °C for 5 min. Cells were left on ice to chill for 5 min, then washed once with 1 ml of freshly prepared antibody diluting buffer (PBS, 0.1% Triton X-100, 1% (w/v) BSA) and collected by centrifugation at 3000 × g RT for 10 min. Cells were stained with 200 ng of FITC-conjugated anti-BrdU antibody (diluted to 100 μl with antibody diluting buffer) at RT for 30min in the absence of light and analyzed on FACSCalibur (BD Biosciences).
Mass Spectrometry
FLAG-RBP1 was expressed in HEK293 cells after transfection with pCMV Tag2A-RBP1 plasmid using FuGENE HD, immunoprecipitated, and separated by SDS-PAGE, and gel slices containing FLAG-RBP1 were treated with 0.5 μg of trypsin (Promega) (51). Tryptic digests were dried under vacuum, resuspended in 60% (v/v) acetonitrile, 5% (v/v) trifluoroacetic acid, and purified on TiO2 beads (GL Sciences) using micro-columns. The TiO2 beads were washed twice with 60 μl of 60% (v/v) acetonitrile, 5% (v/v) trifluoroacetic acid and eluted with 120 μl of 1.25% (w/v) NH4OH (BDH). Purified phosphopeptides were dried under vacuum, resuspended in 20 μl of 2% (v/v) formic acid (Sigma), and analyzed by LC/MS. Chromatography was performed on a Dionex Ultimate 3000 nano-LC at a flow rate of 500 nl/min. Peptides were resolved on a 20-cm × 75-μm C18 column (LC Packings Pepmap100 C18 resin) using a 3–35% acetonitrile gradient. Mass spectrometry was performed on a QSTAR-pulsar i (Applied Biosystems) using a micro-ion spray source (Applied Biosystems). Data were acquired using a data-dependent acquisition program and analyzed by MASCOT software (Matrix Science, supplied by the Australian Proteomics Computational Facility and funded by the Australian National Health and Medical Research Council Grant 381413). MS/MS spectra were manually validated to confirm the accuracy of assigned phosphorylation sites.
In Vitro RBP1 Binding Studies
10 μg of MBP-pRb279–928 was immobilized on 20 μl of packed amylose resin in the presence of 3 mg of BSA in buffer W in a total volume of 400 μl. Immobilized MBP-pRb279–928 was either unphosphorylated or phosphorylated with cyclin A/CDK2 and extensively washed with cold buffer W. Immobilized MBP-pRb279–928 was then mixed with 2.5 μg of either unphosphorylated or cyclin A/CDK2 prephosphorylated GST-RBP1 (purified from E. coli) in a total volume of 30 μl at 4 °C for 5 h. As control, ATP was omitted from the kinase reactions. To assess the effect of cyclin A/CDK2-mediated phosphorylation on the association of pre-assembled RBP1 and pRb, 20 μg of MBP-pRb279–928 was immobilized on 40 μl of packed amylose resin as described above and mixed with 5 μg of GST-RBP1 to allow binding. After extensive washing with 1 ml of buffer W, the amylose resin with MBP-pRb279–928·GST-RBP1 was divided into two equal volumes and was either phosphorylated with cyclin A/CDK2 in a total volume of 30 μl or left unphosphorylated. After washing with buffer W, proteins were separated by 9% SDS-PAGE and transferred to nitrocellulose for analysis of the level of bound GST-RBP1 by immunoblotting using anti-GST antibody.
For GST-RBP1784–1257 binding studies with SAP30, E. coli lysate expressing GST-RBP1784–1257 was mixed with 15 μl of packed glutathione-agarose. After extensive washing with buffer W, the glutathione-agarose conjugated GST-RBP1784–1257 was either left unphosphorylated or phosphorylated with cyclin A/CDK2, washed again, and then mixed with 10 μg of His6-SAP30 in a total volume of 30 μl at 4 °C for 5 h. The samples were then extensively washed with buffer W and subjected to immunoblotting using anti-penta-His antibody to detect His6-SAP30. To assess if CDK-mediated phosphorylation affects the association of pre-assembled RBP1·SAP30 complex, GST-RBP1784–1257 was immobilized on glutathione-agarose as described and then mixed with 10 μg of His6-SAP30 in a total volume of 30 μl at 4 °C for 5 h. After extensive washing with buffer W, the samples were divided into two equal volumes and were either incubated with or without cyclin A/CDK2 in a total volume of 30 μl under phosphorylation conditions. The samples were then washed extensively with buffer W and then subjected to immunoblotting with anti-penta-His antibody to detect His6-SAP30.
Analysis of Cell Cycle-dependent Association of RBP1 with pRb, SAP30, and mSin3A
MCF-7 cells, transfected with either pCMV Tag2A or pCMV Tag2A-RBP1, were arrested in G0/G1 phase and stimulated to synchronously re-enter the cell cycle as described under cell cycle studies. FLAG-RBP1 was immunoprecipitated with anti-FLAG M2-agarose from lysates prepared from MCF-7 cells arrested in G0/G1 phase or 27 h after E2 addition when cells peaked in S phase. The samples were separated by SDS-PAGE and subjected to immunoblotting with the appropriate antibodies to detect pRb, SAP30, and mSin3A. All densitometry analyses were performed using the ImageQuant TL software (GE Healthcare).
HDAC Assay
[3H]Acetylated histones were prepared as described previously (52). To measure HDAC activity, FLAG-RBP1 immunoprecipitates were incubated in 100 μl of HDAC assay buffer (100 mm HEPES pH7.5, 100 mm NaCl, 0.1 mm EDTA, 1 mm PMSF) with 1 μg of [3H]acetylated histones (∼21,400 cpm) in the presence or absence of 3 μm HDAC inhibitor trichostatin A (Cell Signaling) at 37 °C for 1 h. The reaction was stopped by adding 100 μl of 0.1 n HCl, 0.16 n CH3COOH solution and mixed with 900 μl of ethyl acetate. After centrifugation, the upper, organic layer containing [3H]acetyl was removed and measured by scintillation counting in a Tri-Carb 2900TR (PerkinElmer Life Sciences).
RESULTS
RBP1 Is a CDK Substrate, Which Is Phosphorylated in a Cell Cycle-dependent Manner
To identify novel CDK substrates, we performed phosphorylation screening of a λ-phage cDNA expression library with cyclin A/CDK2 in vitro, as described previously (37). One clone identified in this screen encoded a partial sequence corresponding to SAP180 (Sin3-associated protein of 180 kDa). As noted in the introduction, SAP180 is related to RBP1 (27, 32), which plays a critical role in pRb-mediated E2F repression by recruiting the mSin3·HDAC transcriptional repressor complex (22, 24, 28). Because CDK-mediated phosphorylation of pRb is known to inactivate its inhibitory function toward E2Fs, we investigated if CDK-mediated phosphorylation of RBP1 also contributes to the regulation of this complex.
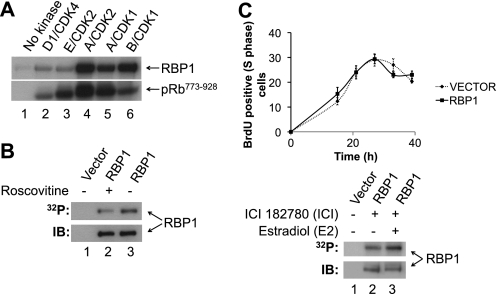
Sequence analysis revealed that RBP1 contains 16 CDK consensus phosphorylation sites, which consist of a phosphorylated serine or threonine followed by an obligatory C-terminal proline ((S/T)P) (53–55). To investigate if RBP1 is a CDK substrate, full-length purified recombinant GST-RBP1 was phosphorylated with purified recombinant cyclin/CDKs in vitro. GST-RBP1 was phosphorylated by cyclin D1/CDK4, cyclin E/CDK2, cyclin A/CDK2, cyclin A/CDK1, and cyclin B/CDK1, as was GST-pRb773–928, which was included as a positive control (Fig. 1A). We next evaluated if RBP1 is phosphorylated in vivo by expressing FLAG-RBP1 in HEK293 cells in the presence of [32P]orthophosphate. FLAG-RBP1 was readily phosphorylated in vivo (Fig. 1B). Importantly, the level of phosphorylation was reduced by ∼40% in the presence of CDK1 and CDK2 inhibitor Roscovitine (50 μm) (49), indicating that RBP1 phosphorylation is dependent on cyclin/CDK activity in vivo.
FIGURE 1.
RBP1 is phosphorylated by CDKs in vitro and in vivo in a cell cycle-dependent manner. A, recombinant GST-RBP1 or GST-pRb773–928 were incubated with the indicated cyclin/CDKs in the presence of [γ-32P]ATP, separated by SDS-PAGE, and analyzed by autoradiography. B, FLAG-RBP1 was immunoprecipitated from [32P]orthophosphate labeled cells, incubated in the presence (lane 2) or absence (lane 3) of the CDK1/2 inhibitor Roscovitine (50 μm), separated by SDS-PAGE, and subjected to autoradiography (32P) or immunoblotting (IB) with anti-FLAG antibody. Immunoprecipitation of cells transfected with empty vector was performed as control (lane 1). C, upper panel, shown is the kinetics of G1-S phase progression of MCF-7 cells transfected with empty vector (dashed black line) or expressing wild-type RBP1 (solid black line) after cells were arrested in G0/G1 phase by ICI 182780 treatment and stimulated with estradiol to synchronously re-enter cell cycle. Cells were stained with BrdU and harvested at the indicated times to determine the percentage of BrdU-positive cells. Error bars represent ± S.E of three independent experiments. Lower panel, FLAG-RBP1 expressed in [32P]orthophosphate-labeled MCF-7 cells, subjected to the same treatment as in the upper panel, was immunoprecipitated from G0/G1 phase-arrested cells (time = 0 h; lane 2, ICI) or at 27 h after estradiol addition when cells were in S phase (time = 27 h, lane 3, ICI + E2). The immunoprecipitates were separated by SDS-PAGE and subjected to autoradiography (32P) or immunoblotting with anti-FLAG antibody (IB) to determine loading. Immunoprecipitation of cells transfected with empty vector was performed as control (lane 1). This experiment is representative of two independent experiments.
To investigate if RBP1 is phosphorylated in a cell cycle-dependent manner, FLAG-RBP1 was expressed in MCF-7 cells, which were metabolically labeled with [32P]orthophosphate. MCF-7 cells are dependent on estrogen for growth and, therefore, can be arrested in G0/G1 phase of the cell cycle by the addition of the estrogen receptor antagonist ICI 182780 (ICI). The addition of excess of E2 can then be used to stimulate cells to synchronously reenter the cell cycle (41, 50). To quantify the kinetics of G0/G1-S-phase cell cycle progression by determining the proportion of cells in S phase, cells were labeled with BrdU and analyzed by flow cytometry. Treatment of cells with ICI 182780 resulted in the arrest of most MCF-7 cells in G0/G1 phase, with only 12% remaining in S phase (Fig. 1C, upper panel), compared with asynchronous cells, which contained ∼40% of cells in S-phase (data not shown). After estradiol treatment, cells progressed from G1 into S phase and peaked at ∼42% S phase 27 h after re-entry into the cell cycle, and thereafter, the proportion of cells in S phase decreased as cells progressed into G2/M phase (Fig. 1C, upper panel), similar to previous reports with this model (50). MCF-7 cells transfected with empty vector or those expressing ectopic FLAG-RBP1 displayed similar kinetics of cell cycle progression (Fig. 1C, upper panel). We repeated this experiment after labeling cells in the presence of [32P]orthophosphate and immunoprecipitated FLAG-RBP1 from cells arrested in G0/G1 phase and 27 h after estradiol addition, when cells peaked in S phase. Phosphorylation of FLAG-RBP1 increased 2-fold when cells were in S phase compared with G0/G1 phase (Fig. 1C, lower panel).
RBP1 Is Phosphorylated on CDK Consensus Site Serines 864 and 1007 in Vivo and in Vitro
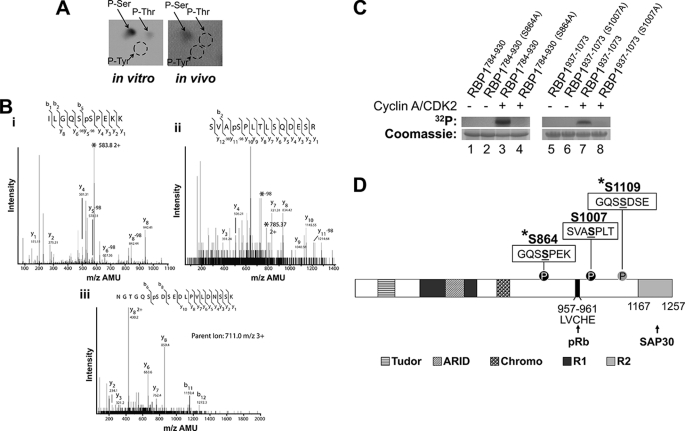
We next performed studies to identify the sites on RBP1 phosphorylated in vitro and in vivo. For in vitro phosphorylation, we utilized cyclin A/CDK2 because this cyclin/CDK robustly phosphorylated purified GST-RBP1 (Fig. 1A), and RBP1 phosphorylation increased as cells progressed through S phase (Fig. 1C), when A/CDK2 is active. Furthermore, our previous studies demonstrate that A/CDK2 phosphorylates nearly all the substrates phosphorylated by E/CDK2, which is the other major cyclin/CDK active during G1-S phase (8). Phosphoamino acid analysis revealed that RBP1 was predominantly phosphorylated on the serine residue(s) in vitro by cyclin A/CDK2, whereas 32P-labeled RBP1 purified from HEK293 cells was exclusively phosphorylated on serine residue(s) (Fig. 2A).
FIGURE 2.
RBP1 is phosphorylated on serines 864 and 1007 in vivo and in vitro by cyclin A/CDK2. A, shown is phosphoamino acid analysis of GST-RBP1 phosphorylated in vitro by cyclin A/CDK2 in the presence of [γ-32P]ATP (left panel) or immunoprecipitated FLAG-RBP1 metabolically labeled with [32P]orthophosphate in HEK293 cells in vivo (right panel). The positions of phosphoserine (p-Ser), phosphothreonine (p-Thr), and phosphotyrosine (p-Tyr) are marked by arrows. B, tandem mass spectrometry spectra of FLAG-RBP1 immunoprecipitated from HEK293 cell lysates is shown. (i) RBP1 859–867 (parent ion, 583.8 m/z 2+) shows phosphorylation at S864. (ii) RBP1 1004–1017 (parent ion, 785.37 m/z) shows phosphorylation at S1007. (iii) RBP1 1103–1122 (parent ion, 711.0 m/z 3+) shows phosphorylation at Ser-1109. AMU, atomic mass units. C, purified recombinant His6-RBP1784–930 (left panel) and His6-RBP1937–1073 (right panel) and their corresponding alanine mutants His6-RBP1784–930 (S864A) and His6-RBP1937–1073 (S1007A) were incubated in the absence (−) or presence (+) of cyclin A/CDK2 and [γ-32P]ATP. After SDS-PAGE, the proteins were visualized by autoradiography (upper panels) and Coomassie Blue (lower panels). D, shown is a schematic representation of the domain organization of RBP1 and the position of the three phosphorylation sites (underlined) identified by mass spectrometry. Ser-864 and -1007, which conform to CDK consensus phosphorylation sites, are marked by black circles, whereas a gray circle marks Ser-1109. The asterisk denotes phosphorylation sites identified on endogenous RBP1 in phosphoproteomic studies (56, 57). The LVCHE motif (amino acids 957–961) responsible for binding to the pocket of pRb is indicated. The Tudor, ARID, and Chromo domains and the repressor regions 1 and 2 of RBP1 are indicated. Repressor region 2 binds to SAP30.
To identify the site(s) on RBP1 phosphorylated in vivo, we performed tandem mass spectrometry on FLAG-RBP1 immunoprecipitated from HEK293 cells. FLAG-RBP1 was phosphorylated on three sites, at serine residues 864, 1007, and 1109. Sites 864 with the sequence GQSpSPEK and 1007 with the sequence SVApSPLT (Fig. 2B) conform to the CDK consensus phosphorylation motif ((S/T)P). Site 1109 with the sequence GQSpSDSE (Fig. 2B) does not conform to a CDK consensus phosphorylation site, suggesting that another kinase(s) is responsible for phosphorylation of this site. In agreement with our findings, phosphorylation of serines 864 and 1109 on endogenous RBP1 has previously been reported in large-scale phosphoproteomics screens (56, 57). Furthermore, RBP1 serine 864 shows increased phosphorylation during G1/S phase and decreases during mitosis, indicating that phosphorylation of this site is cell cycle-regulated (57), consistent with our findings that phosphorylation of this site is regulated by CDKs.
Mass spectrometry analysis demonstrated that GST-RBP1 serines 864 and 1007 were also phosphorylated by A/CDK2 in vitro (data not shown). In addition, cyclin A/CDK2 phosphorylated threonine 1124 and serines 1140 and 1145 in vitro (data not shown). Threonine 1124 likely accounts for the small level of phosphothreonine observed in the phosphoamino acid analysis (Fig. 2A). Some of these extra sites phosphorylated by cyclin A/CDK2 in vitro may be inaccessible for phosphorylation in vivo due to binding to proteins such as SAP30 (22, 27), which may preclude access of cyclin/CDKs. To confirm phosphorylation of serines 864 and 1007 by cyclin A/CDK2, we generated two truncated His6-RBP1 constructs, His6-RBP1784–930 and His6-RBP1937–1073, incorporating these sites and their corresponding mutants, where serines 864 and 1007 were mutated to alanine, to generate His6-RBP1784–930 (S864A) and His6-RBP1937–1073 (S1007A). Both His6-RBP1784–930 and His6-RBP1937–1073 were readily phosphorylated by A/CDK2 in vitro, whereas their respective alanine mutants, His6-RBP1784–930 (S864A) and His6-RBP1937–1073 (S1007A), showed no phosphorylation (Fig. 2C).
CDK-mediated Phosphorylation of RBP1 Does Not Affect Its Binding to SAP30 in Vitro
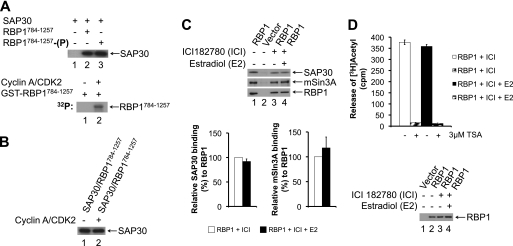
Because RBP1 recruits the mSin3·HDAC complex via the SAP30 subunit to pRb (22, 24, 26–28), we investigated if CDK-mediated phosphorylation of RBP1 might regulate its binding to SAP30. The C-terminal region of RBP1 (1167–1257) interacts with SAP30 (22, 27) (Fig. 2D). We established an in vitro binding assay with recombinant GST-RBP1784–1257 and His6-SAP30. First, GST-RBP1784–1257 was immobilized on glutathione-agarose and either left unphosphorylated or phosphorylated with cyclin A/CDK2 before incubation with His6-SAP30. The agarose beads were then extensively washed before immunoblotting to assess the level of His6-SAP30 bound. Consistent with previous studies (22, 24, 26–28), His6-SAP30 specifically interacted with GST-RBP1784–1257 (Fig. 3A, lanes 1 and 2). Cyclin A/CDK2-mediated phosphorylation of GST-RBP1784–1257 did not alter the level of His6-SAP30 binding compared with unphosphorylated GST-RBP1784–1257 (Fig. 3A, lanes 2 and 3). Similarly, when the GST-RBP1784–1257·His6-SAP30 complex was preassembled and subsequently incubated with cyclin A/CDK2, phosphorylation of GST-RBP1784–1257 did not affect the level of binding of His6-SAP30 (Fig. 3B). Therefore, CDK-mediated phosphorylation of RBP1 did not alter its affinity for SAP30 in vitro.
FIGURE 3.
CDK-mediated phosphorylation of RBP1 does not impact on its interaction with SAP30 in vitro or affect associated SAP30·mSin3A·HDAC activity in vivo. A, cyclin A/CDK2-mediated phosphorylation of GST-RBP1 does not affect binding to SAP30 in vitro. Upper panel, GST-RBP1784–1257 immobilized on glutathione-agarose was either unphosphorylated (lane 2) or phosphorylated with cyclin A/CDK2 (lane 3) and incubated with purified recombinant His6-SAP30 to assess binding, as described under “Experimental Procedures.” The level of His6-SAP30 binding was assessed by immunoblotting with anti-His antibody. As a control, His6-SAP30 was incubated with empty glutathione agarose (lane 1). Lower panel, in vitro kinase assay confirming phosphorylation of purified GST-RBP1784–1257 with cyclin A/CDK2 + [γ-32P]ATP (lane 2). B, preformed GST-RBP1784–1257·His6-SAP30 complex was incubated in the absence (lane 1) or presence of cyclin A/CDK2 (lane 2) under phosphorylation conditions and then washed to remove unbound His6-SAP30. The level of His6-SAP30 bound to GST-RBP1784–1257 was assessed by immunoblotting with anti-His antibody. C, the levels of RBP1-associated SAP30 and mSin3A do not change during G0/G1-S phase cell cycle progression. FLAG-RBP1 was immunoprecipitated from either exponentially growing (lane 1), G0/G1-arrested with ICI 182780 (lane 3, ICI), or S phase (27 h after the addition of estradiol to ICI 182780-arrested cells) (lane 4, ICI + E2) MCF-7 cells. The immunoprecipitates were assessed for the levels of FLAG-RBP1 and co-immunoprecipitating SAP30 and mSin3A by immunoblotting with the appropriate antibodies. Lane 2 represents control immunoprecipitates from cells transfected with empty vector. Lower panel, the histograms represent the relative levels of SAP30 (left) and mSin3A (right) bound to the immunoprecipitated FLAG-RBP1 from G0/G1-arrested (ICI) cells or cells in S phase (ICI + E2). Error bars represent ±S.E of three independent experiments. D, RBP1-associated HDAC activity does not change during progression from G0/G1 (ICI) into S phase (ICI + E2). FLAG-RBP1 was immunoprecipitated from MCF-7 cells arrested in G0/G1 or in S phase, as described in C. Immunoprecipitates were then incubated with [3H]acetylhistones in the absence or presence of the HDAC inhibitor trichostatin A (TSA, 3 μm) to perform HDAC assays, as described under “Experimental Procedures.” The graph represents the means ± S.E. of three independent experiments. Immunoprecipitates were subjected to immunoblotting with anti-FLAG antibody to demonstrate RBP1 loading (lower panel, lanes 2–4).
CDK-mediated Phosphorylation of RBP1 Does Not Impact on Its Interaction with or Alter SAP30·mSin3A·HDAC-associated activity in Vivo
We next assessed if the association or activity of the SAP30·mSin3A·HDAC complex is altered in immunoprecipitates of RBP1 from MCF-7 cells arrested in G0/G1 phase or those in S phase, when RBP1 phosphorylation is increased (Fig. 1C). To perform these studies, MCF-7 cells, which express wild-type pRb (58), were transiently transfected with pCMV Tag2A-RBP1 to ectopically express FLAG-RBP1. MCF-7 cells were arrested in G0/G1 phase of the cell and stimulated to synchronously re-enter the cell cycle, as described in Fig. 1C. Immunoprecipitation of FLAG-RBP1 revealed that the levels of bound SAP30 and mSin3A remained unaltered between cells in G0/G1 (ICI) or S phase (ICI + E2) (Fig. 3C, lanes 3 and 4).
It is possible that CDK-mediated phosphorylation of RBP1 may alter the activity of the SAP30·mSin3·HDAC complex through allosteric mechanisms without altering the subunit composition. To evaluate this possibility and quantitatively assess the levels of RBP1-associated SAP30·mSin3·HDAC activity, RBP1 immunoprecipitates from lysates of G0/G1 or S phase MCF-7 cells were assayed for associated HDAC activity. These results showed the same level of RBP1-associated HDAC activity from MCF-7 cells in G0/G1 or S phase of the cell cycle (Fig. 3D). Altogether, the in vitro and in vivo studies indicate that CDK-mediated phosphorylation of RBP1 does not impact on its interaction with nor alter the activity of the associated SAP30·mSin3·HDAC complex.
CDK-mediated Phosphorylation of RBP1 and pRb Disrupts Their Interaction in Vitro
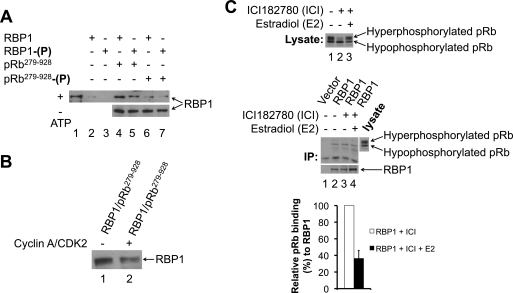
Because CDK-mediated phosphorylation of RBP1 did not alter its interaction with SAP30·mSin3·HDAC, we next tested if the interaction with pRb was affected. RBP1 binds to the pocket region of pRb via an LXCXE motif (23), which is present in many pRb binding proteins (29). The CDK phosphorylation site serines 864 and 1007 flank the LXCXE pRb binding motif (amino acids 957–961) (Fig. 2D). We established an in vitro RBP1-pRb binding assay, where recombinant MBP-pRb279–928 containing the pRb pocket region was incubated with GST-RBP1. Briefly, MBP-pRb was captured on amylose resin, which was washed extensively before detection of bound RBP1 by immunoblotting with anti-GST antibody. GST-RBP1 specifically bound to MBP-pRb279–928 (Fig. 4A, lane 4). To determine whether this interaction is sensitive to phosphorylation, we phosphorylated GST-RBP1 and/or MBP-pRb279–928 with cyclin A/CDK2 before the binding reaction. Phosphorylation of GST-RBP1 with cyclin A/CDK2 reduced its level of binding to unphosphorylated MBP-pRb279–928 by ∼40% compared with the level of binding observed when both proteins were unphosphorylated (Fig. 4A, lanes 4 and 5, upper panel). Conversely, phosphorylation of MBP-pRb279–928 reduced its binding to unphosphorylated GST-RBP1 by ∼75% (Fig. 4A, lanes 4 and 6, upper panel). However, when both proteins were phosphorylated by A/CDK2, the GST-RBP1·MBP-pRb279–928 interaction was maximally reduced (∼85%) (Fig. 4A, lanes 4 and 7, upper panel). Omitting ATP from the phosphorylation reactions before the binding assay did not affect the binding of GST-RBP1 to MBP-pRb279–928, demonstrating that the observed changes were due to phosphorylation by cyclin A/CDK2 (Fig. 4A, lanes 4–7, lower panel).
FIGURE 4.
CDK-mediated phosphorylation of RBP1 and pRb induces their dissociation in vitro and in vivo during G0/G1-S phase cell cycle progression. A, cyclin A/CDK2-mediated phosphorylation of RBP1 and pRb disrupts their association in vitro. Purified recombinant MBP-pRb279–928 immobilized on amylose resin was either unphosphorylated (pRb279–928, lanes 4 and 5) or phosphorylated with cyclin A/CDK2 (pRb279–928-(P), lanes 6 and 7). The samples were then incubated with either unphosphorylated GST-RBP1 (RBP1, lanes 4 and 6) or cyclin A/CDK2-phosphorylated GST-RBP1 (RBP1-(P), lanes 5 and 7), and binding was performed as described under “Experimental Procedures.” The level of GST-RBP1 binding was assessed by immunoblotting with anti-GST antibody (upper panel). As controls, unphosphorylated (lane 2) or phosphorylated GST-RBP1 (lane 3) was incubated with empty amylose resin. To demonstrate that changes in association between RBP1 and pRb were phosphorylation-dependent, ATP was omitted from the phosphorylation reactions (lanes 4–7, lower panel). Lane 1 represents 10% of the input of GST-RBP1 in the binding reaction. B, cyclin A/CDK2-mediated phosphorylation disrupts preformed RBP1·pRb complex in vitro. Pre-formed MBP-pRb279–928·GST-RBP1 complex immobilized on amylose resin was incubated in the absence (lane 1) or presence of cyclin A/CDK2 (lane 2) under phosphorylation conditions and then washed. Bound GST-RBP1 was assessed by immunoblotting with anti-GST antibody. C, the level of RBP1-associated with pRb decreases as cells progress from G0/G1 to S phase of the cell cycle. Upper panel, lysates from either exponentially growing (lane 1), G0/G1 phase-arrested with ICI 182780 (lane 2, ICI), or S phase (27 h after the addition of estradiol to ICI 182780-arrested cells) (lane 3, ICI + E2) MCF-7 cells were subjected to Western blotting with an anti-pRb antibody to demonstrate relative levels of hypophosphorylated and hyperphosphorylated pRb. Middle panel, FLAG-RBP1 was immunoprecipitated (IP) from either exponentially growing (lane 2), G0/G1 phase arrested with ICI 182780 (lane 3, ICI), or S phase (27 h after the addition of estradiol to ICI 182780-arrested cells) (lane 4, ICI + E2) MCF-7 cells. Lane 1 represents control vector transfected cells. The immunoprecipitates were assessed for the levels of FLAG-RBP1 and co-immunoprecipitating pRb by immunoblotting with the appropriate antibodies. Lower panel, the histograms represent the relative levels of pRb bound to the immunoprecipitated FLAG-RBP1 from G0/G1 arrested (ICI) or cells in S phase (ICI + E2). Error bars represent ± S.E. of three independent experiments.
We next tested if cyclin A/CDK2-mediated phosphorylation disrupts a preassembled GST-RBP1·MBP-pRb279–928 complex. We mixed both unphosphorylated proteins to form a complex as described under “Experimental Procedures.” The amylose resin-bound GST-RBP1·MBP-pRb279–928 complex was then incubated either in the absence or presence of cyclin A/CDK2 under phosphorylation conditions followed by washing of the resin and immunoblotting for bound GST-RBP1 with anti-GST antibody. Compared with the control unphosphorylated GST-RBP1·MBP-pRb279–928, ∼35% less GST-RBP1 was bound to MBP-pRb279–928 when the GST-RBP1·MBP-pRb279–928 complex was phosphorylated by A/CDK2 (Fig. 4B). Altogether, these studies demonstrate that A/CDK2-mediated phosphorylation of both RBP1 and pRb disrupts their association in vitro.
RBP1 Dissociates from pRb as Cells Progress from G0/G1 into S phase of the Cell Cycle
We next assessed if CDK-mediated phosphorylation of RBP1 and pRb leads to their dissociation in vivo. To perform these studies, MCF-7 cells were transiently transfected with pCMV Tag2A-RBP1 and RBP1 immunoprecipitated from cells in either G0/G1 phase (ICI) or S phase (ICI + E2) of the cell cycle, as described in Figs. 1C and 3C. We monitored the phosphorylation status of pRb, which changes substantially when cells progress from G0/G1 phase, where CDK activity is low and pRb is hypophosphorylated, into S phase, where cyclin D·CDK4/6, cyclin E·CDK2, and cyclin A·CDK2 complexes are active and hyperphosphorylate pRb (8). Hyperphosphorylated pRb migrates slower than hypophosphorylated pRb on SDS-PAGE (8). Lysate prepared from an asynchronous culture of exponentially growing MCF-7 cells contains both hypo- and hyperphosphorylated forms of pRb (Fig. 4C, upper panel, lane 1), reflective of cells in various phases of the cell cycle. In G0/G1 phase, pRb was largely hypophosphorylated due to low CDK activity, whereas the majority of pRb was hyperphosphorylated and migrated slower on SDS-PAGE, when the MCF-7 cells were in S phase (Fig. 4C, upper panel, lanes 2 and 3). To assess the RBP1 interaction with pRb in vivo and its dependence on the phosphorylation status of RBP1 and pRb, we immunoprecipitated FLAG-RBP1 from MCF-7 cells that were asynchronous, G0/G1-arrested or in S phase. Immunoblotting the FLAG-RBP1 immunoprecipitates for associated pRb revealed that RBP1 bound almost exclusively to the hypophosphorylated form of pRb, strongly suggesting that the interaction is phosphorylation-sensitive (Fig. 4C, middle panel, lanes 2–4). Furthermore, ∼2.5-fold more pRb was associated with RBP1 in G0/G1 phase-arrested cells when CDK is low, compared with cells in S phase, when CDK activity is high (Fig. 4C, middle panel, lanes 3 and 4). These results are consistent with our in vitro findings showing that cyclin A/CDK2-mediated phosphorylation promotes dissociation of RBP1 and pRb. Altogether, the in vitro and in vivo studies indicate that CDK-mediated phosphorylation of both RBP1 and pRb induces their dissociation during progression from G0/G1 to S phase of the cell cycle.
DISCUSSION
In the present study we identify the pRb-binding protein RBP1 as a novel CDK substrate. Numerous studies have demonstrated that the inhibitory function of pRb on E2F-mediated transcription is inactivated by CDK-mediated phosphorylation of pRb on several sites as cells progress from G1 through to S phase (13, 18, 20, 33, 34). Although the precise mechanism underpinning this process is not entirely understood, it is believed that CDK-mediated phosphorylation of pRb on different sites changes the conformation of this protein to inhibit its binding to the E2F transactivation domain and the SAP30·mSin3·HDAC complex to relieve transcriptional repression. For example, cyclin D/CDK4/6-mediated phosphorylation of serines 788 and 795 at the C terminus is thought to destabilize the interaction with E2F (13, 20), whereas phosphorylation of serine 567 by cyclin E/CDK2 is thought to lead to a conformational change in the pRb pocket to disrupt E2F binding (13). Other studies have shown that phosphorylation of pRb threonines 356 and 373 on the interdomain linker (positioned between the N-terminal region and the pocket) and serine 608 and 612 on the pocket region linker (the spacer region located between the A and B domains) by cyclin A/CDK2 or cyclin K/CDK6 significantly reduces the affinity of pRb for the E2F transactivation domain (18). In addition to disrupting the interaction with E2F, phosphorylation of pRb threonines 821 and 826 by cyclin E/CDK2 and cyclin A/CDK2 can disrupt its interaction with HDACs to relieve transcriptional repression (13, 20, 36).
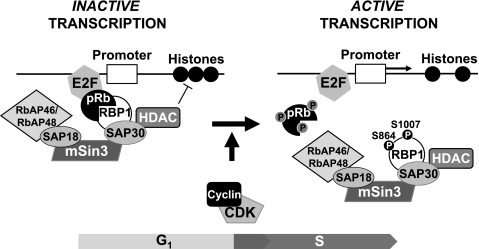
Our results now show that in addition to phosphorylation of pRb, concurrent CDK-mediated phosphorylation of RBP1 is important for efficient dissociation of these proteins, and this mechanism likely underpins dissociation of the SAP30·mSin3·HDAC complex from pRb as cells progress from G1 into S phase of the cell cycle. Hence, CDK-mediated phosphorylation of RBP1 and pRb contributed to their partial dissociation, whereas concurrent phosphorylation of both proteins resulted in their maximal dissociation in vitro (Fig. 4A). Consistent with these observations, examination of RBP1/pRb association in MCF-7 cells revealed maximal association in G0/G1 phase-arrested cells, when CDK activity and pRb and RBP1 phosphorylation is low (Figs. 1C and 4C). Conversely, as cells progressed into S phase when cyclin E/CDK2 and cyclin A/CDK2 are active and RBP1 and pRb phosphorylation is increased, their level of association was significantly reduced (Fig. 4C). In contrast to the disruption of association with pRb, CDK-mediated phosphorylation of RBP1 did not impact on its association with SAP30 in vitro (Figs. 3, A and B) nor did the association of RBP1 with SAP30 and mSin3 or the associated HDAC activity vary as cells progressed from G1 to S phase in MCF-7 cells (Figs. 3, C and D). Altogether, these data indicate that CDK-mediated phosphorylation of RBP1 and pRb disrupts their interaction to dissociate the RBP1·SAP30·mSin3·HDAC complex from E2F, as depicted in Fig. 5.
FIGURE 5.
CDK-mediated cumulative phosphorylation of pRb and concurrent phosphorylation of RBP1 promotes their dissociation to mediate release of the SAP30·mSin3·HDAC complex. During G0/G1 phase, when CDK activity is low, the RBP1/pRb interaction mediates HDAC-dependent pRb inhibition on E2F transcription factors. As cells progress into the cell cycle, G1 and S phase CDKs phosphorylate both pRb and RBP1 to disrupt their interaction, removing SAP30·mSin3·HDACs from pRb and E2F and thereby allowing transcription of genes necessary for S phase cell cycle progression.
How phosphorylation of RBP1 and pRb leads to their dissociation at a molecular level is not clear. RBP1 contains a Tudor domain at the N terminus (amino acids 58–113), an ARID domain (amino acids 314–409), a chromatin organization modifier (Chromo) domain (amino acids 593–634), and the LXCXE pRb binding motif (amino acids 957–961). The RBP1 CDK phosphorylation sites (serines 864 and 1007) are not located within any of these domains; however, they flank the LXCXE pRb binding motif (amino acids 957–961), suggesting that phosphorylation at these sites may reduce binding of the RBP1 LXCXE motif to the pRb pocket (Fig. 2D). Previous studies have shown that CDK-mediated phosphorylation of pRb threonines 821 and 826 disrupts its interaction with HDACs (13, 20, 36), suggesting that phosphorylation of these pRb sites in addition to RBP1 phosphorylation contributes to their dissociation. The importance of concurrent CDK-mediated phosphorylation of RBP1 and pRb is exemplified in preliminary studies in our laboratory, investigating if phosphorylation of RBP1 is important for cell cycle progression. We generated RBP1 phosphosite mutants, where serines 864 and 1007 were mutated to alanine (RBP1-Ala) or aspartate residues (RBP1-Asp) to potentially mimic constitutively unphosphorylated and phosphorylated RBP1, respectively. Studies of the G1-S cell cycle progression in MCF-7 cells ectopically expressing wild-type RBP1, RBP1-Ala, or RBP1-Asp as described in Fig. 1C revealed similar kinetics of cell cycle progression (data not shown). Because maximal RBP1/pRb dissociation and abrogation of pRb inhibition toward E2F is achieved by CDK phosphorylation of both proteins on different sites, perturbation of the cell cycle progression likely requires simultaneous co-expression of phosphosite mutants of both RBP1 and pRb. It will be important to identify the relevant phosphorylation sites on pRb that regulate dissociation from RBP1 to perform these studies. In addition, further studies of the pRb·RBP1 complex will be required to provide specific structural information on this interaction and how phosphorylation of specific sites mediates the dissociation of these proteins. For example, x-ray crystallography studies on the association of the pRb C-terminal region with E2F have provided insights into how CDK-mediated phosphorylation of pRb contributes to E2F release (18, 20).
Our data expand on the existing understanding on the importance of cumulative CDK-mediated phosphorylation in regulating pRb function to control cell cycle progression. Multiple phosphorylations in controlling cell cycle regulators are emerging as a mechanism for setting a threshold for substrate regulation and commitment to cell cycle progression. For example, in Saccharomyces cerevisiae the CDK inhibitor Sic1 must be degraded to allow activation of S phase CDKs and cell cycle progression (59, 60). Efficient Sic1 ubiquitination by the Skp·Cullin·F-box ubiquitin ligase complex leading to its proteasomal degradation and cell cycle progression is dependent on phosphorylation on multiple sites by G1 phase CDKs, which sets a phosphorylation threshold. This ensures establishment of a G1 phase period that can only be overcome when G1-phase CDKs reach a critical threshold leading to rapid Sic1 phosphorylation and degradation (61). Our data suggest a similar paradigm, where the cumulative and concurrent CDK-mediated phosphorylation of pRb and RBP1 sets a higher phosphorylation threshold for efficient dissociation of the SAP30·mSin3·HDAC complex from E2F, thus ensuring cells only commit to cell cycle progression when cyclin/CDK activity is high.
Acknowledgments
We thank Raymond E. Jones, Department of Cancer Research, Merck Research Laboratories, West Point, PA for RBP1 cDNA. SAP30.pET28a(+) plasmid was a kind gift from Danny Reinberg of the Howard Hughes Medical Institute, Department of Biochemistry, New York University School of Medicine, New York. We thank Jon Oakhill of the Protein Chemistry Unit, St. Vincent's Institute of Medical Research, Fitzroy, Victoria, Australia for critical reading of the manuscript.
This work was supported by grants from the Cancer Council Victoria and the Department of Defense Breast Cancer Research Program (BC020800).
- CDK
- cyclin-dependent kinase
- HDAC
- histone deacetylase
- RBP1
- retinoblastoma-binding protein 1
- ARID4
- AT-rich interaction domain 4
- E2
- 17β-estradiol
- ICI
- ICI 182780
- RT
- room temperature.
REFERENCES
- 1. Morgan D. O. (1995) Nature 374, 131–134 [DOI] [PubMed] [Google Scholar]
- 2. Matsushime H., Ewen M. E., Strom D. K., Kato J. Y., Hanks S. K., Roussel M. F., Sherr C. J. (1992) Cell 71, 323–334 [DOI] [PubMed] [Google Scholar]
- 3. Meyerson M., Harlow E. (1994) Mol. Cell. Biol. 14, 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohtsubo M., Theodoras A. M., Schumacher J., Roberts J. M., Pagano M. (1995) Mol. Cell. Biol. 15, 2612–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pagano M., Pepperkok R., Verde F., Ansorge W., Draetta G. (1992) EMBO J. 11, 961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Draetta G., Beach D. (1988) Cell 54, 17–26 [DOI] [PubMed] [Google Scholar]
- 7. Jessus C., Beach D. (1992) Cell 68, 323–332 [DOI] [PubMed] [Google Scholar]
- 8. Sarcevic B., Lilischkis R., Sutherland R. L. (1997) J. Biol. Chem. 272, 33327–33337 [DOI] [PubMed] [Google Scholar]
- 9. Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. (2003) Nature 425, 859–864 [DOI] [PubMed] [Google Scholar]
- 10. Suryadinata R., Sadowski M., Sarcevic B. (2010) Biosci. Rep. 30, 243–255 [DOI] [PubMed] [Google Scholar]
- 11. Mittnacht S. (1998) Curr. Opin. Genet. Dev. 8, 21–27 [DOI] [PubMed] [Google Scholar]
- 12. Suzuki-Takahashi I., Kitagawa M., Saijo M., Higashi H., Ogino H., Matsumoto H., Taya Y., Nishimura S., Okuyama A. (1995) Oncogene 10, 1691–1698 [PubMed] [Google Scholar]
- 13. Harbour J. W., Luo R. X., Dei Santi A., Postigo A. A., Dean D. C. (1999) Cell 98, 859–869 [DOI] [PubMed] [Google Scholar]
- 14. Lee E. Y., Cam H., Ziebold U., Rayman J. B., Lees J. A., Dynlacht B. D. (2002) Cancer Cell 2, 463–472 [DOI] [PubMed] [Google Scholar]
- 15. Brehm A., Miska E. A., McCance D. J., Reid J. L., Bannister A. J., Kouzarides T. (1998) Nature 391, 597–601 [DOI] [PubMed] [Google Scholar]
- 16. Magnaghi-Jaulin L., Groisman R., Naguibneva I., Robin P., Lorain S., Le Villain J. P., Troalen F., Trouche D., Harel-Bellan A. (1998) Nature 391, 601–605 [DOI] [PubMed] [Google Scholar]
- 17. Wu J., Grunstein M. (2000) Trends Biochem. Sci. 25, 619–623 [DOI] [PubMed] [Google Scholar]
- 18. Burke J. R., Deshong A. J., Pelton J. G., Rubin S. M. (2010) J. Biol. Chem. 285, 16286–16293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee J. O., Russo A. A., Pavletich N. P. (1998) Nature 391, 859–865 [DOI] [PubMed] [Google Scholar]
- 20. Rubin S. M., Gall A. L., Zheng N., Pavletich N. P. (2005) Cell 123, 1093–1106 [DOI] [PubMed] [Google Scholar]
- 21. Hassig C. A., Fleischer T. C., Billin A. N., Schreiber S. L., Ayer D. E. (1997) Cell 89, 341–347 [DOI] [PubMed] [Google Scholar]
- 22. Lai A., Kennedy B. K., Barbie D. A., Bertos N. R., Yang X. J., Theberge M. C., Tsai S. C., Seto E., Zhang Y., Kuzmichev A., Lane W. S., Reinberg D., Harlow E., Branton P. E. (2001) Mol. Cell. Biol. 21, 2918–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Defeo-Jones D., Huang P. S., Jones R. E., Haskell K. M., Vuocolo G. A., Hanobik M. G., Huber H. E., Oliff A. (1991) Nature 352, 251–254 [DOI] [PubMed] [Google Scholar]
- 24. Lai A., Lee J. M., Yang W. M., DeCaprio J. A., Kaelin W. G., Jr., Seto E., Branton P. E. (1999) Mol. Cell. Biol. 19, 6632–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilsker D., Patsialou A., Dallas P. B., Moran E. (2002) Cell Growth Differ. 13, 95–106 [PubMed] [Google Scholar]
- 26. Zhang Y., Sun Z. W., Iratni R., Erdjument-Bromage H., Tempst P., Hampsey M., Reinberg D. (1998) Mol. Cell 1, 1021–1031 [DOI] [PubMed] [Google Scholar]
- 27. Binda O., Roy J. S., Branton P. E. (2006) Mol. Cell. Biol. 26, 1917–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lai A., Marcellus R. C., Corbeil H. B., Branton P. E. (1999) Oncogene 18, 2091–2100 [DOI] [PubMed] [Google Scholar]
- 29. Taya Y. (1997) Trends Biochem. Sci. 22, 14–17 [DOI] [PubMed] [Google Scholar]
- 30. Laherty C. D., Yang W. M., Sun J. M., Davie J. R., Seto E., Eisenman R. N. (1997) Cell 89, 349–356 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D. (1997) Cell 89, 357–364 [DOI] [PubMed] [Google Scholar]
- 32. Fleischer T. C., Yun U. J., Ayer D. E. (2003) Mol. Cell. Biol. 23, 3456–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown V. D., Phillips R. A., Gallie B. L. (1999) Mol. Cell. Biol. 19, 3246–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Knudsen E. S., Wang J. Y. (1996) J. Biol. Chem. 271, 8313–8320 [DOI] [PubMed] [Google Scholar]
- 35. Weinberg R. A. (1995) Cell 81, 323–330 [DOI] [PubMed] [Google Scholar]
- 36. Zarkowska T., Mittnacht S. (1997) J. Biol. Chem. 272, 12738–12746 [DOI] [PubMed] [Google Scholar]
- 37. Sarcevic B., Mawson A., Baker R. T., Sutherland R. L. (2002) EMBO J. 21, 2009–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fukunaga R., Hunter T. (1997) EMBO J. 16, 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao J., Dynlacht B., Imai T., Hori T., Harlow E. (1998) Genes Dev. 12, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilsker D., Probst L., Wain H. M., Maltais L., Tucker P. W., Moran E. (2005) Genomics 86, 242–251 [DOI] [PubMed] [Google Scholar]
- 41. Lai A., Sarcevic B., Prall O. W., Sutherland R. L. (2001) J. Biol. Chem. 276, 25823–25833 [DOI] [PubMed] [Google Scholar]
- 42. Huyen Y., Zgheib O., Ditullio R. A., Jr., Gorgoulis V. G., Zacharatos P., Petty T. J., Sheston E. A., Mellert H. S., Stavridi E. S., Halazonetis T. D. (2004) Nature 432, 406–411 [DOI] [PubMed] [Google Scholar]
- 43. Ball L. J., Murzina N. V., Broadhurst R. W., Raine A. R., Archer S. J., Stott F. J., Murzin A. G., Singh P. B., Domaille P. J., Laue E. D. (1997) EMBO J. 16, 2473–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V., Elgin S. C. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 9923–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paro R. (1993) Curr. Opin. Cell Biol. 5, 999–1005 [DOI] [PubMed] [Google Scholar]
- 46. Wu M. Y., Eldin K. W., Beaudet A. L. (2008) J. Natl. Cancer Inst. 100, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sadowski M., Mawson A., Baker R., Sarcevic B. (2007) Biochem. J. 405, 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Campbell D. H., Sutherland R. L., Daly R. J. (1999) Cancer Res. 59, 5376–5385 [PubMed] [Google Scholar]
- 49. Meijer L., Borgne A., Mulner O., Chong J. P., Blow J. J., Inagaki N., Inagaki M., Delcros J. G., Moulinoux J. P. (1997) Eur. J. Biochem. 243, 527–536 [DOI] [PubMed] [Google Scholar]
- 50. Prall O. W., Sarcevic B., Musgrove E. A., Watts C. K., Sutherland R. L. (1997) J. Biol. Chem. 272, 10882–10894 [DOI] [PubMed] [Google Scholar]
- 51. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 52. Yoshida M., Kijima M., Akita M., Beppu T. (1990) J. Biol. Chem. 265, 17174–17179 [PubMed] [Google Scholar]
- 53. Songyang Z., Blechner S., Hoagland N., Hoekstra M. F., Piwnica-Worms H., Cantley L. C. (1994) Curr. Biol. 4, 973–982 [DOI] [PubMed] [Google Scholar]
- 54. Songyang Z., Lu K. P., Kwon Y. T., Tsai L. H., Filhol O., Cochet C., Brickey D. A., Soderling T. R., Bartleson C., Graves D. J., DeMaggio A. J., Hoekstra M. F., Blenis J., Hunter T., Cantley L. C. (1996) Mol. Cell. Biol. 16, 6486–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Srinivasan J., Koszelak M., Mendelow M., Kwon Y. G., Lawrence D. S. (1995) Biochem. J. 309, 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Sci. Signal 3, ra3. [DOI] [PubMed] [Google Scholar]
- 58. Sutherland R. L., Watts C. K. W., Lee C. S. L., Musgrove E. A. (1999) in Human Cell Culture (Masters J. R. W., Palsson B. eds) pp. 79–106, Kluwer Academic Publishers, Bodmin, Cornwall, UK [Google Scholar]
- 59. Schwob E., Böhm T., Mendenhall M. D., Nasmyth K. (1994) Cell 79, 233–244 [DOI] [PubMed] [Google Scholar]
- 60. Sadowski M., Suryadinata R., Lai X., Heierhorst J., Sarcevic B. (2010) Mol. Cell. Biol. 30, 2316–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. (2001) Nature 414, 514–521 [DOI] [PubMed] [Google Scholar]