Abstract
Vesicular transport shuttles cargo among intracellular compartments. Several stages of vesicular transport are mediated by the small GTPase Arf, which is controlled in a cycle of GTP binding and hydrolysis by Arf guanine-nucleotide exchange factors and Arf GTPase-activating proteins (ArfGAPs), respectively. In budding yeast the Age2 + Gcs1 ArfGAP pair facilitates post-Golgi transport. We have found the AGE1 gene, encoding another ArfGAP, can in high gene-copy number alleviate the temperature sensitivity of cells carrying mutations affecting the Age2 + Gcs1 ArfGAP pair. Moreover, increased AGE1 gene dosage compensates for the complete absence of the otherwise essential Age2 + Gcs1 ArfGAP pair. Increased dosage of SFH2, encoding a phosphatidylinositol transfer protein, also allows cell growth in the absence of the Age2 + Gcs1 pair, but good growth in this situation requires Age1. The ability of Age1 to overcome the need for Age2 + Gcs1 depends on phospholipase D activity that regulates lipid composition. We show by direct assessment of Age1 ArfGAP activity that Age1 is regulated by lipid composition and can provide ArfGAP function for post-Golgi transport.
Keywords: Endosomes, G Proteins, Golgi, Membrane Trafficking, Yeast, Arf GTPase-activating Protein, Arf1, Diacylglycerol, Phosphatidylinositol Transfer Protein, Post-Golgi Transport
Introduction
Eukaryotic cells move protein and membrane cargo between the plasma membrane and various organelles by a process termed vesicular transport. Each step in the vesicular-transport process, including vesicle generation, cargo packaging, vesicle targeting, and vesicle fusion, is controlled by a variety of proteins (for review, see Ref. 1). Regulated vesicular transport not only ensures the fidelity of cargo delivery but also maintains the structural and functional integrity of membrane organelles.
Each stage of vesicular transport relies on a specific set of proteins to form the transport vesicle and direct the vesicle to the target compartment. For example, vesicles involved in retrograde transport from the Golgi to the endoplasmic reticulum require coat protein complex I (COPI) and the small GTPase Arf1 (2, 3), whereas transport vesicles originating from the trans-Golgi network also require the Arf1 GTPase but use the clathrin coat complex (4, 5). Arf1 regulates recruitment of coat protein complex I and clathrin coat complexes to the donor membrane, where these coat complexes mediate the generation of transport vesicles by deforming the membrane. Arf regulatory activity depends on a cycle of GTP binding and hydrolysis, which is in turn regulated by two types of proteins, Arf guanine-nucleotide exchange factors and Arf GTPase-activating proteins (ArfGAPs).6 Arf guanine-nucleotide exchange factors mediate the exchange of GDP for GTP on Arf to activate Arf and allow coat recruitment (6), whereas ArfGAPs stimulate hydrolysis of Arf-bound GTP to inactivate Arf and allow release of coat proteins from transport vesicles in preparation for fusion of the transport vesicle with a target membrane (7).
The budding yeast Saccharomyces cerevisiae contains ArfGAP proteins that are characterized by a zinc-binding motif (CXXCX16CXXC, where C is cysteine and X is any amino acid) and a nearby invariant arginine residue, which together constitute a catalytic center known as the ArfGAP domain (4). Four yeast proteins, Gcs1, Glo3, Age2, and Age1, have been shown to exert ArfGAP activity in vitro (8–11), with the Age2 + Gcs1 ArfGAP pair providing essential overlapping function for post-Golgi transport (10).
Vesicular transport is also mediated by membrane lipid composition through the effects of lipid-protein interactions on protein function. The membrane lipid phosphatidylinositol is phosphorylated on the inositol ring to produce distinct phospholipid species that, like Arf, are involved in protein recruitment to membranes for vesicle formation. One family of yeast proteins that affects membrane lipid composition is defined by the Sec14 phosphatidylinositol transfer protein that is involved in vesicle formation for post-Golgi transport, presumably by creating a lipid composition permissive for vesicle formation. Indeed, we and others have shown that ArfGAP activity is modulated by lipid environment (12, 13) and that increased abundance of the Sec14 homologue Sfh2 enhances Gcs1 activity for post-Golgi transport (14).
We have investigated the function of the Age2 + Gcs1 pair for post-Golgi transport by identifying yeast genes that, in increased dosage, alleviate the deleterious effects of impaired ArfGAP function. The AGE1 gene is shown here to alleviate defects resulting from deficient ArfGAP function for post-Golgi transport. The AGE1 gene was initially described as a suppressor of Arf1 temperature sensitivity (SAT1) (15) and was later renamed as encoding an ArfGAP with effector functions (AGE1) (11). Those studies implicated the Age1 protein in Arf1 function because increased AGE1 gene dosage alleviates the temperature sensitivity caused by the arf1-3 mutant allele (15). A more direct role for Age1 in Arf1 function was implied by the finding that the Age1 protein has ArfGAP activity in vitro (11). We show here that deletion of the AGE1 gene compromises the ability of increased SFH2 gene dosage to alleviate post-Golgi transport defects in cells lacking the Age2 + Gcs1 ArfGAP pair. Moreover, we find that the ability of AGE1 to effectively alleviate post-Golgi transport defects depends upon phospholipase D, an effector of an Sfh2-mediated phosphoinositide metabolic pathway (16). In addition, we show that the in vitro ArfGAP activity of Age1 is stimulated by diacylglycerol (DAG), a downstream product of phospholipase D activity. Thus, our analysis highlights the importance of phospholipid metabolism for ArfGAP activity in post-Golgi vesicular transport.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Yeast strains used in this study are described in Table 1. Yeast cells were propagated and transformed using standard techniques. Genes encoding the organelle-specific red fluorescent protein (RFP) markers (17) were backcrossed three times into the W303 genetic background. Plasmids used in this study are described in Table 2. To generate the N-terminal deletion alleles of AGE1 expressed from the endogenous AGE1 promoter, a 600-bp sequence upstream of AGE1 including the first two codons of AGE1 was amplified by PCR and cloned into YEp351 to yield plasmid pSL485. The segments of Age1 encoding the various truncations plus ∼300 bp of downstream sequence were amplified by PCR and cloned downstream of the AGE1 promoter in pSL485 to yield the various high-copy plasmids expressing the AGE1 truncations. The AGE1 sequences from the high-copy plasmids were subcloned into pRS315 to create the low-copy plasmids expressing the AGE1 truncations. For protein expression in bacteria, the entire AGE1 open reading frame and AGE1 codons 169–482 were separately subcloned into pET21b (Novagen) to create plasmids pSL363 and pSL396, respectively. To produce the Age1-GFP and Age1ΔN-GFP fusions, the AGE1 open reading frame and AGE1 sequence from codon 164 to 482 were amplified by PCR and inserted into vector pGREG600 by in vivo ligation in yeast (18), yielding plasmids pJB-Age1-GFP and pPPL149, respectively. A Yep213 yeast genomic library (a gift from D. Thomas) was used to identify dosage suppressors of the temperature sensitivity of age2Δ gcs1-3 mutant cells.
TABLE 1.
Yeast strains used in this study
| Strain | Genotypea | Source |
|---|---|---|
| AAY10 | age2Δ::HIS3/AGE2 gcs1Δ::URA3/GCS1 | This study |
| AAY20 | age2Δ::HIS3/AGE2 gcs1Δ::LEU2/GCS1 | (14) |
| JBY10 | age2Δ::HIS3/AGE2 gcs1Δ::LEU2/GCS1 age1Δ::natMX4/AGE1 | This study |
| JBY12 | age2Δ::HIS3/AGE2 gcs1Δ::URA3/GCS1 sfh2Δ::kanMX4/SFH2 | This study |
| JBY29 | age2Δ::HIS3 gcs1Δ::LEU2 [pMG4-4] | This study |
| JBY65 | age2Δ::HIS3 gcs1Δ::LEU2 [pMG4-4] | This study |
| JBY64 | age2Δ::HIS3 gcs1Δ::LEU2 spo14Δ::kanMX4 [pMG4-4] | This study |
| PPY169 | age2Δ::HIS3 gcs1Δ::URA3 [pLAA314-3] | This study |
| JBY122 | age2Δ::HIS3 gcs1Δ::URA3 [pMG4-4] | This study |
| SL215 | age1Δ::natMX4 | This study |
| PPY203G-Chc1–11A | ADE2 CHC1-mRFP::kanMX6 | This study |
| PPY203G-Snf7–4A | ADE2 SNF7-mRFP::kanMX6 | This study |
| PPY203G-Erg6–7D | ADE2 ERG6-mRFP::kanMX6 | This study |
| PPY203G-Chc1–28B | age2Δ::HIS3 gcs1Δ::LEU2 ADE2 CHC1-mRFP::kanMX6 [pMG4-4] | This study |
| PPY203G-Snf7–5C | age2Δ::HIS3 gcs1Δ::LEU2 ADE2 SNF7-mRFP::kanMX6 [pMG4-4] | This study |
| PPY203G-Erg6–1A | age2Δ::HIS3 gcs1Δ::LEU2 ADE2 ERG6-mRFP::kanMX6 [pMG4-4] | This study |
| PPY205 | age2Δ::HIS3 gcs1Δ::natMX4 ADE2 [pMG4-4 and p416-GFP-Snc1] | This study |
a All strains were derived from diploid W303 (ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) or its isogenic haploid derivatives W303-1A (MATa) and W303-1B (MATα) (30); additional alterations to the W303 genotype are indicated.
TABLE 2.
Plasmids used in this study
| Plasmid | Gene insert | Vector features | Source or reference |
|---|---|---|---|
| pRS314 | TRP1 CEN6a | (31) | |
| pLAA314–3 | gcs1-3 | TRP1 CEN6 | (14) |
| pMG4-4 | gcs1-4 | TRP1 CEN6 | (14) |
| pRS315 | LEU2 CEN6 | (31) | |
| pSH4 | GCS1 | LEU2 CEN6 | This study |
| pSL340 | AGE1 | LEU2 CEN6 | This study |
| pSL494 | AGE1ΔN (167–482)b | LEU2 CEN6 | This study |
| pJB315-AGE1–61 | AGE1 (61–482) | LEU2 CEN6 | This study |
| pJB315-AGE1–71 | AGE1 (71–482) | LEU2 CEN6 | This study |
| pJB315-AGE1–81 | AGE1 (81–482) | LEU2 CEN6 | This study |
| pJB315-AGE1–91 | AGE1 (91–482) | LEU2 CEN6 | This study |
| pJB315-AGE1–101 | AGE1 (101–482) | LEU2 CEN6 | This study |
| pJB315-AGE1–111 | AGE1 (111–482) | LEU2 CEN6 | This study |
| pJB315-AGE1–121 | AGE1 (121–482) | LEU2 CEN6 | This study |
| pJB315-AGE1–131 | AGE1 (131–482) | LEU2 CEN6 | This study |
| pJB315-AGE1–141 | AGE1 (141–482) | LEU2 CEN6 | This study |
| pJB315-AGE1–161 | AGE1 (161–482) | LEU2 CEN6 | This study |
| pRS316 | URA3 CEN6 | (31) | |
| pGCS1–316 | GCS1 | URA3 CEN6 | This study |
| pJB1736 | AGE1ΔN (167–482) | URA3 CEN6 | This study |
| YEp351 | LEU2 2μc | (32) | |
| pRS425 | LEU2 2μ | (33) | |
| pEP1 | GCS1 | LEU2 2μ | This study |
| pSL485 | AGE1prom | LEU2 2μ | This study |
| pLAA-GLL5 | AGE1 | LEU2 2μ | This study |
| pSL377 | AGE1 | LEU2 2μ | This study |
| pSL489 | AGE1ΔN (167–482) | LEU2 2μ | This study |
| pJB351-AGE1–61 | AGE1 (61–482) | LEU2 2μ | This study |
| pJB351-AGE1–71 | AGE1 (71–482) | LEU2 2μ | This study |
| pJB351-AGE1–81 | AGE1 (81–482) | LEU2 2μ | This study |
| pJB351-AGE1–91 | AGE1 (91–482) | LEU2 2μ | This study |
| pJB351-AGE1–101 | AGE1 (101–482) | LEU2 2μ | This study |
| pJB351-AGE1–111 | AGE1 (111–482) | LEU2 2μ | This study |
| pJB351-AGE1–121 | AGE1 (121–482) | LEU2 2μ | This study |
| pJB351-AGE1–131 | AGE1 (131–482) | LEU2 2μ | This study |
| pJB351-AGE1–141 | AGE1 (141–482) | LEU2 2μ | This study |
| pJB351-AGE1–161 | AGE1 (161–482) | LEU2 2μ | This study |
| pSL344 | AGE2 | LEU2 2μ | This study |
| pRS426 | URA3 2μ | (33) | |
| pPP421 | GCS1 | URA3 2μ | This study |
| pSL473 | AGE1 | URA3 2μ | This study |
| pJB1737 | AGE1ΔN (167–482) | URA3 2μ | This study |
| pCTY201 | SFH2 | URA3 2μ | (34) |
| pRS426-STT4 | STT4 | URA3 2μ | Scott Emr |
| pRS426-PIK1 | PIK1 | URA3 2μ | Scott Emr |
| pRS426-MSS4 | MSS4 | URA3 2μ | Scott Emr |
| pRS426-SPO14 | SPO14 | URA3 2μ | This study |
| pET21b | Escherichia coli expression | Novagen | |
| pPPL21 | GCS1 | E. coli expression | (8) |
| pSL363 | AGE1 | E. coli expression | This study |
| pSL396 | AGE1ΔN | E. coli expression | This study |
| pPPL149 | GAL1prom-AGE1ΔN-GFP | URA3 CEN6 | This study |
| pJB-Age1-GFP | GAL1prom -AGE1-GFP | URA3 CEN6 | This study |
| pGREG576 | GAL1prom -GFP | URA3 CEN6 | This study |
| pPPL165 | AGE1prom-AGE1-GFP | LEU2 CEN6 | This study |
| p416-GFP-Snc1 | GFP-SNC1 | URA3 CEN6 | (22) |
| pET-Arf1 | Arf1-His6 | E. coli expression | (8) |
| pACYC/ET3d/yNMT | NMT1 | E. coli expression | (35) |
a Low-copy plasmid.
b Bracketed numbers indicate encoded amino acids; see “Experimental Procedures” for details.
c High-copy plasmid.
Microscopy
Staining with the lipophilic dye FM 4-64 was performed as described (14). For subcellular localization of GFP and RFP protein fusions, cells were concentrated in growth medium by centrifugation just before analysis by fluorescence microscopy.
His6-tagged Age1ΔN and Myristoylated-Arf1
For phospholipid binding and in vitro ArfGAP assays, bacterially expressed Age1ΔN-His6 was purified and eluted from nickel-bead resin under native conditions according to the manufacturer's protocols (Qiagen Inc.).
To isolate myristoylated-Arf1-His6 (myr-Arf1-His6), bacterial cells carrying plasmids pET-Arf1 and pACYC/ET3d/yNMT were first grown at 37 °C to an A600 of 0.5. A 100-fold concentrated solution of 50 μm myristate, 6 μm bovine serum albumin was added to the culture and incubated for 20 min before inducing protein expression with 1 mm IPTG. The cells were then grown overnight at 25 °C, harvested, and frozen at −20 °C. Cell pellets were resuspended in 1 mg/ml lysozyme, 100 mm NaCl, 1 mm MgCl2, 25 mm Tris-HCl, pH 8, and incubated at room temperature for 15 min, and the cells were lysed by adjusting the cell suspension to 0.2% Triton X-100. Lysates were incubated with 50 μg/ml DNase on ice for 1 h and centrifuged at 9000 × g for 30 min to pellet cell debris. Supernatants were adjusted to 300 mm NaCl, 10 mm imidazole, and treated with nickel-nitrilotriacetic acid beads (Qiagen Inc.) for 2 h at 4 °C. The nickel-nitrilotriacetic acid beads were collected in a disposable column and washed twice with 300 mm NaCl, 20 mm imidazole, 25 mm Tris-HCl, pH 8. Myr-Arf1-His6 was eluted with 250 mm imidazole, 300 mm NaCl, 25 mm Tris-HCl, pH 8. Fractions enriched with myr-Arf1-His6, identified by Bradford assay, were washed and concentrated with the use of Centricon filtering systems. Samples of myr-Arf1-His6 were stored at −80 °C as protein preparations in 100 mm NaCl, 1 mm MgCl2, 1 mm dithiothreitol, 20 mm Tris-HCl, pH 8, 10% glycerol.
Phospholipid Binding
Phospholipid strips (Echelon Inc.) were pretreated with 3% bovine serum albumin (BSA) in Tris-buffered saline + 0.1% Tween 20 (TBST) for 2 h. The strips were then incubated overnight at 4 °C in the dark with 0.5 μg/ml purified Age1ΔN-His6 in TBST containing 1% BSA. The strips were washed with several changes of TBST over a 1-h period and then exposed to affinity-purified anti-Age1 antibodies, diluted in TBST containing 1% BSA, for 1.5 h at room temperature. The strips were washed with TBST, treated with horseradish peroxidase-conjugated goat anti-rabbit antibody for 1.5 h, and washed with TBST; strip-bound Age1ΔN-His6 was detected by enhanced chemiluminescence as described by the manufacturer (Pierce). A GST-tagged C-terminal fragment of the Legionella pneumophila SidC protein (PI(4)P Grip, Echelon Inc.) was used as a control for binding specificity to the phospholipid strips. Bound protein was detected with horseradish peroxidase-conjugated anti-GST antibody (GE Healthcare).
GTP Loading
Unilamellar lipid vesicles were prepared essentially as described (20) with minor modifications. Lipid mixtures were prepared in chloroform, evaporated as thin films, and then resuspended in 25 mm MOPS, pH 7.4, as a solution of either 10 mm dimyristoylphosphatidylcholine or 8.5 mm dimyristoylphosphatidylcholine, 1.5 mm dioleoylglycerol (mixture of 1,3 and 1,2 isomers) or 8.5 mm dimyristoylphosphatidylcholine, 1.5 mm phosphatidic acid. The lipid mixtures were then extruded by 20 passages through 100-nm pore filters to produce uniform-sized liposomes.
1-ml loading reactions prepared on ice consisted of 8 μm myr-Arf1-His6, 1 mm ATP, 1 mm dithiothreitol, 1 μm [γ-32P]GTP and unlabeled GTP (10,000–20,000 cpm/pmol), 2 mm EDTA, 1 mm MgCl2, 50 mm NaCl, 25 mm MOPS, pH 7.4, and 100 μl of one of the lipid preparations, which were added after a brief warming of the reaction mixture. Reaction mixtures were incubated at 30 °C for 40 min and terminated by adjusting the MgCl2 concentration to 2 mm and icing the reaction. Unincorporated GTP was removed by harvesting and washing the GTP-loaded myr-Arf1 with Amicon Ultra-2 centrifugal filter units with Ultracel-10 membranes.
GAP Assay
GTP hydrolysis was assayed essentially as previously described (20). 100-μl reactions consisted of varying amounts of Age1ΔN-His6 in 5 mm MgCl2, 1 mm ATP, 100 μg/ml BSA, 25 mm MOPS, pH 7.4, and 10 μl of [γ-32P]GTP-loaded myr-Arf1-His6, added last to initiate the reaction. Assays were incubated at 30 °C for 15 min and terminated with the addition of 500 μl of a cold charcoal suspension and incubation on ice for 30 min. The samples were centrifuged for 1 min, 300 μl of the supernatant was transferred to scintillation mixture, and the amount of radioactivity released was quantified by scintillation counting.
RESULTS
Increased Abundance of the AGE1 Gene Alleviates the Effects of ArfGAP Deficiencies Impairing Post-Golgi Transport
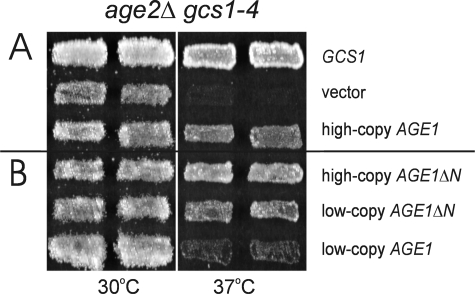
The gcs1-3 or gcs1-4 mutation in combination with the age2Δ gene deletion results in double-mutant cells that exhibit a temperature sensitivity for growth associated with defective post-Golgi transport (10, 14). To further characterize Age2 + Gcs1 ArfGAP function, we sought genes that, in increased gene dosage, alleviate this temperature sensitivity. We introduced a yeast genomic library into age2Δ gcs1-3 double-mutant cells and selected for colony formation at 37 °C. Of the 8 colonies capable of growth at 37 °C, 6 carried genomic inserts containing AGE2, and 2 were found to carry the AGE1 gene (Fig. 1A). AGE1 encodes a protein with 40% identity and 62% similarity to the ArfGAP Gcs1 and that has ArfGAP activity in vitro (11). Thus, increasing the gene dosage for another ArfGAP, Age1, alleviates the growth defect caused by deficient ArfGAP function in post-Golgi transport.
FIGURE 1.
Increased AGE1 dosage alleviates age2Δ gcs1-4 temperature sensitivity. Patches of cells harboring plasmid-borne GCS1 or AGE1 (A) and AGE1ΔN or AGE1 (B) were grown on selective medium at 23 °C, replica-plated to enriched medium, and incubated at 37 °C for 1 day or at 30 °C for 2 days. For each situation, two independent yeast transformants are displayed.
Increased AGE1 Dosage Restores Vesicular Transport in Cells with Impaired ArfGAP Function
The ability of increased AGE1 gene dosage to allow growth by age2Δ gcs1-3 cells probably results from the restoration of effective transport that is compromised in the ArfGAP mutant cells. To address this possibility directly, we monitored the effects of increased AGE1 dosage on the trafficking of two reporters that depend on vesicular transport for proper localization.
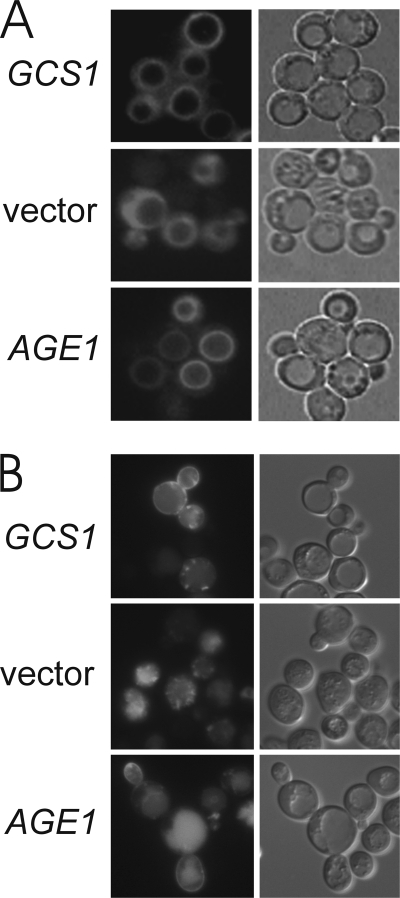
The lipophilic dye FM 4-64 provides a sensitive measure of post-Golgi function for the endocytic transport pathway (21). FM 4-64 is internalized from the cell surface via endocytosis, transported through endosomal compartments, and delivered to the vacuolar periphery. Activity of the Age2 + Gcs1 ArfGAP pair mediates this delivery of FM 4-64 to the vacuole (10). At the restrictive temperature, age2Δ gcs1-3 cells carrying vector alone exhibited impaired endocytic transport such that FM 4-64 extensively stained the cytoplasm (Fig. 2A), indicating that the membrane-bound dye remained trapped in endosomal compartments. In marked contrast, age2Δ gcs1-3 mutant cells carrying either a low-copy GCS1 plasmid or a high-copy AGE1 plasmid effectively transported FM 4-64 to the vacuole periphery, yielding the typical ring staining pattern seen in a cell with intact endocytic transport. Thus, the Age1 ArfGAP can provide function for post-Golgi transport.
FIGURE 2.
Increased AGE1 dosage restores post-Golgi function in age2Δ gcs1 cells. Cells were visualized by fluorescence (left panels) and differential interference contrast microscopy (right panels). A, mutant age2Δ gcs1-3 cells growing at 30 °C and carrying plasmid-borne GCS1 or AGE1 genes were stained with FM 4-64 as described (14) and then incubated in fresh medium at 37 °C for 45 min before visualization. B, mutant age2Δ gcs1-4 cells carrying p416-GFP-Snc1 and plasmid-borne GCS1 or AGE1 genes were grown at 23 °C and then incubated at 37 °C for 3 h before visualization.
Another measure of post-Golgi transport is the recycling of the v-SNARE Snc1 (22). Snc1 mediates the targeting of vesicles from the Golgi to the plasma membrane and is then retrieved from the plasma membrane to the endosomes and subsequently transported to the trans-Golgi network. This retrieval pathway recycles Snc1 for successive rounds of transport to the cell surface. In wild-type cells, green fluorescent protein (GFP) fused to Snc1 is seen at Golgi and endosomal compartments but is enriched at the plasma membrane of the growing bud (22). This localization of GFP-Snc1 was also observed at 37 °C in age2Δ gcs1-4 cells carrying a GCS1 plasmid (Fig. 2B). In contrast, age2Δ gcs1-4 cells carrying empty vector exhibited punctate GFP-Snc1 staining without staining at the plasma membrane, indicative of a block in transport of GFP-Snc1 from internal compartments to the cell surface. Although the majority of age2Δ gcs1-4 cells carrying an AGE1 plasmid displayed GFP-Snc1 localized at internal structures, a portion of the population exhibited GFP-Snc1 at the plasma membrane, a situation not observed in mutant cells carrying vector alone. Thus, an increased dosage of AGE1 can restore (albeit partially) the ability of age2Δ gcs1-4 mutant cells to properly traffic GFP-Snc1.
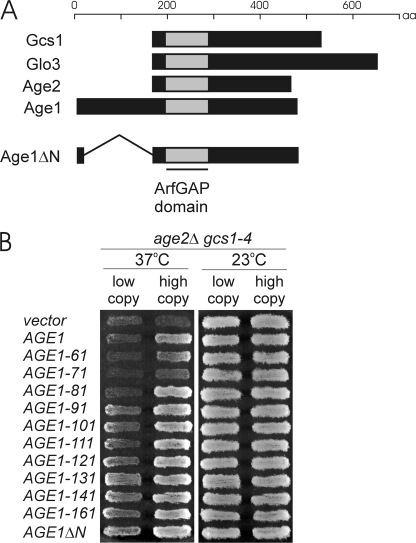
N-terminal Sequences of the Age1 Protein Restrict Its Post-Golgi Activity
The Age1 protein differs from other members of the yeast ArfGAP family by possessing a long N-terminal extension (Fig. 3A). To assess the involvement of these N-terminal sequences in Age1 function, we created the AGE1ΔN allele, which is expressed from the endogenous AGE1 promoter but lacks the sequences encoding codons 3–166 of the Age1 protein (Fig. 3A). A similarly truncated Age1 protein retains ArfGAP activity in vitro (11).
FIGURE 3.
The N terminus of Age1 inhibits the ability of Age1 to alleviate age2Δ gcs1-4 temperature sensitivity. A, schematic alignment of four yeast proteins with ArfGAP activity and the Age1ΔN protein is shown. B, patches of cells harboring plasmid-borne N-terminal truncations of Age1 were grown on selective medium at 23 °C, replica-plated to fresh medium, and incubated at 37 °C for 1 day or 23 °C for 2 days. The allele number refers to the codon that is fused downstream of the promoter and the first two codons of AGE1.
Temperature-sensitive age2Δ gcs1-4 mutant cells carrying a high-copy AGE1ΔN plasmid grew at 37 °C and actually grew better than cells carrying the high-copy intact AGE1 gene (Fig. 1B). When these AGE1 alleles were on low-copy plasmids, the difference was even more pronounced; cells carrying a low-copy AGE1ΔN plasmid grew well at 37 °C, whereas cells harboring a low-copy AGE1 plasmid did not (Fig. 1B). The absence of residues 3–166 from the N terminus of Age1, therefore, results in enhanced ability to alleviate the growth defects of mutant cells, suggesting that N-terminal sequences have a pronounced effect upon Age1 activity.
To localize potential regulatory regions within the Age1 N terminus, we assessed various AGE1 truncation alleles for alleviation of growth defects of mutant cells. The truncation alleles were constructed in a manner similar to AGE1ΔN, with the deletion of sequences from codon 3 to downstream codons ranging from codon 60 to 160. When expressed from low-copy plasmids, truncated proteins lacking the first 80 residues or fewer failed, like intact Age1, to alleviate the temperature sensitivity of age2Δ gcs1-4 cells (Fig. 3B). These truncations did, however, alleviate growth defects when expressed from high-copy plasmids. In contrast, truncations lacking the first 90 or more residues of Age1 were effective in alleviating the growth defects even at low plasmid copy number.
Increased Age1 Abundance Can “Bypass” the Need for an Otherwise Essential ArfGAP Pair
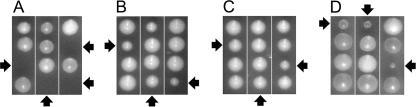
The alleviation by AGE1 (or AGE1 truncations) of temperature sensitivity in the situations described above took place in cells with mutant Gcs1 proteins that may provide residual Gcs1 activity. Therefore, we determined the ability of increased Age1 abundance to compensate for (bypass) the complete absence of the Age2 + Gcs1 essential protein pair. Diploid cells heterozygous for age2Δ and gcs1Δ deletion mutations were transformed with a high-copy AGE1 or AGE1ΔN plasmid or empty vector and then induced to undergo meiosis (sporulation). Resulting haploid segregants lacking both the Age2 and Gcs1 proteins were recovered when the AGE1 or AGE1ΔN plasmid was present (Fig. 4, B and C), an effect not seen for empty vector (Fig. 4A). These results indicate that the lethality due to the absence of Age2 + Gcs1 can be circumvented by increased abundance of the Age1 ArfGAP or its N-terminal-truncated derivative.
FIGURE 4.
Increased abundance of Age1, Age1ΔN, or Sfh2 bypasses the requirement for the essential Age2 + Gcs1 ArfGAP pair. Diploid cells heterozygous for age2Δ and gcs1Δ deletion mutations and harboring high-copy plasmids carrying vector (A), AGE1 (B), AGE1ΔN (C), or SFH2 (D) were sporulated, and the resulting haploid segregants were incubated on solid selective medium at 30 °C for 5 days. Arrows indicate age2Δ gcs1Δ double-mutant segregants that are either nonviable (A) or kept alive by the plasmid-borne gene (B–D).
Increased SFH2 Gene Dosage Also Bypasses the Need for the Age2 + Gcs1 Pair
Previously we showed that increased dosage of SFH2, encoding a member of the Sec14 family of phosphatidylinositol transfer proteins, alleviates the temperature sensitivity of age2Δ gcs1-4 double-mutant cells (14). Here we found that haploid cells harboring a high-copy SFH2 plasmid and both age2Δ and gcs1Δ deletion mutations were viable (Fig. 4D). Therefore, increased SFH2 gene dosage, like increased AGE1 dosage, also bypasses the need for the Age2 + Gcs1 pair.
Age1 Is Needed for Effective SFH2 Bypass of the Age2 + Gcs1 Pair
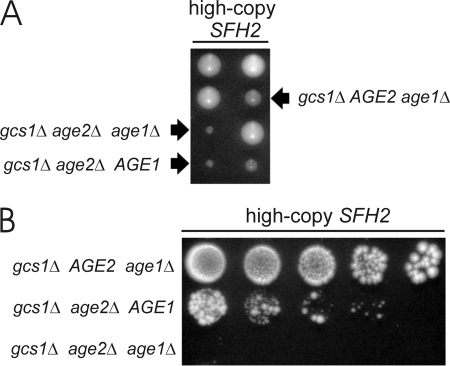
Sfh2 activity may influence ArfGAP activity through modification of the lipid environment (13, 14). The finding that increased dosage of the SFH2 gene bypasses the need for the Age2 + Gcs1 ArfGAP pair may reflect an altered lipid environment that permits vesicular transport in the absence of ArfGAP activity or that allows another ArfGAP to substitute for the missing Age2 and Gcs1. We, therefore, assessed the involvement of Age1 in this SFH2 effect. Diploid cells heterozygous for gcs1Δ, age2Δ, and age1Δ deletion mutations and harboring a high-copy SFH2 plasmid were induced to undergo meiosis. Both gcs1Δ age2Δ AGE1 (double-mutant) and gcs1Δ age2Δ age1Δ (triple-mutant) haploid segregants with high-copy SFH2 grew to form small colonies on synthetic complete medium (Fig. 5A), but a significant difference in growth was evident when the cells were grown on enriched medium (Fig. 5B). In total we assessed 11 double-mutant segregants and 6 triple-mutant segregants, and all exhibited consistent behaviors; that is, double-mutant cells harboring the high-copy SFH2 plasmid grew better than triple-mutant cells (lacking the AGE1 gene) harboring the SFH2 plasmid. Thus, Age1 is required for efficient bypass, by increased SFH2 dosage, of the need for the Age2 + Gcs1 pair.
FIGURE 5.
Effective SFH2 bypass depends on Age1. A, diploid cells heterozygous for gcs1Δ, age2Δ, and age1Δ deletion mutations and carrying a high-copy SFH2 plasmid were sporulated, and the resulting haploid segregants were incubated on solid synthetic complete medium for 5 days at 30 °C. B, equal numbers of cells from colonies identified in panel A were serially diluted (5-fold dilutions), spotted onto solid enriched medium, and incubated for 5 days at 30 °C.
Phospholipase D Mediates AGE1 Alleviation of age2Δ gcs1-4 Temperature Sensitivity
Sfh2 activity is suggested to affect post-Golgi transport by indirectly influencing phospholipase D activity, an enzyme activated by phosphoinositides (16). Sfh2 is thought to mediate phosphoinositide synthesis by delivering substrate to lipid kinases, such as Stt4, that in turn provide stimulatory phosphoinositides for phospholipase D (16).
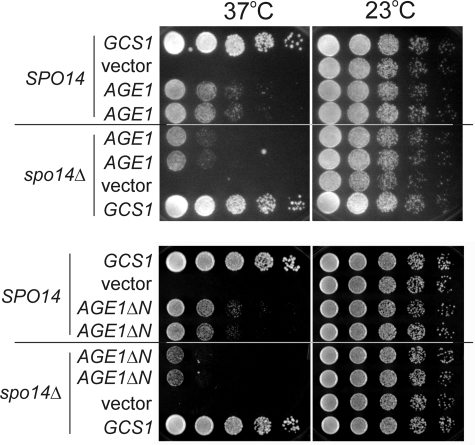
If increased SFH2 gene dosage results in a lipid environment that permits Age1 to functionally replace Age2 + Gcs1, then the absence of Sfh2 or downstream effectors of Sfh2 may compromise the ability of Age1 to replace the essential Age2 + Gcs1 ArfGAP pair. Cells deleted for the lipid kinase genes are inviable, preventing assessment of genetic interactions in cells lacking these genes. Deletion of the SFH2 gene did not impair the ability of AGE1 to bypass the lethality of age2Δ gcs1Δ cells (data not shown). In contrast, deletion of the phospholipase D gene, SPO14, did compromise the ability of AGE1 and AGE1ΔN to alleviate the temperature sensitivity of age2Δ gcs1-4 cells (Fig. 6). These results indicate that phospholipase D, a downstream target of Sfh2 activity, is required for aspects of Age1 function.
FIGURE 6.
Effective alleviation by AGE1 and AGE1ΔN of the post-Golgi ArfGAP defect depends on SPO14. age2Δ gcs1-4 cells with the wild-type SPO14 gene or a spo14Δ deletion mutation and carrying either GCS1, high-copy AGE1, or low-copy AGE1ΔN plasmids were grown in selective medium at 23 °C. Equal numbers of cells were then serially diluted (5-fold dilutions), spotted onto solid enriched medium, and incubated at 37 and 23 °C for 2 days. Two independent transformants are shown for cells carrying the AGE1 or AGE1ΔN plasmid.
Increased Gene Dosage for Phosphatidylinositol Kinases Fails to Compensate for a Post-Golgi ArfGAP Defect
To determine if potential downstream effectors of Sfh2 activity could mimic the SFH2 relief of growth inhibition for mutant cells with inadequate post-Golgi ArfGAP activity, we tested high-copy plasmids encoding the lipid kinases Stt4, Mss4, and Pik1 for the ability to support 37 °C growth of age2Δ gcs1-4 cells. In contrast to what was seen for age2Δ gcs1-4 cells carrying the SFH2, AGE1, or GCS1 plasmids, increased dosage of lipid kinase genes failed to restore high temperature growth (data not shown). Unlike increased SFH2 gene dosage itself, increased dosage of genes whose functions may be affected by Sfh2 activity does not alleviate the temperature sensitivity of age2Δ gcs1-4 cells.
Age1-GFP and Age1ΔN-GFP Colocalize with Golgi and Endosomal Markers
The observations that increased levels of Age1 can provide post-Golgi function suggest that Age1 might be localized to Golgi and/or endosomal compartments. Age1-GFP and Age1ΔN-GFP fusions, expressed under control of the inducible GAL1 promoter, were used to investigate the localization of Age1 protein. The expression of each GFP fusion, like the increased expression of wild-type Age1 and Age1ΔN, alleviated the temperature sensitivity of age2Δ gcs1-4 cells (data not shown), demonstrating that both Age1-GFP and Age1ΔN-GFP retain biological function.
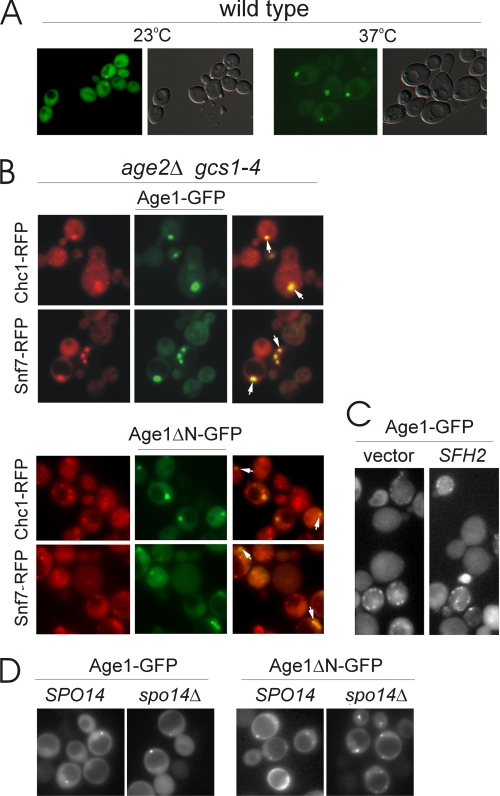
In wild-type cells at 23 °C, Age1ΔN-GFP was found throughout the cytoplasm and in punctate dots (Fig. 7A). At 37 °C, Age1ΔN-GFP (Fig. 7A) and Age1-GFP (data not shown) were in large punctate dots with minimal cytoplasmic staining. These dots were typically present once or twice per cell and were often localized near the vacuole. Under the same growth conditions and temperatures, wild-type cells expressing GFP itself failed to exhibit any punctate staining (data not shown).
FIGURE 7.
Age1-GFP localization at the trans-Golgi and endosome. A, with galactose as the sole carbon source, cells were grown at 23 °C, diluted in fresh medium, and grown for an additional 16 h at 23 or 37 °C before visualization. The localization of Age1ΔN-GFP, expressed under control of the GAL1 promoter, was assessed in wild-type cells at 23 °C and 37 °C. Cells were visualized by fluorescence (left panels) and differential interference contrast microscopy (right panels). B, the colocalization of Age1ΔN-GFP and Age1-GFP, expressed under control of the GAL1 promoter, with RFP organelle markers was assessed in age2Δ gcs1-4 cells at 37 °C. Arrows indicate representative colocalization. C, the localization of Age1-GFP, expressed under control of the AGE1 promoter, was monitored in proliferating age2Δ gcs1-4 cells also carrying an empty vector or a high-copy SFH2 plasmid and incubated for 2 h at 37 °C. D, the localization of Age1ΔN-GFP and Age1-GFP, expressed under control of the GAL1 promoter, was analyzed at 37 °C in age2Δ gcs1-4 cells with the wild-type SPO14 gene or a spo14Δ deletion mutation.
In age2Δ gcs1-4 cells grown at the restrictive temperature of 37 °C, both Age1-GFP and Age1ΔN-GFP exhibited punctate staining similar to that seen in wild-type cells (Fig. 7B). To determine whether the punctate staining corresponded to Golgi or endosomal compartments, we assessed the colocalization of the GFP fusions with organelle-specific RFP fusion proteins (17). Both Age1-GFP and Age1ΔN-GFP localization coincided with the compartments visualized by Chc1-RFP and Snf7-RFP, markers localized at trans-Golgi and endosomal compartments, respectively. The localization of Age1-GFP and Age1ΔN-GFP to Golgi/endosomal compartments is consistent with the ability of Age1 to alleviate deleterious effects caused by the absence of post-Golgi ArfGAPs.
Altered Gene Dosage of SFH2 or SPO14 Does Not Alter Age1-GFP Distribution
The ability of Sfh2 to bypass the need for the essential Age2 + Gcs1 ArfGAP pair in an Age1-dependent manner raises the possibility that increased Sfh2 abundance leads to the recruitment or localization of Age1 to Golgi/endosomal compartments. To address this possibility, we monitored the localization of Age1-GFP, whose expression was controlled by the native AGE1 promoter. A low-copy plasmid harboring AGE1pr-AGE1-GFP, like a low-copy AGE1 plasmid, was unable to alleviate the temperature sensitivity of age2Δ gcs1-4 cells unless a high-copy SFH2 plasmid was also present (data not shown). Irrespective of the presence or absence of a high-copy SFH2 plasmid, Age1-GFP exhibited a punctate staining pattern in age2Δ gcs1-4 cells incubated for a short duration at 37 °C, consistent with Golgi/endosomal localization (Fig. 7C). Thus, an increased abundance of Sfh2 does not appear to alter the localization of Age1.
The ability of increased Age1 abundance to alleviate the temperature sensitivity of age2Δ gcs1-4 cells is compromised in the absence of Spo14, suggesting the possibility that Age1 fails to localize properly in cells lacking Spo14. To test this, we assessed the localization of GFP fusions in spo14Δ cells. Age1-GFP and Age1ΔN-GFP were localized to punctate dots in both wild-type and spo14Δ cells (Fig. 7D), indicating that Spo14 is not required for the localization of Age1 to Golgi/endosomal compartments.
Age1ΔN Binds Phospholipids
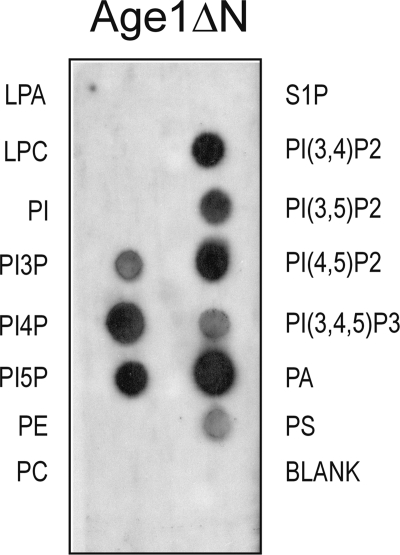
Gcs1 binds to a subset of phospholipids in vitro (14). The finding that SFH2-mediated bypass may act through Age1 led us to ask whether Age1, like Gcs1, binds phospholipids. The Age1ΔN protein and intact Age1 were expressed in bacteria as His6-tagged proteins; only the Age1ΔN protein could be isolated in soluble form under non-denaturing conditions, so intact Age1 protein was not tested. Using commercial phospholipid strips, affinity-purified soluble Age1ΔN-His6 avidly bound phosphatidic acid (PA), phosphatidylinositol 4,5-diphosphate (PI(4,5)P2), and phosphatidylinositol 3,4-diphosphate (PI(3,4)P2)and moderately bound phosphatidylserine (PS) and phosphatidylinositol 3-phosphate (PI3P) (Fig. 8). This pattern of lipid binding suggests that Age1 may be regulated by the phospholipid content of membranes.
FIGURE 8.
Phospholipid binding by Age1ΔN. Phospholipid membrane strips were incubated overnight at 4 °C with proteins (0.5 μg/ml), affinity-purified under native conditions from bacterial cells, and then washed and treated with Age1-specific affinity-purified antibodies; proteins bound to the phospholipid membrane strip were detected by enhanced chemiluminescence. No signal was detected when a membrane strip was treated with proteins isolated from bacteria carrying empty vector and probed with Age1-specific antibodies (data not shown). Incubation with 2.5 μg of C-terminal fragment of SidC, a phosphatidylinositol (PI) 4-phosphate (PI(4)P)-binding protein (29), demonstrated preferential binding of the protein to phosphatidylinositol 4-phosphate and to a much lesser extent to phosphatidylinositol 3,4-diphosphate (PI(3,4)P2) and phosphatidylinositol 4,5-diphosphate (PI(4,5)P2) (data not shown). The different pattern of binding for this phosphatidylinositol 4-phosphate (PI(4)P)-binding protein versus Age1ΔN-His6 demonstrates that phospholipid binding is protein-specific. PE, phosphatidylethanolamine; PC, phosphatidylcholine; PA, phosphatidic acid; PS, phosphatidylserine; PI, phosphatidylinositol; LPA, lysophosphatidic acid; LPC, lysophosphocholine; S1P, sphingosine-1-phosphate.
Age1 ArfGAP Activity in Vitro Is Enhanced by DAG
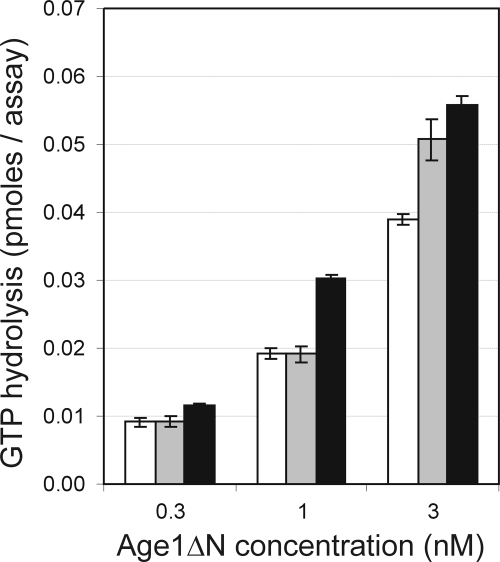
The in vitro activation of GTP-bound Arf1 hydrolysis by Gcs1 and by the rat ArfGAP1 is enhanced by long-chain forms of DAG (23). Phospholipase D produces phosphatidic acid with DAG as a secondary product. Our finding that phospholipase D may influence Age1 function to alleviate the defect in age2Δ gcs1-4 cells suggests that the ArfGAP activity of Age1 may be stimulated by phosphatidic acid or DAG. To address this possibility directly, we employed an in vitro assay to assess the stimulation by Age1ΔN-His6 of Arf1-bound GTP hydrolysis in the presence or absence of phosphatidic acid or DAG. In our assay, GTP-Arf1 is bound to unilamellar lipid vesicles of uniform size. Substituting 15% of the lipid content with DAG resulted in increased stimulation of GTP hydrolysis, which was greater than that observed when the lipid content was substituted with 15% phosphatidic acid (Fig. 9). At 1 nm Age1ΔΝ-His6, the presence of DAG enhanced GTP hydrolysis by 50% compared with what was seen in the absence of DAG. DAG, a downstream product of phospholipase D activity, stimulates the in vitro ArfGAP activity of Age1.
FIGURE 9.
DAG stimulates in vitro ArfGAP activity of Age1ΔN. Activation of hydrolysis of GTP-loaded myristoylated-Arf1 by Age1ΔN-His6 was assessed in assays in which the lipid content was either 100% dimyristoylphosphatidylcholine (white bar), 85% dimyristoylphosphatidylcholine, 15% phosphatidic acid (gray bar), or 85% dimyristoylphosphatidylcholine, 15% dioleoylglycerol (black bar). The low level of background radioactivity observed in the absence of Age1 protein was subtracted from all data values. S.D. were calculated from triplicate samples.
DISCUSSION
We show here that an increased abundance of Age1 and various Age1 truncations alleviates the deleterious effects caused by deficient Age2 + Gcs1 activity for post-Golgi transport. These effects of increased Age1 ArfGAP abundance are consistent with previous observations that endocytic transport is delayed in both age1Δ arf1Δ and age1Δ age2Δ double-mutant cells (11), suggesting an Age1 involvement in post-Golgi function. Elimination of the N-terminal 90 residues of Age1, external to the ArfGAP domain, improved the ability of the Age1 protein to alleviate the temperature sensitivity of age2Δ gcs1-4 double-mutant cells. Hence, the N-terminal region of Age1, specifically the first 90 residues, has a negative effect on Age1 function. Sequence analysis of the N-terminal region of Age1 did not reveal a motif that was conserved in the protein sequences of other eukaryotes.
The ability to compensate for the complete absence of Age2 + Gcs1 implies that Age1ΔN and Age1 provide essential functions that are normally supplied by the missing ArfGAP pair. Because Arf activity is essential in yeast (24), one function most likely supplied by Age1 and Age1ΔN is ArfGAP activity to regulate the GTPase cycle of Arf for post-Golgi transport. The Gcs1 ArfGAP also stimulates the interactions between v-SNAREs and Arf1/coatomer as determined by in vitro assays (25). These interactions are thought to promote vesicle “priming” to produce v-SNARE-equipped vesicles competent to deliver cargo to target compartments. In the absence of the proteins that normally stimulate incorporation of v-SNAREs into transport vesicles, Age1 and Age1ΔN may be capable of fulfilling this function. Thus, Age1 may support vesicular transport through ArfGAP activity and/or as a priming component for vesicle maturation.
A particularly informative finding here is that increased dosage of the SFH2 gene, encoding a phosphatidylinositol transfer protein, restores growth to cells lacking the otherwise essential Age2 + Gcs1 ArfGAP pair. Routt et al. (16) characterized a phosphoinositide metabolic pathway fed by increased SFH2 dosage that augments the delivery of substrates to lipid kinases, which in turn produce phosphoinositides stimulatory for phospholipase D activity. The alleviating effect of increased SFH2 dosage on age2Δ gcs1-4 temperature sensitivity depends on the phospholipase D gene SPO14 (Ref. 14 and data not shown). Phospholipase D cleavage of phosphatidylcholine yields phosphatidic acid, which is then hydrolyzed to DAG, and we found that alleviation of age2Δ gcs1-4 temperature sensitivity can also be achieved by supplying DAG exogenously (14). Therefore, the mechanism by which SFH2 bypasses the need for the Age2 + Gcs1 ArfGAP pair is likely through Sfh2 enhancement of a phospholipid metabolic pathway leading to phospholipase D.
Surprisingly, increasing the dosage of genes encoding downstream effectors of Sfh2 did not alleviate the temperature sensitivity of age2Δ gcs1-4 double mutants. In contrast to our earlier observation (14), a multicopy SPO14 plasmid failed to relieve the temperature sensitivity of age2Δ gcs1-4 cells. Further analysis indicated that our initial observation was due to a second-site spontaneous mutation independent of the SPO14 plasmid. Our findings, therefore, suggest that alleviation of age2Δ gcs1-4 temperature sensitivity is due to increased flux through the Sfh2-mediated phosphoinositide metabolic pathway rather than increased activity of pathway proteins downstream of Sfh2.
Effective SFH2-mediated relief of the lethality of the age2Δ gcs1Δ combination depends on the chromosomal copy of AGE1, implying that SFH2 bypass acts through Age1. Furthermore, this bypass raises the possibility that the alleviation of age2Δ gcs1-4 growth defect by a multicopy SFH2 plasmid is independent of the gcs1-4 protein. Instead, the alleviation by SFH2 may operate mainly, if not exclusively, through Age1. We found that purified Age1ΔN protein binds to several phospholipids and Age1-GFP, expressed at high or low levels, localized to Golgi and endosomal compartments. Therefore, Age1 may be regulated by phospholipid composition at the membranes of post-Golgi compartments.
A multicopy AGE1 plasmid was found to overcome the lethality of the age2Δ gcs1Δ combination even in cells lacking the SFH2 gene. This finding suggests that the remaining Sfh proteins, known to have overlapping function with Sfh2 (16), may provide compensatory activity. Consistent with this idea, increasing SFH3, SFH4, and SFH5 dosage also alleviates the temperature sensitivity of age2Δ gcs1-4 cells, albeit poorly in comparison to SFH2 (14).
We show here that the phospholipase D enzyme Spo14, which is required for the alleviation of age2Δ gcs1-4 temperature sensitivity by increased SFH2 gene dosage (14), is also required for the alleviation of that temperature sensitivity by AGE1 dosage, reinforcing the idea that phosphatidic acid and/or DAG, the immediate and downstream products of phospholipase D activity, enhance Age1 activity. Interactions between ArfGAPs, specifically Gcs1, and phospholipase D have previously been described for sporulating cells, where Gcs1 is required for phospholipase D function during formation of the prospore membrane (26). The in vitro GAP activities of Gcs1 and its rat ArfGAP1 ortholog are stimulated by phosphatidylinositol 4,5-diphosphate, phosphatidic acid, and most effectively, DAG (12, 23). In similar fashion, we show here that long-chain DAG also stimulates the in vitro ArfGAP activity of Age1. The genetic interactions identified here among SFH2, SPO14, and AGE1 and the stimulation of Age1 in vitro ArfGAP activity by DAG suggest a mechanism for Age1-mediated effects on post-Golgi vesicular transport; an Sfh2-mediated phosphoinositide pathway increases the abundance of phospholipids to activate phospholipase D activity, which in turn augments the pool of DAG to stimulate the ArfGAP activity of Age1 at post-Golgi membranes.
Our data suggest that Age1, like Gcs1, may be recruited to membranes through recognition of localized lipid packing and the resulting membrane topology. Gcs1 protein has an ArfGAP1 lipid packing sensor motif that regulates recruitment to membranes of high curvature (13). Although Age1 lacks an obvious ArfGAP1 lipid packing sensor motif, the involvement of phospholipase D and DAG in Age1 activity suggests that Age1 might also be recruited to highly curved membranes; the products of phospholipase D activity contribute to high membrane curvature (27). Because the overexpression of SFH2 or the deletion of SPO14 did not alter the Golgi/endosomal punctate staining of Age1-GFP, changes in membrane lipid content may affect the putative recruitment of Age1 at a local level that cannot be discerned by fluorescence microscopy.
The effect of depleting the cellular pool of DAG in mammalians cells has been reported (28). A diminished DAG pool results in the inhibition of Golgi-to-endoplasmic reticulum retrograde transport as a consequence of decreased abundance of ArfGAP1 at Golgi membranes. This decrease in Golgi-localized ArfGAP1 is associated with an accumulation of coat protein complex I-coated buds, implying that ArfGAP1 is required to complete membrane fission as part of the process of transport-vesicle production. The consequences of depleted DAG pools in mammalian cells raises the possibility that DAG pools in yeast regulate the recruitment of Age1 ArfGAP to participate in the production of transport vesicles at post-Golgi membranes.
In summary, increasing the activity of a phosphoinositide pathway can restore viability to age2Δ gcs1Δ mutant cells lacking the essential Age2 + Gcs1 post-Golgi ArfGAP pair. The phospholipids generated lead to the activation of phospholipase D, and the lipid products then stimulate Age1 activity to provide essential post-Golgi ArfGAP function.
Acknowledgments
We thank Vytas Bankaitis, Scott Emr, Joanne Engebrecht, and Joel Moss for plasmids and Roberto de Antueno for technical assistance in lipid preparations. We also thank Dan Cassel, Ameer Jarrar, Christopher McMaster, and Anne Spang for critical reading of the manuscript and two anonymous reviewers for their constructive suggestions.
This work was supported by Grant MOP-64293 from the Canadian Institutes of Health Research (to G. C. J. and R. A. S.).
- ArfGAP
- Arf GTPase-activating protein
- DAG
- diacylglycerol
- myr
- myristoylated
- RFP
- red fluorescent protein.
REFERENCES
- 1. Bonifacino J. S., Glick B. S. (2004) Cell 116, 153–166 [DOI] [PubMed] [Google Scholar]
- 2. Letourneur F., Gaynor E. C., Hennecke S., Démollière C., Duden R., Emr S. D., Riezman H., Cosson P. (1994) Cell 79, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 3. Serafini T., Orci L., Amherdt M., Brunner M., Kahn R. A., Rothman J. E. (1991) Cell 67, 239–253 [DOI] [PubMed] [Google Scholar]
- 4. Goldberg J. (1999) Cell 96, 893–902 [DOI] [PubMed] [Google Scholar]
- 5. Stamnes M. A., Rothman J. E. (1993) Cell 73, 999–1005 [DOI] [PubMed] [Google Scholar]
- 6. Peyroche A., Paris S., Jackson C. L. (1996) Nature 384, 479–481 [DOI] [PubMed] [Google Scholar]
- 7. Cukierman E., Huber I., Rotman M., Cassel D. (1995) Science 270, 1999–2002 [DOI] [PubMed] [Google Scholar]
- 8. Poon P. P., Wang X., Rotman M., Huber I., Cukierman E., Cassel D., Singer R. A., Johnston G. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10074–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poon P. P., Cassel D., Spang A., Rotman M., Pick E., Singer R. A., Johnston G. C. (1999) EMBO J. 18, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poon P. P., Nothwehr S. F., Singer R. A., Johnston G. C. (2001) J. Cell Biol. 155, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang C. J., Bowzard J. B., Anido A., Kahn R. A. (2003) Yeast 20, 315–330 [DOI] [PubMed] [Google Scholar]
- 12. Yanagisawa L. L., Marchena J., Xie Z., Li X., Poon P. P., Singer R. A., Johnston G. C., Randazzo P. A., Bankaitis V. A. (2002) Mol. Biol. Cell 13, 2193–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bigay J., Casella J. F., Drin G., Mesmin B., Antonny B. (2005) EMBO J. 24, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wong T. A., Fairn G. D., Poon P. P., Shmulevitz M., McMaster C. R., Singer R. A., Johnston G. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12777–12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang C. J., Cavenagh M. M., Kahn R. A. (1998) J. Biol. Chem. 273, 19792–19796 [DOI] [PubMed] [Google Scholar]
- 16. Routt S. M., Ryan M. M., Tyeryar K., Rizzieri K. E., Mousley C., Roumanie O., Brennwald P. J., Bankaitis V. A. (2005) Traffic 6, 1157–1172 [DOI] [PubMed] [Google Scholar]
- 17. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 18. Jansen G., Wu C., Schade B., Thomas D. Y., Whiteway M. (2005) Gene 344, 43–51 [DOI] [PubMed] [Google Scholar]
- 19. Deleted in proof.
- 20. Huber I., Rotman M., Pick E., Makler V., Rothem L., Cukierman E., Cassel D. (2001) Methods Enzymol. 329, 307–316 [DOI] [PubMed] [Google Scholar]
- 21. Vida T. A., Emr S. D. (1995) J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lewis M. J., Nichols B. J., Prescianotto-Baschong C., Riezman H., Pelham H. R. (2000) Mol. Biol. Cell 11, 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Antonny B., Huber I., Paris S., Chabre M., Cassel D. (1997) J. Biol. Chem. 272, 30848–30851 [DOI] [PubMed] [Google Scholar]
- 24. Stearns T., Kahn R. A., Botstein D., Hoyt M. A. (1990) Mol. Cell. Biol. 10, 6690–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson M., Poon P. P., Schindler C., Murray L. E., Kama R., Gabriely G., Singer R. A., Spang A., Johnston G. C., Gerst J. E. (2006) Mol. Biol. Cell 17, 1845–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Connolly J. E., Engebrecht J. (2006) Eukaryot. Cell. 5, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Corda D., Hidalgo Carcedo C., Bonazzi M., Luini A., Spanò S. (2002) Cell. Mol. Life Sci. 59, 1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fernández-Ulibarri I., Vilella M., Lázaro-Diéguez F., Sarri E., Martínez S. E., Jiménez N., Claro E., Mérida I., Burger K. N., Egea G. (2007) Mol. Biol. Cell 18, 3250–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weber S. S., Ragaz C., Reus K., Nyfeler Y., Hilbi H. (2006) PLoS Pathog. 2, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Archambault J., Drebot M. A., Stone J. C., Friesen J. D. (1992) Mol. Gen. Genet. 232, 408–414 [DOI] [PubMed] [Google Scholar]
- 31. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. (1986) Yeast 2, 163–167 [DOI] [PubMed] [Google Scholar]
- 33. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 34. Li X., Routt S. M., Xie Z., Cui X., Fang M., Kearns M. A., Bard M., Kirsch D. R., Bankaitis V. A. (2000) Mol. Biol. Cell 11, 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haun R. S., Tsai S. C., Adamik R., Moss J., Vaughan M. (1993) J. Biol. Chem. 268, 7064–7068 [PubMed] [Google Scholar]