Abstract
Tumor-associated cell surface antigens and tumor-associated vascular markers have been used as a target for cancer intervention strategies. However, both types of targets have limitations due to accessibility, low and/or heterogeneous expression, and presence of tumor-associated serum antigen. It has been previously reported that a mitochondrial/cell surface protein, p32/gC1qR, is the receptor for a tumor-homing peptide, LyP-1, which specifically recognizes an epitope in tumor cells, tumor lymphatics, and tumor-associated macrophages/myeloid cells. Using antibody phage technology, we have generated an anti-p32 human monoclonal antibody (2.15). The 2.15 antibody, expressed in single-chain fragment variable and in trimerbody format, was then characterized in vivo using mice grafted subcutaneously with MDA-MB-231 human breast cancers cells, revealing a highly selective tumor uptake. The intratumoral distribution of the antibody was consistent with the expression pattern of p32 in the surface of some clusters of cells. These results demonstrate the potential of p32 for antibody-based tumor targeting strategies and the utility of the 2.15 antibody as targeting moiety for the selective delivery of imaging and therapeutic agents to tumors.
Keywords: Antibodies, Cell Surface Protein, Tumor, Tumor Marker, Tumor Therapy, Globular Domain of C1q Receptor, Imaging, Lymphatic, Macrophages, Molecular Targeting
Introduction
The localization of tumors may be accomplished by any of several combinations including computed tomography, ultrasonography, gamma camera examination, and glucose consumption (1, 2). However, targeted localization of the tumors is preferred, mainly using specific probes that bind to tumor-associated cell surface antigens or to markers of angiogenesis expressed by endothelial cells or present in the surrounding extracellular matrix (3–6). Probes that bind to tumor-associated cell surface antigens have some drawbacks (7) such as the heterogeneous expression on the cell surface or the increased serum levels of the antigen as tumors grow, which may act as a trap for the targeting agent. Angiogenesis related targets are readily accessible; however, the relatively low abundance of endothelial cells in tumor tissue makes the molecular imaging of tumor neovessels more challenging. Furthermore, angiogenesis may occur also in a physiological context, thus adding more complexity to the targeting.
With these limitations in mind, we hypothesized as an alternative target a marker selectively expressed in different compartments in the tumor area. One targeting agent specific for the tumor but not restricted to the tumor cells is the tumor homing peptide (LyP-1), which strongly and specifically accumulates in the tumor after systemic administration, localizing preferentially associated to lymphatic markers (8–10). LyP-1-binding protein was characterized as p32 (10), a multiligand and multicompartmental protein that has been independently identified in several contexts and has been named accordingly as SF2P32 (splicing factor SF2-associated protein; 11), HABP-1 (hyaluronic acid binding protein-1; 12), gC1qR (globular domain of C1q receptor; Ref. 13), or HIV TAP (Tat-associated protein; 14). Although p32 is primarily present in the mitochondria, it has been, under certain conditions (15), detected in different cellular compartments (nucleus, cellular surface, endoplasmic reticulum (13, 16–20)) and in different cell types (B lymphocyte (13)), platelets (21), neutrophils (22), eosinophils (23), endothelial cells (24), macrophages and dendritic cells (25, 26), or fibroblasts (27)). p32 has also been recently reported in the surface of tumor cells in hypoxic/nutrient-deprived areas as well as in the cell surface of a tumor-associated macrophage/myeloid cell subpopulation closely linked to tumor lymphatics (10).
In this work, we take advantage of the over-expression of the multicompartmental p32/gClqR (hereafter referred to as p32) associated to tumors (in tumor cells, tumor lymphatics, and tumor-associated macrophages) to generate a human anti-p32 single-chain Fv (scFv)4 antibody (2.15). This antibody has shown to selectively target solid tumors in vivo both as a monovalent and trivalent antibody fragment.
EXPERIMENTAL PROCEDURES
Cells and Culture Conditions
All cells were from the ATCC. HEK-293 cells (human embryonic kidney epithelia; CRL-1573), and MDA-MB-231 (human breast adenocarcinoma; HTB-26) were grown in DMEM supplemented with 10% heat-inactivated FCS (all from Invitrogen) in humidified CO2 (5%) incubator at 37 °C. U-937 cells (human histiocytic lymphoma; CRL-1593.2) and 4T1 cells (mouse breast tumor; CRL-2539) were maintained in RPMI supplemented with 10% FCS. Differentiation of the U-937 cells was induced for the indicated time intervals in fresh culture medium containing 5 nm phorbol myristic acid (Sigma-Aldrich).
Recombinant Proteins, Antibodies, Peptides, and Reactives
Recombinant human p32 (rhp32) was obtained from bacteria and purified by immobilized metal ion affinity chromatography. Recombinant mouse p32 was purchased from United States Biological (USBio). Purified rabbit polyclonal anti-full-length p32 was directed against the N terminus (amino acids 76–93). The mAbs used included mouse anti-p32 (60.11 and 74.5.2), anti-human c-Myc 9.E10, FITC-conjugated anti-human c-Myc 9.E10 (Abcam, Cambridge, UK); anti-human MHC class I molecules W6/32 (eBioscience, San Diego, CA); rat anti-mouse CD31 (BD Biosciences); HRP-conjugated anti-human c-Myc (Invitrogen); and HRP-conjugated anti-M13 bacteriophage (GE Healthcare). The polyclonal antibodies used included an Alexa Fluor 546-conjugated anti-rat IgG (Invitrogen); a phycoerythrin-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Europe, Suffolk, UK); an HRP-conjugated donkey anti-rabbit IgG; and an HRP-conjugated sheep anti-mouse IgG (GE Healthcare). Trypsin, BSA, o-phenylenediamine dihydrochloride, and isopropyl-beta-d-thiogalactopyranoside were from Sigma-Aldrich. BSA was conjugated with 4-hydroxy-5-iodo-3-nitrophenyl (NIP; Sigma-Aldrich) in a molar ratio of 10:1 (NIP10-BSA) as described (28). Mouse EHS-laminin (LM111) was from (BD Biosciences).
Selection of scFv Phage Library on rhp32
Recombinant scFv phages (Griffin.1 library, Medical Research Council Cambridge; total diversity, ∼1.2 × 109) (29) were panned for binding on purified antigen (rhp32) as described (30) with slight modifications: immunotubes (Maxisorp, Nunc, Roskilde, Denmark) were coated overnight at 4 °C with 4 ml of rhp32 at a concentration of 10 μg/ml in PBS. After washing twice with PBS, the tubes were blocked for 2 h at 37 °C with 4% BSA in PBS. Meanwhile, 1013 phages were blocked with 1 ml 4% BSA in PBS. Preblocked phages were added to the immunotube and incubated at room temperature with continuous rotation for 30 min, followed by 90 min of stationary incubation. The tubes were washed 10 times (in the first round of selection, 20 in the subsequent selections) with PBS containing 0.05% Tween 20 and then with PBS. Bound phages were eluted with 1 ml of trypsin (1 mg/ml in 50 mm Tris-HCl, pH 7.4, 1 mm CaCl2) at room temperature with continuous rotation for 20 min. Eluted phages were recovered by infecting logarithmically growing (A600 = 0.5) Escherichia coli TG1 (K12, Δ(lac-pro), supE, thi, hsdD5/F′traD36, proA+B+, lacIq, lacZΔM15 (31)) at 37 °C for 30 min. The infected cells were plated on LB agar supplemented with 100 μg/ml ampicillin and 1% glucose and incubated overnight at 37 °C. This enriched library was grown on E. coli TG1 and rescued upon infection with the helper phage KM13 (32). Phages displaying scFv fragments were purified from the culture supernatant by precipitation with 20% PEG 6000 and 2.5 m NaCl and were resuspended in sterile cold PBS with 15% glycerol for long term storage at −80 °C and for subsequent rounds of selection.
Screening of Selected Phages by ELISA
Single colonies were screened by ELISA to evaluate the frequency of phage displaying rhp32-binding scFv fragments as described (33). rhp32-binding phages were fingerprinted by amplifying the scFv using primers LMB3 and FdSeq1 (LMB3, 5′-CAG GAA ACA GCT ATG AC-3′; FdSeq1, 5′-GAA TTT TCT GTA TGA GG-3′) followed by digestion with the frequent cutting enzyme BstN-I (New England Biolabs). Molecular characterization was completed by sequencing the variable regions using primers FOR_LinkSeq (VH; 5′-GCC ACC TCC GCC TGA ACC-3′) and pHEN_Seq (VL; 5′-CTA TGC GGC CCC ATT CA-3′). Sequences were analyzed and aligned to the VBASE2 database (34) to learn the amino acids forming the loops in the complementarity-determining regions used and type of chains present.
Soluble Antibody Expression and Purification
Phage particles from selected clones were used to infect logarithmically growing (A600 = 0.5) E. coli HB2151 (nonsuppresser strain (K12, ara, Δ(lac-pro), thi/F′ proA+B+, lacIqZΔM15 (35)), and soluble scFv fragments were obtained as described (33). Purification was performed using the ÄKTAprime plus system (affinity step: HisTrap or HiTrap rProtein A FF columns (GE Healthcare) according to the manufacturer's protocol followed by gel filtration HiPrep 16/60 Sephacryl S100-HR) and checked by ELISA and SDS-PAGE. Either supernatant from isopropyl-beta-d-thiogalactopyranoside-induced HB2151 or purified scFv was used. Competition ELISA was performed as a standard ELISA but with a previous step of blockade using mAb; after blocking with 300 μl 4% BSA in PBS at 37 °C for 1 h, wells were incubated with 100 μl of a 20 μg/ml solution of the appropriate reagent (mAb 60.11, mAb 74.5.2, or control mouse IgG1) for 1 h at room temperature and 30 rpm.
Flow Cytometry
To study the ability of the scFv to detect p32 on the cell surface, unstimulated mouse 4T1 cells and phorbol myristic acid-stimulated human U-937 cells (5 nm for 3, 6, or 12 h prior to the staining) were incubated with anti-p32 mAb (5 μg/ml) or purified scFv (10 μg/ml) and mAb 9E10 (4 μg/ml) in 100 μl for 45 min. After washing, the cells were treated with appropriate dilutions of phycoerythrin-conjugated goat anti-mouse IgG. The samples were analyzed with an EPICS XL (Coulter Electronics, Hialeah, FL).
Construction of Expression Vectors and Purification of Recombinant Multivalent Antibodies
The coding sequence of the scFv 2.15 was amplified using primers ClaI-2.15 (5′-TCA TCG ATG GAG GTG CAG CTG GTG GAG-3′) and FdSeq1 and ligated into pCR2.1 TOPO. The ClaI/NotI-digested fragment was ligated into the ClaI/NotI pCR3.1-L36-NC1ES−-digested plasmid (6) to obtained the pCR3.1–2.15-NC1ES− plasmid. All constructs were verified by sequencing. The details about the plasmid pCEP4-B1.8-NC1ES− containing the B1.8 (anti-NIP) trimerbody and the procedure to obtain purified trimerbodies can be found elsewhere (6).
Antibody Labeling with Cyanine 5
Purified antibodies (scFvs and trimerbodies) were labeled with the near-infrared cyanine 5 (Cy5) N-hydroxysuccinimide (NHS) esters (GE Healthcare) according to the manufacturer's recommendations. One milliliter of the antibody solution (1 mg/ml) was conjugated with 0.1 ml of a 2 mg/ml Cy5 solution for 1 h in the dark at room temperature. Cy5-labeled recombinant antibodies were separated from unconjugated Cy5 dye by gel filtration on Sephadex G-25 Superfine HiTrap Desalting columns (PD-10 columns, GE Healthcare), and concentrated in 10,000 molecular weight cutoff (MWCO) Vivaspin 500 filter (Vivascience) to 1 mg/ml. The labeling ratio of Cy5 to antibody (Cy5:antibody) was calculated as described (36) and was close to 1:(1–2). The functionality of Cy5-labeled antibodies was verified by ELISA against specific antigen.
Infrared Immunophotodetection in Tumor-bearing Mice
Imaging was performed as described (6) with slight modifications. Briefly, wild-type MDA-MB-231 cells (2 × 106) were implanted into the mammary fat pad of 6-week-old female Hsd:athymic nude-Foxn1nu mice (Harlan Ibérica, Barcelona, Spain) maintained with a low manganese diet (ssniff Spezialdiäten GMBH, Soest, Germany). Nodule dimensions were used to calculate tumor volume using the formula: width2 × length × 0.52. When tumors reached a volume of 0.2–0.4 cm3, mice were injected in the tail vein with 100 μl Cy5-labeled antibody solution in PBS. Mice were imaged using the high resolution charge-coupled device cooled digital camera ORCA-2BT and Hokawo software (Hamamatsu Photonics France, Massy, France) under anesthesia. Three images were acquired for each experiment: a bright field image, a Cy5-specific image (emission, red light filter centered at 632.8 nm; optical filter, 665–680 nm), and an autofluorescence reference image (emission, blue light filtered at 470 nm; optical filter, 665–680 nm). Normalized reference autofluorescence was subtracted from the Cy5-specific image, and the resultant was tinted and merged with the bright-field image (tinted in the GFP blue-shifted spectral (448 nm) for better contrast) using the Hokawo software. Further editing included only cropping, resizing, and rotating the image for a better view of the picture. All mice were handled in accordance with the guidelines of the Hospital Universitario Puerta de Hierro Animal Care and Use Committee and performed in accordance with Spanish legislation.
Immunohistology
Tumors were removed after infrared imaging (2.5 h after i.v. injection), frozen in optimal cutting temperature (OCT) embedding medium (Sakura Tissue Tek, Alphen aan den Rijn, The Netherlands), and sectioned (4–7-μm thickness) using the Leica CM1850 cryostat. Sections were incubated overnight with the primary antibodies (anti-Myc:FITC antibody (1:200) and rat anti-mouse CD31 (1:100)), followed by anti-rat secondary reagents (1:1000), and mounted by using VectaShield mounting media with 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Images were acquired using a confocal scanning inverted Leica AOBS/SP2-microscope (Leica Microsystems).
RESULTS
Isolation of Human Anti-rhp32 Antibodies by Panning a scFv Library
The Griffin.1 library was panned against affinity purified recombinant human p32 (rhp32) immobilized in Nunc immunotubes. The frequency of binding clones was studied by ELISA after each round of selection. The percentage of binders was 20% after the first round and 60% after the second. BstN1 fingerprinting of 24 selected clones (by rhp32/BSA ratio (in preliminary 96-well ELISA) > 3) indicated 16 different restriction patterns, which were later confirmed by DNA sequencing.
Biochemical Characterization of scFv Fragments
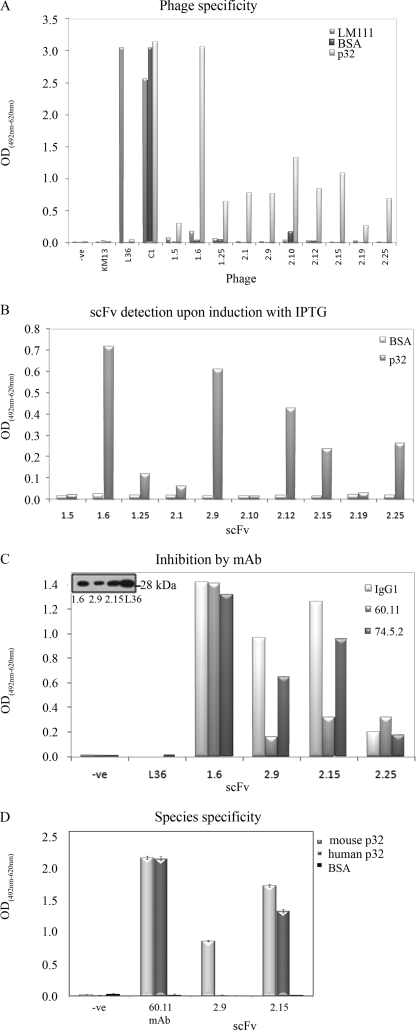
Ten different clones that were consistent binders were used in a phage-ELISA assay against rhp32 and other unrelated proteins (Fig. 1A). Most of the clones were highly specific for rhp32, showing almost no reactivity against other elements present in the selection process (plastic and BSA) or an unrelated protein (LM111). After expression as soluble scFv, although different from clone to clone, there was a significant reactivity of most of the scFvs against rhp32 (Fig. 1B). The binding of LyP-1 phage to p32 is inhibited by monoclonal antibody 60.11, which is directed against the C1q binding domain of p32 (10). To identify scFv sharing the epitope with mAb 60.11, four scFv fragments (1.6, 2.9, 2.15, and 2.25) were expressed using the nonsuppressor host E. coli HB2151 and purified from the supernatant by standard affinity chromatography procedures. The purified scFv fragments gave rise to a single protein band of expected mobility (Fig. 1C, inset); a competition ELISA was designed using either mAb 60.11 directed against p32 N-terminal amino acids 76–93 or 74.5.2 directed against amino acids 204–218 from the C terminus of p32. Preincubation with 60.11 but not 74.5.2 greatly diminished the binding of scFv 2.9 and 2.15, indicating overlapping, if not identical, epitopes (Fig. 1C). Due to the degree of cross-species conservation between rodents and humans, both in sequence and in the ability to bind C1q (37), we further investigated whether the selected scFv fragments were able to bind immobilized purified recombinant mouse p32. It was found that 2.9 gave no signal on recombinant mouse p32, whereas 2.15 gave comparable signals on both mouse and human p32 immobilized on plastic (Fig. 1D).
FIGURE 1.
Characterization of anti-p32 phage-derived scFvs. A, specificity of phage scFv fragments from selected clones by ELISA. Reactivity was assessed against the following: elements present in the selection process (BSA); murine laminin-1 (LM111); and the target protein rhp32 (p32). Internal controls were as follows; L36 binds to LM111, but not other proteins; KM13, empty phage, does not bind any protein, whereas C1-phage binds BSA). Phage input was essentially the same for each clone and each ELISA as assessed by phage-ELISA. The y axis represents A492 nm corrected with A620 nm except otherwise stated. B, soluble scFv fragments retained their specificity after cloning in HB2151 and expression after induction with isopropyl-β-d-thiogalactopyranoside (IPTG). C, mononclonal antibody competition analyses for p32 binding. Binding of 2.9 and 2.15 to rhp32 was diminished after incubation with mAb 60.11, suggesting a common epitope for the scFv and mAb. Inset, Western blotting of the purified scFv. D, 2.15, but not 2.9, was able to bind to human and mouse p32. Error bars represent the standard error of the mean of three different experiments.
Reactivity of scFv Fragments to Cell Surface-expressed p32
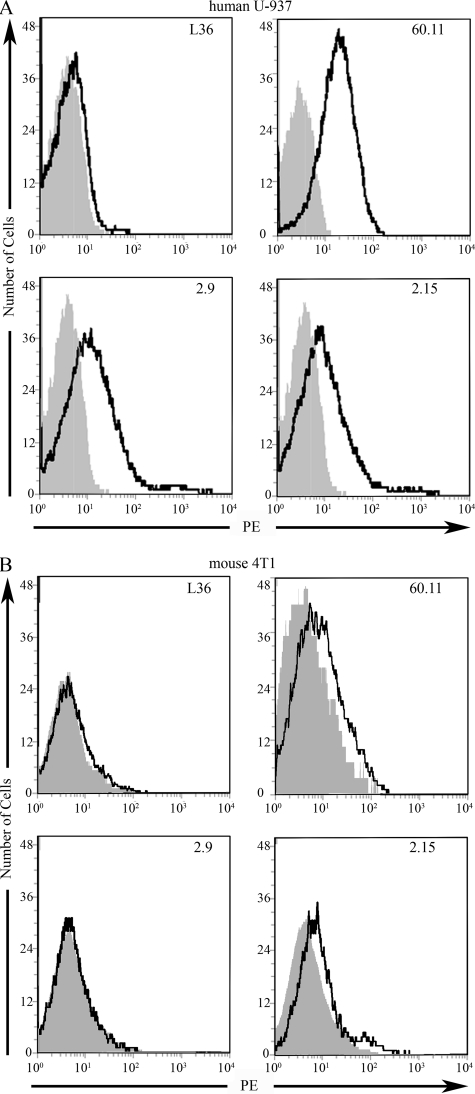
The ability of the scFv fragments to recognize cell surface-expressed p32 was assessed by flow cytometry using freshly purified soluble scFv. Nonstimulated human U-937 cells reacted minimally with either anti-p32 mAbs (60.11 or 74.5.2) or scFv (2.9 or 2.15) (data not shown). In contrast, U-937 activation with phorbol myristic acid produced a consistently increased p32 expression as reflected by mAbs and scFv fragments. Staining with mAb 60.11 showed that ∼50% of stimulated U-937 cells expressed p32 in the surface (Fig. 2A). Purified scFv fragments corresponding to 2.9 and 2.15, also stained the U-937 cells, and a slightly higher percentage of the cells were stained than with the 60.11 antibody (Fig. 2A). Surface staining of mouse 4T1 cells demonstrated the presence of p32 on the cell surface, in agreement with previous studies (10). The mAb 60.11 produced a small but consistent shift in flow cytometry analysis of live 4T1 cells (Fig. 2B). A similar staining pattern was observed in 4T1 cells incubated with the 2.15 scFv. In contrast, incubation of 4T1 cells with 2.9 scFv revealed no staining (Fig. 2B). Thus, further corroborating the ability of 2.15 to detect the mouse cell surface p32 to a extent similar to the mAb 60.11.
FIGURE 2.
Reactivity of scFv fragments to cell surface expressed p32. A, binding of anti-p32 mAb and purified scFv fragments to phorbol myristic acid-stimulated human U-937 cells by flow cytometry. B, binding of anti-p32 mAb and purified scFv fragments to mouse 4T1 cells by flow cytometry. For purified scFv fragments, the bound scFv was detected with sequential incubations with 9E10 anti-Myc mAb and phycoerythrin (PE)-labeled goat anti-mouse IgG. FACScan histograms show the binding of each scFv clone (bold line) and the backgrounds of phycoerythrin-conjugated secondary antibodies (gray).
Tumor Targeting with Mono- and Trivalent Anti-p32 Antibodies
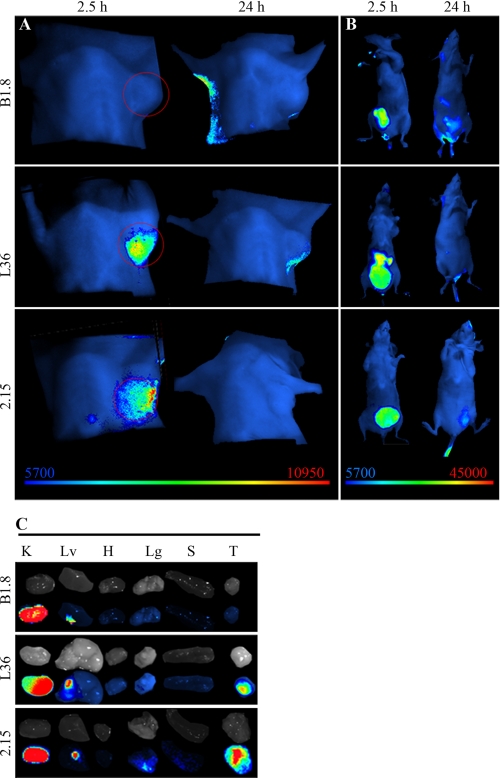
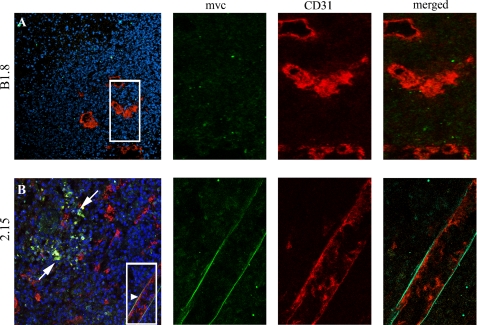
We selected the scFv 2.15 for in vivo targeting assays based on its inhibition by the 60.11 mAb as shown by ELISA on its ability to detect p32 expressed in the surface of both human and mouse cells as shown by flow cytometry. Control scFv antibodies included the following: B1.8 (anti-hapten NIP) and L36 (recognizes a conformational epitope of LM111 exposed in several solid tumor models (6)). Antibodies were labeled with the near-infrared fluorochrome Cy5 and injected in the tail vein of nude mice bearing MDA-MB-231 human xenografts. All of the antibodies showed a rapid renal clearance after i.v. injection, with peak signal intensity at 1–2 h and no detectable bladder signal at 24 h post-injection (Fig. 3B). The control scFv-B1.8 showed no detectable localization, whereas both 2.15 and L36 localized in the tumors (Fig. 3A). Maximum resolution was achieved at ∼2h (Fig. 3A), when the ratio of the signal of the 2.15 and that of the B1.8 was near 7 (supplemental Fig. 1). Ex vivo imaging of the organs further confirms the specific accumulation of 2.15 and L36 in the tumors, while showing a similar uptake by the kidneys for all the antibodies (Fig. 3C). Staining of the tumors from mice that received the scFvs with FITC-conjugated anti-Myc mAb (to detect the injected scFv) and anti-CD31 mAb showed that most of the endothelial cells were negative for the scFv, and most of the scFv staining was dispersed in clusters of cells (Fig. 4, white arrow). However, some of the main vessels showed a distinct co-staining in the basal side of the cells (Fig. 4, arrowhead).
FIGURE 3.
Targeting of fluorescently labeled anti-p32 scFv to human tumors. Near-infrared fluorescence imaging of nude mice bearing MDA-MB-231 human breast tumor xenografts. A, anti-hapten B1.8 did not localize in the tumor, whereas L36 and 2.15 showed a specific signal. A representative image of four mice is shown (2.15 scFv) and three mice (B1.8 and L36). B, ventral view shows similar accumulation of the scFv in the bladder. C, ex vivo imaging of the organs (kidney (K), liver (Lv), heart (H), lung (Lg), spleen (S), and tumor (T)) shows similar uptake of the Cy5-labeled reagent in the kidneys.
FIGURE 4.
2.15 scFv recognizes dispersed clusters of cells and the basal side of endothelial cells. After i.v. administration of the scFv, tumors were removed, embedded in OCT, and stained for myc (green) or for an endothelial marker (CD31, red). A, B1.8 does not localize in the tumor stroma. It does not localize either associated with vessels (right panel). B, 2.15 localizes in the tumor in dispersed clusters of cells (white arrow). A distinct co-staining occurred in the basal side of endothelial cells (arrowhead and right panel).
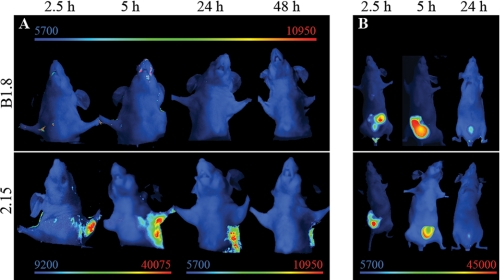
We have previously reported that the trimerbody format offers advantages over scFv molecules for tumor-targeting applications in vivo against a cell surface antigen or an extracellular matrix antigen (LM111) (6). We hypothesized that an anti-p32 trimerbody would also surpass its scFv counterpart for in vivo imaging. For this reason, the 2.15 scFv was assembled in the trimerbody format and expressed as a soluble secreted protein in human HEK-293 cells and purified from conditioned medium. The purification scheme yielded antibodies that were >95% pure by SDS-PAGE. The functionality of purified 2.15 trimerbody was demonstrated by ELISA against rhp32 (supplemental Fig. 2). Trimerbodies (2.15 and B1.8) were labeled with Cy5 and injected in the tail vein of nude mice bearing MDA-MB-231 human xenografts. Trimerbodies showed slower clearance than the corresponding scFvs (Fig. 5B). The control B1.8 trimerbody showed no detectable localization in the tumor, whereas a strong and selective accumulation was observed in the case of the 2.15 trimerbody. Maximum tumor uptake was detected at 2.5 and 5 h; the signal intensity decreased at 24 h although remained detectable for at least 48 h (Fig. 5A), whereas most of the systemic protein was eliminated at 24h (Fig. 5B).
FIGURE 5.
Targeting of fluorescently labeled anti-p32 trimerbody to human tumors. Near-infrared fluorescence imaging of nude mice bearing MDA-MB-231 human breast tumor xenografts. A, an anti-hapten B1.8 trimerbody did not localize in the tumor, whereas the 2.15 trimerbody showed a rapid (2.5 h) and sustained (up to 48 h) localization. Note the different scale bar for the early images. B, ventral view shows similar trimerbody accumulation in the bladder, as well as some elimination through the liver.
DISCUSSION
We have generated a human recombinant antibody against the multicompartmental protein p32/gClqR. We have demonstrated the ability of different formats of this antibody (scFv and trimerbody) to target solid tumors in vivo. These results represent a new concept in tumor targeting. So far, antibody-based tumor targeting strategies have been based on tumor-associated cell surface antigens or tumor-associated vascular markers. Antibodies specific to tumor surface antigens, such as HER2-neu (38), carcinoembryonic antigen (39), or prostate-specific antigen (40), among others, have proven useful for in vivo localization of solid tumors. However, tumor surface antigens exhibit a high shedding profile and are dependent on tumor dedifferentiation or clonal proliferation. Antigens that are preferentially expressed in the tumor extracellular matrix may be better suited for tumor-targeting applications. In fact, several groups have demonstrated that antibodies specific to components of the extracellular matrix (EDB domain of fibronectin (41), domain C of tenascin C (42), and laminin (6, 33)) were capable of selective targeting of neovascular structures in solid tumors. However, it has been shown that antibodies against tumor-associated vascular markers fail to accumulate in nonproliferating tumors (43). It is tempting to speculate that this can be a major problem in a proportion of patients as well as certain types of tumors with a very low growth rate.
Although p32 has been reported in the surface of several other cell types under certain circunstances (fibroblasts, neutrophils, endothelial cells, platelets, etc. (15)), the majority of the protein is cytoplasmic and is detectable after permeabilization of the cell membrane (44). Nonetheless, certain cells are able to translocate and release the protein, which can result in the modification of a number of cellular and vascular protein responses (15). The role of cell surface p32 in the tumor remains unclear; however, the unique expression pattern of p32 in tumor cells, tumor lymphatics, and tumor-associated macrophages/myeloid cells makes p32 an ideal target for the diagnosis and therapy of cancer. In fact, it has been reported that p32/gC1qR is the receptor for a tumor-homing peptide, LyP-1. The LyP-1 peptide has been shown to localize in the tumor lymphatics (9), and to effectively target nanoparticles to the tumor (45). In this work, we confirmed that systemically administered 2.15 scFv penetrates the tumor covering a broad population of cells but in a pattern distinct from that of the LyP-1 peptide (8, 9). Although LyP-1 is probably processed as a CendR peptide upon binding to p32 and thus can actively penetrate the tissue (46, 47), the antibody may remain partially bound to the p32 readily accessible from the lumen of the blood vessels and also reach some clusters of cells (probably macrophages) in the stroma nearby.
Multimerization of scFv constructs has important advantages for tumor-targeting applications. Trimerbodies are intermediate-sized molecules that exhibit high stability under physiological conditions and enhanced avidity for the target owing to the trivalent structure (6, 48). In fact, we report that the 2.15 antibody in a trimerbody format stained the tumor more brightly than the 2.15 scFv at all time points. Anti-p32 trimerbody localized rapidly and specifically in the tumors. The tumor uptake reached a maximum at 2.5–5 h post injection and slowly washed out over time. Fluorescence was still detectable in the tumor 48 h after the trimerbody inoculation.
In summary, we have demonstrated that the human anti-p32 antibody 2.15 can selectively localize tumors in vivo. These results illustrate the potential of this new antibody for imaging and therapeutic applications and suggest that p32 might be universal target for cancer targeting.
Supplementary Material
Acknowledgments
We thank Drs. Paloma Sánchez-Mateos and Rafael Samaniego for help with the confocal microscope.
This work was supported by grants from the Ministerio de Ciencia e Innovación (BIO2008-03233), the Comunidad Autónoma de Madrid (S-BIO-0236-2006), the European Union (SUDOE-FEDER (IMMUNONET-SOE1/P1/E014; to L. A.-V.), and a grant from the U. S. Department of Defense Breast Cancer Program (to E. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- scFv
- single-chain fragment variable
- NIP
- 4-hydroxy-5-iodo-3-nitrophenyl
- rhp32
- recombinant human p32.
REFERENCES
- 1. Jacobsson H., Wallin G., Werner S., Larsson S. A. (1994) Eur. J. Nucl. Med. 21, 582–586 [DOI] [PubMed] [Google Scholar]
- 2. Nguyen B. D., Roarke M. C., Karstaedt P. J., Ingui C. J., Ram P. C. (2009) Curr. Probl. Diagn. Radiol. 38, 68–83 [DOI] [PubMed] [Google Scholar]
- 3. Vogel C. A., Bischof-Delaloye A., Mach J. P., Pèlegrin A., Hardman N., Delaloye B., Buchegger F. (1993) Br. J. Cancer 68, 684–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sanz L., Kristensen P., Blanco B., Facteau S., Russell S. J., Winter G., Alvarez-Vallina L. (2002) Gene Ther. 9, 1049–1053 [DOI] [PubMed] [Google Scholar]
- 5. Steffen A. C., Orlova A., Wikman M., Nilsson F. Y., Ståhl S., Adams G. P., Tolmachev V., Carlsson J. (2006) Eur. J. Nucl. Med. Mol. Imaging 33, 631–638 [DOI] [PubMed] [Google Scholar]
- 6. Cuesta A. M., Sánchez-Martín D., Sanz L., Bonet J., Compte M., Kremer L., Blanco F. J., Oliva B., Alvarez-Vallina L. (2009) PLoS ONE 4, e5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buchsbaum G. M., Moll C., Duecy E. E. (2004) Int. Urogynecol. J. Pelvic. Floor Dysfunct. 15, 432–433 [DOI] [PubMed] [Google Scholar]
- 8. Laakkonen P., Akerman M. E., Biliran H., Yang M., Ferrer F., Karpanen T., Hoffman R. M., Ruoslahti E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9381–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laakkonen P., Porkka K., Hoffman J. A., Ruoslahti E. (2002) Nat. Med. 8, 751–755 [DOI] [PubMed] [Google Scholar]
- 10. Fogal V., Zhang L., Krajewski S., Ruoslahti E. (2008) Cancer Res. 68, 7210–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krainer A. R., Mayeda A., Kozak D., Binns G. (1991) Cell 66, 383–394 [DOI] [PubMed] [Google Scholar]
- 12. Gupta S., Batchu R. B., Datta K. (1991) Eur. J. Cell Biol. 56, 58–67 [PubMed] [Google Scholar]
- 13. Ghebrehiwet B., Lim B. L., Peerschke E. I., Willis A. C., Reid K. B. (1994) J. Exp. Med. 179, 1809–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu L., Loewenstein P. M., Zhang Z., Green M. (1995) J. Virol. 69, 3017–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eggleton P., Tenner A. J., Reid K. B. (2000) Clin. Exp. Immunol. 120, 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dedio J., Müller-Esterl W. (1996) FEBS Lett. 399, 255–258 [DOI] [PubMed] [Google Scholar]
- 17. Braun L., Ghebrehiwet B., Cossart P. (2000) EMBO J. 19, 1458–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kittlesen D. J., Chianese-Bullock K. A., Yao Z. Q., Braciale T. J., Hahn Y. S. (2000) J. Clin. Invest. 106, 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mahdi F., Shariat-Madar Z., Todd R. F., 3rd, Figueroa C. D., Schmaier A. H. (2001) Blood 97, 2342–2350 [DOI] [PubMed] [Google Scholar]
- 20. Mahdi F., Madar Z. S., Figueroa C. D., Schmaier A. H. (2002) Blood 99, 3585–3596 [DOI] [PubMed] [Google Scholar]
- 21. Peerschke E. I., Reid K. B., Ghebrehiwet B. (1994) J. Immunol. 152, 5896–5901 [PubMed] [Google Scholar]
- 22. Eggleton P., Ghebrehiwet B., Sastry K. N., Coburn J. P., Zaner K. S., Reid K. B., Tauber A. I. (1995) J. Clin. Invest. 95, 1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuna P., Iyer M., Peerschke E. I., Kaplan A. P., Reid K. B., Ghebrehiwet B. (1996) Clin. Immunol. Immunopathol. 81, 48–54 [DOI] [PubMed] [Google Scholar]
- 24. Peerschke E. I., Smyth S. S., Teng E. I., Dalzell M., Ghebrehiwet B. (1996) J. Immunol. 157, 4154–4158 [PubMed] [Google Scholar]
- 25. Steinberger P., Szekeres A., Wille S., Stöckl J., Selenko N., Prager E., Staffler G., Madic O., Stockinger H., Knapp W. (2002) J. Leukocyte Biol. 71, 133–140 [PubMed] [Google Scholar]
- 26. Vegh Z., Goyarts E. C., Rozengarten K., Mazumder A., Ghebrehiwet B. (2003) International Immunopharmacol. 3, 345–357 [DOI] [PubMed] [Google Scholar]
- 27. Oiki S., Okada Y. (1988) J. Immunol. 141, 3177–3185 [PubMed] [Google Scholar]
- 28. Hawkins R. E., Russell S. J., Winter G. (1992) J. Mol. Biol. 226, 889–896 [DOI] [PubMed] [Google Scholar]
- 29. Griffiths A. D., Williams S. C., Hartley O., Tomlinson I. M., Waterhouse P., Crosby W. L., Kontermann R. E., Jones P. T., Low N. M., Allison T. J. (1994) EMBO J. 13, 3245–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nissim A., Hoogenboom H. R., Tomlinson I. M., Flynn G., Midgley C., Lane D., Winter G. (1994) EMBO J. 13, 692–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gibson T. (1984) Studies in the Epstein-Barr Virus Genome, Ph.D. thesis, MRC Laboratory of Molecular Biology, University of Cambridge, Cambridge, UK [Google Scholar]
- 32. Kristensen P., Winter G. (1998) Folding Des. 3, 321–328 [DOI] [PubMed] [Google Scholar]
- 33. Sanz L., Kristensen P., Russell S. J., Ramirez García J. R., Alvarez-Vallina L. (2001) Cancer Immunol. Immunother. 50, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Retter I., Althaus H. H., Münch R., Müller W. (2005) Nucleic Acids Res. 33, D671–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carter P., Bedouelle H., Winter G. (1985) Nucleic Acids Res. 13, 4431–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Birchler M., Neri G., Tarli L., Halin C., Viti F., Neri D. (1999) J. Immunol. Methods 231, 239–248 [DOI] [PubMed] [Google Scholar]
- 37. Lynch N. J., Reid K. B., van den Berg R. H., Daha M. R., Leigh L. A., Ghebrehiwet B., Lim W. B., Schwaeble W. J. (1997) FEBS Lett. 418, 111–114 [DOI] [PubMed] [Google Scholar]
- 38. Harries M., Smith I. (2002) Endocr. Relat. Cancer 9, 75–85 [DOI] [PubMed] [Google Scholar]
- 39. Begent R. H., Verhaar M. J., Chester K. A., Casey J. L., Green A. J., Napier M. P., Hope-Stone L. D., Cushen N., Keep P. A., Johnson C. J., Hawkins R. E., Hilson A. J., Robson L. (1996) Nat. Med. 2, 979–984 [DOI] [PubMed] [Google Scholar]
- 40. McDevitt M. R., Barendswaard E., Ma D., Lai L., Curcio M. J., Sgouros G., Ballangrud A. M., Yang W. H., Finn R. D., Pellegrini V., Geerlings M. W., Jr., Lee M., Brechbiel M. W., Bander N. H., Cordon-Cardo C., Scheinberg D. A. (2000) Cancer Res. 60, 6095–6100 [PubMed] [Google Scholar]
- 41. Neri D., Carnemolla B., Nissim A., Leprini A., Querzè G., Balza E., Pini A., Tarli L., Halin C., Neri P., Zardi L., Winter G. (1997) Nat. Biotechnol. 15, 1271–1275 [DOI] [PubMed] [Google Scholar]
- 42. Silacci M., Brack S. S., Späth N., Buck A., Hillinger S., Arni S., Weder W., Zardi L., Neri D. (2006) Protein Eng. Des. Sel. 19, 471–478 [DOI] [PubMed] [Google Scholar]
- 43. Birchler M. T., Thuerl C., Schmid D., Neri D., Waibel R., Schubiger A., Stoeckli S. J., Schmid S., Goerres G. W. (2007) Otolaryngol. Head Neck. Surg. 136, 543–548 [DOI] [PubMed] [Google Scholar]
- 44. van den Berg R. H., Prins F., Faber-Krol M. C., Lynch N. J., Schwaeble W., van Es L. A., Daha M. R. (1997) J. Immunol. 158, 3909–3916 [PubMed] [Google Scholar]
- 45. Karmali P. P., Kotamraju V. R., Kastantin M., Black M., Missirlis D., Tirrell M., Ruoslahti E. (2009) Nanomedicine 5, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruoslahti E., Bhatia S. N., Sailor M. J. (2010) J. Cell Biol. 188, 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Teesalu T., Sugahara K. N., Kotamraju V. R., Ruoslahti E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16157–16162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cuesta A. M., Sainz-Pastor N., Bonet J., Oliva B., Alvarez-Vallina L. (2010) Trends Biotechnol. 28, 355–362 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.