Abstract
The essential and evolutionarily conserved Smc5-Smc6 complex (Smc5/6) is critical for the maintenance of genome stability. Partial loss of Smc5/6 function yields several defects in DNA repair, which are rescued by inactivation of the homologous recombination (HR) machinery. Thus HR is thought to be toxic to cells with defective Smc5/6. Recent work has highlighted a role for Smc5/6 and the Sgs1 DNA helicase in preventing the accumulation of unresolved HR intermediates. Here we investigate how deletion of MPH1, encoding the orthologue of the human FANCM DNA helicase, rescues the DNA damage sensitivity of smc5/6 but not sgs1Δ mutants. We find that MPH1 deletion diminishes accumulation of HR intermediates within both smc5/6 and sgs1Δ cells, suggesting that MPH1 deletion is sufficient to decrease the use of template switch recombination (TSR) to bypass DNA lesions. We further explain how avoidance of TSR is nonetheless insufficient to rescue defects in sgs1Δ mutants, by demonstrating a requirement for Sgs1, along with the post-replicative repair (PRR) and HR machinery, in a pathway that operates in mph1Δ mutants. In addition, we map the region of Mph1 that binds Smc5, and describe a novel allele of MPH1 encoding a protein unable to bind Smc5 (mph1-Δ60). Remarkably, mph1-Δ60 supports normal growth and responses to DNA damaging agents, indicating that Smc5/6 does not simply restrain the recombinogenic activity of Mph1 via direct binding. These data as a whole highlight a role for Smc5/6 and Sgs1 in the resolution of Mph1-dependent HR intermediates.
Keywords: DNA Helicase, DNA Recombination, DNA Repair, DNA Replication, DNA Topology, Mph1, Sgs1, Smc5/6
Introduction
The Smc5/6 complex is one of three structural maintenance of chromosome (SMC)2 complexes within eukaryotic cells, the two others being the Smc1/3 (cohesin) and Smc2/4 (condensin) complexes (1–3). Cohesin, along with playing transcriptional regulatory roles, serves to maintain cohesion between sister chromatids during DNA replication, ensuring their proper mitotic segregation and also facilitating HR-based repair of DNA double strand breaks. Condensin promotes chromosome compaction, enabling proper chromatid segregation. Despite a detailed understanding of cohesion and condensin, the essential biochemical role(s) played by the Smc5/6 complex in genome integrity remains relatively unclear (4).
The Smc5/6 complex is well conserved among higher eukaryotes and contains six non-SMC subunits including the SUMO E3 ligase, Mms21 (5, 6). Cells containing hypomorphic alleles of Smc5/6 complex members, from here on referred to as smc5/6 mutants, exhibit defects in DNA metabolism. Similar deficiencies in DNA repair as those in smc5/6 mutants are also observed in cells globally deficient for protein sumoylation (the conjugation of the small protein modifier SUMO to target proteins) or selectively defective for Mms21 SUMO ligase activity, such as the Saccharomyces cerevisiae mms21-sp allele (C200S, H202A).
Cells with hypomorphic alleles of Smc5/6 complex members are sensitive to a variety of genotoxic stressors such as the DNA alkylating agent methyl methanesulfonate (MMS) and the ribonucleotide reductase inhibitor hydroxyurea (HU) (7, 8). Seminal work within yeast characterizing the role of Smc5/6 during DNA repair determined that the complex functions within an HR-dependent repair pathway, because mutations disabling HR-based genome repair after UV or γ-irradiation are epistatic to Smc5/6 deficiency (9). However, subsequent analysis has revealed that, depending on the genotoxic stressor, inactivation of HR can actually alleviate smc5/6 mutant phenotypes. These findings suggest the existence of different subpathways of HR, some of which are detrimental to the cell when the Smc5/6 complex is not fully functional (10–13). These genetic findings, together with physical observations of accumulated recombination intermediates within smc5/6 mutants, have led to a model implicating the Smc5/6 complex in the resolution of a particular HR-mediated sister chromatid linkage termed a Rec-X. In the absence of a properly functioning Smc5/6 complex, these HR intermediates are thought to form a physical linkage between chromatids, preventing their separation and promoting lethal nondisjunction events (10, 14–19). Previous work by several groups has found the accumulation of biochemically similar Rec-X intermediates within cells defective for the DNA helicase Sgs1 (15, 16). These data have led us and others to suggest cooperativity between the Smc5/6 and Sgs1 complexes in the resolution of HR-generated Rec-X intermediates (14–16).
During the course of our work characterizing a novel role for the DNA helicase Mph1 in promoting Rec-X formation, a report emerged demonstrating a role for Mph1 in promoting a detrimental form of homologous recombination-based repair within cells defective for the Smc5/6 complex but not cells mutant for the DNA helicase Sgs1 (17). Mph1 is the orthologue of human FANCM, a component of the Fanconi Anemia protein complex, which is critical for repair of interstrand DNA crosslinks (20). This report uncovered a direct physical interaction between the Smc5/6 complex and the Mph1 helicase, and documented robust rescue of DNA damage sensitivity upon deletion of Mph1 from smc5/6 but not sgs1Δ mutant cells. Two models explaining these results were proposed, one suggesting that the Smc5/6 complex serves as a negative regulator of Mph1 recombinogenic activity and the other proposing the existence of two distinct DNA repair pathways with their own unique recombination intermediates that are dependent upon either the Smc5/6 or Sgs1 protein complexes for their resolution.
Similar to this report, we had found that removal of Mph1 within smc5/6 but not sgs1Δ mutant cells restored resistance to the DNA damaging agents MMS and HU. We further investigated whether the difference in rescue between smc5/6 and sgs1Δ mutants might be due to the Smc5/6 complex directly binding the Mph1 helicase to prevent Mph1-mediated aberrant recombinogenic activity, which would otherwise promote DNA damage sensitivity. To test this, we generated a novel allele of Mph1 (mph1-Δ60) that is unable to bind Smc5. The mph1-Δ60 allele behaved similarly to wild-type, arguing against direct Smc5 repression of Mph1 activity. We next sought to determine if Mph1 was involved only in the generation of recombination intermediates within smc5/6 but not sgs1Δ mutants as recently reported (17). In contrast to this report, but consistent with a different report (46), we demonstrate a clear role for the DNA helicase Mph1 in promoting the formation of recombination intermediates within both smc5/6 and sgs1Δ mutants. Finally, to explain the lack of rescue within sgs1Δ cells, we conducted a series of epistasis experiments and uncovered an alternative repair pathway utilized by cells upon MPH1 deletion. This alternative repair pathway is dependent upon the post-replicative repair and HR machinery, including the DNA helicase Sgs1, thus explaining why sgs1Δ mutants are not rescued from DNA damage sensitivity upon MPH1 deletion. Our findings provide greater insight into DNA repair pathways that are critically dependent upon the Smc5/6 complex along with the interplay between this complex and the DNA helicases Mph1 and Sgs1.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Complete strain and plasmid details are described in supplemental Table S2. The BY4741 yeast background was utilized for the majority of experiments. Gateway-compatible pAG CEN/ARS destination vectors were utilized for complementation experiments (22). mph1-DE and mph1-Δ60 were generated through site-directed mutagenesis or sewing PCR of the corresponding wild-type entry clones and verified by sequencing. SGS1 alleles were PCR amplified from preexisting vectors and Gateway recombination was utilized to generate the corresponding entry vectors. Yeast-two-hybrid experiments were performed utilizing the PJ694 strain and gateway compatible activation and DNA binding domain vectors.
Analysis of Recombination Intermediates (X-structures)
Preparation of DNA and probing for ARS305 were performed as described (23). Nocodazole at a concentration of 10 μg/ml was added to cultures for 2.5 h to synchronize heterogeneously cycling populations. Cells were then released into liquid YPAD media containing 0.016% or 0.033% MMS and incubated at 30 °C. We note that the majority of our experiments are performed using 0.016% MMS during release after synchronization. This concentration was chosen because it represents a level of MMS closer to that at which the majority of spot assays were performed but was also of a sufficient level to readily observe X-structures. All comparisons were performed with samples grown and prepared in parallel to remove interexperimental variability.
Spot Assays
Yeast were grown overnight at 23 °C in liquid YPAD or, when required, selective media to assure plasmid maintenance. Cells were then pelleted and resuspended into 1× PBS and counted. 105 cells, and serial 10-fold dilutions, were spotted onto YPAD plates or YPAD plates containing MMS or HU, as indicated. Plates were incubated at 30 °C and photographed 2–3 days later.
RESULTS
MPH1 Deletion Rescues the DNA Damage Sensitivity of smc5/6 Mutants
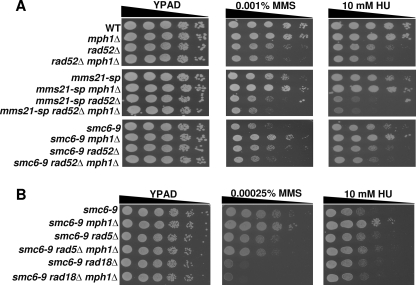
Using Synthetic Genetic Array analysis (24), we screened 26 deletion mutations in loci encoding factors important in DNA metabolism for their ability to suppress the temperature sensitive phenotype of smc6-9 mutants (supplemental Table S1) (13). Only deletion of MPH1, which encodes the Mph1 DNA helicase, suppressed the temperature sensitivity of smc6-9 mutants (data not shown). Given its ability to suppress the temperature sensitivity of smc6-9 mutants, we asked if deletion of MPH1 could also rescue the DNA damage sensitivity of cells defective for Smc5/6 function. mms21-sp and smc6-9 mutants, (henceforth both will be referred to as smc5/6 mutants) deleted for MPH1, exhibited an increased resistance to the DNA-damaging agents, MMS and HU, as compared with the single mms21-sp and smc6-9 controls (Fig. 1A). Though this is consistent with our hypothesis that MPH1 deletion can rescue phenotypes caused by loss of Smc5/6 function, it is somewhat surprising given that MPH1 deletion in wild-type cells leads to mild MMS sensitivity (Fig. 1A) (25). As previously noted, a similar rescue upon MPH1 deletion was reported while our studies were under review (17).
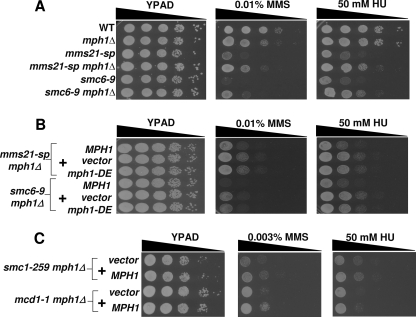
FIGURE 1.
Deletion of MPH1 rescues the DNA damage-sensitivity of smc5/6 mutants. A, spot assay comparing the relative growth rates of various single and double mutants between mph1Δ and mms21-sp or smc6–9 grown on YPAD alone or with the indicated genotoxins. B, Mph1 helicase-dependent functions are necessary for sensitizing smc5/6 mutants to DNA-damaging agents. mms21-sp mph1Δ and smc6–9 mph1Δ cells were transformed with either wild-type MPH1, empty vector, or mph1-DE (helicase dead) containing plasmids and spotted to the indicated media. C, deletion of MPH1 confers no DNA damage resistance to cells with mutant forms of Smc1/3 complex members. Hypomorphic alleles encoding the Smc1/3 complex members Smc1 and Mcd1 were used. The indicated double mutants were transformed with either the vector control or MPH1-containing plasmid, and spotted to the indicated media.
To better understand how loss of MPH1 leads to the observed rescue in smc5/6 mutants, we sought to determine whether the helicase activity of Mph1 was essential for promoting DNA damage sensitivity or if Mph1 has helicase-independent activities similar to its mammalian counterpart, FANCM (26, 27). To create a helicase-impaired allele of MPH1, two residues predicted to be essential for the hydrolysis of ATP were mutated within the conserved helicase motif II (D209N, E210Q), generating the mph1-DE allele (28). mms21-sp mph1Δ and smc6-9 mph1Δ cells were then transformed with single copy plasmids containing either wild-type MPH1, vector or the mph1-DE allele. In contrast to wild-type Mph1, the predicted helicase-dead form of Mph1 did not restore the DNA damage sensitivity of smc5/6 mph1Δ mutants even though it did show a similar level of protein expression (Fig. 1B and supplemental Fig. S1).
Given the functional cooperativity between Smc5/6 and Smc1/3 complexes during DNA repair, and the overlap in their genomic binding sites (29–32), we asked if MPH1 might also play a role in the DNA damage-sensitivity of smc1/3 mutants. Expression of MPH1 in a smc1/3 mph1Δ double mutant had no effect on HU-sensitivity compared with the vector control, and rather than sensitizing smc1/3 mutants to MMS, Mph1 increased their resistance, contrary to the result observed in smc5/6 mutants (Fig. 1C). These data demonstrate that the rescue seen upon MPH1 deletion is specific to the Smc5/6 complex and further suggest that the molecular mechanisms governing the DNA damage sensitivity of smc5/6 mutants are distinct from those of smc1/3 mutants.
Removal of Mph1 Causes a Decrease in the Levels of Unresolved Recombination Intermediates within smc5/6 Mutants
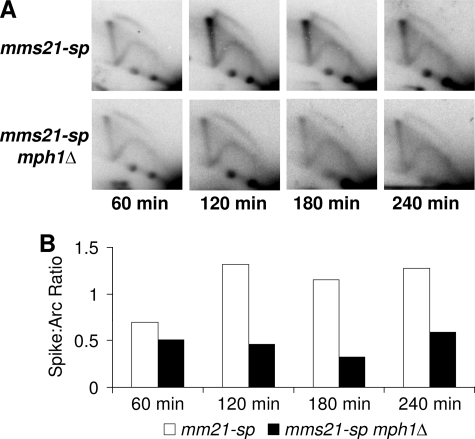
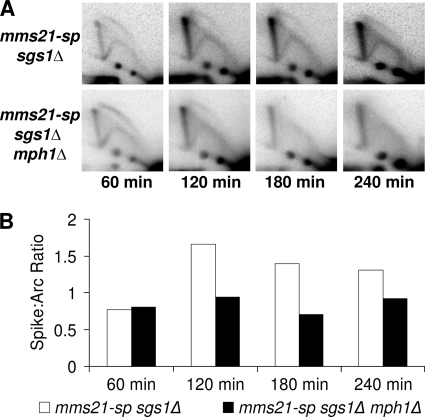
Several laboratories, including our own, have shown that mutants within the Smc5/6 complex accumulate abundant levels of recombination-dependent sister chromatid linkages when replicating through damaged DNA templates (10, 14–19). Efficient resolution of these linkages requires the activity of the Smc5/6 complex, as well as the Sgs1 helicase complex consisting of the proteins Sgs1, Top3, and Rmi1 (hereon referred to as the Sgs1 complex) (21, 33). In the absence of a properly functioning Smc5/6 or Sgs1 complex, unresolved recombination intermediates accumulate and can be visualized as a linear “X-spike” when analyzed by two-dimensional gel electrophoresis (2DGE) followed by Southern blotting. These observed recombination-dependent linkages are thought to prevent the proper segregation of sister chromatids at mitosis and promote lethal nondisjunction events. Because deletion of MPH1 rescues the DNA damage sensitivity of smc5/6 mutants (which accumulate unresolved recombination intermediates), we hypothesized that diminished recombination-dependent chromatid linkage underlies the observed rescue. To test this, mms21-sp cells with and without MPH1 were synchronized with nocodazole and released into media containing 0.016% MMS. Samples were then taken each hour for a total of 4 hours and analyzed by 2DGE followed by Southern blotting with probes specific to the ARS305 region. As predicted, a robust decrease in X-shaped molecules was seen in mms21-sp mph1Δ cells as compared with mms21-sp controls. Similar results were also observed when comparing smc6-9 mph1Δ mutants to smc6-9 controls (Fig. 2 and supplemental Fig. S2).
FIGURE 2.
Deletion of MPH1 reduces accumulation of recombination intermediates in mms21-sp mutants. A, representative time course of two-dimensional gel electrophoresis (2DGE) followed by Southern blotting examining replication intermediates near ARS305 in mms21-sp and mms21-sp mph1Δ mutants at each time point after release into 0.016% MMS. B, quantification of the ratio of X-shaped molecules to structures running within the replication arc.
Sgs1 Mutants Are Not Rescued by MPH1 Deletion
Because deletion of MPH1 led to increased DNA damage resistance and decreased unresolved recombination intermediates within smc5/6 mutants, we asked if the DNA damage sensitivity of sgs1Δ cells, which also show a similar accumulation of unresolved recombination intermediates, would be rescued by removal of MPH1. To our surprise but in agreement with recent reports (17, 19), there was little difference in the DNA damage-sensitivity between sgs1Δ and sgs1Δ mph1Δ mutants (a slight unexplained increase in growth in HU was seen, but it is important to point out that the level of rescue is clearly much less than observed upon MPH1 deletion in smc5/6 mutants) (supplemental Fig. S3). We tested three possible interpretations for the failure of MPH1 deletion to rescue sgs1Δ mutants despite its ability to rescue smc5/6 mutants. 1) The Smc5/6 complex functions to restrain aberrant Mph1 recombinogenic activity (which would otherwise promote DNA damage sensitivity) via direct binding of Smc5 to Mph1 (17), 2) The Smc5/6 and Sgs1 complexes function within two different DNA repair pathways (i.e. the observed recombination intermediates seen within smc5/6 and sgs1Δ mutant are derived from different DNA repair pathways), 3) the DNA repair pathway employed in cells lacking MPH1 is dependent upon Sgs1 for its function.
The Interaction between Smc5 and Mph1 Is Not Essential for Efficient DNA Repair
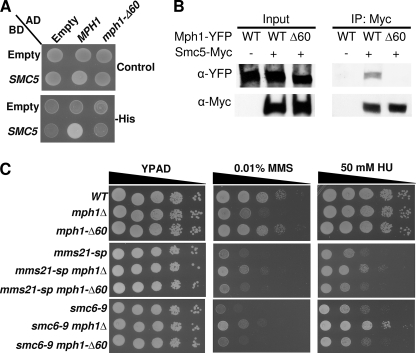
Recent work identified a physical interaction between Smc5 and Mph1 and proposed the interesting possibility that Smc5 directly regulates Mph1 helicase activity (17). To test if the interaction between Smc5 and Mph1 functions to prevent aberrant Mph1 activity, we mapped the interaction using a series of MPH1 deletion mutants. We found a stretch of amino acids from 751–810 within Mph1 that were sufficient to mediate binding to Smc5 (supplemental Fig. S4 and data not shown). This stretch is evolutionarily conserved as similar residues are also present in the Mph1 orthologues of Homo sapiens, Danio rerio, and Xenopus laevis (supplemental Fig. S5). We next asked if these same amino acids were not only sufficient but also necessary for the interaction between Smc5 and Mph1. An allele of MPH1, mph1-Δ60, lacking amino acids 751–810 was tested within a yeast-two-hybrid system and found to be defective in binding Smc5 (Fig. 3A). To verify our two-hybrid results we next generated C-terminally tagged alleles of Mph1 and performed immunoprecipitation experiments. In agreement with our two-hybrid results, full-length Mph1, but not the Mph1-Δ60 protein, co-immunoprecipitated with Smc5 (Fig. 3B). Having established that the Mph1-Δ60 protein could not bind Smc5 and was expressed at similar levels as wild type Mph1 (supplemental Fig. S6), we tested its effect on in vivo growth and DNA repair by integrating it into the endogenous genomic locus. mph1-Δ60 mutants grew normally on media with or without genotoxic agents. Similar results were obtained within the smc5/6 mutant background, with cells expressing mph1-Δ60 behaving similarly to cells expressing wild-type MPH1 (Fig. 3C). We interpret these data to suggest that the interaction between Smc5 and Mph1 is not necessary for Mph1 to promote its DNA repair roles nor is the interaction essential to prevent aberrant Mph1 recombinogenic activity.
FIGURE 3.
The interaction between Smc5 and Mph1 is dispensable with regard to MMS and HU sensitivity. A, two-hybrid analysis comparing the DBD alone or fused to the N terminus of Smc5 with regard to its ability to interact with either AD alone or AD fused to full-length Mph1 or Mph1-Δ60 (lacking amino acids 751–810). Control selects for the presence of both the DBD and AD containing plasmids, -His media selects for the presence of an interaction between the expressed DBD and AD containing proteins. B, co-IP performed by immunoprecipitating Smc5-Myc and probing for the presence of the C-terminal YFP tag on the various Mph1 proteins. C, spot assays comparing the growth of MPH1, mph1Δ, or mph1-Δ60 cells in SMC5/6 wild-type or smc5/6 mutant backgrounds on various media with or without genotoxic agents. The MPH1 and mph1-Δ60 alleles are integrated at the native MPH1 locus.
Sgs1 Is Required for the Rescue Observed upon Deletion of MPH1
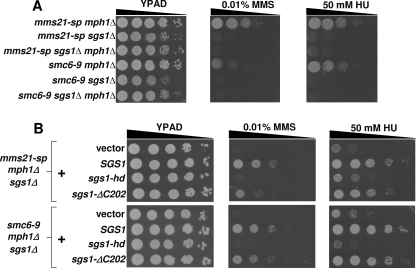
Previous models have suggested that Smc5/6 and Sgs1 might function within different DNA repair pathways, thus explaining the lack of rescue upon MPH1 deletion in sgs1 mutant cells (17). If this were indeed the case, one would expect that smc5/6 sgs1Δ double mutants would benefit from deletion of MPH1, as it should rescue the smc5/6 defect within the double mutant. To test this possibility, we compared the DNA damage sensitivities of mms21-sp sgs1Δ and smc6–9 sgs1Δ mutants with or without MPH1 (Fig. 4A and supplemental Fig. S7). Remarkably, MPH1 deletion did not promote DNA damage resistance in smc5/6 mutants lacking Sgs1, inconsistent with independent functions of Smc5/6 and Sgs1, but instead suggesting that the MPH1 deletion-mediated rescue of smc5/6 mutants is dependent upon functional Sgs1. We next asked if the helicase or checkpoint activities of Sgs1 were important in this setting (34, 35). For rescue, Sgs1 required its helicase activity and was only partially dependent upon its intra-S-phase checkpoint function (Fig. 4B). We found that the wild type and mutant Sgs1 proteins were expressed at similar levels, as previously reported (36, 37) (supplemental Fig. S8).
FIGURE 4.
Sgs1 is essential for rescue of the DNA damage sensitivity mediated by deletion of MPH1. A, Sgs1 is required for deletion of MPH1 to rescue the DNA damage-sensitivity of smc5/6 mutants. Spot assay comparing the growth rates of the various mutant strains. B, Sgs1 requires its helicase and S-phase checkpoint functions for full rescue of the DNA damage sensitivity within smc5/6 mph1Δ cells. mms21-sp mph1Δ sgs1Δ, and smc6–9 mph1Δ sgs1Δ cells were transformed with either empty vector, wild-type SGS1, sgs1-hd (helicase dead allele of SGS1), or sgs1-ΔC202 (allele of SGS1 lacking the C-terminal 202 amino-acids and defective in intra-S-phase checkpoint activity)-containing plasmids and spotted to the indicated media.
The requirement for Sgs1 helicase activity for full mph1Δ-mediated rescue of smc5/6 deficiency was of particular interest given that Sgs1, presumably through its helicase activity, is needed to remove the recombination intermediates that form during replication through damaged DNA templates (33). Thus the lack of rescue observed within smc5/6 sgs1Δ cells might have been due to an inability of MPH1 deletion to lower levels of unresolved recombination intermediates. Contrary to this prediction, MPH1 deletion reduced levels of unresolved recombination intermediates in mms21-sp sgs1Δ mutants (Fig. 5). The effect of MPH1 deletion in sgs1Δ single mutants was also examined and found to cause a marked decrease in the level of unresolved recombination intermediates (supplemental Fig. S9). Based on these results we conclude that Mph1 promotes the formation of recombination intermediates that are dependent upon both Smc5/6 and Sgs1 for their timely resolution. Furthermore, these data demonstrate that the function of Sgs1 in the observed mph1Δ-mediated rescue was not solely through its role in X-structure resolution since these structures were decreased in a similar fashion in mms21-sp and mms21-sp sgs1Δ mutants upon MPH1 deletion. Instead, our data suggest that Sgs1 may play a critical role in an alternative DNA repair pathway that functions in the absence of the Mph1 helicase.
FIGURE 5.
Sgs1 is not required for the decreased levels of recombination intermediates seen upon MPH1 deletion. A, representative time course of 2DGE Southern blots examining replication intermediates near ARS305 in mms21-sp sgs1Δ and mms21-sp sgs1Δ mph1Δ mutants at the indicated times after release into 0.016% MMS. B, quantification of the ratio of X-shaped molecules to structures running within the replication arc.
A Mph1-independent Repair Pathway Provides DNA Damage Tolerance within smc5/6 Mutants
To better understand the Sgs1-dependent alternative DNA repair pathway, which we propose functions upon MPH1 deletion, we performed a series of targeted epistasis experiments. Our analysis revealed that deletion of MPH1 promoted DNA damage tolerance in smc5/6 mutants through a HR- and post-replicative repair (PRR)-dependent alternative repair pathway, because it required RAD51 and RAD52, as well as RAD5 and RAD18, respectively (Fig. 6, A and B and supplemental Fig. S10) (38, 39). Notably, our analysis also revealed that rescue of both MMS and HU sensitivity by removal of MPH1 was independent of various checkpoint proteins as well as factors required for nucleotide excision repair, non-homologous end joining, and single strand annealing repair pathways (supplemental Fig. S11).
FIGURE 6.
MPH1 deletion rescues the DNA damage-sensitivity of smc5/6 mutants through a HR- and PRR-dependent pathway. A, spot assays comparing the growth rate of strains mutant for members of the Smc5/6 complex along with deletions in MPH1 and RAD52. B, spot assays comparing the growth of smc6–9 mutants with and without MPH1 and members of the PRR machinery.
DISCUSSION
Smc5/6 mutants show a dramatic growth defect in the presence of DNA damaging agents; this growth is strongly rescued when the DNA helicase, Mph1, is no longer expressed. Data from our laboratory as well as other groups (40, 41) indicate that cells defective in HR-dependent repair suffer no further increase in DNA damage-sensitivity when MPH1 is also deleted, arguing that Mph1 mediates DNA damage tolerance through a HR-dependent pathway. Biochemical analyses of Mph1 homologues from P. furiosus, S. pombe, and H. sapiens have demonstrated a robust ability for Mph1 family members to act on DNA substrates which typify stalled replication forks (42–44). MPH1 deletion reduces the levels of recombination intermediates in mutants unable to resolve these structures (Figs. 2 and 5 and supplemental Figs. S3 and S10), suggesting that Mph1 acts early in the repair process, likely at the stalled replication fork, to influence the chosen DNA repair pathway (supplemental Fig. S12A). Furthermore, because the rescue that occurs when MPH1 is deleted from smc5/6 mutants is dependent upon PRR, we speculate that the PRR machinery offers an alternative repair pathway for the stalled replication fork (supplemental Fig. S12, A and D). Once engaged, the PRR machinery reverses the replication fork into a “chicken-foot” intermediate, allowing for the continuation of DNA synthesis using the complementary newly synthesized sister and thus bypassing the stall-inducing lesion (supplemental Fig. S12D). We propose that the resumption of DNA replication off of parental templates then occurs via a double Holliday junction (HJ) intermediate, which would require HR proteins for its biogenesis and would also employ the Sgs1 helicase for its seamless resolution (supplemental Fig. S12, E, F, and C) (45). This alternative repair pathway, occurring in the absence of Mph1 in smc5/6 mutants, would explain our observations that mph1Δ-mediated rescue requires the PRR and HR machinery, including the Sgs1 helicase. Interestingly a report characterizing the S. pombe Mph1 orthologue, Fml1, described rescue of HU sensitivity upon deletion of Fml1 from both smc6 and rqh1 (Sgs1 orthologue) mutants (44). The authors proposed that Fml1 initiates replication fork reversal that in smc6 and rqh1 mutants would be deleterious due to defect(s) in fork stabilization or downstream recombination intermediate resolution. We speculate that their findings together with our own suggest the ability of rqh1 mutants to benefit from fml1 deletion may arise from an additional mechanism in S. pombe that compensates for loss of Rqh1 function within the equivalent of our proposed Mph1/Fml1-independent repair pathway. Further work will be necessary to understand the basis for this difference in rescue between yeast species.
Our results demonstrating that the observed rescue upon MPH1 deletion is dependent on the HR machinery is intriguing given previous reports suggesting that the utilization of HR repair within smc5/6 mutants can be a deleterious event (10–13). Our data suggest that HR, as a whole, does not cause the DNA damage sensitivity of Smc5/6 mutants, but rather, a sub-pathway of HR-mediated repair under the control of the Mph1 helicase is detrimental when undertaken by cells defective for Smc5/6 function.
Similar to our findings, two previous reports examining sgs1Δ cells demonstrated no DNA damage resistance when MPH1 was deleted. In addition, both studies used 2DGE to examine the levels of unresolved X-shaped recombination intermediates in sgs1Δ versus sgs1Δ mph1Δ mutants, and both concluded that an Mph1-independent pathway of X-shaped intermediate generation explained the lack of rescue when MPH1 was deleted in sgs1Δ mutants (17, 46). In contrast to these studies, we observed a ∼50% decrease in X-shaped molecules upon removal of Mph1 from sgs1Δ cells. Because only one of the two previous studies (46) provided quantification of their 2D gel analysis (which, furthermore, involved only a single time point), where they note a ∼30% decrease in X-shaped structures upon deletion of MPH1 from sgs1Δ cells, it is difficult to determine if differences truly exist between our data and previous results.
It has been suggested that the observed X-shaped recombination intermediates that accumulate within smc5/6 and sgs1Δ mutants arise from independent DNA repair pathways. We instead propose that the X-shaped molecules found within smc5/6 and sgs1Δ mutants are derived from a single DNA repair pathway (template switch recombination), which is initiated by Mph1 and depends upon the Smc5/6 and Sgs1 complexes for its completion (supplemental Fig. S12, A–C). Several observations support this hypothesis 1) Mph1 promotes the formation of recombination intermediates within smc5/6 and sgs1Δ mutants, 2) the X-shaped molecules found within smc5/6 and sgs1Δ mutants have identical biochemical characteristics (15, 16), and 3) the level of unresolved recombination intermediates are not increased in sgs1Δ mms21-sp mutants as compared with sgs1Δ cells alone (14).
A recent report looking at stalled replication forks revealed that the Smc5/6 complex plays an important role in the recruitment of Rad52 to stalled replication forks (47). This recruitment was independent of Rad52 focus formation and was attributed to a role for Rad52 in maintaining the stalled fork in a recombination-competent position. While it is possible that the requirement we see for Rad52 in mph1Δ-mediated rescue can be partly attributed to its role in replication fork stability, we also find that the rescue depends upon Rad51 (which is not recruited to stably stalled forks), suggesting that Rad52 most likely has other roles in the observed rescue aside from stabilizing replication forks (supplemental Fig. S10).
In this study, we identify an important functional link between the Smc5/6 complex and the Mph1 and Sgs1 helicases. These results expand upon previous studies showing increased DNA damage sensitivity and accumulation of unresolved recombination intermediates within smc5/6 mutants. Our results suggest that one of the critical mechanisms mediating DNA damage sensitivity within smc5/6 mutants is the utilization of a HR-based DNA repair pathway that is promoted by Mph1.
Additional work is needed to know if our new model for how Mph1 initiates template switch recombination is conserved. The human Mph1 orthologue, FANCM, functions in conjunction with a multi-subunit Fanconi Anemia (FA) core complex. Defective proteins within this complex cause a variety of congenital abnormalities along with bone marrow failure and cancer predisposition (48). Interestingly, unlike other mutations within FA core complex members only a single patient has been found with biallelic mutations within FANCM (50). Surprisingly, this same patient also contained additional pathogenic mutations in each FANCA allele (26). How combined FANCA and FANCM mutations interact is not fully understood, but the rarity of observed FANCM patients suggests that deficiency within FANCM may yield phenotypes distinct from those caused by deficiencies in other FA complex members. Indeed, mice deleted for FANCM show more pronounced elevation in levels of sister-chromatid exchanges, reduction of life-span and tumor free survival when compared with mice mutant in other FA core complex members (49). A role for FANCM in template switch recombination may be more important than the role of the broader complex in crosslink repair, and explain these more severe consequences of FANCM deficiency.
Supplementary Material
Acknowledgments
We thank Amy Tsou, Amar Majmundar, Andrew Lippa, Avinash Bhandoola, Brian Keith, Xiaolan Zhao, and members of the Brown, Greenberg, and Johnson laboratories for their thoughtful discussions and/or comments on the manuscript. Charles Boone, Aaron Gitler, Luis Aragón, Vincent Guacci, Rodney Rothstein, and Kim Nasmyth are acknowledged for their generous gifts of yeast strains or plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AG021521 and P01-AG031862 (to F. B. J.) and T32-AG000255 (to A. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S12 and Table S1.
- SMC
- structural maintenance of chromosome
- Smc5/6
- Smc5-Smc6
- HR
- homologous recombination
- MMS
- methyl methanesulfonate
- HU
- hydroxyurea
- PRR
- post-replicative repair.
REFERENCES
- 1. Losada A., Hirano T. (2005) Genes Dev. 19, 1269–1287 [DOI] [PubMed] [Google Scholar]
- 2. Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., Ebmeier C. C., Goossens J., Rahl P. B., Levine S. S., Taatjes D. J., Dekker J., Young R. A. (2010) Nature 467, 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., Yahata K., Imamoto F., Aburatani H., Nakao M., Imamoto N., Maeshima K., Shirahige K., Peters J. M. (2008) Nature 451, 796–801 [DOI] [PubMed] [Google Scholar]
- 4. De Piccoli G., Torres-Rosell J., Aragón L. (2009) Chromosome Res. 17, 251–263 [DOI] [PubMed] [Google Scholar]
- 5. Murray J. M., Carr A. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 177–182 [DOI] [PubMed] [Google Scholar]
- 6. Potts P. R. (2009) DNA Repair. 8, 499–506 [DOI] [PubMed] [Google Scholar]
- 7. Prakash S., Prakash L. (1977) Genetics 87, 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Verkade H. M., Bugg S. J., Lindsay H. D., Carr A. M., O'Connell M. J. (1999) Mol. Biol. Cell 10, 2905–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lehmann A. R., Walicka M., Griffiths D. J., Murray J. M., Watts F. Z., McCready S., Carr A. M. (1995) Mol. Cell Biol. 15, 7067–7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ampatzidou E., Irmisch A., O'Connell M. J., Murray J. M. (2006) Mol. Cell Biol. 26, 9387–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cost G. J., Cozzarelli N. R. (2006) Genetics 172, 2185–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pebernard S., Wohlschlegel J., McDonald W. H., Yates J. R., 3rd, Boddy M. N. (2006) Mol. Cell Biol. 26, 1617–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Torres-Rosell J., Machín F., Farmer S., Jarmuz A., Eydmann T., Dalgaard J. Z., Aragón L. (2005) Nat. Cell Biol. 7, 412–419 [DOI] [PubMed] [Google Scholar]
- 14. Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. (2006) Cell 127, 509–522 [DOI] [PubMed] [Google Scholar]
- 15. Branzei D., Vanoli F., Foiani M. (2008) Nature 456, 915–920 [DOI] [PubMed] [Google Scholar]
- 16. Chavez A., George V., Agrawal V., Johnson F. B. (2010) J. Biol. Chem. 285, 11922–11930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Y. H., Choi K., Szakal B., Arenz J., Duan X., Ye H., Branzei D., Zhao X. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21252–21257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi K., Szakal B., Chen Y. H., Branzei D., Zhao X. (2010) Mol. Biol. Cell 21, 2306–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sollier J., Driscoll R., Castellucci F., Foiani M., Jackson S. P., Branzei D. (2009) Mol. Biol. Cell 20, 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kee Y., D'Andrea A. D. (2010) Genes Dev. 24, 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mankouri H. W., Ngo H. P., Hickson I. D. (2007) Mol. Biol. Cell 18, 4062–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alberti S., Gitler A. D., Lindquist S. (2007) Yeast 24, 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liberi G., Cotta-Ramusino C., Lopes M., Sogo J., Conti C., Bensimon A., Foiani M. (2006) Methods Enzymol. 409, 442–462 [DOI] [PubMed] [Google Scholar]
- 24. Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 25. Scheller J., Schürer A., Rudolph C., Hettwer S., Kramer W. (2000) Genetics 155, 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singh T. R., Bakker S. T., Agarwal S., Jansen M., Grassman E., Godthelp B. C., Ali A. M., Du C. H., Rooimans M. A., Fan Q., Wahengbam K., Steltenpool J., Andreassen P. R., Williams D. A., Joenje H., de Winter J. P., Meetei A. R. (2009) Blood 114, 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xue Y., Li Y., Guo R., Ling C., Wang W. (2008) Hum. Mol. Genet. 17, 1641–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pyle A. M. (2008) Annu. Rev. Biophys. 37, 317–336 [DOI] [PubMed] [Google Scholar]
- 29. Birkenbihl R. P., Subramani S. (1992) Nucleic Acids Res. 20, 6605–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim S. T., Xu B., Kastan M. B. (2002) Genes Dev. 16, 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindroos H. B., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. (2006) Mol. Cell 22, 755–767 [DOI] [PubMed] [Google Scholar]
- 32. Potts P. R., Porteus M. H., Yu H. (2006) EMBO J. 25, 3377–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J. E., Foiani M. (2005) Genes Dev. 19, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frei C., Gasser S. M. (2000) Genes Dev. 14, 81–96 [PMC free article] [PubMed] [Google Scholar]
- 35. Lu J., Mullen J. R., Brill S. J., Kleff S., Romeo A. M., Sternglanz R. (1996) Nature 383, 678–679 [DOI] [PubMed] [Google Scholar]
- 36. Bernstein K. A., Shor E., Sunjevaric I., Fumasoni M., Burgess R. C., Foiani M., Branzei D., Rothstein R. (2009) EMBO J. 28, 915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mullen J. R., Kaliraman V., Brill S. J. (2000) Genetics 154, 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krogh B. O., Symington L. S. (2004) Annu. Rev. Genet. 38, 233–271 [DOI] [PubMed] [Google Scholar]
- 39. Lee K. Y., Myung K. (2008) Molecules and Cells 26, 5–11 [PMC free article] [PubMed] [Google Scholar]
- 40. Lee W., St Onge R. P., Proctor M., Flaherty P., Jordan M. I., Arkin A. P., Davis R. W., Nislow C., Giaever G. (2005) PLoS Genet. 1, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schürer K. A., Rudolph C., Ulrich H. D., Kramer W. (2004) Genetics 166, 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gari K., Décaillet C., Stasiak A. Z., Stasiak A., Constantinou A. (2008) Mol. Cell 29, 141–148 [DOI] [PubMed] [Google Scholar]
- 43. Komori K., Hidaka M., Horiuchi T., Fujikane R., Shinagawa H., Ishino Y. (2004) J. Biol. Chem. 279, 53175–53185 [DOI] [PubMed] [Google Scholar]
- 44. Sun W., Nandi S., Osman F., Ahn J. S., Jakovleska J., Lorenz A., Whitby M. C. (2008) Mol. Cell 32, 118–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu L., Hickson I. D. (2003) Nature 426, 870–874 [DOI] [PubMed] [Google Scholar]
- 46. Mankouri H. W., Ngo H. P., Hickson I. D. (2009) Mol. Biol. Cell 20, 1683–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Irmisch A., Ampatzidou E., Mizuno K., O'Connell M. J., Murray J. M. (2009) EMBO J. 28, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. D'Andrea A. D. (2010) N. Engl. J. Med. 362, 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bakker S. T., van de Vrugt H. J., Rooimans M. A., Oostra A. B., Steltenpool J., Delzenne-Goette E., van der Wal A., van der Valk M., Joenje H., te Riele H., de Winter J. P. (2009) Hum. Mol. Genet. 18, 3484–3495 [DOI] [PubMed] [Google Scholar]
- 50. Meetei A. R., Medhurst A. L., Ling C., Xue Y., Singh T. R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C. G., Hoatlin M., Joenje H., de Winter J. P., Wang W. (2005) Nat. Genet. 37, 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.