Abstract
Members of the perilipin family of lipid droplet scaffold proteins are thought to play important roles in tissue-specific regulation of triglyceride metabolism, but the mechanisms involved are not fully understood. Present results indicate that adipose triglyceride lipase (Atgl) interacts with perilipin-5 (Plin5) but not perilipin-1 (Plin1). Protein interaction assays in live cells and in situ binding experiments showed that Atgl and its protein activator, α-β-hydrolase domain-containing 5 (Abhd5), each bind Plin5. Surprisingly, competition experiments indicated that individual Plin5 molecules bind Atgl or Abhd5 but not both simultaneously. Thus, the ability of Plin5 to concentrate these proteins at droplet surfaces involves binding to different Plin5 molecules, possibly in an oligomeric complex. The association of Plin5-Abhd5 complexes on lipid droplet surfaces was more stable than Plin5-Atgl complexes, and oleic acid treatment selectively promoted the interaction of Plin5 and Abhd5. Analysis of chimeric and mutant perilipin proteins demonstrated that amino acids 200–463 are necessary and sufficient to bind both Atgl and Abhd5 and that the C-terminal 64 amino acids of Plin5 are critical for the differential binding of Atgl to Plin5 and Plin1. Mutant Plin5 that binds Abhd5 but not Atgl was defective in preventing neutral lipid accumulation compared with wild type Plin5, indicating that the ability of Plin5 to concentrate these proteins on lipid droplets is critical to functional Atgl activity in cells.
Keywords: Fluorescence, Fluorescence Resonance Energy Transfer (FRET), Lipase, Lipid Droplet, Lipolysis, Protein Chimeras, Protein Targeting
Introduction
Regulation of triglyceride storage and mobilization is critically dependent on the subcellular targeting and trafficking of specific proteins. Recent work demonstrates that this trafficking involves scaffold proteins of the perilipin (Plin)2 family, including those that are ubiquitously expressed, such as Plin2 (adipose differentiation-related protein) and Plin3 (tail-interacting protein 47, TIP47), and those with restricted expression, such as Plin1 (perilipin) and Plin5 (muscle lipid droplet protein) that appear to have specialized functions (1). Although each Plin homolog has a conserved Plin domain (pfam 03036), amino acid sequences of family members diverge widely outside of this domain (2). Nonetheless, the sequences of individual orthologs are well conserved in mammals and suggest that important functions might be mediated by sequences outside of the Plin domain.
We have been investigating how Plin family members organize and regulate the trafficking of lipolytic effector proteins and have focused on Plin1 and Plin5 (3–6). Plin1 is expressed almost exclusively in adipose tissues and plays a central role in the storage of triglyceride and in the rapid mobilization of fatty acids by activators of protein kinase A (1). Recent work indicates that one means by which Plin1 regulates triglyceride storage and mobilization is by controlling the availability of α-β-hydrolase domain-containing 5 (Abhd5), a potent activator of adipose triglyceride lipase (Atgl) (4).
In contrast, Plin5 is highly expressed in tissues that have high rates of fatty acid oxidation, such as heart, skeletal muscle, and liver (7, 8). Interestingly, expression of Plin5 promotes both triglyceride storage and fatty acid oxidation. Plin5 expression is up-regulated by peroxisome proliferator-activated receptor α, and this regulation appears to be part of an expression program that shifts the metabolism of cells from fatty acid storage to oxidation (7).
Lipolysis occurs on the surface of intracellular lipid droplets, and several lines of evidence indicate that droplet targeting is critical to the cellular function of Abhd5 and Atgl (9, 10). Abhd5 is targeted to lipid droplets via direct interactions with Plin1 and Plin5 (5, 10, 11). However, unlike Plin1, Plin5 expression promotes the colocalization and interaction of Abhd5 and Atgl in unstimulated cells, which facilitates lipolysis (3–5). It is not known how Plin5 coordinates the interaction of Atgl and Abhd5. On the one hand, Plin5 could bind Atgl5 directly. Alternatively, Atgl might be recruited to the Plin5-containing lipid droplets by virtue of its interaction with Abhd5.
In the experiments below, we determined that Atgl interacts with Plin5 but not Plin1. Interestingly, although Plin5 binds both Abhd5 and Atgl, the same Plin5 molecule does not bind both at the same time. Protein complementation experiments, however, indicate that Plin5 forms homo-oligomers and suggest that Abhd5 and Atgl interact as part of this oligomeric structure. Analysis of chimeric and truncated Plin proteins demonstrates that binding of both Atgl and Abhd5 occurs outside of the Plin domain and that the C terminus of Plin5 is critical for the binding of Atgl. Comparison of wild type Plin5 with a Plin5/Plin1 chimera that does not bind Atgl confirmed that Plin5 coordinates the interaction of Atgl and Abhd5, thus preventing accumulation of intracellular triglyceride.
EXPERIMENTAL PROCEDURES
Immunohistochemistry and Biochemical Fractionation of Steatotic Mouse Livers
Male 129S1/SvImJ or C57BL/6J mice were untreated or infused with the β3-adrenergic receptor agonist CL 316,243 (CL, 0.75 nmol/h) for 24 h to mobilize fatty acids from adipose tissue and produce triglyceride accumulation in liver, as described previously (12). Livers of 129S1/SvImJ control and CL-treated were fixed in methacarn, and 5-μm-thick paraffin sections were processed for immunofluorescence histochemistry. Briefly, sections were blocked with 5% normal goat serum and then incubated with affinity-purified antibodies for Plin5 (5) or Atgl (Proteintech) (1.5 μg/ml). Control sections were incubated with the same concentration of rabbit IgG. Primary antibodies were detected with Cy5-conjugated goat anti-rabbit secondary antibodies, diluted 1:1000. For tissue fractionation, livers of control and CL-treated C57BL/6 mice were homogenized in 25 mm HEPES (pH 7.5), 20% sucrose buffer containing protease inhibitors. Post-nuclear supernatants were centrifuged at 16,000 × g for 15 min in a fixed angle rotor to obtain a crude lipid droplet fraction. Lipid droplet fractions were overlaid with HEPES buffer containing 10 and 0% sucrose and then centrifuged at 100,000 × g for 15 min in a swinging bucket rotor. The centrifuge tubes were then frozen, and the lipid droplet fractions were obtained by scraping the top 2 mm of the tubes. Proteins in the droplet fractions were precipitated with acetone and equal fraction volumes subjected to immunoblot analysis. All animal experiments were approved by the Institutional Animal Use and Care Committee of Wayne State University.
Generation of Fluorescent Fusion Proteins and Protein Complementation Constructs
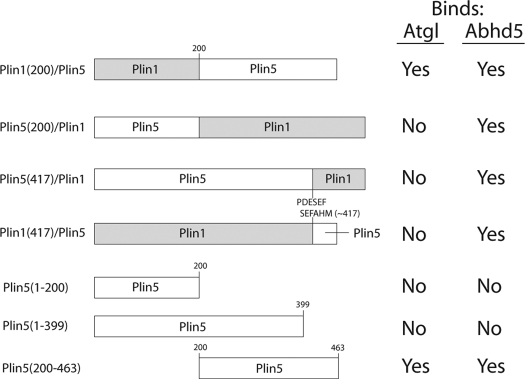
Construction and validation of fluorescently tagged proteins and Gaussia princeps luciferase protein complementation constructs (13) have been described (5). All constructs are based on mouse sequences. DNA encoding chimeric and truncated Plin proteins was generated by PCR. The primary structure and nomenclature of constructs used in this study are detailed in Table 1. All PCR-generated constructs were verified by DNA sequencing.
TABLE 1.
Schematic representation of the design, nomenclature, and binding results of chimeric and mutant Plin constructs used in this study
Cell Transfection for Subcellular Colocalization of Fluorescently Tagged Proteins, Fluorescence Resonance Energy Transfer (FRET), and Fluorescence Recovery after Photobleaching (FRAP)
COS7 or H4IIe cells were transfected with Plin1-EYFP or Plin5-EYFP and ECFP-Atgl or ECFP-Abhd5, incubated with oleic acid (400 μm complexed to BSA) overnight to promote lipid droplet formation, and imaged live the next day.
Luciferase Protein Complementation Analysis
COS7 or 293T cells were grown in 24- or 48-well plates and were transfected in triplicate or quadruplicate with appropriate N- and C-luciferase fragments fused to Atgl, Abhd5, wild type Plin1 and Plin5, and mutant Plin molecules, as specified under “Results.” Cells were cultured for 18–24 h in media containing oleic acid, and luciferase activity was determined as described previously (5).
Binding of Recombinant ECFP-Atgl and Abhd5-Cherry to Plin5-EYFP in Permeabilized Cells
Recombinant ECFP-Atgl and Abhd5-Cherry were prepared from lysates of transiently transfected 293T cells, as described previously (5). Briefly, transfected 293T cells were washed in PBS, collected by centrifugation, and suspended in intracellular buffer (IB: 10 mm HEPES (pH 7.3), 140 mm KCl, 6 mm NaCl, 1 mm MgCl2, 2 mm EGTA) at a concentration of 0.5 ml/10-cm plate. Cells were frozen and thawed, then passed 15 times through a 26-gauge needle, and centrifuged for 10 min at 16,000 × g, and the fluorescence of supernatants was determined. COS7 cells were transfected with Plin5-EYFP, Plin1-EYFP, or fluorescent Plin1/5 chimeras as above. After 24 h, cells were lightly fixed in 1% fresh paraformaldehyde at 4 °C for 30 min, washed with PBS, and then permeabilized for 30 min at room temperature in IB containing 0.02% saponin and 1% BSA. Permeabilized cells were washed once in IB and incubated with ECFP/Atgl extracts for 45 min at room temperature. Binding reactions were washed rapidly 2–3 times with IB and then postfixed with 1% paraformaldehyde prior to confocal microscopic analysis.
Two additional experiments were performed to examine the ability of Plin5 to bind both Abhd5 and Atgl simultaneously. In one experiment, permeabilized COS7 cells expressing Plin5-EYFP were incubated with ECFP-Atgl alone or with an excess of Abhd5-Cherry. In a second experiment, binding of Abhd5-Cherry was performed on fixed permeabilized cells that had been cotransfected with ECFP-Atgl and Plin5-EYFP.
Microscopy
Images for colocalization, binding, and fluorescence resonance energy transfer (FRET) were acquired with an Olympus IX-81 microscope equipped with a spinning disc confocal unit. Microscope control and data acquisition were performed using IPlabs (Scanalytics, BD Biosciences) software. Images were captured using ×40 (0.9 NA) or ×60 (1.2 NA) plan apo water immersion lens and a Hamamatsu ORCA cooled CCD camera. The filter combinations used are listed below according to the format fluorophore, exciter(bandwidth)/dichroic/emitter(bandwidth) as follows: mCherry, 575(50)/610LP/640(50); ECFP, 436(20)/455LP/480(40); EYFP, 500(20)/515LP/535(30); Nile Red, 535(50)/556LP/610(75); LipidTox Deep Red; and Cy5, 628(40)/660BP/692(40). EYFP FRET used the ECFP exciter and the EYFP dichroic/emission filters. FRAP experiments were performed using a Leica TCS SP5 laser-scanning microscope with a ×63 (1.4 NA) oil objective. FRAP was performed using the Leica FRAP wizard module. Briefly, a region of interest (ROI) on a patch of lipid droplets was bleached by 1–2 scans (2.6 s/scan) with full laser power from the 405, 458, and 496 nm lasers. ECFP-Abhd5 and ECFP-Atgl fluorescence was monitored by 5% 405 nm laser excitation and 454–493 nm emission.
Image Analysis
FRET was performed using the three filter method, and net FRET was calculated using the FRET extension of IPlabs software, as described previously (3). For FRAP experiments, fluorescence intensity was monitored in the bleached ROI and in an unbleached ROI on droplets in the same focal plane. Average fluorescence intensity of the bleached ROI was compared with pre-bleach level (with 100% bleach, 0% recovery = first post-bleach scan), with correction for bleaching during acquisition. Rate and extent of recovery were estimated by fitting recovery values to a single exponential association function by nonlinear regression (GraphPad software).
Binding of fluorescent Atgl and Abhd5 to fluorescent Plin variants was assessed by linear regression of line scans. Briefly, the fluorescence channels corresponding to fluorescent proteins were merged, and fluorescence intensities were collected pixel by pixel over cellular regions that included both lipid droplet and cytosolic regions of transfected cells. Fluorescence intensities (>500 pixels/cell) were evaluated by Pearson's linear regression (GraphPad), with the regression slope indicating the amount of Atgl or Abhd5 bound per unit Plin over a range of Plin, and the y intercept estimating nonspecific binding. The binding of Abhd5-Cherry to cells coexpressing ECFP-Atgl and Plin5-EYFP was evaluated by multiple linear regression (Microsoft Excel), with Plin5-EYFP and ECFP-Atgl fluorescence intensities as independent predictors of Abhd5-Cherry binding.
Lipid Droplet Formation and Accumulation of Neutral Lipids
COS7 cells were transfected with Plin5-EYFP or Plin5/Plin1-EYFP chimera with ECFP-Atgl and Abhd5-Cherry at a plasmid ratio of 1:1:1. Following transfection, cells were incubated with media supplemented with 400 μm oleic acid complexed to BSA and fixed in 1% paraformaldehyde after 18 h. Cells were localized using EYFP fluorescence and scored as to the presence or absence of lipid droplet clusters by an analyst who was blind to the transfection conditions. Three coverslips were evaluated per condition per experiment, and the results of four independent experiments (>1600 cells/condition) were combined for presentation.
Statistical Analyses
Quantitative data were evaluated by analysis of variance, and group means were compared by Bonferroni's test. Nominative data (cell frequencies) were assessed by χ2 statistic.
RESULTS
Plin5, Atgl, and Abhd5 Colocalize on Lipid Droplets of Steatotic Mouse Livers
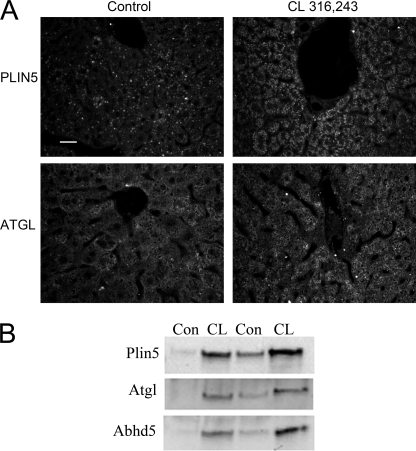
The localization of Plin5, Atgl, and Abhd5 was determined in a model of acute hepatic steatosis that we have developed. In this model, fatty acids are mobilized from adipose tissue by activating β3-adrenergic receptors with CL and deposited in the liver as triglycerides. As shown in Fig. 1A, control livers had relatively few intracellular lipid droplets that contained Plin5, whereas CL treatment led to pronounced hepatic steatosis in which numerous intracellular lipid droplets were heavily stained with antibodies to Plin5. In control livers, Atgl immunofluorescence appeared to be mostly cytosolic, with occasionally targeting to lipid droplets, whereas the lipase was strongly targeted to hepatic lipid droplets of CL-treated mice. Biochemical fractionation experiments confirmed the colocalization of Plin5, Atgl, and Abhd5 in hepatic lipid droplet fractions of CL-treated mice (Fig. 1B).
FIGURE 1.
Colocalization of Plin5, Atgl, and Abhd5 on lipid droplets of steatotic liver. A, immunofluorescence localization of Plin5 and Atgl was performed on liver sections of control mice and mice treated with CL to produce hepatic steatosis. B, immunoblot detection of Plin5, Atgl, and Abhd5 in lipid droplet fractions of control (Con) and CL-treated mice. Bar, 20 μm.
Atgl Colocalizes and Interacts with Plin5 but Not Plin1
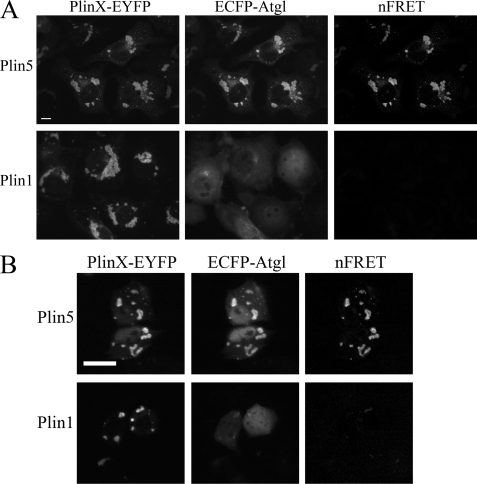
We previously reported that Plin5 expression supports the colocalization of Abhd5 and Atgl in COS7 cells, whereas Plin1 expression does not (5). These observations raised the possibility that Plin5 and Plin1 might differentially interact with Atgl and provide a mechanism for the colocalization of Plin5 Abhd5 and Atgl on lipid droplets. We examined the subcellular distribution of ECFP-Atgl in COS7 cells that were cotransfected with Plin1-EYFP or Plin5-EYFP and found that ECFP-Atgl was precisely colocalized to lipid droplets containing Plin5-EYFP in greater than 97% of transfected cells (Fig. 2A). In contrast, ECFP-Atgl was colocalized to lipid droplets containing Plin1-EYFP in fewer than 1% of cells (p < 0.001). The molecular proximity of ECFP-Atgl and Plin5-EYFP permitted fluorescence energy transfer between the fluorescent reporters, indicating a close (<8 nm) and likely direct interaction. Similar results were obtained in H4IIe hepatoma cells, in which the fluorescent reporters were expressed at much lower levels (∼10%) compared with COS7 cells (Fig. 2B).
FIGURE 2.
ECFP-Atgl colocalizes and interacts with Plin5 but not Plin1. A, interaction of ECFP-Atgl and Plin5-EYFP in COS7 cells. COS7 cells were transiently transfected with ECFP-Atgl and EYFP-tagged Plin5 or Plin1 and imaged by confocal microscopy. ECFP-Atgl was highly targeted to lipid droplets containing Plin5-EYFP but not to droplets containing Plin1. The colocalization of ECFP-Atgl and Plin5-EYFP supported FRET on lipid droplets. Bar, 10 μm. B, interaction of ECFP-Atgl and Plin5-EYFP in H4IIe hepatoma cells. Cotransfection of Plin5-EYFP and ECFP-Atgl leads to colocalization on lipid droplets and FRET between fluorescent proteins. In contrast, Plin1-EYFP did not support droplet targeting of ECFP-Atgl or FRET. Bar, 10 μm.
Atgl Binds to Plin5 in Permeabilized Cells
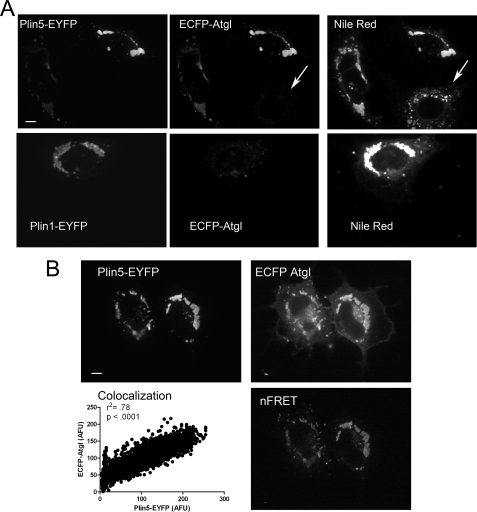
We next determined whether ECFP-Atgl, prepared from lysates from 293T cells, would differentially bind Plin5- and Plin1-EYFP in permeabilized COS7 cells. As shown in Fig. 3A, ECFP-Atgl bound to lipid droplets containing Plin5-EYFP but not to lipid droplets of adjacent untransfected cells (arrow). Furthermore, no specific binding of ECFP-Atgl was detected on lipid droplets of cells expressing Plin1-EYFP. The binding of ECFP-Atgl occurred in direct proportion to Plin5-EYFP concentration and supported energy transfer between the fluorescent proteins (Fig. 3B).
FIGURE 3.
ECFP-Atgl selectively binds Plin5-EYFP in permeabilized cells. A, binding of recombinant ECFP-Atgl in permeabilized COS7 cells expressing EYFP-tagged Plin5 or Plin1. Permeabilized cells were incubated with the same preparation of ECFP-Atgl, and all capture parameters were the same for each fluorescence channel. ECFP-Atgl bound to lipid droplets (post-stained with Nile Red) of cells transfected with Plin5, but not to untransfected cells (arrow) or cells expressing Plin1. B, cells expressing Plin5-EYFP were permeabilized and bound with recombinant ECFP-Atgl as detailed under “Experimental Procedures.” Atgl bound in direct proportion to Plin5 concentration as demonstrated by linear regression of pixel intensities (see also A). In addition, bound ECFP-Atgl supported FRET with droplet-associated Plin5, indicating close interaction. Bar, 10 μm.
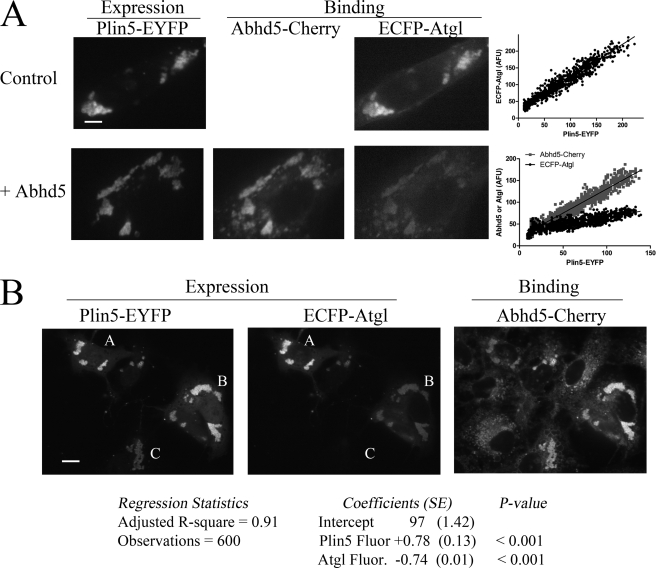
Interaction of Atgl and Abhd5 with Plin5 Is Mutually Exclusive
Abhd5 binds Plin5 (4), and cells that express Plin5 often express both Abhd5 and Atgl (5, 7, 15–17). These observations raised the question of the mode of interaction among Plin5, Abhd5, and Atgl in cells expressing all three proteins. We previously reported that Plin5 supports the colocalization of Abhd5 and Atgl in live COS7 cells, whereas Plin1 does not (5). Retrospective analysis of these experiments surprisingly indicated cells having high levels of Abhd5 bound to Plin5 had relatively low levels of bound Atgl, suggesting mutually exclusive interactions with Plin5. However, no firm conclusion could be drawn because the concentrations of free Atgl and Abhd5 were not directly controlled in these experiments. Therefore, we examined the binding of a fixed concentration of ECFP-Atgl to Plin5 in the absence and presence of excess Abhd5-Cherry. Fig. 4A shows the results of a typical experiment. In the absence of Abhd5, Atgl bound lipid droplets in direct proportion to Plin5 concentration (Fig. 4A, top panel). The presence of excess Abhd5-Cherry greatly reduced ECFP-Atgl binding at the expense of strong Abhd5-Cherry binding to Plin5-containing lipid droplets (Fig. 4A, bottom panel). The amount of ECFP-Atgl bound per unit Plin5-EYFP was estimated by the regression slope and was found to be reduced by Abhd5-Cherry from 0.99 ± 0.013 to 0.34 ± 0.007. In two independent experiments, competition with Abhd5-Cherry reduced binding of ECFP-Atgl to Plin5 by 58 ± 12% (p = 0.0082).
FIGURE 4.
Mutually exclusive binding of Atgl and Abdh5 to Plin5. A, binding of Abhd5-Cherry suppresses binding of ECFP-Atgl to Plin5-EYFP. COS7 cells expressing Plin5-EYFP were fixed and permeabilized and then bound with ECFP-Atgl alone (top panel) or with Abhd5-Cherry (bottom panel). Images are from a representative experiment, and all capture parameters were equal for each fluorescence channel. Coincubation with Abhd5-Cherry significantly reduced binding of ECFP-Atgl as assessed by the regression analysis (p < 0.001). B, binding of ECFP-Atgl to Plin5-EYFP prevents binding of Abhd5-Cherry. COS7 cells coexpressing variable amounts of ECFP-Atgl and Plin5-EYFP were fixed and permeabilized and then bound with Abhd5-Cherry. A, B, and C are cells with variable amounts of bound ECFP-Atgl. Binding was assessed by the intensity of bound Abhd5-Cherry fluorescence as a function of Plin5 fluorescence (Plin5 Fluor.) and Atgl fluorescence (Atgl Fluor.) intensities. Binding was directly proportional to Plin 5 fluorescence (slope = +0.78) and inversely proportional to Atgl fluorescence (slope = −0.74). Bar, 10 μm.
Because Abhd5 and Atgl interact (4, 14), it is conceivable that free Abhd5 might have acted as a “decoy” to suppress Atgl binding to Plin5. To address this possibility, we examined the binding of Abhd5-Cherry to permeabilized cells expressing variable levels of ECFP-Atgl that had been cross-linked to Plin5-EYFP. Fig. 4B shows a typical result in which cells marked A–C had high, medium, or low levels of ECFP-Atgl bound to Plin5-EYFP. We found that the binding of Abhd5-Cherry to Plin5 was systematically reduced in cells with high levels of bound Atgl. For example, cells A and B have similar levels of Plin5-EYFP, and the reduced binding of Abhd5-Cherry in cell A is associated with high levels of bound ECFP-Atgl. The impact of Plin5 and Atgl concentration on Abhd5 binding across all transfected cells was evaluated by multiple regression of pixel intensities. This analysis confirmed ECFP-Abhd5 binding is directly proportional to Plin5-EYFP concentration and inversely proportional to bound Atgl concentration. Multiple and partial regression coefficients were >0.9, strongly indicating that binding of Atgl and Abhd5 to Plin5 is mutually exclusive.
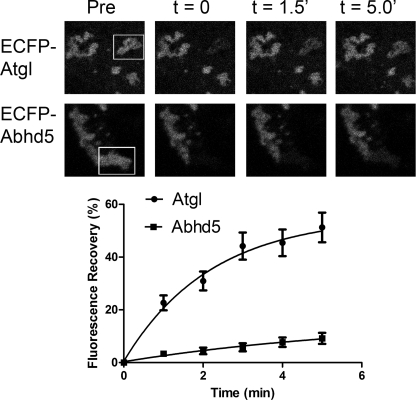
Plin5-Abhd5 Complexes on Lipid Droplets Are More Stable Than Plin5-Atgl Complexes
The experiments above establish that Plin5 increases the steady-state concentration of Abhd5 and Atgl on lipid droplets. We next examined fluorescence recovery after photobleaching to determine the relative stability of Plin5 complexes on lipid droplets. As shown in Fig. 5, fluorescence of ECFP-tagged Atgl recovered with an estimated half-time of 1.60 ± 0.18 min. The maximal recovery of Atgl fluorescence was 52 ± 4%, indicating that bound Atgl may also exist in a pool that exchanges much more slowly. ECFP-Abhd5 fluorescence also clearly recovered after photobleaching, but the magnitude of recovery was significantly less than ECFP-Atgl during the 5-min recording period, indicating that Abhd5-Plin5 complexes are much more stable. In most cells, the kinetics of recovery of ECFP-Abhd5 fluorescence were too slow to be accurately estimated; however, in those cells exhibiting substantial recovery (>20%), recovery half-times were estimated to be between 2 and 5 min.
FIGURE 5.
Complexes of ECFP-Abhd5 and Plin5-EYFP on lipid droplets are more stable than complexes of ECFP-Atgl and Plin5-EYFP. Top panel, confocal micrographs of cells before (Pre) and after photobleaching of indicated region of interest (box). Bottom panel, summary of recovery in 10–14 bleaching experiments. Composite data were fit to a single exponential association function by nonlinear regression.
Differential Interactions among Plin Homologs and Lipolytic Effectors
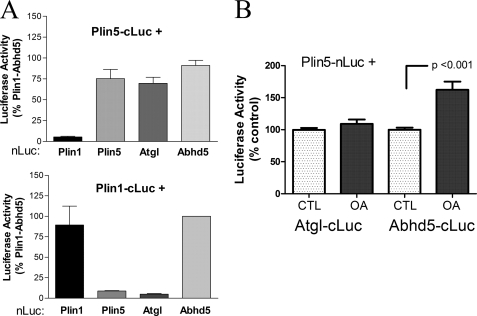
We next evaluated protein-protein interaction among Plin homologs and potential binding partners with luciferase complementation assays in 293T cells. As shown in Fig. 6A, Plin5 produced strong luciferase complementation when coexpressed with Atgl or Abhd5. In contrast, Plin1 did not interact with Atgl but did interact with Abhd5, as expected (5). Interestingly, Plin5 interacted with itself, indicating the formation of higher order structures. Plin1 also interacted with itself, as expected (3). Importantly, Plin1 and Plin5 did not form hetero-oligomers, supporting the specificity of the interaction.
FIGURE 6.
Protein-protein interaction of Atgl with Plin5 and Plin1 assessed by luciferase complementation assays. A, Plin5 interacts with Atgl, but Plin1 does not. 293T cells were transiently transfected with Plin5 or Plin1 tagged with the C-terminal fragment of Gaussia princeps luciferase (cLuc) and the complementary N-terminal luciferase (nLuc) fragment fused to potential binding partners. Complementation of luciferase activity was used to evaluate protein-protein interaction. Plin5 interacted with Atgl, Abhd5, and itself but not with Plin1. Plin1 interacted with Abhd5 and itself but not with Atgl or Plin5. Results are from four independent experiments. B, oleic acid (OA) loading promotes the interaction of Plin5 with Abhd5 but not with Atgl. COS7 cells were cotransfected with complementary fragments of luciferase and then exposed to 400 μm oleic acid or BSA (CTL) for 2 h prior to determining luciferase activity. Oleic acid loading selectively increased the interaction of Plin5 with Abhd5. Results are from four independent experiments.
Brief treatment of cells with oleic acid has been shown to promote the interaction of Abhd5 with Plin5 in a manner that is dependent upon triglyceride synthesis (5), so it was of interest to determine whether oleic acid affects the interaction of Plin5 with Atgl. As shown in Fig. 6B, a 2-h oleic acid treatment increased the interaction of Plin5 with Abhd5 by 62% (p < 0.001) but had no effect on the interaction of Plin5 and Atgl.
C Terminus of Plin5 Specifies Its Interactions with Atgl: Analysis of Plin1/Plin5 Chimeras, and Truncation Mutants of Plin5
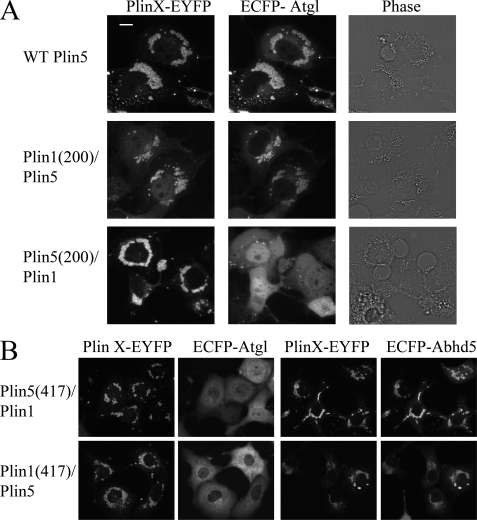
Plin family members share a protein domain (perilipin domain, pfam number = 03036) that defines its members. In the case of Plin1 and Plin5, the greatest region of conservation is in the N-terminal 200 amino acids, whereas sequences outside of this domain are far more divergent. We explored the interaction of Atgl to a series of mutant and chimeric Plin polypeptides by subcellular colocalization, FRET, protein complementation, and binding of recombinant proteins in permeabilized cells. The nomenclature of the mutant constructs and results are summarized in Table 1. We first evaluated binding to chimeric proteins which, unlike truncation mutants, maintained proper targeting to lipid droplets. Our results demonstrated that Atgl binding was conferred by the sequences in the C-terminal half of Plin5 (Fig. 7A). Thus, ECFP-Atgl colocalized with Plin1(1–200)/Plin5 in 89% of transfected cells, whereas ECFP-Atgl failed to associate with Plin5(1–200)/Plin1(0/108 cells; p < 0.0001). Atgl selectively interacted with Plin1(1–200)/Plin5 in protein complementation assay and in binding assays in permeabilized cells (data not shown). Swapping the C termini of Plin homologs (amino acids 417–463 of Plin5 and amino acids 417–517 of Plin1) effectively eliminated Plin interactions with Atgl but allowed proper targeting to lipid droplets and interaction with Abhd5 (Fig. 7B).
FIGURE 7.
Colocalization of ECFP-Atgl with wild type Plin5 and Plin1/5 chimeras. A, colocalization of ECFP-Atgl with Plin requires expression of Plin5 amino acids 200–463. Bar, 10 μm. B, swapping the C terminus of Plin5 with sequence from Plin1 abolished interaction with Atgl but not with Abhd5 (top panel). However, exchanging the C terminus of Plin1 with that of Plin5 did not allow interaction of ECFP-Atgl with Plin1 (bottom panel).
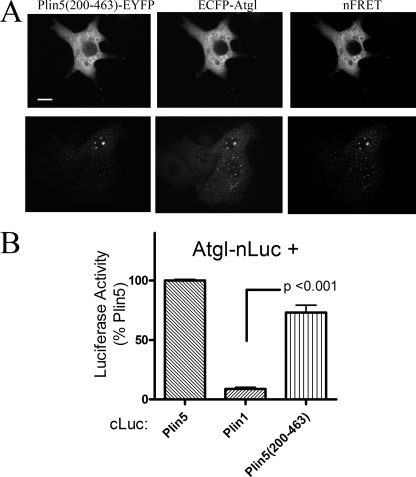
The results with chimeric proteins localized the determinants of Atgl binding to the non-Plin domain of Plin5 and demonstrated the requirement for the C-terminal 46 amino acids. We therefore examined a truncation mutant, Plin5(200–463), that eliminated the conserved N-terminal perilipin domain while retaining the critical elements identified above (Fig. 8A). When expressed alone or with ECFP-Atgl, Plin5(200–463) appeared cytosolic in the majority of transfected cells. Plin5(200–463)-EYFP and ECFP-Atgl were highly colocalized within the cytoplasm of these cells as evidenced by strong FRET between the fluorescent proteins (Fig. 8A, right panel). In about 25% of transfected cells, Plin5(200–463)-EYFP accumulated on small (<1 μm) punctate vesicle-like structures (Fig. 8A, bottom panel). ECFP-Atgl was precisely targeted to these structures and exhibited strong FRET with Plin5(200–463)-EYFP. It is important to note that ECFP-Atgl was never observed on such punctate structures in the absence of mutant Plin5 and that Plin5(200–463) expression alone was sufficient to induce the appearance of these structures. Interestingly, the structures are not typical lipid droplets because they were not stained by neutral lipid dyes (data not shown). Luciferase complementation assays confirmed the interaction of Atgl with Plin5(200–463) (Fig. 8B). Evaluation of cellular colocalization, FRET, and protein complementation assays leads us to conclude that the determinants of Atgl binding reside exclusively in the C-terminal half of Plin5. In further support of this conclusion, Plin5 truncation mutants encompassing amino acids 1–200 and 1–399 failed to colocalize with ECFP-Atgl or target endogenous lipid droplets (Table 1, data not shown).
FIGURE 8.
C-terminal half of Plin5 mediates interactions with Plin5. A, targeting and colocalization of Plin5(200–463)-EYFP in COS7 cells. Plin5(200–463)-EYFP was targeted to cytosol (top panel) and to punctate vesicle-like structures that lack a neutral lipid core (bottom panel). Regardless, ECFP-Atgl was highly colocalized with truncated Plin5, as shown by strong FRET. Bar, 10 μm. B, Atgl interacted with Plin5(200–463) in luciferase complementation assay. Values are from four independent assays.
Plin5 Coordinates the Functional Interaction of Atgl and Abhd5
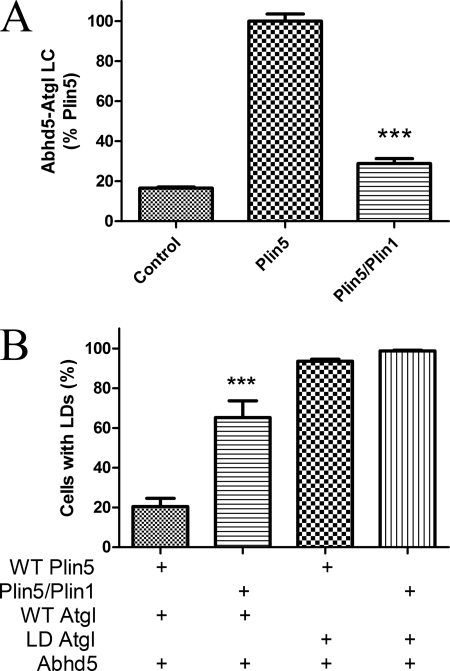
We previously reported that Plin5 expression concentrates Abhd5 and Atgl on lipid droplets and thereby prevents accumulation of intracellular lipid in cells challenged with oleic acid (5). In those experiments, we found that mutant Abhd5 that does not bind Plin5 does not promote Atgl activity. To extend these findings, we compared wild type Plin5 with chimeric Plin5(417)/Plin1 that does not bind Atgl because of replacement of the C-terminal 46 amino acids with a sequence derived from Plin1. Both Plin proteins were targeted to lipid droplets and bound Abhd5 similarly (Fig. 7B). As expected from targeting data, expression of Plin5 greatly increased the interaction of Abhd5 and Atgl, as detected by luciferase complementation assay, and this effect was significantly reduced in cells expressing mutant Plin5 (Fig. 9A). The functional effect of this interaction was investigated by examining lipid droplet accumulation in transfected cells. As shown in Fig. 9B, few cells expressing wild type Plin5, Atgl, and Abhd5 had few visible lipid droplets when challenged with oleic acid, whereas a significantly higher percentage of cells expressing mutant Plin5 accumulated lipid droplets. This large difference in the percentage of cells with lipid droplets was not observed in cells expressing lipase-dead Atgl, indicating that the interaction of Plin5 with Atgl facilitates its activity.
FIGURE 9.
Interaction of Atgl with Plin5 facilitates Atgl activity. A, COS7 cells were transfected with Abhd5 and Atgl fused to complementary fragments of luciferase along with wild type Plin5-EYFP or Plin5(417)/Plin1-EYFP (Plin5/Plin1) or control (ECFP) vectors. Coexpression of Plin5 greatly increased luciferase complementation (LC), and this effect was significantly impaired in cells expressing Plin5/1, which does not bind Atgl. Graph is summary of three independent experiments performed in quadruplicate. ***, p < 0.001. B, COS7 cells were transfected with plasmids encoding wild type (WT) Plin5-EYFP or Plin5/Plin1-EYFP, along with Abhd5-Cherry and wild type or lipase-dead (LD) ECFP-Atgl. Transfected cells were scored as to the presence of lipid droplet clusters by an analyst that was blind to transfection conditions. The graph is a summary of four independent experiments. Significantly more cells expressing Plin5/Plin1 accumulated lipid droplets versus WT Plin5, and this effect required active Atgl. ***, p < 0.001.
DISCUSSION
Growing evidence indicates that the Plin family members play a central role in the organization and trafficking of lipolytic effectors to lipid droplet surfaces. Although Plin family members share a domain on the N terminus, the C-half of the proteins is not well conserved. Furthermore, some members like Plin1 and Plin5 have restricted tissue distribution and are subject to distinct physiological regulation. Together, these observations suggest that various Plin paralogs serve distinct functions that are mediated by divergent sequences in the proteins.
Plin1 and Plin5 both bind Abhd5; however, Plin5 promotes the colocalization and functional interaction of Abhd5 and Atgl in the basal state (5). In contrast, Abhd5 and Atgl do not colocalize in cells expressing Plin1 in the absence of stimulation. Indeed, Plin1 expression suppresses Abhd5-Atgl interactions in the basal state and promotes their interaction upon phosphorylation of Plin1 by protein kinase A (4). These observations suggest that Atgl might interact differentially with Plin1 and Plin5.
We investigated the mechanism by which Plin5 concentrates Atgl on lipid droplets in colocalization, in situ binding, and protein interaction assays. Results from these experiments demonstrate that Atgl interacts with Plin5 and not with Plin1. Because Plin5 binds both Abhd5 and Atgl, it was of interest to examine the mode of binding when all three proteins are present. Binding of Atgl to Plin5 was not mediated by Abhd5 bound to Plin5 because Atgl bound Plin5 in the absence of Abhd5 and did not bind Plin1, which also binds Abhd5. Furthermore, binding experiments demonstrated that Atgl and Abhd5 compete for Plin5-binding sites and, once bound, Atgl potently suppresses binding of Abhd5. Thus, individual Plin5 molecules bind Abhd5 or Atgl, but not both at the same time. These data indicate that there is some overlap in the interaction sites or that the binding of one partner produces a conformational change that prevents binding of the other. In support of the former possibility, we note that all mutations that disrupt Abhd5 binding also disrupt Atgl binding.
The interaction assays used in this study relied on recombinant expression in mammalian cells. Because we have not yet demonstrated the interaction of Plin5 and Atgl with purified proteins, we cannot conclusively exclude the possibility that the interaction involves an unknown cellular component. However, we note that the interaction of Atgl with Plin5 is directly proportional to Plin5 concentration over a large range of Plin5 expression and occurs in (and between) at least four diverse cell types. Moreover, Atgl interacts with Plin5(200–463) in the cytoplasm and on intracellular structures lacking neutral lipid, indicating that the interaction does not require lipid droplets.
The observation that Abhd5 and Atgl interact separately with Plin5 was surprising and raises questions as to how these proteins interact on the lipid droplet surface. Plin5 clearly concentrates Atgl and Abhd5 on intracellular lipid droplets and facilitates their interaction (5). Furthermore, this concentrating effect is important for promoting lipolysis. Thus, targeting of Atgl to Plin5 alone is insufficient to promote cellular lipolysis in the absence of Abhd5, and mutations that prevent proper targeting of Abhd5 (5) or Atgl (this work) greatly reduce lipolysis when both proteins are present. The simplest explanation is that Abhd5 and Atgl interact while associated with Plin5. Protein complementation data indicate that Plin5 forms oligomers that may serve to coordinate the interaction and facilitate lipolysis. It is possible that Plin5 exerts conformational effects on Atgl or Abhd5 that influence their interaction. For example, Abdh5 may preferentially interact with bound Atgl.
The interactions among Plin5, Atgl, and Abhd5 in the regulation of triglyceride metabolism are likely to be complex and may depend on the metabolic status of the cell. Plin5 itself traffics between cytosol and lipid droplets. Plin5 and Atgl are both up-regulated by fasting (7, 18), which increases basal lipolysis. Under these conditions, Plin5 may concentrate Atgl on lipid droplets which, along with increased availability of Abhd5, elevates basal lipolysis. In addition, it should be recognized that Abhd5 and Atgl have multiple independent activities (14, 19–21), and it is conceivable that the ability of Plin5 to concentrate these proteins at droplet surfaces influences those activities as well.
Analysis of chimeric and mutant proteins indicates that the binding of both Atgl and Abhd5 is conferred by amino acids 200–463 of Plin5, which is outside of the conserved PAT1 domain (after Perilipin-Adipophilin-TIP47). This sequence is most similar to amino acids 191–437 of Plin3 (also known as TIP47), for which the crystal structure is known (22). The domain is thought to fold into a single structural unit containing four helix bundles and a compact α/β domain that together form an L-shape. The four-bundle α-helices are structurally similar to the N-terminal region of apoE and may be involved in lipid interactions (22). It is not presently known whether Abhd5 or Atgl bind Plin3. However, Plin3 has been found to suppress the retinyl ester hydrolase activity of PNPLA5 (GS2), presumably by binding, and this effect requires intact four-helix bundles of Plin3. Atgl (PNPLA2) and PNPLA5 are paralogs of the patatin protein family of esterase/phospholipases (23). Taken together, these observations suggest possible coevolution of Plin and patatin family members.
Additional studies will be required to determine the role that Plin5 plays in coordinating Atgl and Abhd5 activity in vivo. In this regard, targeting of Abhd5 and Atgl to lipid droplet fractions is strongly correlated with expression of Plin5, but not Plin2 or Plin3, in mice fed a high fat diet (24), similar to the present results with drug-induced fatty liver. Proper targeting appears to be critical for the functioning of Atgl and Abhd5 in vivo, as it is in vitro, because certain mutations of these proteins that produce disease in humans interfere with lipid droplet targeting, rather than abrogating enzymatic activity per se (5, 9, 10). Finally, our FRAP and protein complementation data indicate that the interactions of Plin5 with Abhd5 and Atgl are dynamic, which may allow further regulation by nutritional and hormonal signals.
Acknowledgments
We thank R. Granneman and A. Petkova for technical assistance and Drs. T. Leff, R. MacKenzie, and V. Kimler for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK62292 and DK76629. This work was also supported by the Department of Veterans Affairs.
- Plin
- perilipin
- Atgl
- adipose triglyceride lipase
- FRAP
- fluorescence recovery after photobleaching
- ROI
- region of interest
- CL
- CL 316,243
- EYFP
- enhanced YFP
- ECFP
- enhanced cyan fluorescent protein.
REFERENCES
- 1. Brasaemle D. L. (2007) J. Lipid Res. 48, 2547–2559 [DOI] [PubMed] [Google Scholar]
- 2. Londos C., Brasaemle D. L., Schultz C. J., Segrest J. P., Kimmel A. R. (1999) Semin. Cell Dev. Biol. 10, 51–58 [DOI] [PubMed] [Google Scholar]
- 3. Granneman J. G., Moore H. P., Granneman R. L., Greenberg A. S., Obin M. S., Zhu Z. (2007) J. Biol. Chem. 282, 5726–5735 [DOI] [PubMed] [Google Scholar]
- 4. Granneman J. G., Moore H. P., Krishnamoorthy R., Rathod M. (2009) J. Biol. Chem. 284, 34538–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Granneman J. G., Moore H. P., Mottillo E. P., Zhu Z. (2009) J. Biol. Chem. 284, 3049–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moore H. P., Silver R. B., Mottillo E. P., Bernlohr D. A., Granneman J. G. (2005) J. Biol. Chem. 280, 43109–43120 [DOI] [PubMed] [Google Scholar]
- 7. Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Croce M. A., Gropler M. C., Varma V., Yao-Borengasser A., Rasouli N., Kern P. A., Finck B. N., Bickel P. E. (2006) Diabetes 55, 3418–3428 [DOI] [PubMed] [Google Scholar]
- 8. Yamaguchi T., Matsushita S., Motojima K., Hirose F., Osumi T. (2006) J. Biol. Chem. 281, 14232–14240 [DOI] [PubMed] [Google Scholar]
- 9. Schweiger M., Schoiswohl G., Lass A., Radner F. P., Haemmerle G., Malli R., Graier W., Cornaciu I., Oberer M., Salvayre R., Fischer J., Zechner R., Zimmermann R. (2008) J. Biol. Chem. 283, 17211–17220 [DOI] [PubMed] [Google Scholar]
- 10. Yamaguchi T., Omatsu N., Matsushita S., Osumi T. (2004) J. Biol. Chem. 279, 30490–30497 [DOI] [PubMed] [Google Scholar]
- 11. Subramanian V., Rothenberg A., Gomez C., Cohen A. W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M. P., Brasaemle D. L. (2004) J. Biol. Chem. 279, 42062–42071 [DOI] [PubMed] [Google Scholar]
- 12. Granneman J. G., Li P., Zhu Z., Lu Y. (2005) Am. J. Physiol. Endocrinol. Metab. 289, E608–E616 [DOI] [PubMed] [Google Scholar]
- 13. Remy I., Michnick S. W. (2006) Nat. Methods 3, 977–979 [DOI] [PubMed] [Google Scholar]
- 14. Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. (2006) Cell Metab. 3, 309–319 [DOI] [PubMed] [Google Scholar]
- 15. Haemmerle G., Zimmermann R., Hayn M., Theussl C., Waeg G., Wagner E., Sattler W., Magin T. M., Wagner E. F., Zechner R. (2002) J. Biol. Chem. 277, 4806–4815 [DOI] [PubMed] [Google Scholar]
- 16. Schoiswohl G., Schweiger M., Schreiber R., Gorkiewicz G., Preiss-Landl K., Taschler U., Zierler K. A., Radner F. P., Eichmann T. O., Kienesberger P. C., Eder S., Lass A., Haemmerle G., Alsted T. J., Kiens B., Hoefler G., Zechner R., Zimmermann R. (2010) J. Lipid Res. 51, 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., Kratky D., Wagner E. F., Klingenspor M., Hoefler G., Zechner R. (2006) Science 312, 734–737 [DOI] [PubMed] [Google Scholar]
- 18. Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S. (2004) J. Biol. Chem. 279, 47066–47075 [DOI] [PubMed] [Google Scholar]
- 19. Caviglia J. M., Sparks J. D., Toraskar N., Brinker A. M., Yin T. C., Dixon J. L., Brasaemle D. L. (2009) Biochim. Biophys. Acta 1791, 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghosh A. K., Ramakrishnan G., Chandramohan C., Rajasekharan R. (2008) J. Biol. Chem. 283, 24525–24533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. (2004) J. Biol. Chem. 279, 48968–48975 [DOI] [PubMed] [Google Scholar]
- 22. Hickenbottom S. J., Kimmel A. R., Londos C., Hurley J. H. (2004) Structure 12, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 23. Kienesberger P. C., Oberer M., Lass A., Zechner R. (2009) J. Lipid Res. 50, S63–S68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bell M., Wang H., Chen H., McLenithan J. C., Gong D. W., Yang R. Z., Yu D., Fried S. K., Quon M. J., Londos C., Sztalryd C. (2008) Diabetes 57, 2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]