Abstract
Apolipoprotein (apo) E4 is the major genetic risk factor for late-onset Alzheimer disease (AD). ApoE4 assumes a pathological conformation through an intramolecular interaction mediated by Arg-61 in the amino-terminal domain and Glu-255 in the carboxyl-terminal domain, referred to as apoE4 domain interaction. Because AD is associated with mitochondrial dysfunction, we examined the effect of apoE4 domain interaction on mitochondrial respiratory function. Steady-state amounts of mitochondrial respiratory complexes were examined in neurons cultured from brain cortices of neuron-specific enolase promoter-driven apoE3 (NSE-apoE3) or apoE4 (NSE-apoE4) transgenic mice. All subunits of mitochondrial respiratory complexes assessed were significantly lower in NSE-apoE4 neurons compared with NSE-apoE3 neurons. However, no significant differences in levels of mitochondrial complexes were detected between astrocytes expressing different apoE isoforms driven by the glial fibrillary acidic protein promoter, leading to our conclusion that the effect of apoE4 is neuron specific. In neuroblastoma Neuro-2A (N2A) cells, apoE4 expression reduced the levels of mitochondrial respiratory complexes I, IV, and V. Complex IV enzymatic activity was also decreased, lowering mitochondrial respiratory capacity. Mutant apoE4 (apoE4-Thr-61) lacking domain interaction did not induce mitochondrial dysfunction in N2A cells, indicating that the effect is specific to apoE4-expressing cells and dependent on domain interaction. Consistent with this finding, treatment of apoE4-expressing N2A cells with a small molecule that disrupts apoE4 domain interaction restored mitochondrial respiratory complex IV levels. These results suggest that pharmacological intervention with small molecules that disrupt apoE4 domain interaction is a potential therapeutic approach for apoE4-carrying AD subjects.
Keywords: Alzheimer Disease, Apolipoproteins, Mitochondria, Neurodegeneration, Protein Structure, Apolipoprotein E
Introduction
Apolipoprotein (apo)2 E4 is the major genetic risk factor for late-onset Alzheimer disease (AD) (1–6). The protein sequence of apoE4 (299 amino acids) differs from the other two common isoforms (apoE2, apoE3) at residues 112 or 158; apoE3 has a cysteine at residue 112 and an arginine at residue 158, whereas apoE4 has arginines at both positions and apoE2 has cysteines (3, 4). These differences profoundly affect the tertiary protein structure (3–5). ApoE4 displays an intramolecular interaction between its amino-terminal and carboxyl-terminal domains (7, 8), referred to as apoE4 domain interaction. The domain interaction is mediated by Arg-112 in apoE4, which induces a salt bridge between Arg-61 in the amino-terminal domain and Glu-255 in the carboxyl-terminal domain (7, 8). This interaction is decreased in apoE2 and apoE3 due to the presence of Cys-112 (8). Substitution of a threonine for Arg-61 in apoE4 disrupts the domain interaction, converting apoE4 to an apoE3-like conformation (7–9).
It is postulated that domain interaction is primarily responsible for the pathogenic effects of apoE4 in AD (3–5). Because AD is known to be associated with mitochondrial dysfunction (10–13), apoE4 domain interaction might cause mitochondrial dysfunction. Patients in the early stages of AD have abnormally low glucose metabolism in brain regions susceptible to AD pathology, as shown by PET studies (14, 15). Because the brain is highly dependent on mitochondrial activity for energy production from glucose (12), cerebral glucose hypometabolism may indicate perturbed mitochondrial function in AD brains. Consistent with this notion, gene expression (16, 17) and metabolic activity (12, 18) of mitochondrial respiratory complexes are decreased in AD brains. Most protein subunits of mitochondrial respiratory complexes are expressed from the nuclear genome, but 13 of the complex subunits are encoded by mtDNA (19). Mutations in mtDNA are more frequently detected in the brains of AD patients than those of age-matched controls, which may also contribute to the mitochondrial dysfunction in AD patients (10, 19). Importantly, PET studies have also detected an AD-like regional pattern of glucose hypometabolism in the brains of cognitively normal apoE4 carriers decades before the age of onset of clinical AD (20–22). This result raises the possibility that apoE4 may perturb mitochondrial respiratory function in the brain, rendering subjects with apoE4 more susceptible to AD neuropathology.
In the nervous system, apoE functions as a major carrier and distributor of cholesterol and other lipids, which are essential components of neuronal membranes and myelin sheaths (6, 23). In an unstressed brain, apoE is predominantly synthesized and secreted by astrocytes (6, 24). However, neurons in the central nervous system synthesize apoE under various pathophysiological conditions (6, 24, 25). The stress-induced apoE synthesis by neurons is thought to protect neurons against injury or to promote neuronal regeneration (23, 24). However, neuronal expression of apoE4 is neurotoxic due to domain interaction (3–5). Transgenic expression of apoE4, but not apoE3, in neurons using a neuron-specific enolase (NSE) promoter, induces age-dependent learning and memory deficits and neurodegenerative changes in mice (26, 27). Neuropathogenic effects of apoE4 are more pronounced when apoE4 is expressed in neurons compared with glial cells. No significant neuropathological changes were found in transgenic mice expressing apoE4 in glial cells driven by the glial fibrillary acidic protein (GFAP) promoter (28, 29). It is postulated that a neuron-specific apoE proteolytic machinery may enhance the neurotoxic effect of apoE4 (29, 30).
To determine whether neuronal expression of apoE4 and apoE4 domain interaction cause mitochondrial dysfunction, we compared the apoE isoform-dependent effects on mitochondria in both neurons and astrocytes. Our findings show that apoE4 expressed in neurons exerts a deleterious effect on mitochondrial protein expression and function. Importantly, the detrimental effects of apoE4 on mitochondria can be mitigated by genetic and pharmacologic interference with apoE4 domain interaction.
EXPERIMENTAL PROCEDURES
Materials
All of the chemicals were of analytical grade and were obtained from Sigma unless stated otherwise.
Cell Culture and Cell Transfection
Primary neuronal cultures were prepared from NSE-apoE3 or NSE-apoE4 mice on a mouse apoE knock-out background on postnatal day 0 as described (31). Primary astrocytes were prepared from GFAP-apoE3 or GFAP-apoE4 mice (29) on a mouse apoE-null background on postnatal day 0 as described (31). After 10–15 days of in vitro culture, cortical neurons or astrocytes were harvested into PhosphoSafe extraction buffer (Novagen) containing mixtures of protease inhibitors and phosphatase inhibitors (Roche Applied Science). Neuroblastoma Neuro-2A (N2A) cells stably expressing apoE3, apoE4, or a control vector were described previously (31). An R61T mutation was made on apoE4 plasmid DNA by site-directed mutagenesis. Cell transfection and selection of stably transfected cells were done as described (31). Similar phenotypes were observed from several individually established clones. For treatment with GIND25 (9), cells were incubated in OptiMEM (Invitrogen) with 0, 10, or 20 μm of GIND25 (dissolved in dimethyl sulfoxide) for 24 h; the dimethyl sulfoxide concentration was 0.5%. N2A cell extracts were prepared in radioimmune precipitation assay buffer (Pierce) containing protease inhibitors and phosphatase inhibitors.
Western Blot Analysis and Confocal Microscopy
Western blotting was performed by standard methods using MitoProfile total OXPHOS rodent WB antibody cocktail (MitoSciences) or individual antibodies of cytochrome c oxidase subunit 1 (Mitosciences), ATP synthase subunit α (Invitrogen), mitochondrial heat shock protein 70 (MtHsp70, Affinity BioReagents), voltage-dependent anion channels (VDACs, MitoSciences), apoE (Calbiochem), actin (Sigma), and tubulin (Novus Biologicals). Indirect immunofluorescence staining was performed as described (32). DNA was stained with TOTO-3 (Invitrogen). Images from the same confocal plane were captured on a Leica TCS SP5 confocal microscope.
Real-time Quantitative PCR
mRNA was isolated using an AllPrep DNA/RNA/protein mini kit (Qiagen), and reverse transcription was performed with a Superscript III kit (Invitrogen). Real-time quantitative PCR was performed with a HotSybr PCR kit (Molecular Cloning Laboratories) and a 7900HT fast PCR system (Applied Biosystems). The primer pairs were 5′-GGTCAACCAGGTGCACTTTT, and 5′-TGGGGCTCCGATTATTAGTG (cytochrome c oxidase subunit 1), 5′-TTATCCCCCGAATCTCTGTG and 5′-GCAATCGATGTTTTCCCAGT (ATP synthase subunit α), 5′-CTGAGTATGGGCTGACGTTTAC and 5′-GGTGAGCTTCAGTCCACGAG (VDAC1), 5′-GGCTGTATTCCCTCCATCG, and 5′-CCAGTTGGTAACAATGCCATGT (β-actin). The relative amounts of mRNA of different genes were compared after normalization to β-actin mRNA levels.
Cytochrome c Oxidase and Citrate Synthase Assays
Spectrophotometric assays for cytochrome c oxidase and citrate synthase were performed as described (33) on mitochondria prepared with a Qproteome mitochondria isolation kit (Qiagen). Oxidation of reduced cytochrome c by cytochrome c oxidase in the first 45 s was measured at 550 nm to calculate the fast initial rate. For the citrate synthase assay, the reaction was initiated by adding 0.1 mm oxaloacetate to the reaction mixture at 30 °C. The formation of 5-thio-2-nitrobenzoate was measured at 412 nm for 5 min.
Oxygen Consumption Measurements
Cellular oxygen consumption rate (OCR) was measured with an XF24 extracellular flux analyzer (Seahorse Biosciences) in 24-well plates at 37 °C as described with modifications (34). N2A-apoE3, N2A-apoE4, and control cells were seeded at 30,000 cells/well, 4 h before analysis. Before the measurement, the culture medium was removed and replaced with nonbuffered Dulbecco's minimum essential medium (Invitrogen), pH 7.4, prewarmed to 37 °C. After a 17-min equilibration period, four measurements of OCR were recorded in the first 60 min in each well to establish a baseline OCR. After the baseline measurement, carbonylcyanide-3-chlorophenylhydrazone (CCCP, final concentration, 5 μm) was added, and OCR was measured five times. The maximal uncoupled OCR was expressed as a percent change in OCR over baseline.
Statistical Analysis
Statistical analyses were performed by two-tailed t test or one-way ANOVA followed by Tukey's post hoc comparisons. p < 0.05 was considered statistically significant.
RESULTS
ApoE4 Reduces Steady-state Levels of Mitochondrial Respiratory Complexes in Neuronal Cells
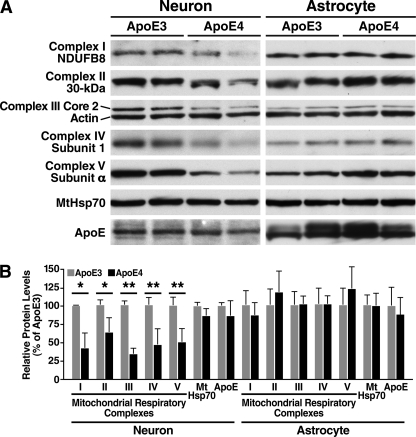
To assess the potential role of apoE4 in mitochondrial dysfunction, we first determined whether apoE4 reduces the steady-state levels of mitochondrial respiratory complexes in primary neurons. We performed Western blot analyses on lysates from cultured brain cortical neurons from transgenic mice that had NSE promoter-driven expression of either apoE3 (NSE-apoE3) or apoE4 (NSE-apoE4) on a mouse apoE knock-out background. The NSE-apoE3 and NSE-apoE4 neurons expressed apoE and MtHsp70 at comparable levels (Fig. 1, A and B). However, steady-state levels of all five subunits of mitochondrial respiratory complexes (I–V) assessed were significantly lower in NSE-apoE4 than NSE-apoE3 neurons: complex I subunit NDUFB8, 43 ± 20% of that in NSE-apoE3 neurons (p < 0.01 by two-tailed t test, n = 3); complex II 30-kDa subunit, 64 ± 19% (p < 0.05, n = 3); complex III subunit Core 2, 35 ± 7% (p < 0.001, n = 3); complex IV subunit 1, 46 ± 23% (p < 0.001, n = 6); and complex V subunit α, 51 ± 18% (p < 0.001, n = 6). Thus, neuronal expression of apoE4 is associated with a reduced level of several different subunits of mitochondrial respiratory complexes.
FIGURE 1.
ApoE4 reduces steady-state levels of mitochondrial respiratory complexes in cultured brain cortical neurons but not in astrocytes. A, Western blot analysis of mitochondrial respiratory complexes either in cultured cortical neurons from NSE-apoE3 or NSE-apoE4 transgenic mice or in primary cortical astrocytes from GFAP-apoE3 or GFAP-apoE4 mice. Two separate samples are shown for each cell type. B, mitochondrial protein levels relative to those in apoE3-expressing neurons or astrocytes. Values are mean ± S.D. after normalization to actin levels. (n ≥ 3 for neurons; n ≥ 6 for astrocytes). *, p < 0.05; **, p < 0.001 versus apoE3 by two-tailed t test.
To determine whether expression of apoE4 in astrocytes has a similar effect on mitochondria, we performed Western blot analyses on lysates from primary cortical astrocytes from transgenic mice with GFAP promoter-driven expression of apoE3 (GFAP-apoE3) or apoE4 (GFAP-apoE4) on a mouse apoE-null background (29). In contrast to its effect in neurons, expression of apoE4 in astrocytes did not reduce the levels of mitochondrial respiratory complexes (n ≥ 6, Fig. 1, A and B). These results are consistent with a more pronounced pathogenic effect of apoE4 than apoE3 when expressed in neuronal cells (26, 28, 29).
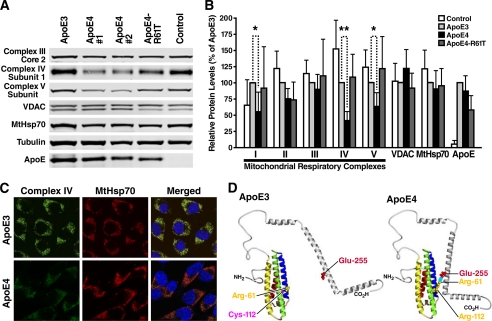
To confirm the effect observed in primary cultured neurons, we examined mitochondrial respiratory complexes in N2A cells stably expressing either apoE3 (N2A-apoE3) or apoE4 (N2A-apoE4). Again, the steady-state levels of complex IV (cytochrome c oxidase) subunit 1 and complex V (ATP synthase) subunit α in N2A-apoE4 cells were lower in two different lines of N2A-apoE4 cells than in N2A-apoE3 cells or control cells expressing vector alone (Fig. 2A). Statistical analysis showed that amounts of mitochondrial respiratory complexes I, IV, and V were significantly lower in N2A-apoE4 cells than in N2A-apoE3 cells or control cells (Fig. 2B), suggesting an apoE4-specific effect. Steady-state levels of apoE and mitochondrial VDAC and MtHsp70 did not differ significantly between N2A-apoE3 and N2A-apoE4, even though amounts of VDAC were occasionally slightly increased in N2A-apoE4 cells (Fig. 2, A and B). In contrast to the effects of apoE4 on complexes I, IV, and V, levels of complexes II and III were not changed significantly in N2A cells (Fig. 2B). These results indicate that a subset of mitochondrial respiratory complexes is more responsive to the adverse effects of apoE4 in N2A cells.
FIGURE 2.
ApoE4 induces deficiency of mitochondrial respiratory complexes through intramolecular domain interaction. A, Western blot analysis of mitochondrial respiratory complexes in N2A cells expressing apoE3, apoE4, apoE4-R61T, or vector alone (control). Levels of complex IV subunit 1 and complex V subunit α were lower in cells expressing apoE4 (clones 1 and 2), but not in cells expressing apoE3 or apoE4-R61T. B, mitochondrial protein levels relative to those in apoE3-expressing cells after normalization to tubulin levels. Values are mean ± S.D. (n = 6). *, p < 0.05; **, p < 0.001 versus apoE3 by Tukey's test. C, confocal microscopy revealed lower immunoreactivity of mitochondrial complex IV subunit 1 (green) in N2A-apoE4 than in N2A-apoE3 cells. Mitochondria were detected by immunofluorescence of MtHsp70 (red). Nuclear staining is shown in blue in merged images. D, a model of apoE4 domain interaction. The residues critical for domain interaction are shown.
To further investigate the adverse effects of apoE4 in N2A cells, we performed confocal microscopy. In most, if not all, mitochondria (labeled by MtHsp70), immunofluorescence of complex IV subunit 1 (mitochondrially encoded) was lower in N2A-apoE4 cells than in N2A-apoE3 cells (Fig. 2C). This finding indicates that apoE4 may have a widespread effect on mitochondrial expression of complex IV subunit 1. Immunofluorescence of MtHsp70 displayed a typical mitochondrial staining pattern in both N2A-apoE3 and N2A-apoE4 cells (Fig. 2C). N2A-apoE3 and N2A-apoE4 cells did not differ in gross morphology or number of mitochondria. These results suggest that apoE4 has differential effects on the expression of different mitochondrial proteins.
ApoE4 Reduces mRNA Expression in Subunits of Mitochondrial Complexes IV and V Encoded by mtDNA and Nuclear Genome
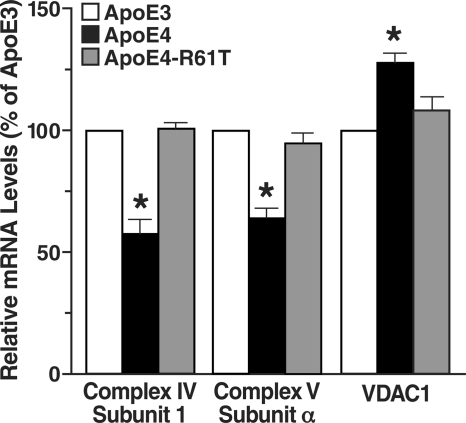
To test whether apoE4 alters the expression of mitochondrial genes encoded by both mtDNA and the nuclear genome, we analyzed cDNA from N2A-apoE3 and N2A-apoE4 cells by quantitative real-time PCR (Fig. 3). Relative mRNA levels for complex IV subunit 1 (mitochondrially encoded) and complex V subunit α (encoded by the nuclear genome) in apoE4-expressing cells were significantly decreased (to 57 ± 6% and 64 ± 4%, respectively, of that in apoE3-expressing cells) (Fig. 3). On the other hand, mRNA levels of VDAC isoform 1 (VDAC1, a nuclear gene) were increased in apoE4-expressing cells (to 127 ± 4% of that in apoE3-expressing cells), possibly reflecting a cellular mechanism to compensate for the reduction of mitochondrial respiratory complexes. Alternatively, apoE4 may have different effects on expression of individual mitochondrial genes. These findings indicate that apoE4 perturbs expression of at least a subset of mitochondrial genes, including those encoded by both mtDNA and the nuclear genome.
FIGURE 3.
ApoE4 alters expression of mitochondrial genes in N2A cells. Relative mRNA levels of mitochondrial complex IV subunit 1, complex V subunit α, and VDAC1 in N2A-apoE3, N2A-apoE4, and N2A-apoE4-R61T cells were determined by quantitative real-time PCR. β-Actin levels were used for normalization. Values are mean ± S.D. (n = 3). *, p < 0.001 versus apoE3, two-tailed t test.
ApoE4 Reduces Mitochondrial Respiratory Function
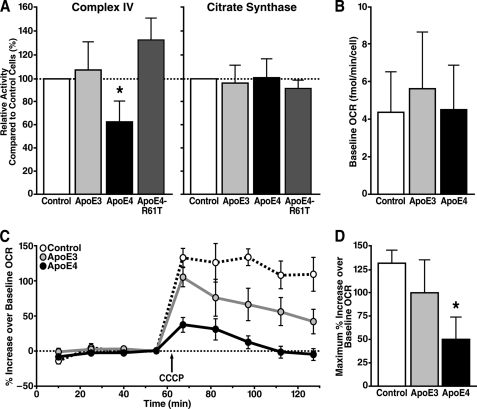
To investigate the effect of apoE4 on mitochondrial metabolic activity, we performed spectrophotometric activity assays on mitochondria isolated from N2A cells stably expressing various forms of apoE. The activity of complex IV in mitochondria was significantly lower in N2A-apoE4 cells than in N2A-apoE3 or control cells (Fig. 4A). This finding suggests that apoE4 not only reduces the expression of complex IV subunit 1 but also decreases activity of the holoenzyme. In a parallel experiment, we found that activity of mitochondrial citrate synthase, a Krebs cycle enzyme, was unchanged. Thus, apoE4 appears to have differential effects on enzymes involved in mitochondrial energy metabolism (Fig. 4A).
FIGURE 4.
ApoE4 causes mitochondrial respiratory dysfunction. A, activities of mitochondrial complex IV and citrate synthase in mitochondria isolated from N2A cells stably expressing apoE3, apoE4, or apoE4-R61T and control cells were determined by spectrophotometric assays. Values are mean ± S.D. (complex IV, n = 4; citrate synthase, n = 3). *, p < 0.05, ANOVA with Tukey's test. B, no significant differences were observed at baseline OCR between N2A cells expressing different apoE isoforms. Values are mean ± S.D. (n = 6). C, changes in OCR after uncoupling in N2A-apoE3, N2A-apoE4, and control cells. After baseline OCR measurements, CCCP was added to stimulate maximal OCR. A representative experiment using the XF24 extracellular flux analyzer is shown. Values are mean ± S.D. of quadruplicate wells. D, maximal uncoupled OCR was significantly lower in apoE4-expressing cells. Values are mean ± S.D. of six experiments. *, p < 0.05 by ANOVA with Tukey's test.
To evaluate apoE4-induced mitochondrial respiratory dysfunction in living cells, we performed cellular oxygen consumption assays using an XF24 extracellular flux analyzer (Seahorse Biosciences) (34). The OCR reflects mitochondrial respiratory function, as mitochondria consume >95% of oxygen uptake in aerobically growing higher organisms (35). We did not detect significant OCR differences at baseline between N2A cells expressing different apoE isoforms (Fig. 4B). This observation was not surprising because mitochondria are known to have high reserve capacity of respiratory complexes. Typically, the activities of such complexes need to be reduced by >60% before major changes in basal oxygen consumption occur (36). To determine the total capacity of mitochondrial respiratory function, we induced maximum OCR with an uncoupling agent, CCCP (37). Shortly after addition of CCCP, OCR increased by 98% in N2A-apoE3 cells and 130% in control cells over the initial basal rate (Fig. 4, C and D). The CCCP-induced OCR increased by only 50% in N2A-apoE4 cells, which was significantly less than in N2A-apoE3 cells (p < 0.05) or control cells (p < 0.001). Our results indicate that functional measures of mitochondrial respiratory capacity were decreased by apoE4.
Domain Interaction Mediates ApoE4-induced Mitochondrial Respiratory Dysfunction
ApoE4 domain interaction (Fig. 2D) is thought to mediate the detrimental effects of apoE4 (3, 5, 9, 38). To assess its potential role in apoE4-induced mitochondrial dysfunction, we generated N2A cells with stable expression of mutant apoE4-R61T, which lacks domain interaction (7–9). Unlike apoE4, apoE4-R61T did not reduce expression of complex I, IV, or V (Fig. 2, A and B, and Fig. 3) or complex IV activity (Fig. 4A) in neuronal cells. Therefore, apoE4 domain interaction underlies the ability of apoE4 to induce mitochondrial respiratory dysfunction.
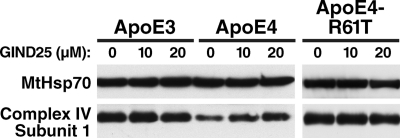
If apoE4 domain interaction is essential to induce mitochondrial respiratory dysfunction, the adverse effects of apoE4 may be mitigated by small molecules that disrupt domain interaction, i.e. apoE structural correctors (3, 9). To test this possibility, we incubated N2A-apoE3, N2A-apoE4, and N2A-apoE4-R61T cells with a known apoE structural corrector, GIND25 (9). Mitochondria from GIND25-treated N2A-apoE4 cells showed a dose-dependent increase in the levels of complex IV subunit 1 (n = 4, p < 0.05 by ANOVA with Tukey's test), whereas MtHsp70 levels were unaltered (Fig. 5). Mitochondria from the mock-treated N2A-apoE3 or N2A-apoE4-R61T cells had higher levels of complex IV subunit 1 than the mock-treated N2A-apoE4 cells but were unaffected by GIND25 (n = 3), suggesting an apoE4 domain interaction-specific effect of GIND25. These results confirm that neuronal apoE4-induced mitochondrial respiratory dysfunction is mediated by domain interaction and is reversible by interventions that disrupt that interaction.
FIGURE 5.
Disrupting apoE4 domain interaction with a small molecule, GIND25, restores mitochondrial complex IV levels. Western blot analysis of mitochondria from GIND25-treated N2A-apoE3, N2A-apoE4, and N2A-apoE4-R61T cells. MtHsp70 served as an internal control.
DISCUSSION
This study shows for the first time that apoE4 causes mitochondrial respiratory dysfunction in neuronal cells through apoE4 domain interaction. ApoE4, but not apoE3, reduced expression of mitochondrial respiratory complexes and perturbed mitochondrial respiratory function in neuronal cells. Mutant apoE4-R61T lacking domain interaction did not induce mitochondrial dysfunction. In addition, disrupting domain interaction with a small molecule (GIND25) reversed the apoE4-induced deficiency of mitochondrial respiratory complex IV. Thus, the isoform-specific effects of apoE4 on mitochondrial function are mediated by its domain interaction.
Our findings suggest that the structure of apoE4 is a potential therapeutic target for apoE4-related neurodegeneration. ApoE4 domain interaction mediates a broad spectrum of isoform-specific pathophysiological properties of apoE4 (3, 5). First, apoE4 in plasma binds preferentially to very low density lipoproteins, whereas apoE3 binds preferentially to HDL (5, 7). Abolishing domain interaction with the R61T mutation changes the binding preference of apoE4 to HDL (7, 8). Second, apoE4 enhances amyloid β production in rat neuroblastoma cells stably expressing human wild type amyloid precursor protein to a greater extent than apoE3 (9). The effect of apoE4 is abolished by disrupting apoE4 domain interaction with either small molecules or the R61T mutation (9). Finally, unlike human apoE4, mouse apoE does not display domain interaction owing to the presence of a Thr-61 residue at the critical position for domain interaction to occur (39). Introducing an apoE4-like domain interaction into the mouse apoE gene induces neurodegenerative pathology and a mild memory deficit (40, 41). These apoE4-specific effects, which are mediated by domain interaction, likely contribute to the pathogenesis of AD (3–5, 38). Thus, disrupting apoE4 domain interaction with small molecules may be beneficial for apoE4-carrying subjects, which make up 65–80% of all patients with AD (5).
Mitochondrial dysfunction is thought to be an early event in the disease progression of AD (10–13, 16). Previous studies have implicated mitochondria as a potential target for apoE or its proteolytic fragments (32, 42, 43). When synthesized in neurons, apoE (amino acids 1–299) can be cleaved by a neuron-specific protease to produce carboxyl-terminal-truncated apoE fragments (29, 30). The apoE4 (1–272) (amino acids 1–272) fragment mislocalized to mitochondria when transiently expressed in N2A cells (32). In addition, this apoE4 (1–272) fragment can bind to subunits of mitochondrial respiratory complexes III, IV, and V and perturb the activities of complexes III and IV (43). Notably, apoE4 (1–272) appears to associate with these mitochondrial proteins more robustly than full-length apoE4 (32, 43). It remains unanswered whether full-length apoE4 also perturbs mitochondrial function. In this study, we demonstrate that expression of full-length apoE4 induces mitochondrial dysfunction in both N2A cells and cultured cortical neurons but not in astrocytes. In our N2A stable transfectants of full-length apoE4, most of the intracellular apoE4 was localized to the endoplasmic reticulum and Golgi apparatus (32); however, it remains possible that trace amounts of apoE4 proteolytic fragments may be targeted to mitochondria and perturb mitochondrial function.
The mechanism of apoE4-induced mitochondrial dysfunction is not known in detail. One possibility is that apoE4 perturbs expression of mitochondrial energy metabolism genes. In a gene expression profiling study of postmortem human hippocampus, apoE4 gene expression was found to be associated with down-regulation of gene transcripts of mitochondrial respiratory complexes I, IV, and V from the nuclear genome, compared with apoE3 (44). In our study, expression of apoE4 in N2A cells reduced the levels of mitochondrial gene transcripts from both the nuclear genome (complex V subunit α) and mtDNA (complex IV subunit 1) (Fig. 3). These findings are relevant to AD pathogenesis because gene transcripts for mitochondrial respiratory complexes I–V are also reduced in AD brains (12, 16, 17, 45), especially in the same regions that have reduced glucose metabolism in AD patients and cognitively normal apoE4 carriers (17). Thus, alterations in mitochondrial gene expression may serve as markers for the disease progression of AD. It would be of interest to determine whether up-regulating the expression of mitochondrial energy metabolism genes can protect against apoE4-induced mitochondrial dysfunction and mitigate AD pathology.
Mitochondria are emerging as a primary site of action for pathogenic proteins involved in AD pathology including amyloid β, presenilin, and Tau (10, 11, 13, 46). In addition, amyloid β is physically present in mitochondria (46) and is associated with impaired activity of mitochondrial respiratory complex IV, both in vitro and in vivo (47–49). We found that apoE4 induces mitochondrial respiratory dysfunction independently of human amyloid β. Because apoE4 carriers develop AD-like cerebral glucose hypometabolism decades before the onset of clinical features of AD (21), apoE4-induced mitochondrial dysfunction is likely an early pathogenic event. ApoE4-induced mitochondrial dysfunction may lower the threshold for other mitochondrial stressors such as amyloid β and mutant Tau to induce AD pathology. More importantly, because apoE4 causes mitochondrial dysfunction through its domain interaction, small molecules that interfere with this interaction hold great therapeutic potential for the majority of late-onset AD patients (65–80%) who carry at least one apoE4 allele.
Acknowledgments
We thank James McGuire, Sandra Almeida, and Aurawan Vongs for technical assistance; Sylvia Richmond, Mimi Zeiger, and Stephen Ordway for manuscript preparation and editorial assistance; and John C. W. Carroll for graphics.
This work was supported in part by Merck Research Laboratories.
- apo
- apolipoprotein
- AD
- Alzheimer disease
- CCCP
- carbonylcyanide-3-chlorophenylhydrazone
- GFAP
- glial fibrillary acidic protein
- MtHsp70
- mitochondrial heat shock protein 70
- N2A
- Neuro-2A
- NSE
- neuron-specific enolase
- OCR
- oxygen consumption rate
- VDAC
- voltage-dependent anion channels
- ANOVA
- analysis of variance.
REFERENCES
- 1. Corder E. H., Saunders A. M., Strittmatter W. J., Schmechel D. E., Gaskell P. C., Small G. W., Roses A. D., Haines J. L., Pericak-Vance M. A. (1993) Science 261, 921–923 [DOI] [PubMed] [Google Scholar]
- 2. Strittmatter W. J., Saunders A. M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G. S., Roses A. D. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1977–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mahley R. W., Weisgraber K. H., Huang Y. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5644–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mahley R. W., Huang Y. (2009) Ann. Neurol. 65, 623–625 [DOI] [PubMed] [Google Scholar]
- 5. Mahley R. W., Weisgraber K. H., Huang Y. (2009) J. Lipid Res. 50, S183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim J., Basak J. M., Holtzman D. M. (2009) Neuron 63, 287–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dong L. M., Wilson C., Wardell M. R., Simmons T., Mahley R. W., Weisgraber K. H., Agard D. A. (1994) J. Biol. Chem. 269, 22358–22365 [PubMed] [Google Scholar]
- 8. Dong L. M., Weisgraber K. H. (1996) J. Biol. Chem. 271, 19053–19057 [DOI] [PubMed] [Google Scholar]
- 9. Ye S., Huang Y., Müllendorff K., Dong L., Giedt G., Meng E. C., Cohen F. E., Kuntz I. D., Weisgraber K. H., Mahley R. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18700–18705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin M. T., Beal M. F. (2006) Nature 443, 787–795 [DOI] [PubMed] [Google Scholar]
- 11. Hauptmann S., Keil U., Scherping I., Bonert A., Eckert A., Müller W. E. (2006) Exp. Gerontol. 41, 668–673 [DOI] [PubMed] [Google Scholar]
- 12. Parihar M. S., Brewer G. J. (2007) Am. J. Physiol. Cell Physiol. 292, C8–23 [DOI] [PubMed] [Google Scholar]
- 13. Atamna H., Frey W. H., 2nd (2007) Mitochondrion 7, 297–310 [DOI] [PubMed] [Google Scholar]
- 14. Minoshima S., Giordani B., Berent S., Frey K. A., Foster N. L., Kuhl D. E. (1997) Annals Neurol. 42, 85–94 [DOI] [PubMed] [Google Scholar]
- 15. Mosconi L., Pupi A., De Leon M. J. (2008) Ann. N.Y. Acad. Sci. 1147, 180–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de la Monte S. M., Wands J. R. (2006) J. Alzheimers Dis. 9, 167–181 [DOI] [PubMed] [Google Scholar]
- 17. Liang W. S., Reiman E. M., Valla J., Dunckley T., Beach T. G., Grover A., Niedzielko T. L., Schneider L. E., Mastroeni D., Caselli R., Kukull W., Morris J. C., Hulette C. M., Schmechel D., Rogers J., Stephan D. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4441–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Valla J., Berndt J. D., Gonzalez-Lima F. (2001) J. Neurosci. 21, 4923–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wallace D. C. (2005) Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Small G. W., Mazziotta J. C., Collins M. T., Baxter L. R., Phelps M. E., Mandelkern M. A., Kaplan A., La Rue A., Adamson C. F., Chang L., Guze B. H., Corder E. H., Saunders A. M., Haines J. L., Pericak-Vance M. A., Roses A. D. (1995) J. Am. Med. Assoc. 273, 942–947 [PubMed] [Google Scholar]
- 21. Reiman E. M., Chen K., Alexander G. E., Caselli R. J., Bandy D., Osborne D., Saunders A. M., Hardy J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 284–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reiman E. M., Chen K., Alexander G. E., Caselli R. J., Bandy D., Osborne D., Saunders A. M., Hardy J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8299–8302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mahley R. W. (1988) Science 240, 622–630 [DOI] [PubMed] [Google Scholar]
- 24. Huang Y., Weisgraber K. H., Mucke L., Mahley R. W. (2004) J. Mol. Neurosci. 23, 189–204 [DOI] [PubMed] [Google Scholar]
- 25. Xu Q., Bernardo A., Walker D., Kanegawa T., Mahley R. W., Huang Y. (2006) J. Neurosci. 26, 4985–4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buttini M., Orth M., Bellosta S., Akeefe H., Pitas R. E., Wyss-Coray T., Mucke L., Mahley R. W. (1999) J. Neurosci. 19, 4867–4880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raber J., Wong D., Buttini M., Orth M., Bellosta S., Pitas R. E., Mahley R. W., Mucke L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10914–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hartman R. E., Wozniak D. F., Nardi A., Olney J. W., Sartorius L., Holtzman D. M. (2001) Exp. Neurol. 170, 326–344 [DOI] [PubMed] [Google Scholar]
- 29. Brecht W. J., Harris F. M., Chang S., Tesseur I., Yu G.-Q., Xu Q., Fish J. D., Wyss-Coray T., Buttini M., Mucke L., Mahley R. W., Huang Y. (2004) J. Neurosci. 24, 2527–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang Y., Liu X. Q., Wyss-Coray T., Brecht W. J., Sanan D. A., Mahley R. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8838–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harris F. M., Tesseur I., Brecht W. J., Xu Q., Mullendorff K., Chang S., Wyss-Coray T., Mahley R. W., Huang Y. (2004) J. Biol. Chem. 279, 3862–3868 [DOI] [PubMed] [Google Scholar]
- 32. Chang S., ran Ma T., Miranda R. D., Balestra M. E., Mahley R. W., Huang Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18694–18699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cardoso S. M., Proença M. T., Santos S., Santana I., Oliveira C. R. (2004) Neurobiol. Aging 25, 105–110 [DOI] [PubMed] [Google Scholar]
- 34. Wu M., Neilson A., Swift A. L., Moran R., Tamagnine J., Parslow D., Armistead S., Lemire K., Orrell J., Teich J., Chomicz S., Ferrick D. A. (2007) Am. J. Physiol. Cell Physiol. 292, C125-C136 [DOI] [PubMed] [Google Scholar]
- 35. Richter O. M., Ludwig B. (2003) Rev. Physiol. Biochem. Pharmacol. 147, 47–74 [DOI] [PubMed] [Google Scholar]
- 36. Davey G. P., Peuchen S., Clark J. B. (1998) J. Biol. Chem. 273, 12753–12757 [DOI] [PubMed] [Google Scholar]
- 37. Mozo J., Ferry G., Studeny A., Pecqueur C., Rodriguez M., Boutin J. A., Bouillaud F. (2006) Biochem. J. 393, 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhong N., Weisgraber K. H. (2009) J. Biol. Chem. 284, 6027–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weisgraber K. H. (1994) Adv. Protein Chem. 45, 249–302 [DOI] [PubMed] [Google Scholar]
- 40. Raffaï R. L., Dong L. M., Farese R. V., Jr., Weisgraber K. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11587–11591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhong N., Scearce-Levie K., Ramaswamy G., Weisgraber K. H. (2008) Alzheimer Dementia 4, 179–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mahley R. W., Hui D. Y., Innerarity T. L., Beisiegel U. (1989) Arteriosclerosis 9, I-14–I-18 [PubMed] [Google Scholar]
- 43. Nakamura T., Watanabe A., Fujino T., Hosono T., Michikawa M. (2009) Mol. Neurodegener. 4, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu P.-T., Li Y.-J., Qin X. J., Scherzer C. R., Xu H., Schmechel D. E., Hulette C. M., Ervin J., Gullans S. R., Haines J., Pericak-Vance M. A., Gilbert J. R. (2006) Neurobiol. Dis. 21, 256–275 [DOI] [PubMed] [Google Scholar]
- 45. Bonilla E., Tanji K., Hirano M., Vu T. H., DiMauro S., Schon E. A. (1999) Biochim. Biophys. Acta 1410, 171–182 [DOI] [PubMed] [Google Scholar]
- 46. LaFerla F. M., Green K. N., Oddo S. (2007) Nat. Rev. Neurosci. 8, 499–509 [DOI] [PubMed] [Google Scholar]
- 47. Keil U., Bonert A., Marques C. A., Scherping I., Weyermann J., Strosznajder J. B., Müller-Spahn F., Haass C., Czech C., Pradier L., Müller W. E., Eckert A. (2004) J. Biol. Chem. 279, 50310–50320 [DOI] [PubMed] [Google Scholar]
- 48. Devi L., Prabhu B. M., Galati D. F., Avadhani N. G., Anandatheerthavarada H. K. (2006) J. Neurosci. 26, 9057–9068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rhein V., Song X., Wiesner A., Ittner L. M., Baysang G., Meier F., Ozmen L., Bluethmann H., Dröse S., Brandt U., Savaskan E., Czech C., Götz J., Eckert A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]