Abstract
Yeast cells begin to bud and enter the S phase when growth conditions are favorable during the G1 phase. When subjected to some oxidative stresses, cells delay entry at G1, allowing repair of cellular damage. Hence, oxidative stress sensing is coordinated with the regulation of cell cycle. We identified a novel function of the cell cycle regulator of Saccharomyces cerevisiae, Swi6p, as a redox sensor through its cysteine residue at position 404. When alanine was substituted at this position, the resultant mutant, C404A, was sensitive to several reactive oxygen species and oxidants including linoleic acid hydroperoxide, the superoxide anion, and diamide. This mutant lost the ability to arrest in G1 phase upon treatment with lipid hydroperoxide. The Cys-404 residue of Swi6p in wild-type cells was oxidized to a sulfenic acid when cells were subjected to linoleic acid hydroperoxide. Mutation of Cys-404 to Ala abolished the down-regulation of expression of the G1 cyclin genes CLN1, CLN2, PCL1, and PCL2 that occurred when cells of the wild type were exposed to the lipid hydroperoxide. In conclusion, oxidative stress signaling for cell cycle regulation occurs through oxidation of the G1/S-speicific transcription factor Swi6p and consequently leads to suppression of the expression of G1 cyclins and a delay in cells entering the cell cycle.
Keywords: Cell Cycle, Cellular Regulation, Oxidative Stress, Thiol, Yeast, Cell Cycle Delay, Cysteine Oxidation, Sulfenic Acid
Introduction
The cell cycle consists of a series of coordinated events that ensure duplication of genetic material, chromosome segregation, cell growth, and cytokinesis, producing two daughter cells. In eukaryotes, cell cycle events are governed by phase-specific cyclins that complex and activate cyclin-dependent kinase (CDK)2 for activation of phase-specific events. Cell division is regulated in part by the periodic expression of genes specific to each of the four phases (G1, S, G2, and M) (1, 2). In Saccharomyces cerevisiae, the single CDK, Cdc28, can complex with nine cyclins that are transcribed distinctly in the four stages of the cell cycle. Cells enter the cell cycle to undergo division when both intrinsic and extrinsic requirements are met. In S. cerevisiae, when cells attain a critical size in the presence of sufficient nutrients in the late G1 phase, they reach an interval called “Start,” at which the bud begins to emerge, DNA replication is initiated, and cells duplicate their spindle pole body preparing for mitosis and cytokinesis (3).
Cell cycle initiation and progression are tightly regulated to ensure that cell division does not take place under unfavorable conditions (4, 5). In proliferating cells, oxidative stress can arise from reactive oxygen species that are generated from incomplete reduction of oxygen from the electron transport chain. Environmental factors, including heat and other stresses, ionizing radiation, metal ions, herbicides such as paraquat, and the vitamin K precursor menadione, can also induce reactive oxygen species production in cells (6). Reactive oxygen species are highly toxic because of their ability to directly damage nucleic acids, proteins, and lipids (6). In particular, oxidation of membrane lipids generates lipid hydroperoxides, which may be involved in the pathogenesis of human diseases such as atherosclerosis and neurodegenerative disorders (7, 8). Upon lipid peroxidation and oxidative stress, cells respond by delaying exit from G1 so that cellular damage can be removed or repaired to avoid irreparable damage being passed on to daughter cells (9, 10). Some of these G1 arrest responses are conserved from yeast to human (11, 12).
At cell cycle checkpoints, cells monitor the internal and environmental redox conditions to initiate the appropriate responses to oxidative stress. In mammalian cells, p53 participates in major checkpoints for DNA damage (10). In S. cerevisiae, DNA damage caused by hydrogen peroxide activates the Rad53p checkpoint via the RAD9 signaling pathway in response to DNA damage (9). However, G1 delay in response to lipid hydroperoxides operates independently of the RAD9 pathway (13, 14). From a genome-wide screen of S. cerevisiae deletion mutants to identify genes needed to maintain resistance to a range of different oxidative stresses, 259 deletants were found to be sensitive to the lipid hydroperoxide, linoleic acid hydroperoxide (LoaOOH) (15). Because LoaOOH induces G1 arrest in yeast, the LoaOOH-sensitive mutants were subjected to a second screen to identify genetic factors that may be involved in signaling and regulation of G1 arrest in response to LoaOOH (16). Deletion of SWI6, which encodes the G1/S phase-specific transcription factor, resulted in the loss of the G1 delay phenotype in response to LoaOOH (16).
Swi6p is a cell cycle transcription factor that activates the periodic expression of G1 cyclins required for the G1 to S phase transition. Swi6p forms two heterodimeric complexes, SBF and MBF, with Swi4p and Mbp1p, respectively (17, 18) and relies on its partner protein for DNA interaction (19, 20). Swi6p has also been shown to regulate cell cycle arrest upon DNA damage through phosphorylation by Rad53p (21). However, the mechanism of its involvement in regulating cell cycle arrest in response to oxidative stress is largely unknown and is not dependent on the DNA repair pathway (9). Here, we report how Swi6p can regulate cell cycle delay by functioning as a sensor for oxidative stress through an intrinsically reactive cysteine residue.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
Yeast strains and plasmids are listed in supplemental Table S1. All of the strains are isogenic with the haploid BY4741 background and were cultured in synthetic complete medium (SC) with the omission of leucine where necessary for plasmid maintenance. SWI6 coding sequence and its flanking sequences (1 kb upstream and 0.5 kb downstream) were cloned into the centromeric plasmid pRS415, and the resultant plasmid pSWI6 was transformed into the swi6Δ mutant (16). This complemented wild-type strain was designated SWI6 to differentiate it from the wild-type strain BY4741 carrying the empty vector pRS415. The Cys-404 in Swi6p was mutated to alanine using pSWI6 (16) as a template by site-directed, ligase-independent mutagenesis (22, 23). Primers used for site-directed mutagenesis are listed in supplemental Table S2. For immunoprecipitation of Swi6p or its C404Ap variant, the N-terminal His9 tag was added to the SWI6 or C404A coding sequence by PCR and cloned into pAG416GPD-ccdB from the S. cerevisiae Gateway Cloning vector suite (24, 25) for expression under the glyceraldehyde-3-phosphate dehydrogenase (TDK3) promoter. The resulting plasmids were pGPD-9HisSWI6 and pGPD-9HisC404A (supplemental Table S1).
Analysis of Swi6p Structure
The Protein Data Bank (26) was mined to identify proteins with potentially reactive surface-exposed cysteine residues that form covalent dimers during crystallization.3 The Protein Quaternary State Web server (27) was used to determine whether the reactive Sw6p Cys residue might be involved in physiological dimer formation or be susceptible to other forms of modification such as glutathionylation or nitrosylation. Protein Quaternary State determines whether dimer formation is likely to be physiological or an artifact of crystallization by assessing the amount of solvent-accessible surface buried during covalent dimer formation. For Protein Data Bank entries where the quaternary state is known, the solvent-accessible surface buried per chain ranges from ∼370–4750 Å2 for homodimers and from ∼640–3230 Å2 for heterodimers (28). A threshold of 400 Å2 of buried solvent-accessible surface/chain was used to predict physiological oligomers. Oligomers with less than this value are unlikely to be physiologically relevant. Protein Quaternary State also identifies residues buried at the interface.
Sensitivity to Oxidants
Linoleic acid hydroperoxide was synthesized as described (29) and stored at −20 °C for up to 4 months. LoaOOH (in HPLC grade methanol) was added to molten agar medium to the required concentrations, and the plates were poured. Other oxidant/reductant stocks (1 m menadione, 0.8 m H2O2, 0.5 m diamide, and 1 m dithiothreitol) were freshly prepared in water prior to addition to molten agar. The plates were left to dry for 1 h. Overnight cultures were serially diluted using fresh medium, and 5 μl of each dilution was spotted onto the LoaOOH-containing plates. The plates were dried and incubated at 30 °C in the dark for 3 days.
Cell Cycle Analysis
The strains were cultured in SC-Leu medium at 30 °C to A600 0.2. α-Mating factor (Zymo Research, Orange, CA) was added to a final concentration of 3 μg/ml, and cultures were synchronized at 30 °C for 2 h. The cells were washed once, resuspended in PBS, and split into equal portions. LoaOOH (15–20 mm in methanol) was added to a final concentration of 30 μm in one culture, and an equal volume of methanol was added to the other culture as control. The cultures were incubated with shaking at 30 °C for 45 min and were washed once with PBS. The cells were released into fresh SC-Leu medium, and 1 ml of culture was sampled at the indicated time intervals. The cell samples were centrifuged, fixed with 1 ml of ice-cold 70% (v/v) ethanol, and resuspended in 1 ml of water. The cells were subsequently sonicated in a M250 Branson digital sonifier (Branson, Danbury, CT) for 20 s at 30% amplitude to disperse clumps, washed once with 1 ml of 50 mm Tris-HCl, pH 7.5, and resuspended in 20 μl of the same buffer. The budding index was determined by fluorescence microscopy as described (11). To examine the cellular DNA content by flow cytometry, the samples were fixed with 70% (v/v) ice-cold ethanol and resuspended in 1 ml of water. The samples were subjected to sonication to disperse clumps, centrifuged, and resuspended in 100 μl of Tris-HCl, pH 7.5, containing RNaseA (100 μg/ml) for digestion for 2 h at 37 °C. The samples were stained overnight in 10 μg/ml propidium iodide as described previously (16). DNA content of samples was analyzed as described by Haase and Lew (30) on a Cell Lab QuantaTM SC flow cytometer (Beckman Coulter Australia, Gladesville, Australia).
Immunoprecipitation and Detection of Sulfenic Acid in Proteins
To detect the presence of cysteines converted to sulfenic acid residues, cells expressing Swi6p from the glyceraldehyde-3-phosphate dehydrogenase (TDK3) promoter were grown in SC-Ura to A600 of 0.4 and split into equal aliquots for treatment with and without LoaOOH (30 μm) for 5 min. Cells were collected by centrifugation at 4 °C and were resuspended in PBS. Each culture was divided into two equal aliquots, and dimedone (1 m stock in dimethyl sulfoxide) was added to a final concentration of 50 mm to one aliquot. The cells treated with dimedone were incubated with shaking at 30 °C for 15 min and collected by centrifugation at 4 °C. The cell pellets were stored at −80 °C until lysis. For detection of sulfenic acid formation in Swi6p during the cell cycle, the cells were grown in 500 ml of SC-Ura to A600 of 0.2 and synchronized as described above. The cells were resuspended in 500 ml of PBS buffer, and 50 ml was collected as zero time. LoaOOH was then added to the culture to final concentration of 30 μm, and 50 ml of cells was collected at intervals. The collected cells were immediately subjected to dimedone treatment as described above. The remaining 250-ml culture from LoaOOH treatment was centrifuged, washed once in PBS, and released into 250 ml of fresh SC-Ura medium. The cells (50 ml) were collected at indicated intervals and subjected to dimedone treatment as described above.
The cells were lysed using a bead beater with acid-washed glass beads in lysis buffer containing 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 1% (v/v) Triton X-100, 5 mm PMSF, fungal protease inhibitor mixture (Sigma-Aldrich), phosphatase inhibitor mixture (Roche Applied Science), and 70 mm iodoacetamide. Cell debris was removed by centrifugation, and supernatant-containing soluble protein was transferred to fresh tubes. Immunoprecipitation was performed using 500 μg of soluble protein and diluted 5-fold (final volume of 1 ml) in dilution buffer containing 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 5 mm PMSF, fungal protease inhibitor mixture, phosphatase inhibitor mixture, and 70 mm iodoacetamide. The proteins were precleared by adding 10 μl of Dynabeads (Invitrogen), and the mixture was incubated with rotation at 4 °C for 1 h prior to separation of Dynabeads from the protein solution using a magnet. The protein solution was transferred to new tubes, and 1 μg of polyclonal anti-Swi6 yN-19 antibody was added (Santa Cruz Biotechnology, Santa Cruz, CA). The mixture was incubated with rotation at 4 °C for 1 h before the addition of fresh Dynabeads to the antibody-protein mixture and incubation with rotation at 4 °C overnight. The protein was separated from the Dynabeads using a magnet, and the beads were washed three times in 1 ml of wash buffer containing 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, and 0.1% (v/v) Triton X-100. Bound proteins were eluted using 2× SDS sample loading buffer containing 0.1 m DTT at 75 °C for 10 min. The protein eluates were subjected to SDS-PAGE, and the proteins were subsequently transferred on to a 0.45-μm nitrocellulose membrane. Immunoblotting was carried out using anti-cysteine sulfenic acid antibody (Millipore, Billerica, MA) to detect sulfenic acid, and Swi6p was detected by anti-Swi6p antibody.
RNA Extraction and Microarray Analysis
For microarray analysis, asynchronous populations were used. The cultures (200 ml) were grown to A600 of 0.4 at 30 °C and split into equal aliquots for treatment with and without LoaOOH (30 μm) for 45 min. Each culture was collected in tubes containing 20 g of −80 °C crushed ice, and the cells were pelleted by centrifugation (4000 × g, 3 min) and washed once in PBS before storage at −80 °C. Frozen cell pellets were resuspended in 1 ml of TRIzol® reagent (Invitrogen) and lysed in a bead beater with acid-washed glass beads. RNA was extracted as described by the RNA labeling, and hybridization according to the Affymetrix® GeneChip® yeast genome 2.0 array protocol (Affymetrix, Santa Clara, CA) was carried out at the Ramaciotti Centre for Gene Function Analysis (University of New South Wales, Sydney, Australia). Data analysis was performed with Partek Genomics suite software (version 6.4). Probe set data were normalized using the robust multi-array average method and were analyzed as described previously (32). For identification of differentially expressed genes, two-way analysis of variance and a false discovery rate with a 0.05 threshold was used. The microarray data set is deposited at Gene Expression Omnibus under accession number GSE18334. Hierarchical clustering for potentially co-regulated genes was performed using the open source TM4 microarray software suite (33, 34). The web-based program FunSpec was used for analysis of functional enrichment of genes in the gene clusters (35), and the YEASTRACT database was used for search of genes and transcription factors association (36, 37). Upstream sequences (−1000 to −1 bp) of genes were retrieved and subjected to motif search using the oligo-analysis tool from regulatory sequence analysis tools (38).
Real Time PCR
For analysis of transcripts in asynchronous populations, cultures were grown in 60 ml of SC-Leu to A600 of 0.4 at 30 °C and treated with 30 μm LoaOOH as described above. For analysis of changes in G1 cyclin transcripts in the cell cycle, the cultures were grown in 400 ml of SC-Leu medium to A600 of 0.2, synchronized, and split into equal portions with one portion treated with LoaOOH treatment as described above. The cells were then released in fresh SC-Leu medium (200 ml), and 20-ml aliquots were collected in 5 g of −80 °C crushed ice at indicated intervals. The cells were collected by centrifugation (4000 × g, 3 min) and washed once in PBS before storage at −80 °C. RNA was extracted as described above. For RT-PCR, 1 μg of RNA was taken for cDNA synthesis using the SuperScriptTM III first strand cDNA synthesis kit (Invitrogen). Gene-specific primers used in RT-PCR assays are listed in supplemental Table S3. PCR was carried out using the Platinum® SYBR® Green RT-PCR SuperMix-UDG kit (Invitrogen) on a Rotor-Gene RG-3000A (Qiagen) with the following parameters: 50 °C for 2 min, 95 °C for 2 min, and 40 cycles of 95 °C for 15 s and 60 °C for 30 s. The threshold cycle (Ct) value for each gene at the log phase was determined, and normalization of specific genes against reference genes (ΔCt values) was performed according to Pfaffl et al. (39).
RESULTS
Swi6p Has a Potentially Reactive Cys-404 Residue
The involvement of Swi6p in the cell cycle delay in response to oxidative stress was initially identified from a screen of mutants sensitive to LoaOOH (15). It was therefore of interest to identify the mechanism whereby the Swi6p transcription factor is involved in sensing oxidative damage. Swi6p could respond to activation of the cell integrity signaling pathway via the protein kinase Slt2p, which phosphorylates Swi6p directly (40). Although deletion of either SLT2 or SWI6 led to sensitivity to LoaOOH, only the swi6Δ mutant lacked the ability to delay in G1, indicating that transduction of oxidative stress signal is likely to occur via Swi6p and that the MAPK cell integrity signaling cascade (16, 41) is unlikely to be involved. Deletion of either SWI4 or MBP1 did not result in a loss of cell cycle delay (16), indicating the critical role of Swi6p in oxidative stress signaling. Preliminary analysis of the protein was therefore performed to examine whether stress signal transduction could result from its structure. Swi6p contains an ankyrin repeat motif and on closer examination of its crystal structure (42), a potentially reactive cysteine residue at position 404 was identified (supplemental Fig. S1A).
The ankyrin motif in Swi6p is also found in its partner proteins Swi4p and Mbp1p (42); however, Cys-404 is a nonconserved residue found only in Swi6p (supplemental Fig. S1B). Therefore, Swi6p may function as the cellular oxidative stress sensor that detects oxidative stress signals via its potentially reactive Cys-404 residue for regulation of cell cycle delay. Therefore, Cys-404 was mutated to an alanine to assess its role in the cell cycle response to redox stress.
The C404A Mutation Affects Cellular Responses to Some Oxidants and Dithiothreitol
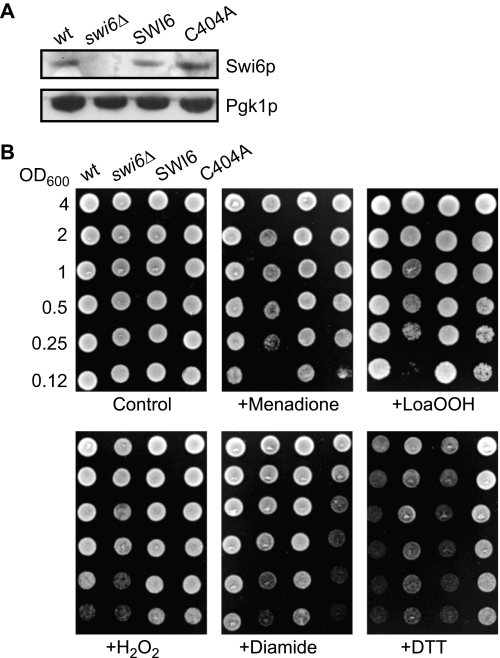
To test whether mutation of the Cys-404 residue led to any phenotype under redox stress, the SWI6 gene or its C404A mutant copy was expressed from its own promoter from a centromeric plasmid in the swi6Δ mutant; the resulting strains were denoted SWI6 and C404A, respectively. The control strain was the wild type transformed with the vector lacking an insert. Although the swi6Δ deletant grew, it had enlarged cells with irregular shape (supplemental Fig. S2) like some other cell cycle mutants (43). When pSWI6 or pC404A was transformed into the swi6Δ mutant, the wild-type morphology was restored completely with the wild-type gene and partially with the C404A variant. Moreover, the SWI6 and C404A strains expressed levels of Swi6p that were comparable with that of the wild-type strain (Fig. 1A).
FIGURE 1.
Sensitivity of the wild-type and swi6Δ and C404A mutant strains to LoaOOH. A, Swi6p and Swi6 C404A mutant protein expression. The wild-type SWI6 and mutant C404A sequences under the control of SWI6 promoter and downstream regulatory elements were expressed in the swi6Δ strain. Cell extracts were prepared, and the Swi6p and Pgk1p (as loading control) levels were determined by Western blotting. B, spot test of sensitivity to LoaOOH. Cultures of BY4741, swi6Δ, and swi6Δ carrying SWI6 or its Ala mutant copy on a centromeric plasmid were grown to stationary phase, and 2-fold serial dilutions from cultures with the same cell density were spotted on media containing oxidants (2 mm menadione, 0.1 mm LoaOOH, 1.2 mm H2O2, 1.5 mm diamide) or reducing agent (2 mm dithiothreitol).
The wild-type, swi6Δ, SWI6, and C404A strains were grown to stationary phase, and appropriate dilutions were tested for sensitivity to LoaOOH, hydrogen peroxide, menadione (which leads to generation of superoxide anion), the thiol oxidant diamide, and dithiothreitol. The swi6Δ mutant was sensitive to all of the compounds except dithiothreitol (Fig. 1B), but its resistance was restored on introducing the SWI6 gene. The C404A strain grew as well as the wild type in the absence of redox stress but was more sensitive to LoaOOH, diamide, and menadione, indicating that the Cys-404 residue is not important for growth under normal aerobic conditions but is functionally important for protection against certain reactive oxygen species. The C404A strain was very resistant to dithiothreitol compared with the other strains and was not sensitive to H2O2. Therefore, the C404A mutant Swi6p protein is active in the absence of redox stress but leads to an altered cellular response to particular reactive oxygen species, including LoaOOH and the superoxide anion, both of which are known to cause cell cycle delay at G1 (9, 11). Because the effect of diamide on cell cycle progression is not known, we studied the response of wild-type (SWI6), swi6Δ, and C404A strains to diamide by flow cytometry. After diamide treatment of cells synchronized with α-factor, all three strains entered S phase as shown by an increase in the 2N DNA peak, and there was no indication of cell cycle delay at G1 (supplemental Fig. S3). In all of the strains, progression of the S phase was found to be slower than in the untreated control, indicating that diamide may induce a delay in the S phase rather than in the G1 phase. Despite the more pronounced sensitivity of C404A strain to diamide, LoaOOH was used in subsequent studies for oxidant treatment because lipid hydroperoxides cause G1 arrest and are products generated from lipid peroxidation that normally occurs in cells, whereas diamide is a synthetic thiol oxidant not usually encountered by cells.
The C404A Mutant Did Not Delay Cell Division on LoaOOH Treatment
To test whether Cys-404 is involved in cell cycle regulation, cells of SWI6, swi6Δ, and C404A were synchronized by arresting at the G1 phase with α-mating factor. After washing with buffer, the cells were treated with LoaOOH or methanol as control before a further wash and resuspension in fresh medium. Because bud emergence and DNA replication take place when cells enter the S phase, the timing of these events post release from pheromone (control) or LoaOOH treatment was compared in the strains to determine whether there was delay in passage to the S phase in response to LoaOOH.
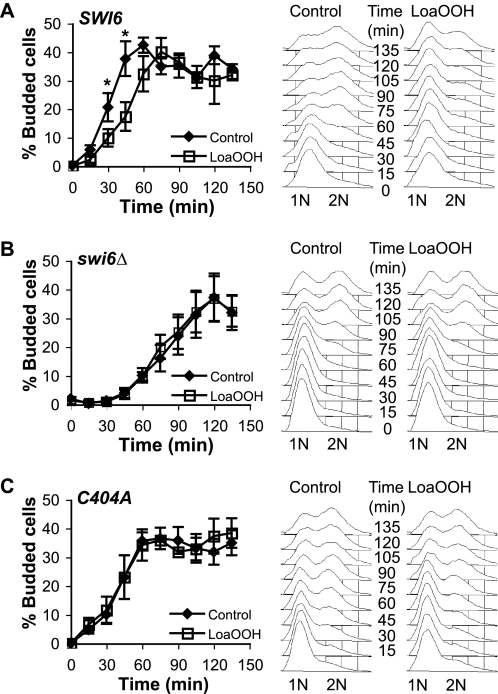
In the SWI6 strain expressing the wild-type gene, bud emergence commenced 10 min post release in the absence of LoaOOH but was delayed by a further 15–20 min after treatment with LoaOOH, as found previously (11, 16) (Fig. 2A). DNA replication was also delayed in the SWI6 strain in response to LoaOOH; compared with the control in which 2N cells appeared 45 min post release from pheromone, treatment with LoaOOH delayed the appearance of 2N cells until 75 min (Fig. 2A). Deletion of SWI6 led to an extended G1 phase, and bud emergence was only observed 45 min after release from pheromone (Fig. 2B). When treated with LoaOOH, swi6Δ cells entered the S phase without further delay at G1 at the same time as the control (45 min after release). This was confirmed by measuring cellular DNA content: 2N cells appeared after an extended 90-min delay regardless of whether the cells were treated with LoaOOH or not (Fig. 2B).
FIGURE 2.
Cell cycle delay phenotypes of the wild-type and swi6Δ and C404A mutant strains in response to LoaOOH. Cultures were synchronized with α-factor and were treated with LoaOOH for 45 min. Aliquots were sampled every 15 min post treatment, and the cells were fixed for bud counting or for FACS analysis of DNA content. For bud counts, 400–600 cells/sample were counted under a fluorescent microscope. A, SWI6 wild type; B, swi6Δ mutant; C, swi6Δ mutant with C404A construct. The data plotted are the averages of three independent experiments, and the error bars represent ± S.E. *, p value < 0.05, determined using the paired, two-tailed Student's t test. For FACS analysis, 25,000 cells were analyzed per sample. 1N represents a single copy of DNA per cell, and 2N represents two copies of DNA. A representative example of three independent experiments is shown.
The C404A strain also entered the S phase at the same time as the wild-type SWI6 strain, but its cell cycle response to LoaOOH treatment differed markedly (Fig. 2C). On LoaOOH treatment, bud emergence in SWI6 cells was delayed, but C404A cells began budding 10 min after LoaOOH treatment and progressed into the S phase at the same time as the control with no LoaOOH treatment. There was a short delay in DNA replication upon LoaOOH treatment but not to the same extent as seen in SWI6. This indicated that regulation of the S phase events of bud emergence and initiation of DNA replication may have been partially uncoupled because of the C404A mutation.
These results show that deletion of SWI6 led to a delay in cells entering the S phase, but there was no further delay following LoaOOH treatment. The C404A mutation did not affect the timing of entry into S phase (determined by bud emergence) under normal conditions but abolishes the ability of the strain to respond to LoaOOH by delaying entry into the S phase. The Cys-404 residue in Swi6p is therefore critical in the cell response to this oxidant, and it might act as a sensor of oxidant damage through its modification by oxidants such as LoaOOH.
Cys-404 of Swi6p Is Oxidized to a Sulfenic Acid upon LoaOOH Treatment
Because the cell cycle delay required the presence of the thiol group at position 404 in Swi6p, we sought to identify how Swi6p is modified in response to LoaOOH. Although Swi6p crystallizes as a dimer via disulfides at Cys-404 residues, only 425 Å is buried per chain during dimer formation, which is above the theoretical cut-off for a likely physiological dimer. Moreover, the energy gain upon complex formation is only −4.5 kcal/mol, and no residues buried by dimer formation were detected during the analysis, whereas a true oligomer would be expected to have at least some residues of this type. Thus, it is unlikely that Swi6p forms a homodimer under physiological conditions, and the reactive cysteine may be oxidatively modified in some other way. Lack of dimer formation was shown by subjecting cell extracts of S. cerevisiae exposed to LoaOOH to nonreducing SDS-PAGE and immunoblotting using anti-Swi6p antibody. No dimer was detected on LoaOOH treatment (supplemental Fig. S4A). Because cysteine residues can be glutathionylated to protect against further cysteine oxidation (44), Swi6p was precipitated from extracts of LoaOOH-treated cells using anti-Swi6p antibody and tested for glutathionylation using the anti-GSH antibody. No evidence for glutathionylation of Swi6p was found (supplemental Fig. S4B).
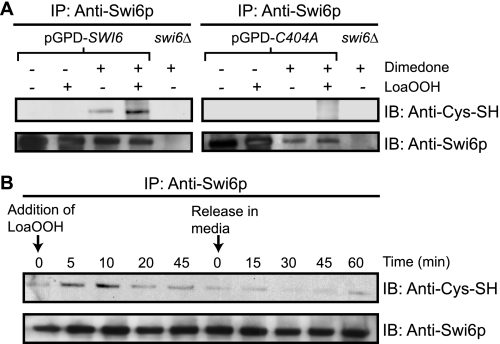
Protein thiols can also be serially oxidized to form sulfenic, sulfinic, and sulfonic acid residues, and oxidation to sulfenic acid in particular can serve as a signal for oxidative stress and damage (45). Hence, the cells were treated with dimedone, which reacts specifically with sulfenic acids, and the extracts were examined using an antibody that recognizes the adduct of dimedone (46). Because the level of oxidized protein was in low abundance compared with the reduced form, a construct was used in which Swi6p was expressed at higher levels from the glyceraldehyde-3-phosphate dehydrogenase (TDK3) promoter. The cells were subjected to LoaOOH treatment followed by the addition of dimedone, and Swi6p was immunoprecipitated from cell extracts and immunoblotted to detect the presence of sulfenic acid residues. Under normal growth conditions, a low level of sulfenic acid could be detected in Swi6p in an asynchronous cell population, indicating that the transcription factor had a basal level of oxidation to the sulfenic acid. Within 5 min of LoaOOH treatment, substantially more Swi6p became oxidized to sulfenic acid. No sulfenic acid was detected in C404A cells grown normally or after exposure to LoaOOH, showing that it was the Cys-404 in Swi6p that was oxidized (Fig. 3A).
FIGURE 3.
Oxidation of the Cys-404 residue to sulfenic acid in LoaOOH-treated cells. A, detection of sulfenic acid in Swi6p in LoaOOH-treated cells. The cells were exposed to LoaOOH (30 μm, 5 min) and were subsequently treated with dimedone as under “Experimental Procedures.” The cells were lysed, and Swi6p or C404Ap was immunoprecipitated (IP) from cell extracts and subjected to immunoblotting (IB) for the presence of sulfenic acid. B, time course of sulfenic acid formation in Swi6p in a synchronous population on LoaOOH treatment and post release from treatment. The cells were arrested at G1 using α-factor, washed, and resuspended in PBS followed by treatment with 30 μm LoaOOH. The samples were taken at the indicated time intervals and treated with dimedone. Additionally LoaOOH-treated cells were washed and released in fresh medium to initiate cell division, and the samples were taken again at intervals as indicated. The cell extracts were prepared, and Swi6p was immunoprecipitated for detection of sulfenic acid as described above.
The above data were obtained with asynchronous cultures. To determine whether formation of the sulfenic acid in Swi6p in response to LoaOOH was relevant to cell cycle delay, α-factor-synchronized cells were treated with LoaOOH, and culture samples were collected during the 45-min treatment and for 60 min post treatment. Swi6p was then precipitated and tested for sulfenic acid formation (Fig. 3B). A low basal level of sulfenic acid in Swi6p was present at zero time, but this increased markedly after 5 min of treatment with LoaOOH. Sulfenic acid formation peaked at 10 min and then decreased gradually after 20 min of treatment. This decrease could be due to further oxidation to the sulfinic or sulfonic acid or to reduction of the sulfenic acid residue by a redox repair system induced by the oxidant (31). After transfer to fresh medium without LoaOOH, the level of sulfenic acid decreased further, and by 30 min, which corresponded to when buds began to emerge, sulfenic acid was barely detectable in Swi6p. At 60 min post treatment when budding was maximal, sulfenic acid was at a similar basal level as the zero time control. The timing of changes in sulfenic acid formation in Swi6p was therefore consistent with the view that transition to the S phase is dependent on the oxidation status of the Cys-404 residue of Swi6p.
The C404A Mutation Affects Expression of Many Genes, Including Those of G1 Cyclins
Entry into the S phase requires transcription of cell cycle genes by the Swi6p/Mbp1p (MBF) and Swi6p/Swi4p (SBF) complexes (18). The transcriptional response of the SWI6, C404A, and swi6Δ strains to LoaOOH was determined by microarray analysis to investigate the effects of Cys-404 oxidation on Swi6p activity. SWI6, C404A, and swi6Δ strains were grown to mid-exponential phase and exposed in parallel to control and LoaOOH-treated conditions. RNA samples were prepared from biological triplicates and subjected to microarray analysis.
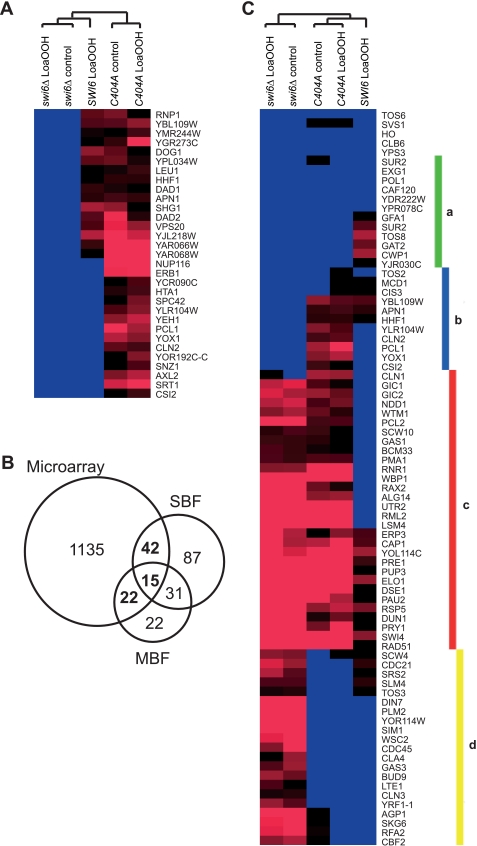
Transcript levels for genes under test conditions were expressed relative to those of the wild type (SWI6) in the absence of LoaOOH. From two-way analysis of variance, 1214 genes had significantly altered expression (p < 0.05) across the six different conditions, and the main sources of variation came from the test conditions, genotype (39.95%) and LoaOOH treatment (5.81%). Differentially expressed genes were clustered hierarchically, resulting in 14 clusters (supplemental Fig. S5), and the functional groupings that were over-represented in each gene cluster are given in supplemental Table S4. Principal component analysis indicated that the transcriptomic profile of the C404A mutant was closer to that of the swi6Δ deletant than to the wild type, with the C404A mutant and swi6Δ deletant showing similarly altered expression of 931 genes.
Four of the 14 gene clusters were enriched in genes related to cell division (clusters 4, 9, 10, and 11; supplemental Table S4). The C404A mutant had a similar expression pattern to the wild type in response to LoaOOH treatment in three of these clusters (clusters 9, 10, and 11), but cluster 4 comprised genes whose expression was higher only in the C404A mutant and not the wild type or swi6Δ deletant (Fig. 4A). Cluster 4 contained cell cycle genes (CLN2, PCL1, and YOX1) that are periodically expressed at late G1 phase, which are necessary for passage from G1 to S phase (47). Despite the use of asynchronous cells in the microarray study, these genes were up-regulated in the C404A mutant with or without LoaOOH treatment. Genes in this cluster were grouped according to their known transcriptional regulators to identify the transcription factor(s) that may account for their regulation. Both SBF and MBF complexes had an equal number of target genes, indicating that Swi6p activity is likely to be important to the expression of this gene cluster (supplemental Table S5). When promoters of these genes were analyzed for potential transcription factor binding motif(s), the motif CGCGAAA corresponding to the SCB motif (CGCSAAA) (48) was over-represented (p value: 1.7 × 10−5), indicating a possible bias of SBF-regulated genes in relation to the function of the Cys-404 residue in Swi6p. To examine this possibility, all known targets of SBF, MBF, or both (49) were identified from the 1214 differentially expressed genes, and 79 genes were identified as targets of Swi6p transcriptional complexes (Fig. 4B). When expression levels of these 79 genes were hierarchically clustered, there was no general pattern that explicitly determined which of the two complexes was affected because of the C404A mutation (Fig. 4C). Genes in each cluster were analyzed for functional enrichment, and their functional groupings revealed important changes in the transcriptional response to LoaOOH because of loss of the Cys-404 residue (Table 1, clusters a–d).
FIGURE 4.
Microarray analysis of gene expression in the wild type and C404A and swi6Δ mutants subjected to LoaOOH treatment. The cultures of each strain were grown to A600 = 0.4 and treated with 30 μm LoaOOH for 45 min, and cell extracts were prepared for RNA extraction. Three biological replicates of each strain and treatment were performed. Gene expression from other genotypes and conditions was normalized as the intensity ratio relative to the untreated wild-type control. A, differentially expressed genes that were uniquely up-regulated in the C404A mutant with or without treatment from hierarchical clustering. B, genes identified in the microarray study that are known targets of SBF and/or MBF complexes (highlighted in bold). C, hierarchical clustering of the differentially expressed genes that are known targets of SBF, MBF, or both. The resulting clusters were enriched with genes in the functional grouping of cell wall components (cluster a), cell cycle (cluster b), cell shape (cluster c), and DNA replication (cluster d).
TABLE 1.
Functional categories
Shown are the functional categories enriched in gene clusters a–d of Fig. 4C. Functional enrichment was determined using FunSpec (35). The complete list of genes in each cluster and their functional groupings can be found in the supplemental material.
| Cluster | Category | p valuea | Genes | Classification source |
|---|---|---|---|---|
| a | Sphingolipid biosynthetic process | 5.65 × 10−4 | SUR2 SUR1 | GO Biological Process |
| Cell wall | 5.87 × 10−5 | CWP1 GFA1 EXG1 | GO Cellular Component | |
| b | Regulation of cyclin-dependent protein kinase activity | 1.10 × 10−5 | CLN1 PCL1 CLN2 | GO Biological Process |
| Cell cycle | 9.69 × 10−5 | MCD1 YOX1 CLN1 PCL1 | GO Biological Process | |
| Cell division | 3.07 × 10−4 | MCD1 CLN1 PCL1 CLN2 | GO Biological Process | |
| c | Reproduction | 5.23 × 10−4 | PCL2 GIC2 RAX2 | GO Biological Process |
| Regulation of cell shape | 1.52 × 10−3 | GIC2 GIC1 | GO Biological Process | |
| d | Telomere maintenance via recombination | 1.95 × 10−3 | YRF1−1 RFA2 | GO Biological Process |
| DNA replication | 9.56 × 10−3 | PLM2 CDC45 RFA2 | GO Biological Process |
a The p value is the probability of chance enrichment, and only genes with p < 0.005 (except for cluster d in which the cut-off is p < 0.01) are shown.
Genes up-regulated in the wild type on LoaOOH treatment (cluster a) were involved in synthesis of cell wall components. Their induction was probably a consequence of cell wall damage from lipid oxidation, because CWP1, EXG1, and SUR1 are induced by other cell wall damaging compounds (50). However, deletion of SWI6 or mutation of its Cys-404 led to reduced expression of these genes, and on LoaOOH treatment their expression remained unchanged. This indicated that Swi6p and its Cys-404 residue might also be important for activation of cell wall genes in cells under oxidative stress.
Cluster b consisted of genes involved in the cell cycle (CLN2, PCL1, and YOX1) that were uniquely more highly expressed in the C404A mutant. Cluster c contained genes involved in cell shape, which were up-regulated in both swi6Δ and the C404A mutant, which may explain the irregular and elongated cell shape seen in swi6Δ (supplemental Fig. S2). Another two G1 cyclins, CLN1 and PCL2, were in this cluster, indicating that Swi6p and its Cys-404 are important for proper regulation of G1 cyclin expression. Genes in cluster d are involved in DNA replication. As noted above, expression of genes involved in DNA replication was reduced in both the wild type and the C404A mutant (cluster 10). Reduced expression of DNA replication genes is consistent with the finding that DNA replication was delayed on LoaOOH treatment in SWI6 and to a lesser extent in C404A mutant cells.
From the above data, abolishing the thiol of Cys-404 in Swi6p resulted in elevated expression of all four G1 cyclins that are important for passage from the G1 phase to the S phase. However, the C404A mutant showed reduced expression of DNA replication genes, which is consistent with the finding that oxidant-induced delay in DNA replication was less affected than budding by the mutation.
LoaOOH Treatment Reduces Cyclin Transcription in Synchronous Cultures of the Wild Type, but Not the C404A Mutant
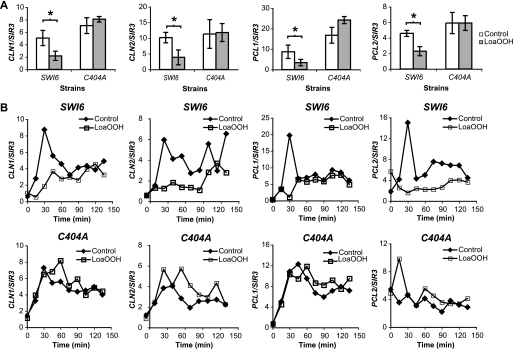
To validate the data from microarray analysis, asynchronous cells were subjected to LoaOOH treatment, and the expression levels of CLN1, CLN2, PCL1, and PCL2 were quantified by RT-PCR. In wild-type cells subjected to LoaOOH, G1 cyclin transcript levels were reduced by 50% compared with the control (Fig. 5A). On the other hand, their expression was higher in the C404A mutant than in the wild type under normal unstressed conditions, and there was little change following exposure of the cells to LoaOOH.
FIGURE 5.
Cyclin expression in LoaOOH-treated cells. A, CLN1, CLN2, PCL1, and PCL2 transcript levels in asynchronous cultures in the presence of LoaOOH by RT-PCR. Cells of the wild-type and C404A mutant strains were grown to A600 = 0.4 and treated with 30 μm LoaOOH for 45 min, and cell extracts were prepared for RNA extraction. Transcript levels were quantified by RT-PCR, and their abundance was expressed relative to SIR3. The average values of four independent experiments are shown, and the error bars represent ± S.D. *, p value <0.05, determined using the paired, two-tailed Student's t test. B, quantification of CLN1, CLN2, PCL1, and PCL2 expression after LoaOOH treatment in α-factor synchronized cultures. Two independent experiments were performed with technical replicates, and a representative experiment is shown.
Because the expression of G1 cyclins is dynamic and fluctuates with the cell cycle (51), expression levels of CLN1, CLN2, PCL1, and PCL2 were monitored using untreated and LoaOOH-treated cells synchronized by α-factor (Fig. 5B). In wild-type cells, in the first 30 min after LoaOOH treatment, CLN1 and PCL1 expression was significantly decreased but subsequently increased slowly to a level similar to that of the untreated control. However, CLN2 and PCL2 expression remained low for 90 min after treatment before increasing. This reduction of G1 cyclin expression in the first 30 min post release correlated with the delay observed in cell cycle progression of the wild-type strain (Fig. 2A). On the other hand, in the C404A mutant, expression of CLN1, CLN2, and PCL1 increased in a similar manner to the control in the first 30 min post LoaOOH treatment. However, expression of these cyclin genes continued to increase, peaking at 60 min post treatment before coming down to a level similar to that of the control. There was a brief induction of PCL2 transcript in the C404A mutant 15 min after LoaOOH treatment, but this was reduced to a similar level to the control by 30 min. Based on these transcript profiles, the inability to oxidize Cys-404 in Swi6p led to up-regulation of G1 cyclins in the C404A mutant, thus driving the cells to progress into S phase despite having been subjected to oxidative stress.
DISCUSSION
Cell division is essential to all living organisms, and the cell cycle is tightly regulated. In response to DNA damage, yeast cells arrest in the G1 phase, initiating repair enzymes to remove damage before resuming cell division (21). Similarly, when yeast cells experience lipid peroxidation, they arrest at G1 delaying budding and S phase progression (11). The signal for each stress or stimulus may differ, but all share a central modulator, Swi6p, for cell cycle regulation. In S. cerevisiae, Rad9p serves as the checkpoint protein for DNA damage and transfers the signals to Swi6p through a phospho-relay (21). SWI6 was identified from a genome-wide screen to be important for cellular defense to oxidative stress induced by LoaOOH, and deletion of SWI6 in cells led to an inability to arrest at G1 despite the presence of LoaOOH. However, oxidative stress induced by lipid hydroperoxide had no apparent sensor pathway (16). Because lipid peroxidation damages cell wall/membranes and activates the cell integrity pathway for cell wall repair (41), its MAPK Slt2p was examined for its role in redox sensing for the cell cycle. Despite its sensitivity to LoaOOH (15, 41), the slt2Δ mutant retained the ability to arrest in G1, hence making it unlikely that SLT2 is involved in cell cycle regulation in response to oxidative stress (41).
Here, we report a novel function of Swi6p that serves as a sensor for oxidative stress through the direct oxidation of its Cys-404 to a sulfenic acid residue, which is therefore not dependent on any signal transduction cascade. This sensing function of Swi6p is unlikely to involve its binding partners Swi4p or Mbp1p because deletion of SWI4 or MBP1 from cells did not abolish their ability to arrest at G1 in response to oxidative stress (16).
In mammalian cells, sensitive protein thiols can be protected from oxidation to sulfenic acid by glutathionylation or other disulfide bond formation (52). These events are reversible by cellular reducing systems when redox homeostasis is restored (52), and this could also occur with the oxidized Swi6p. Neither glutathionylation nor dimerization was detected in Swi6p, and further irreversible thiol oxidation to form sulfinic or sulfonic acids could also occur, probably at higher LoaOOH doses. In mammalian cells, the severity of oxidative stress may be monitored by the extent of oxidation at cysteine residues in proteins (45). Moreover, in S. cerevisiae, a reactive cysteine in the thiol peroxidase (Gpx3p/Orp1p/Hyr1p) that senses hydrogen peroxide is oxidized to the sulfenic acid and transduces the response to the Yap1p transcription factor (53, 54). It is therefore possible that in S. cerevisiae, the oxidation product of the Swi6p Cys-404 may serve as an indicator of oxidative stress level as part of the mechanism used by Swi6p in regulating the cell cycle.
Besides being activated by Slt2p upon cell wall damage signaled via the cell integrity pathway (41, 55), Swi6p is also phosphorylated by Slt2p from the Pkc1p pathway that promotes growth to activate genes for cell cycle progression (56). However, when challenged by oxidative stress, Swi6p can become oxidized at its Cys-404 residue to sulfenic acid, resulting in G1 delay. Hence, oxidation of Cys-404 may serve as a mechanism to antagonize the proliferation signals received. The ability to override one signal by another on the same transcription factor would enable cells to respond rapidly to the change in the environment to elicit appropriate oxidative stress responses rather than initiating cell division.
Upon oxidation of Cys-404 the G1 cyclins, which would normally be required for commitment to cell division, were down-regulated. The mechanism by which this oxidation results in altered transcriptional activity of Swi6p remains to be elucidated. However, because Swi6p is recruited to the Swi4p-Slt2p complex at promoters for activation of transcription (55), one possibility is that formation of sulfenic acid at Cys-404 in the ankyrin motif, which is important for protein-protein interaction, may change the affinity of the transcriptional complex for some promoters. The SBF complex primarily activates the transcription of CLN1 and CLN2, which encode cyclins that bind to and activate Cdc28p kinase activity, as well as PCL1 and PCL2 for binding and activation of Pho85p CDK at late G1 phase (3, 47). G1 cyclin-CDK complexes have long been proposed to be necessary in degradation of the B-type cyclin-CDK inhibitor Sic1p for initiation of S phase events, including DNA replication, bud formation, and spindle body formation (3, 57). However, recent evidence indicates that the maximal cyclin-CDK activity in late G1 phase has a more specific role in the establishment of cell morphogenesis and budding than in the initiation of DNA replication (43). Deletion of all four G1 cyclins causes catastrophic morphogenesis and enormously wide bud necks in the conditional mutant, yet it was able to initiate DNA replication at the proper timing, indicating that these G1 cyclins play no regulatory role in initiation of DNA replication (43). This role for G1 cyclins is in accord with the finding that in the C404A mutant under oxidative stress, there was elevated expression of G1 cyclins, which led to bud formation and expression of genes involved in morphogenesis, yet DNA replication was still partially delayed.
Swi6p has a regulatory role in both morphogenesis (SBF targets) and DNA replication (MBF targets), yet the mutation of Cys-404 to Ala affected one process, budding and morphogenesis involving targets of SBF complex, more than that of DNA replication. Abolishing the Cys-404 thiol may uncouple the regulation of budding and DNA replication by Swi6p. However, this uncoupling is not the case for all SBF targets. Thus, regulation by the redox-sensing residue Cys-404 is more directed to the regulation of G1 cyclins. This perhaps could be a consequence of the timing of events that occur in the S phase. In wild-type cells, DNA replication does not commence until 30–45 min after release from pheromone (43), whereas buds begin to emerge 15 min after release. As a result, in response to certain oxidants (those more likely to damage proteins and lipids than DNA), it may be more critical to prevent the induction of G1 cyclins for morphogenesis.
In summary, we have identified Swi6p as an oxidative stress sensor for the regulation of the cell cycle. This sensing function occurs via direct oxidation of the thiol group of its Cys-404 to sulfenic acid. Oxidation of the Cys-404 residue resulted in down-regulation of G1 cyclins CLN1, CLN2, PCL1, and PCL2, which are required for proper morphogenesis and bud formation. DNA replication is less affected by Cys-404 oxidation, and its initiation in the C404A mutant was delayed upon oxidative stress but not to the same extent as wild-type cells.
Supplementary Material
Acknowledgments
We thank the Ramaciotti Centre for the expertise in microarray and RT-PCR analysis and Geoffrey Kornfeld and Gabriel Perrone for helpful discussion and advice.
This work was supported by discovery grants from Australian Research Council (to I. W. D.) and National Health and Medical Research Council Peter Doherty and University of New South Wales vice-chancellor's postdoctoral fellowships (to R. C. Y. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1–S5, and Figs. S1–S4.
M. A. Wouters, unpublished results.
- CDK
- cyclin-dependent kinase
- LoaOOH
- linoleate hydroperoxide
- MBF
- Swi6p/Mbp1p transcription factor complex
- SBF
- Swi4p/Swi6p transcription factor complex.
REFERENCES
- 1. Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., Eisen M. B., Brown P. O., Botstein D., Futcher B. (1998) Mol. Biol. Cell 9, 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simon I., Barnett J., Hannett N., Harbison C. T., Rinaldi N. J., Volkert T. L., Wyrick J. J., Zeitlinger J., Gifford D. K., Jaakkola T. S., Young R. A. (2001) Cell 106, 697–708 [DOI] [PubMed] [Google Scholar]
- 3. Mendenhall M. D., Hodge A. E. (1998) Microbiol. Mol. Biol. Rev. 62, 1191–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elledge S. J. (1996) Science 274, 1664–1672 [DOI] [PubMed] [Google Scholar]
- 5. Shackelford R. E., Kaufmann W. K., Paules R. S. (1999) Environ Health Perspect 107, (Suppl. 1) 5–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gutteridge J. M., Halliwell B. (1999) Free Radicals in Biology and Medicine, pp. 36–104, Oxford University Press, Oxford [Google Scholar]
- 7. Adibhatla R. M., Hatcher J. F. (2010) Antioxid. Redox. Signal. 12, 125–169 [DOI] [PubMed] [Google Scholar]
- 8. Neely M. D., Boutte A., Milatovic D., Montine T. J. (2005) Brain Res. 1037, 90–98 [DOI] [PubMed] [Google Scholar]
- 9. Flattery-O'Brien J. A., Dawes I. W. (1998) J. Biol. Chem. 273, 8564–8571 [DOI] [PubMed] [Google Scholar]
- 10. Shackelford R. E., Kaufmann W. K., Paules R. S. (2000) Free Rad. Biol. Med. 28, 1387–1404 [DOI] [PubMed] [Google Scholar]
- 11. Alic N., Higgins V. J., Dawes I. W. (2001) Mol. Biol. Cell 12, 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barrera G., Pizzimenti S., Laurora S., Briatore F., Toaldo C., Dianzani M. U. (2005) Biofactors 24, 151–157 [DOI] [PubMed] [Google Scholar]
- 13. Flattery-O'Brien J., Collinson L. P., Dawes I. W. (1993) J. Gen. Microbiol. 139, 501–507 [DOI] [PubMed] [Google Scholar]
- 14. Alic N., Felder T., Temple M. D., Gloeckner C., Higgins V. J., Briza P., Dawes I. W. (2004) Free Rad. Biol. Med. 37, 23–35 [DOI] [PubMed] [Google Scholar]
- 15. Thorpe G. W., Fong C. S., Alic N., Higgins V. J., Dawes I. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6564–6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fong C. S., Temple M. D., Alic N., Chiu J., Durchdewald M., Thorpe G. W., Higgins V. J., Dawes I. W. (2008) FEMS Yeast Res. 8, 386–399 [DOI] [PubMed] [Google Scholar]
- 17. Dirick L., Moll T., Auer H., Nasmyth K. (1992) Nature 357, 508–513 [DOI] [PubMed] [Google Scholar]
- 18. Koch C., Moll T., Neuberg M., Ahorn H., Nasmyth K. (1993) Science 261, 1551–1557 [DOI] [PubMed] [Google Scholar]
- 19. Siegmund R. F., Nasmyth K. A. (1996) Mol. Cell. Biol. 16, 2647–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sedgwick S. G., Taylor I. A., Adam A. C., Spanos A., Howell S., Morgan B. A., Treiber M. K., Kanuga N., Banks G. R., Foord R., Smerdon S. J. (1998) J. Mol. Biol. 281, 763–775 [DOI] [PubMed] [Google Scholar]
- 21. Sidorova J. M., Breeden L. L. (1997) Genes Dev. 11, 3032–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chiu J., March P. E., Lee R., Tillett D. (2004) Nucleic Acids Res. 32, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiu J., Tillett D., Dawes I. W., March P. E. (2008) J. Microbiol. Methods. 73, 195–198 [DOI] [PubMed] [Google Scholar]
- 24. Jansen G., Wu C., Schade B., Thomas D. Y., Whiteway M. (2005) Gene 344, 43–51 [DOI] [PubMed] [Google Scholar]
- 25. Alberti S., Gitler A. D., Lindquist S. (2007) Yeast 24, 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Henrick K., Thornton J. M. (1998) Trends Biochem. Sci. 23, 358–361 [DOI] [PubMed] [Google Scholar]
- 28. Jones S., Thornton J. M. (1995) Prog. Biophys. Mol. Biol. 63, 31–65 [DOI] [PubMed] [Google Scholar]
- 29. Evans M. V., Turton H. E., Grant C. M., Dawes I. W. (1998) J. Bacteriol. 180, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haase S. B., Lew D. J. (1997) Methods Enzymol. 283, 322–332 [DOI] [PubMed] [Google Scholar]
- 31. Vivancos A. P., Castillo E. A., Biteau B., Nicot C., Ayté J., Toledano M. B., Hidalgo E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8875–8880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang B. M., McLean A. S., Dawes I. W., Huang S. J., Lin R. C. (2009) Crit. Care Med. 37, 882–888 [DOI] [PubMed] [Google Scholar]
- 33. Saeed A. I., Bhagabati N. K., Braisted J. C., Liang W., Sharov V., Howe E. A., Li J., Thiagarajan M., White J. A., Quackenbush J. (2006) Methods Enzymol. 411, 134–193 [DOI] [PubMed] [Google Scholar]
- 34. Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. (2003) BioTechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 35. Robinson M. D., Grigull J., Mohammad N., Hughes T. R. (2002) BMC Bioinformatics 3, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monteiro P. T., Mendes N. D., Teixeira M. C., d'Orey S., Tenreiro S., Mira N. P., Pais H., Francisco A. P., Carvalho A. M., Lourenço A. B., Sá-Correia I., Oliveira A. L., Freitas A. T. (2008) Nucleic Acids Res. 36, D132–D136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Teixeira M. C., Monteiro P., Jain P., Tenreiro S., Fernandes A. R., Mira N. P., Alenquer M., Freitas A. T., Oliveira A. L., Sá-Correia I. (2006) Nucleic Acids Res. 34, D446–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Helden J., André B., Collado-Vides J. (1998) J. Mol. Biol. 281, 827–842 [DOI] [PubMed] [Google Scholar]
- 39. Pfaffl M. W., Horgan G. W., Dempfle L. (2002) Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Madden K., Sheu Y. J., Baetz K., Andrews B., Snyder M. (1997) Science 275, 1781–1784 [DOI] [PubMed] [Google Scholar]
- 41. Alic N., Higgins V. J., Pichova A., Breitenbach M., Dawes I. W. (2003) J. Biol. Chem. 278, 41849–41855 [DOI] [PubMed] [Google Scholar]
- 42. Foord R., Taylor I. A., Sedgwick S. G., Smerdon S. J. (1999) Nat. Struct. Biol. 6, 157–165 [DOI] [PubMed] [Google Scholar]
- 43. Moffat J., Andrews B. (2004) Nat. Cell Biol. 6, 59–66 [DOI] [PubMed] [Google Scholar]
- 44. Dalle-Donne I., Rossi R., Colombo G., Giustarini D., Milzani A. (2009) Trends Biochem. Sci. 34, 85–96 [DOI] [PubMed] [Google Scholar]
- 45. Poole L. B., Karplus P. A., Claiborne A. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 325–347 [DOI] [PubMed] [Google Scholar]
- 46. Seo Y. H., Carroll K. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16163–16168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dirick L., Böhm T., Nasmyth K. (1995) EMBO J. 14, 4803–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., Danford T. W., Hannett N. M., Tagne J. B., Reynolds D. B., Yoo J., Jennings E. G., Zeitlinger J., Pokholok D. K., Kellis M., Rolfe P. A., Takusagawa K. T., Lander E. S., Gifford D. K., Fraenkel E., Young R. A. (2004) Nature 431, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Iyer V. R., Horak C. E., Scafe C. S., Botstein D., Snyder M., Brown P. O. (2001) Nature 409, 533–538 [DOI] [PubMed] [Google Scholar]
- 50. García R., Bermejo C., Grau C., Pérez R., Rodríguez-Peña J. M., Francois J., Nombela C., Arroyo J. (2004) J. Biol. Chem. 279, 15183–15195 [DOI] [PubMed] [Google Scholar]
- 51. Breeden L. L. (2003) Curr. Biol. 13, R31–R38 [DOI] [PubMed] [Google Scholar]
- 52. Dalle-Donne I., Rossi R., Giustarini D., Colombo R., Milzani A. (2007) Free Rad. Biol. Med. 43, 883–898 [DOI] [PubMed] [Google Scholar]
- 53. Delaunay A., Pflieger D., Barrault M. B., Vinh J., Toledano M. B. (2002) Cell 111, 471–481 [DOI] [PubMed] [Google Scholar]
- 54. Paulsen C. E., Carroll K. S. (2009) Chem. Biol. 16, 217–225 [DOI] [PubMed] [Google Scholar]
- 55. Kim K. Y., Truman A. W., Levin D. E. (2008) Mol. Cell. Biol. 28, 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gray J. V., Ogas J. P., Kamada Y., Stone M., Levin D. E., Herskowitz I. (1997) EMBO J. 16, 4924–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. (2001) Nature 414, 514–521 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.