Abstract
Previous studies in our laboratory showed that isolated, intact adult rat liver mitochondria are able to oxidize the 3-carbon of serine and the N-methyl carbon of sarcosine to formate without the addition of any other cofactors or substrates. Conversion of these 1-carbon units to formate requires several folate-interconverting enzymes in mitochondria. The enzyme(s) responsible for conversion of 5,10-methylene-tetrahydrofolate (CH2-THF) to 10-formyl-THF in adult mammalian mitochondria are currently unknown. A new mitochondrial CH2-THF dehydrogenase isozyme, encoded by the MTHFD2L gene, has now been identified. The recombinant protein exhibits robust NADP+-dependent CH2-THF dehydrogenase activity when expressed in yeast. The enzyme is localized to mitochondria when expressed in CHO cells and behaves as a peripheral membrane protein, tightly associated with the matrix side of the mitochondrial inner membrane. The MTHFD2L gene is subject to alternative splicing and is expressed in adult tissues in humans and rodents. This CH2-THF dehydrogenase isozyme thus fills the remaining gap in the pathway from CH2-THF to formate in adult mammalian mitochondria.
Keywords: Cell Metabolism, Folate Metabolism, Gene Structure, Membrane Enzymes, Mitochondrial Metabolism, One-carbon Metabolism
Introduction
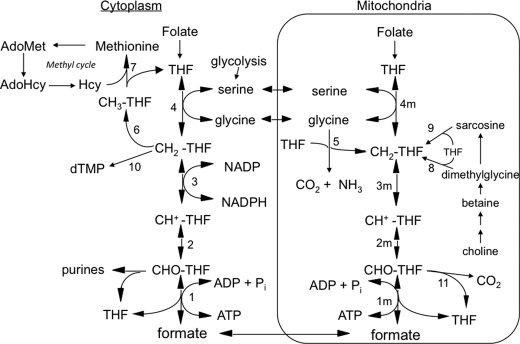
Tetrahydrofolate (THF)3-dependent 1-carbon metabolism is highly compartmentalized in eukaryotes, with THF-dependent enzymes found in mitochondria, cytoplasm, and nuclei (1, 2). The 3-carbon of serine is the major 1-carbon donor in most organisms, including humans (3), and THF can be charged with this 1-carbon unit in both the cytoplasmic and the mitochondrial compartments via serine hydroxymethyltransferase (Fig. 1, reactions 4 and 4m), resulting in the formation of 5,10-methylene-THF (CH2-THF). Cytoplasmic CH2-THF can be reduced to 5-methyl-THF (CH3-THF) (reaction 6) for entry into the methyl cycle, it can be oxidized to 10-formyl-THF (10-CHO-THF) (reactions 3 and 2) for purine synthesis, or it can be used for nuclear thymidylate (dTMP) synthesis (reaction 10) (2). The other product of the serine hydroxymethyltransferase reaction, glycine, can be metabolized by the mitochondrially localized glycine cleavage system (reaction 5), producing CH2-THF from its 2-carbon (4, 5). CH2-THF, from either serine or glycine, can be oxidized to 10-CHO-THF by mitochondrial versions of reactions 3 and 2. 10-CHO-THF can either be converted to formate and THF by 10-formyl-THF synthetase (reaction 1m) or oxidized to form CO2 and THF by 10-formyl-THF dehydrogenase (reaction 11) (6, 7).
FIGURE 1.
Mammalian 1-carbon metabolism. Reactions 1–4 are in both the cytoplasmic and the mitochondrial (m) compartments. Reactions 1, 2, and 3, 10-formyl-THF synthetase, 5,10-methenyl-THF cyclohydrolase, and 5,10-methylene-THF dehydrogenase, respectively, are catalyzed by trifunctional C1-THF synthase in the cytoplasm (MTHFD1). In mammalian mitochondria, reaction 1m is catalyzed by monofunctional MTHFD1L, and reactions 2m and 3m are catalyzed by bifunctional MTHFD2 or MTHFD2L. The other reactions are catalyzed by the following: 4 and 4m, serine hydroxymethyltransferase; 5, glycine cleavage system; 6, 5,10-methylene-THF reductase; 7, methionine synthase; 8, dimethylglycine dehydrogenase; 9, sarcosine dehydrogenase; 10, thymidylate synthase; 11, 10-formyl-THF dehydrogenase (only the mitochondrial activity of this enzyme is shown, but it has been reported in both compartments in mammals). All reactions from choline to sarcosine are mitochondrial except the betaine-to-dimethylglycine conversion, which is cytoplasmic. Hcy, homocysteine; AdoHcy, S-adenosylhomocysteine; AdoMet, S-adenosylmethionine.
The cytoplasmic and mitochondrial compartments are metabolically connected by transport of serine, glycine, and formate across the mitochondrial membranes, supporting a mostly unidirectional flow (clockwise in Fig. 1) of 1-carbon units from serine to formate and on to methionine. In fact, it appears that under most conditions, the majority of 1-carbon units for cytoplasmic processes are derived from mitochondrial formate (6–17).
In eukaryotes, the cytoplasmic activities of CH2-THF dehydrogenase, 5,10-methenyl-THF (CH+-THF) cyclohydrolase, and 10-CHO-THF synthetase (Fig. 1, reactions 1–3) are present on a trifunctional enzyme called C1-THF synthase (18–22). The mammalian version of this trifunctional enzyme is encoded by the MTHFD1 gene (23–25), and its cytoplasmic protein product will herein be designated as MTHFD1.
The enzymes catalyzing reactions 1m–3m (Fig. 1) in mammalian mitochondria are much less clear. MacKenzie and co-workers (26, 27) characterized a bifunctional NAD+-dependent CH2-THF dehydrogenase/CH+-THF cyclohydrolase (reactions 3m and 2m), originally isolated from ascites tumor cells. This enzyme was later shown to be a mitochondrial protein (28, 29), encoded by the nuclear MTHFD2 gene. Notably, this enzyme (MTHFD2 protein) is found only in transformed mammalian cells and embryonic or non-differentiated tissues (26) but is essential during embryonic development (30, 31). The final step in the mammalian mitochondrial pathway to formate (10-CHO-THF synthetase; reaction 1m) is catalyzed by mitochondrial C1-THF synthase, encoded by the MTHFD1L gene (32). This isozyme, herein referred to as MTHFD1L, is a homolog of the cytoplasmic MTHFD1. Unlike MTHFD1, however, MTHFD1L is a monofunctional enzyme, containing only the 10-CHO-THF synthetase activity (33). The lack of CH2-THF dehydrogenase/CH+-THF cyclohydrolase activities (reactions 3m and 2m) in MTHFD1L thus leaves a gap in this pathway in adult mammalian mitochondria.
Here we report the identification and characterization of a new mitochondrial CH2-THF dehydrogenase isozyme, encoded by the MTHFD2L gene. The MTHFD2L gene is expressed in adult tissues, thus completing the pathway from CH2-THF to formate in adult mammalian mitochondria.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Oligonucleotides were synthesized by IDT (Coralville, IA). Nitrocellulose membranes were obtained from Midwest Scientific (Valley Park, MO). The ECL Plus Western blotting detection reagent was from GE Healthcare. T7, T3, and SP6 polymerases were purchased from Epicenter (Madison, WI). Restriction enzymes were purchased from either Invitrogen or Fisher/Promega.
Animals
All study protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of The University of Texas, Austin, TX and conform to the National Research Council Guide for the Care and Use of Laboratory Animals (58). Sprague-Dawley rats, ages 8–12 weeks, were sacrificed by CO2 asphyxiation, and tissues were harvested and stored in RNAlater (Ambion/Applied Biosystems) according to the manufacturer's instructions prior to total RNA isolation.
Cloning of Full-length Rat MTHFD2L cDNA
Total RNA was isolated from adult rat brain by grinding tissue in TRI Reagent (Ambion/Applied Biosystems; 100 mg tissue/ml) and following the manufacturer's instructions for RNA isolation. Contaminating genomic DNA was removed using Turbo DNase (Ambion/Applied Biosystems). Reverse transcription was performed using DNase-treated total rat brain RNA with Moloney murine leukemia virus reverse transcriptase (Ambion/Applied Biosystems) and random hexamer primers. The full-length rat MTHFD2L cDNA was PCR-amplified from the resulting cDNA using oligonucleotide primers rMTHFD2Ls (5′-CCTACATATGGCGACGCGGGCCCGTG-3′; NdeI recognition site underlined; start codon in bold) and rMTHFD2Lr2 (5′-CGCCTCGAGGTAGGTGATATTCTTGGCAGC-3′; XhoI recognition site underlined). Primer design was based on the predicted rat mRNA sequence (National Center for Biotechnology Information (NCBI) Reference Sequence NM_001107211.1). The addition of MasterAmp PCR enhancer with betaine (MasterAmp Tfl DNA polymerase kit; Epicenter Biotechnologies, Madison, WI) was required in this reaction to enhance processivity through the GC-rich 5′ end of the rat cDNA. The resulting PCR-amplified fragment was cleaved with NdeI and XhoI and cloned into the pET22b vector (Novagen/EMD Biosciences). The DNA sequence of the resulting construct was verified at the DNA Core Facilities of the Institute for Cellular and Molecular Biology at the University of Texas, Austin, TX. The full-length rat MTHFD2L cDNA was subsequently cloned into a mammalian expression vector, pcDNA3.1/V5-His-TOPO (Invitrogen), using sequence- and ligation-independent cloning (34). The rat MTHFD2L insert was PCR-amplified from pET22b-rMTHFD2L using the forward primer, 5′-GGCTAGTTAAGCTTGGTACCGAGCTCGGATCATGGCGACGCGGGCCCGT-3′ (start codon in bold), and the reverse primer, 5′-TAG-ACTCGAGCGGCCGCCACTGTGCTGGATGTAGGTGATATTCTTGGC-3′, with underlined portions of the primers complementary to the rat MTHFD2L cDNA sequence. The PCR product contains 1014 bp of MTHFD2L cDNA, excluding the stop codon, flanked by 31 bp (5′) and 30 bp (3′) of the pcDNA3.1 vector. The vector was digested with BamHI and EcoRV and gel-purified prior to ligation-independent cloning with PCR-amplified rat MTHFD2L cDNA. The full-length MTHFD2L protein is expressed with a 14-amino acid V5 epitope and His6 tag fused to its C terminus in this vector. The cloned sequence, confirmed by DNA sequencing, is identical to the predicted mRNA sequence (NCBI Reference Sequence NM_001107211.1).
Transcript Analysis
Total RNA from adult human brain and placenta was obtained from Ambion/Applied Biosystems. Rat RNAs were isolated from various tissues as described above. Transcripts were analyzed by reverse transcription-PCR (RT-PCR). First strand cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase and random decamers. PCR was performed with primers (Table 1) based on the predicted human (NCBI Reference Sequence NM_001144978) or rat mRNA sequence. PCR products were excised and gel-purified using a PCR clean-up kit (Fisher/Promega or Qiagen), ligated into the pGEM-T Easy plasmid (Fisher/Promega), and transformed into Z-Competent (Zymo Research, Orange, CA) XL10 Gold Escherichia coli cells (Agilent Technologies, Santa Clara, CA). Plasmids were isolated using the E.Z.N.A. plasmid mini kit (Omega Bio-Tek, Norcross, GA) or GeneJet plasmid miniprep kit (Fermentas, Glen Burnie, MD), and their inserts were sequenced.
TABLE 1.
Sequences of oligonucleotide primers used to analyze intron-exon structure
| Name | Gene location (exon no.) | Sequence (5′ → 3′) |
|---|---|---|
| hNCRTfor1 | 1 | AGTCCGGAAGCCGGGGAT |
| hNCrace2 | 3 | CCTGACATATGTATGGCTTGCTGG |
| hJunc1to3f2 | 1, 3 | GCGGTGTGAGACATGAAGC |
| hNCRTend1 | 9 | ATAGCATGACTTCAGTTTGC |
| rD2Lexon7 | 7 | CGGTGACCATAGCTCACAGA |
| rD2Lexon9 | 9 | GCTCTCCCCTGCGATCTAGTA |
Mitochondrial Isolation and Submitochondrial Fractionation from CHO/pcDNA3.1-MTHFD2L Cells
The pcDNA3.1-MTHFD2L construct was expressed in the glyB line of CHO cells (35, 36). Cell culture and selection of stable transfectants were carried out as described previously (32, 37). Cells were disrupted and processed to give cytosolic and mitochondrial fractions as described (32). Mitochondria were isolated essentially according to Rickwood et al. (38) and subfractionated by a modified digitonin method (39, 40) using 0.12 mg of digitonin/mg of mitochondria. Digitonin-treated mitochondria were centrifuged at 10,000 × g for 10 min to obtain a pellet containing crude mitoplasts. The supernatant fraction was centrifuged again 10,000 × g for 10 min. The final supernatant (containing the outer membrane and intermembrane space) was centrifuged at 100,000 × g for 1 h at 4 °C in a Beckman L8-70M ultracentrifuge using a 50Ti rotor. The supernatant contained the intermembrane space fraction, and the pellet was resuspended in ∼200 μl of homogenization solution (HMS: 0.25 m sucrose, 1 mm EGTA, 0.5% BSA, 10 mm Hepes-NaOH (pH 7.4)) without BSA to yield the outer membrane fraction. The crude mitoplasts obtained after the first 10,000 × g spin were washed and resuspended in HMS without BSA, mixed with an equal volume of 0.025 m HEPES, pH 7.0, containing 1% Triton X-100 (0.5% final), and incubated on ice for 30 min. Triton-treated mitoplasts were centrifuged at 100,000 × g for 1 h. The supernatant contained the matrix fraction, and the pellet was resuspended in HMS without BSA to yield the inner membrane fraction. Protein concentrations of all fractions were measured using a Bradford assay (41).
Immunoblotting
Mitochondrial subfractions (outer membrane, intermembrane space, inner membrane, and matrix) were separated by SDS-PAGE and immunoblotted as described previously (37). The primary antibodies used were mouse monoclonal anti-V5 (Invitrogen), mouse monoclonal anti-COX I (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal anti-hsp60 (Enzo Life Sciences, Ann Arbor, MI), rabbit polyclonal anti-Bcl-xS/L (Santa Cruz Biotechnology), rabbit polyclonal anti-manganese superoxide dismutase (StressGen Biotechnologies, San Diego, CA), and rabbit polyclonal anti-porin (Calbiochem). Secondary antibodies were HRP-conjugated goat anti-mouse and goat anti-rabbit IgGs from Zymed Laboratories Inc. (San Francisco, CA). Reacting bands were visualized by enhanced chemiluminescence detection.
Alkaline Carbonate Extraction and Protease Treatment of Mitochondria
Alkaline carbonate extraction and protease treatment of mitochondria were performed as described previously (37). The soluble and membrane fractions from each treatment were subjected to SDS-PAGE and immunoblotting as described above.
Expression in Yeast and Enzyme Assay
The full-length rat cDNA was subcloned from pET22b-rMTHFD2L using sequence- and ligation-independent cloning (34) into a YEp24-based yeast expression vector containing the Saccharomyces cerevisiae MET6 promoter (42). This vector (Yep24ES) was modified by Everett Stone (University of Texas, Austin, TX) to include a multiple cloning site and an N-terminal His6 tag. The rat MTHFD2L insert was PCR-amplified from pET22b-rMTHFD2L using the forward primer, 5′-GTCATCACCATCACCATCACGGATCCATGGCGACGCGGGCC-3′ (start codon in bold), and the reverse primer, 5′-ACCTTAGCGGCCGCAGATCTGGTACCCTAGTAGGTGATATTCT-3′, with underlined portions of the primers complementary to the rat MTHFD2L cDNA sequence. In this construct, YEp-rMTHFD2L, the entire rat mitochondrial ORF, including the mitochondrial presequence, is expressed from the strong constitutive MET6 promoter of the vector. Yeast strain MWY4.4 (ser1 ura3-52 trp1 his4 leu2 ade3-65 Δmtd1) (9) was transformed with YEp-rMTHFD2L using the high efficiency lithium acetate yeast transformation protocol from Gietz and Woods (43). Transformed cells were grown in selective synthetic minimal medium and harvested at an A600 of ∼4, and the wet weight of each cell pellet was determined. Cells were disrupted with glass beads using a FastPrep FP120 cell disrupter (MP Biomedicals, Solon, OH) to yield the whole lysate. A cleared lysate was obtained from the whole lysate by centrifugation at 30,000 × g for 30 min. Aliquots (50 μl) of whole and cleared lysates were assayed for NAD+- and NADP+-dependent CH2-THF dehydrogenase activity as described (44). Enzyme activity in each fraction was normalized to the wet weight of the cell pellet. YEp-rMTHFD2L and Yep24ES were also transformed into yeast strain MWY4.5 (ser1 ura3-52 trp1 his4 leu2 ade3-30/65 Δmtd1) (45) for in vivo complementation tests.
RESULTS
Gene Identification and cDNA Cloning
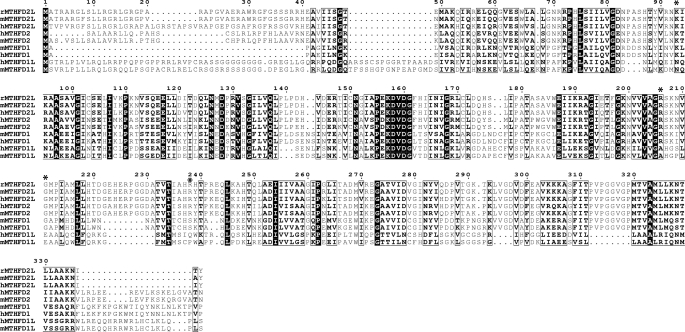
A fourth C1-THF synthase homolog was identified on human chromosome 4 at 4q13.3 by a BLAST search using the human MTHFD1 cDNA sequence. This gene, designated MTHFD2L (MTHFD2-Like), is present in all sequenced vertebrate genomes, including mouse and rat. The mammalian MTHFD2L ORF is homologous to the mitochondrial bifunctional dehydrogenase/cyclohydrolase (MTHFD2 gene) and the N-terminal dehydrogenase/cyclohydrolase domains of cytoplasmic and mitochondrial C1-THF synthase (MTHFD1 and MTHFD1L genes) (Fig. 2). The predicted initiator codon sits within a near perfect expanded Kozak consensus sequence in the first exon (46), and the N terminus has the characteristics of a mitochondrial leader sequence, including the potential to form a positively charged amphipathic α helix. Several expressed sequence tags and cDNAs from the MTHFD2L locus have been sequenced from adult tissues in human, mouse, and rat, indicating that this gene is expressed in adult tissues. Importantly, the predicted MTHFD2L proteins possess four amino acids (Lys-93, Arg-206, Gly-211, and Arg-238 in Fig. 2) shown to be critical for dehydrogenase and/or cyclohydrolase activity (24). These four residues have been substituted in all MTHFD1L-encoded monofunctional mitochondrial C1-THF synthases (33). Based on these observations, we propose that the MTHFD2L gene encodes the enzyme responsible for the CH2-THF dehydrogenase activity observed in adult mammalian mitochondria (6, 7).
FIGURE 2.
Sequence alignment of MTHFD2L, MTHFD1L, MTHFD2, and MTHFD1 proteins. Full-length MTHFD2L and MTHFD2 are aligned with the N-terminal dehydrogenase/cyclohydrolase domains of MTHFD1 and MTHFD1L. The leading lowercase letter in each protein name indicates the source (h, human; m, mouse; r, rat). Black boxes denote identity, and white boxes denote conservative substitutions. Residues shown to be critical to dehydrogenase/cyclohydrolase activity (positions 93, 206, 211, and 238 in the alignment) are indicated by asterisks. The alignment was produced using the CLC Sequence Viewer (version 6.3), and the output was generated by the ESPript 2.2 web server.
CHO Cell Expression and Subcellular Localization
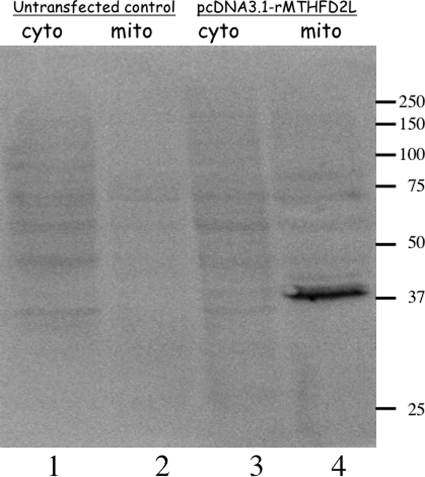
To determine whether the protein encoded by this cDNA was, in fact, mitochondrial, we expressed the cDNA in CHO cells. The C terminus of the full-length rat MTHFD2L cDNA was fused to the 14-amino acid V5 epitope and a His6 tag. This plasmid, pcDNA/rMTHFD2L, was transfected into CHO cells, and G418-resistant colonies were selected and grown. The cytosolic and mitochondrial fractions from transfected and untransfected (control) cells were isolated and subjected to SDS-PAGE and immunoblotting using antibodies against the V5 epitope (Fig. 3). A clear signal at ∼37 kDa was detected in the mitochondrial fraction (lane 4), but not the cytoplasmic fraction (lane 3), of the transfected CHO cell line. This mobility is consistent with the expected size of the epitope-tagged construct. No signal was seen in either fraction of the untransfected CHO cell line (lanes 1 and 2). These results confirm that the MTHFD2L gene encodes a protein that localizes exclusively to mitochondria in a mammalian cell line.
FIGURE 3.
Subcellular localization of epitope-tagged rat MTHFD2L expressed in CHO cells. Cytoplasmic (cyto) and mitochondrial (mito) fractions from untransfected control CHO cells (lanes 1 and 2) or cells transfected with pcDNA3.1-rMTHFD2L (lanes 3 and 4) were fractionated on a 10% SDS-polyacrylamide gel and immunoblotted with anti-V5 antibodies (1:1000 dilution). Each lane contains 50 μg of total protein. Sizes of molecular mass markers in kDa are shown on the right.
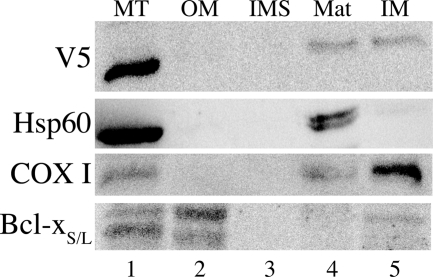
To determine the submitochondrial localization of the rMTHFD2L protein, mitochondria were isolated from transfected CHO cells and subfractionated as described under “Experimental Procedures.” These submitochondrial fractions were subjected to SDS-PAGE and immunoblotting using antibodies against the V5 epitope and against various mitochondrial marker proteins. COX I was found predominantly in the inner membrane fraction (47) as expected (Fig. 4, lane 5). Hsp60, an inner membrane/matrix marker (48), was found predominantly in the matrix fraction (Fig. 4, lane 4), and Bcl-xS/L, an outer membrane marker (49), was found predominantly in the outer membrane fraction (Fig. 4, lane 2). As observed previously (37, 50), anti-Bcl-xS/L antibodies detected a doublet in CHO cell mitochondria. MTHFD2L, detected by the anti-V5 antibodies, was distributed between the inner membrane and matrix fractions (Fig. 4, lanes 4 and 5), suggesting that the protein is partly associated with the mitochondrial inner membrane.
FIGURE 4.
Submitochondrial localization of epitope-tagged rat MTHFD2L expressed in CHO cells. Mitochondria were subfractionated as described under “Experimental Procedures.” Whole mitochondria (MT), outer membrane (OM), intermembrane space (IMS), matrix (Mat), and inner membrane (IM) fractions were resolved on 10% SDS-polyacrylamide gels and subjected to immunoblotting with anti-V5 (1:1500 dilution), anti-Hsp60 (inner membrane/matrix marker; 1:1200 dilution), anti-COX I (inner membrane marker; 1:1000 dilution), or anti-Bcl-xS/L (outer membrane marker; 1:750 dilution) antibodies. MTHFD2L migrated at an apparent molecular mass of 37 kDa, Hsp60 migrated at 60 kDa, COX I migrated at an apparent molecular mass of 35 kDa, and Bcl-xS/L migrated at an apparent molecular mass of 32 and 35 kDa.
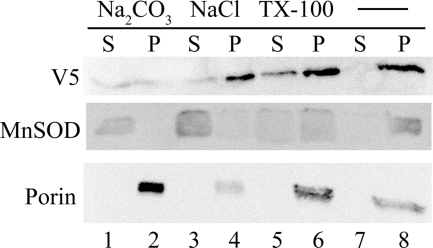
To determine whether the inner membrane-associated MTHFD2L is a peripheral or an integral membrane protein, alkaline (pH 11.5) sodium carbonate extraction was performed on whole mitochondria. Sodium carbonate releases peripheral membrane proteins into the supernatant, whereas integral membrane proteins are retained in the insoluble membrane pellet (51). The integral membrane marker protein, porin (voltage-dependent anion channel (VDAC)), was observed in the pellet after sodium carbonate treatment (membrane fraction; Fig. 5, lane 2), whereas the peripheral membrane marker protein, manganese superoxide dismutase (MnSOD), was released into the supernatant (soluble fraction; Fig. 5, lane 1). In contrast to both porin and manganese superoxide dismutase, V5-tagged MTHFD2L was distributed equally between the membrane and soluble fractions (Fig. 5, lanes 1 and 2). Mitochondria treated with NaCl of the same ionic strength as Na2CO3 failed to release MTHFD2L efficiently from the membrane fraction (Fig. 5, lane 3 versus 4). Triton X-100 treatment also released less than half of the MTHFD2L from the membrane (Fig. 5, lanes 5 versus 6). Manganese superoxide dismutase, although a matrix marker, has been reported to behave partly as an insoluble membrane protein (52), even in the presence of detergent (37, 47), explaining its presence in the membrane fraction even after treatment with Triton X-100 (Fig. 5, lane 6). Thus, rMTHFD2L behaves as a tightly associated peripheral membrane protein of the mitochondrial inner membrane.
FIGURE 5.
Alkaline carbonate extraction of CHO cell mitochondria expressing epitope-tagged rat MTHFD2L. CHO cell mitochondria were prepared and subjected to Na2CO3 or NaCl or Triton X-100 extraction as described under “Experimental Procedures.” The insoluble integral membrane proteins (pellet; P) were separated from the soluble and peripheral membrane proteins (supernatant; S) by ultracentrifugation. Equal concentrations of pellet and supernatant proteins were fractionated on a 10% SDS-polyacrylamide gel and subjected to immunoblotting with anti-V5 (1:4000 dilution), anti-manganese superoxide dismutase (MnSOD) (peripheral membrane protein marker; 1:2000 dilution), or anti-porin (integral membrane protein marker; 1:2000 dilution) antibodies. MTHFD2L migrated at an apparent molecular mass of 37 kDa, manganese superoxide dismutase migrated at an apparent molecular mass of 25 kDa, and porin migrated at an apparent molecular mass of 31 kDa.
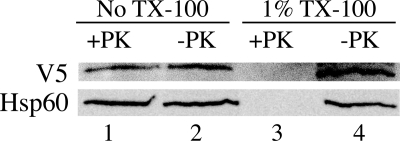
To determine the topology of MTHFD2L within the mitochondrial inner membrane, mitoplasts were treated with proteinase K in the presence or absence of Triton X-100. V5-tagged MTHFD2L behaved identically to the inner membrane/matrix marker, Hsp60. MTHFD2L was completely resistant to digestion in the absence of detergent (Fig. 6, lane 1 versus lane 2) but was completely degraded when the membranes were solubilized by 1% Triton X-100 (Fig. 6, lane 3 versus lane 4). Thus, the inner membrane protected against proteolysis, indicating that rMTHFD2L is located on the matrix side of the inner mitochondrial membrane.
FIGURE 6.
Proteinase K digestion of CHO cell mitochondria expressing epitope-tagged rat MTHFD2L. Mitoplasts were prepared by the digitonin method and treated with 100 μg/ml proteinase K (PK) as described under “Experimental Procedures.” Protease treatment was stopped by the addition of PMSF to 1 mm, and the mitoplast suspensions were centrifuged through sucrose cushions. The resulting pellets were resuspended in equal volumes of HMS without BSA, fractionated on a 10% SDS-polyacrylamide gel, and subjected to immunoblotting with anti-V5 (1:1500 dilution) or anti-Hsp60 (inner membrane/matrix marker; 1:1200 dilution) antibodies. Mitoplasts incubated with 1% Triton X-100 (TX-100) in the presence or absence of proteinase K were used as controls. These Triton X-100-treated controls were analyzed directly without centrifugation through sucrose cushions. MTHFD2L migrated at an apparent molecular mass of 37 kDa, and Hsp60 migrated at an apparent molecular mass of 60 kDa.
Expression in Yeast
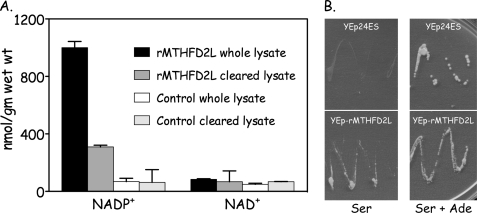
The full-length rat MTHFD2L cDNA fused to a C-terminal His tag was subcloned into a yeast expression vector (Yep24-rMTHFD2L) and transformed into strain MWY4.4. This strain carries a point mutation in the ADE3 gene that inactivates the NADP+-dependent CH2-THF dehydrogenase activity of the cytoplasmic trifunctional C1-THF synthase (8), and it carries a deletion of the MTD1 gene encoding a monofunctional cytoplasmic NAD+-dependent CH2-THF dehydrogenase (44). The low residual CH2-THF dehydrogenase activity in this strain is NADP+-dependent due to the mitochondrial C1-THF synthase isozyme (8). MWY4.4 cells transformed with Yep24-rMTHFD2L overexpressed NADP+-dependent CH2-THF dehydrogenase activity ∼12-fold when compared with control cells (Fig. 7A). NAD+-dependent activity was not detected above background, suggesting that the rat MTHFD2L is specific for NADP+. The recombinant enzyme appears to be partially membrane-associated as ∼70% of the activity pelleted in the particulate fraction after a 30,000 × g centrifugation (Fig. 7A, compare whole versus cleared lysate).
FIGURE 7.
Expression of recombinant rat MTHFD2L in yeast. A, S. cerevisiae strain MWY4.4 (ser1 ura3 trp1 leu2 his4 ade3-65 Δmtd1) harboring YEp-rMTHFD2L and untransformed control cells were grown in selective medium, harvested, and lysed as described under “Experimental Procedures.” Lysates were assayed for NAD+- and NADP+-dependent CH2-THF dehydrogenase activity before (whole lysate) and after a 30,000 × g centrifugation (cleared lysate). Enzyme activity is expressed as nmol product per gm of wet weight (nmol/gm wet wt) of cells. Each column represents the average ± S.D. of duplicate determinations. B, strain MWY4.5 (ser1 ura3 trp1 leu2 his4 ade3-30/65 Δmtd1) was transformed to uracil prototrophy with either YEp-rMTHFD2L or empty plasmid (Yep24ES). Ura+ transformants were streaked onto yeast minimal plates containing serine (left) or serine + adenine (Ser + Ade, right) and incubated at 30 °C for 4 days. Both plates also contained leucine, tryptophan, and histidine to support the other auxotrophic requirements of MWY4.5.
We next asked whether this recombinant MTHFD2L was active in vivo by testing its ability to complement the adenine requirement of yeast strain MWY4.5. Yeast produce 10-formyl-THF for de novo purine biosynthesis from serine, using either the cytoplasmic or the mitochondrial 1-carbon pathway (Fig. 1) (8). MWY4.5 lacks the 10-formyl-THF synthetase activity of the cytoplasmic trifunctional C1-THF synthase and lacks both cytoplasmic CH2-THF dehydrogenase activities (45). Thus, both the cytoplasmic and the mitochondrial pathways to 10-formyl-THF are blocked in MWY4.5; this strain thus requires adenine in the medium for growth (45). If rMTHFD2L is catalytically active in vivo and if at least some of the overexpressed enzyme localizes to the cytoplasm, it should rescue the adenine requirement of MWY4.5. Transformants of MWY4.5 harboring either YEp-rMTHFD2L or empty plasmid (Yep24ES) were streaked onto yeast minimal plates containing serine as 1-carbon donor or serine + adenine and incubated at 30 °C. As shown in Fig. 7B, rMTHFD2L fully complemented the adenine requirement of MWY4.5, indicating that this enzyme functions as a CH2-THF dehydrogenase in vivo as well as in vitro.
Gene and mRNA Structure and Alternative Splicing
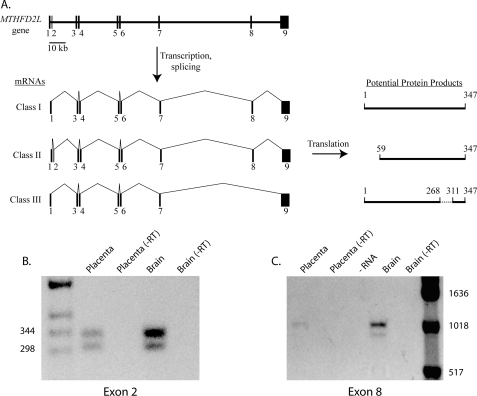
The human MTHFD2L gene spans 145 kbp and consists of nine exons interrupted by eight introns of length ranging from 590 to 56,030 bp (Fig. 8A). The start codon is present in the first exon, and the 5′ end of exon 1 extends at least 31 bp upstream of the ATG start codon, based on NCBI EST database expressed sequence tags containing exon 1. The stop codon is present in exon 9, which also encodes 1283 nucleotides of 3′-UTR, including several potential polyadenylation signals (AATAAA). Exon 1 is very GC-rich (74% G+C), contributing to the difficulty of obtaining full-length cDNAs. All of the intron-exon splice sites follow the GT/AG rule (53), except after the terminal exon.
FIGURE 8.
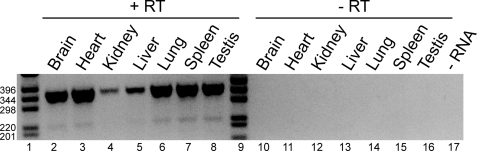
Human MTHFD2L gene structure and alternative transcript splicing. A, gene structure, alternative splicing, and potential protein products. The entire gene spans 145 kbp on chromosome 4 at 4q13.3. Exons are shown as numbered black bars; introns are shown as thin horizontal lines. Exons are not drawn to scale. Three alternative splicing patterns due to skipping of exon 2 and/or 8 are observed in human brain and placenta (see under “Results” for details). B, RT-PCR analysis of exon 2 splicing using primers binding in exons 1 and 3 (hNCRTfor1 and hNCrace2, respectively; Table 1). The expected sizes of the two products are 335 bp (including exon 2) and 301 bp (excluding exon 2). −RT, minus reverse transcriptase control. C, RT-PCR analysis of exon 8 splicing using primers binding in exons 1 and 9 (hJunc1to3f2 and hNCRTend1, respectively; Table 1). The expected sizes of the two products are 980 bp (including exon 8) and 852 bp (excluding exon 8). DNA size markers are indicated.
MTHFD2L transcripts were analyzed by RT-PCR on RNA from human brain and placenta as described under “Experimental Procedures.” These analyses revealed alternative splicing of two exons in the human MTHFD2L gene. Doublets were observed with primers that flanked exon 2 or exon 8 in both brain and placenta RNA (Fig. 8, B and C). The band intensity of the exon 8 doublet was always weak with placenta RNA. Sequence analysis of the cloned PCR products confirmed that the larger product of each doublet included the alternate exon (2 or 8), and the smaller product of each doublet lacked the alternate exon. This alternative splicing thus produces at least three classes of transcripts from the human MTHFD2L gene (Fig. 8A). In class I transcripts, the 34-bp exon 2 is skipped, giving an mRNA that encodes the 347-amino acid protein shown in the alignment in Fig. 2. In class II transcripts, exon 2 is retained; translation of this mRNA from the first AUG codon in exon 1 would produce an early termination product of only 60 amino acids. However, translation of the class II mRNA from an AUG codon in exon 3 would give a protein identical to the last 289 amino acids of the class I protein. In class III transcripts, both exon 2 and exon 8 are skipped. Based on sequenced RT-PCR clones and expressed sequence tags in the NCBI EST databases, all three transcript classes are expressed in both brain and placenta from adult humans.
RT-PCR analysis of RNA isolated from adult rat tissues revealed that the MTHFD2L gene is widely expressed (Fig. 9). The 34-bp exon 2 is also found in the mouse and rat MTHFD2L genes. Although the majority of mouse and rat expressed sequence tags in the NCBI EST database lack exon 2, there are several that contain this exon. RT-PCR on rat brain RNA using primers in exons 1 and 3 produced two products, although the longer product (containing exon 2) was the minor species (data not shown). RT-PCR using primers in exons 7 and 9 also produced two products with rat RNAs, a 338-bp product containing exon 8 and a 212-bp product lacking exon 8 (Fig. 9). Sequence analysis of the 212-bp product indicated precise splicing of exon 7 to exon 9 with the reading frame intact. In all tissues examined, the exon 8 skipped transcript is the minor species.
FIGURE 9.
MTHFD2L expression in adult rat tissues. Total RNA isolated from the indicated adult rat tissues was analyzed by RT-PCR as described under “Experimental Procedures” using primers binding in exons 7 and 9 (rD2Lexon7 and rD2Lexon9, respectively; Table 1). The expected sizes of the two products are 338 bp (including exon 8) and 212 bp (excluding exon 8). Lanes 1 and 9 contain DNA size markers (sizes indicated on left). Lanes 2–8, reverse transcriptase samples (+ RT); lanes 10–16, minus reverse transcriptase controls (− RT); lane 17, minus RNA control (− RNA; contained reverse transcriptase). The image is overexposed to reveal the smaller band.
DISCUSSION
The experiments described here confirm that adult mammals express a CH2-THF dehydrogenase isozyme in mitochondria, encoded by the MTHFD2L gene. This isozyme fills the gap in the adult mammalian mitochondrial 1-carbon pathway left by the lack of CH2-THF dehydrogenase activity in the monofunctional MTHFD1L isozyme (Fig. 1). The MTHFD2L gene encodes a protein of about 340 amino acids that is homologous to the mitochondrial bifunctional dehydrogenase/cyclohydrolase (MTHFD2 gene product) and the N-terminal dehydrogenase/cyclohydrolase domains of cytoplasmic and mitochondrial C1-THF synthase (MTHFD1 and MTHFD1L gene products) (Fig. 2). The recombinant rat protein exhibits robust NADP+-dependent CH2-THF dehydrogenase activity in vitro and can replace the yeast cytoplasmic CH2-THF dehydrogenase activities in vivo (Fig. 7). 5,10-Methenyl-THF cyclohydrolase assays were not performed. The high backgrounds in the cyclohydrolase spectrophotometric assay make it unreliable in crude extracts. Thus, we will have to await purification of the enzyme to determine whether it possesses cyclohydrolase activity. The MTHFD2L proteins have a predicted N-terminal mitochondrial targeting sequence, and when the full-length rat cDNA was expressed in CHO cells, the targeting sequence directed the protein exclusively to mitochondria (Fig. 3).
Evidence from both in vivo (9, 10, 45, 54) and in vitro (6, 7) experiments supports a matrix localization for mitochondrial 1-carbon metabolism. Recently, we found that the MTHFD1L protein is tightly associated with the inner mitochondrial membrane, facing the matrix (37). The sublocalization experiments described here indicated that MTHFD2L also behaves as a peripheral membrane protein tightly associated with the matrix side of the inner mitochondrial membrane (Figs. 4–6). Given the membrane association of MTHFD1L and MTHFD2L, it is likely that folate-dependent 1-carbon metabolism occurs at the inner mitochondrial membrane. Mitochondrial serine hydroxymethyltransferase, glycine cleavage system, sarcosine dehydrogenase, and dimethylglycine dehydrogenase have all been reported to be associated with the inner membrane in rat liver mitochondria (55–57). These enzymes each produce CH2-THF from their respective substrates, which can be oxidized by the CH2-THF dehydrogenase activity of MTHFD2L. In addition, human mitochondrial folylpolyglutamate synthetase, which adds glutamate residues to the mitochondrial folate pool, has also been reported to be tightly associated with the inner membrane (52). It was suggested that the inner membrane localization of mitochondrial folylpolyglutamate synthetase would enable efficient polyglutamylation of folates by promoting substrate channeling between the inner membrane folate carrier and folylpolyglutamate synthetase. It is thus tempting to hypothesize the existence of a large folate-dependent 1-carbon-metabolizing complex at the inner mitochondrial membrane, perhaps including membrane carriers for substrates (e.g. serine, glycine, formate, folate) and/or components of the mitochondrial respiratory chain where the redox cofactors are reoxidized.
The MTHFD2L gene structure is conserved among human, mouse, and rat, consisting of nine exons (Fig. 8A). RT-PCR analysis revealed three classes of MTHFD2L transcripts in adult human brain and placental RNA, potentially encoding three different protein products (Fig. 8A). Although all three transcript classes are approximately equally represented, at least in human brain and placenta, we do not know whether all three are translated into stable proteins in vivo. RT-PCR analysis of rat RNAs also revealed evidence of alternative splicing of both exon 2 and exon 8.
RT-PCR analysis of RNA isolated from adult rat tissues revealed that the MTHFD2L gene is widely expressed, with the highest signals observed in brain, heart, lung, spleen, and testes and lower signals in liver and kidney (Fig. 9). We also detected MTHFD2L expression in mouse embryos as early as 9.5 days after fertilization.4 The MTHFD2 gene, encoding a mitochondrial NAD+-dependent CH2-THF dehydrogenase, is also expressed early in mouse embryos, but its expression decreases as the embryos approach birth (31, 54). In adults, MTHFD2 expression is restricted to tissues that contain differentiating cells such as bone marrow (26). Thus, after birth, the NADP+-dependent MTHFD2L appears to be the only mitochondrial CH2-THF dehydrogenase expressed in differentiated tissues. Experiments to determine when the switch between MTHFD2 and MTHFD2L occurs, and why, are currently underway.
This work was supported, in whole or in part, by National Institutes of Health Grants DK61425 and GM086856 (to D. R. A.).
J. Bryant, J. Lewandowski, and S. Vokes, unpublished data.
- THF
- tetrahydrofolate
- CH+-THF
- 5,10-methenyl-tetrahydrofolate
- CH2-THF
- 5,10-methylene-tetrahydrofolate
- 10-CHO-THF
- 10-formyl-tetrahydrofolate
- MTHFD1
- cytoplasmic C1-tetrahydrofolate synthase
- MTHFD1L
- mitochondrial C1-tetrahydrofolate synthase
- MTHFD2
- NAD+-dependent 5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methenyl-tetrahydrofolate cyclohydrolase
- h
- human
- r
- rat.
REFERENCES
- 1. Appling D. R. (1991) FASEB J. 5, 2645–2651 [DOI] [PubMed] [Google Scholar]
- 2. Woeller C. F., Anderson D. D., Szebenyi D. M., Stover P. J. (2007) J. Biol. Chem. 282, 17623–17631 [DOI] [PubMed] [Google Scholar]
- 3. Davis S. R., Stacpoole P. W., Williamson J., Kick L. S., Quinlivan E. P., Coats B. S., Shane B., Bailey L. B., Gregory J. F., 3rd. (2004) Am. J. Physiol. Endocrinol. Metab. 286, E272–EE279 [DOI] [PubMed] [Google Scholar]
- 4. Fu T. F., Rife J. P., Schirch V. (2001) Arch. Biochem. Biophys. 393, 42–50 [DOI] [PubMed] [Google Scholar]
- 5. Lamers Y., Williamson J., Theriaque D. W., Shuster J. J., Gilbert L. R., Keeling C., Stacpoole P. W., Gregory J. F., 3rd (2009) J. Nutr. 139, 666–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barlowe C. K., Appling D. R. (1988) Biofactors 1, 171–176 [PubMed] [Google Scholar]
- 7. García-Martínez L. F., Appling D. R. (1993) Biochemistry 32, 4671–4676 [DOI] [PubMed] [Google Scholar]
- 8. Barlowe C. K., Appling D. R. (1990) Mol. Cell. Biol. 10, 5679–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pasternack L. B., Laude D. A., Jr., Appling D. R. (1994) Biochemistry 33, 74–82 [DOI] [PubMed] [Google Scholar]
- 10. Pasternack L. B., Littlepage L. E., Laude D. A., Jr., Appling D. R. (1996) Arch. Biochem. Biophys. 326, 158–165 [DOI] [PubMed] [Google Scholar]
- 11. Kastanos E. K., Woldman Y. Y., Appling D. R. (1997) Biochemistry 36, 14956–14964 [DOI] [PubMed] [Google Scholar]
- 12. Gregory J. F., 3rd, Cuskelly G. J., Shane B., Toth J. P., Baumgartner T. G., Stacpoole P. W. (2000) Am. J. Clin. Nutr. 72, 1535–1541 [DOI] [PubMed] [Google Scholar]
- 13. Herbig K., Chiang E. P., Lee L. R., Hills J., Shane B., Stover P. J. (2002) J. Biol. Chem. 277, 38381–38389 [DOI] [PubMed] [Google Scholar]
- 14. Patel H., Pietro E. D., MacKenzie R. E. (2003) J. Biol. Chem. 278, 19436–19441 [DOI] [PubMed] [Google Scholar]
- 15. Quinlivan E. P., Davis S. R., Shelnutt K. P., Henderson G. N., Ghandour H., Shane B., Selhub J., Bailey L. B., Stacpoole P. W., Gregory J. F., 3rd. (2005) J. Nutr. 135, 389–396 [DOI] [PubMed] [Google Scholar]
- 16. Anguera M. C., Field M. S., Perry C., Ghandour H., Chiang E. P., Selhub J., Shane B., Stover P. J. (2006) J. Biol. Chem. 281, 18335–18342 [DOI] [PubMed] [Google Scholar]
- 17. MacFarlane A. J., Liu X., Perry C. A., Flodby P., Allen R. H., Stabler S. P., Stover P. J. (2008) J. Biol. Chem. 283, 25846–25853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paukert J. L., Williams G. R., Rabinowitz J. C. (1977) Biochem. Biophys. Res. Comm. 77, 147–154 [DOI] [PubMed] [Google Scholar]
- 19. Schirch L. (1978) Arch. Biochem. Biophys. 189, 283–290 [DOI] [PubMed] [Google Scholar]
- 20. Smith G. K., Mueller W. T., Wasserman G. F., Taylor W. D., Benkovic S. J. (1980) Biochemistry 19, 4313–4321 [DOI] [PubMed] [Google Scholar]
- 21. Hum D. W., Bell A. W., Rozen R., MacKenzie R. E. (1988) J. Biol. Chem. 263, 15946–15950 [PubMed] [Google Scholar]
- 22. Thigpen A. E., West M. G., Appling D. R. (1990) J. Biol. Chem. 265, 7907–7913 [PubMed] [Google Scholar]
- 23. Howard K. M., Muga S. J., Zhang L., Thigpen A. E., Appling D. R. (2003) Gene 319, 85–97 [DOI] [PubMed] [Google Scholar]
- 24. Christensen K. E., Patel H., Kuzmanov U., Mejia N. R., MacKenzie R. E. (2005) J. Biol. Chem. 280, 7597–7602 [DOI] [PubMed] [Google Scholar]
- 25. MacFarlane A. J., Perry C. A., Girnary H. H., Gao D., Allen R. H., Stabler S. P., Shane B., Stover P. J. (2009) J. Biol. Chem. 284, 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mejia N. R., MacKenzie R. E. (1985) J. Biol. Chem. 260, 14616–14620 [PubMed] [Google Scholar]
- 27. Mejia N. R., Rios-Orlandi E. M., MacKenzie R. E. (1986) J. Biol. Chem. 261, 9509–9513 [PubMed] [Google Scholar]
- 28. Mejia N. R., MacKenzie R. E. (1988) Biochem. Biophys. Res. Comm. 155, 1–6 [DOI] [PubMed] [Google Scholar]
- 29. Bélanger C., MacKenzie R. E. (1989) J. Biol. Chem. 264, 4837–4843 [PubMed] [Google Scholar]
- 30. Di Pietro E., Sirois J., Tremblay M. L., MacKenzie R. E. (2002) Mol. Cell. Biol. 22, 4158–4166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Di Pietro E., Wang X. L., MacKenzie R. E. (2004) Biochim. Biophys. Acta. 1674, 78–84 [DOI] [PubMed] [Google Scholar]
- 32. Prasannan P., Pike S., Peng K., Shane B., Appling D. R. (2003) J. Biol. Chem. 278, 43178–43187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walkup A. S., Appling D. R. (2005) Arch. Biochem. Biophys. 442, 196–205 [DOI] [PubMed] [Google Scholar]
- 34. Li M. Z., Elledge S. J. (2007) Nat. Methods 4, 251–256 [DOI] [PubMed] [Google Scholar]
- 35. Kao F. T., Puck T. (1975) Genetics 79, 343–352 [PubMed] [Google Scholar]
- 36. Kao F., Chasin L., Puck T. T. (1969) Proc. Natl. Acad. Sci. U.S.A. 64, 1284–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prasannan P., Appling D. R. (2009) Arch. Biochem. Biophys. 481, 86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rickwood D., Wilson M. T., Darley-Usmar V. M. (1987) in Mitochondria: A Practical Approach (Darley-Usmar V. M., Rickwood D., Wilson M. T. eds) pp. 1–16, IRL Press, Washington, D. C. [Google Scholar]
- 39. Greenawalt J. W. (1974) Methods Enzymol. 31, 310–323 [DOI] [PubMed] [Google Scholar]
- 40. Pedersen P. L., Greenawalt J. W., Reynafarje B., Hullihen J., Decker G. L., Soper J. W., Bustamente E. (1978) Methods Cell Biol. 20, 411–481 [DOI] [PubMed] [Google Scholar]
- 41. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 42. Suliman H. S., Sawyer G. M., Appling D. R., Robertus J. D. (2005) Arch. Biochem. Biophys. 441, 56–63 [DOI] [PubMed] [Google Scholar]
- 43. Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 44. West M. G., Barlowe C. K., Appling D. R. (1993) J. Biol. Chem. 268, 153–160 [PubMed] [Google Scholar]
- 45. West M. G., Horne D. W., Appling D. R. (1996) Biochemistry 35, 3122–3132 [DOI] [PubMed] [Google Scholar]
- 46. Kozak M. (1987) J. Mol. Biol. 196, 947–950 [DOI] [PubMed] [Google Scholar]
- 47. Vijayvergiya C., Beal M. F., Buck J., Manfredi G. (2005) J. Neurosci. 25, 2463–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garesse R., Vallejo C. G. (2001) Gene 263, 1–16 [DOI] [PubMed] [Google Scholar]
- 49. Boise L. H., González-García M., Postema C. E., Ding L., Lindsten T., Turka L. A., Mao X., Nuñez G., Thompson C. B. (1993) Cell 74, 597–608 [DOI] [PubMed] [Google Scholar]
- 50. Alonso G., Guillemain I., Dumoulin A., Privat A., Patey G. (1997) Cell Tissue Res. 288, 59–68 [DOI] [PubMed] [Google Scholar]
- 51. Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. (1982) J. Cell Biol. 93, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nair J. R., McGuire J. J. (2005) Biochim. Biophys. Acta 1746, 38–44 [DOI] [PubMed] [Google Scholar]
- 53. Breathnach R., Chambon P. (1981) Annu. Rev. Biochem. 50, 349–383 [DOI] [PubMed] [Google Scholar]
- 54. Pike S. T., Rajendra R., Artzt K., Appling D. R. (2010) J. Biol. Chem. 285, 4612–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Motokawa Y., Kikuchi G. (1971) Arch. Biochem. Biophys. 146, 461–464 [DOI] [PubMed] [Google Scholar]
- 56. Da Cruz S., Xenarios I., Langridge J., Vilbois F., Parone P. A., Martinou J. C. (2003) J. Biol. Chem. 278, 41566–41571 [DOI] [PubMed] [Google Scholar]
- 57. Bergeron F., Otto A., Blache P., Day R., Denoroy L., Brandsch R., Bataille D. (1998) Eur. J. Biochem. 257, 556–561 [DOI] [PubMed] [Google Scholar]
- 58. National Research Council (1996) Guide for the Care and Use of Laboratory Animals, National Academy Press, Washington, D.C. [Google Scholar]