Abstract
The therapeutic and diagnostic efficiency of engineered small proteins, peptides, and chemical drug candidates is hampered by short in vivo serum half-life. Thus, strategies to tailor their biodistribution and serum persistence are highly needed. An attractive approach is to take advantage of the exceptionally long circulation half-life of serum albumin or IgG, which is attributed to a pH-dependent interaction with the neonatal Fc receptor (FcRn) rescuing these proteins from intracellular degradation. Here, we present molecular evidence that a minimal albumin binding domain (ABD) derived from streptococcal protein G can be used for efficient half-life extension by indirect targeting of FcRn. We show that ABD, and ABD recombinantly fused to an Affibody molecule, in complex with albumin does not interfere with the strictly pH-dependent FcRn-albumin binding kinetics. The same result was obtained in the presence of IgG. An in vivo study performed in rat confirmed that the clinically relevant human epidermal growth factor 2 (HER2)-targeting Affibody molecule fused to ABD has a similar half-life and biodistribution profile as serum albumin. The proof-of-concept described may be broadly applicable to extend the in vivo half-life of short lived biological or chemical drugs ultimately resulting in enhanced therapeutic or diagnostic efficiency, a more favorable dosing regimen, and improved patient compliance.
Keywords: Drug Design, Drug Metabolism, Endosomal pH Function, Immunology, Ligand-binding Protein, Membrane Recycling, Protein Degradation, Protein Drug Interactions, Protein Targeting, Receptor Recycling
Introduction
Molecular in vitro selection technologies such as phage display and ribosome display have generated an array of novel therapeutically promising small scaffold proteins and peptides with specificity toward signaling molecules as well as tumor surface antigens. Despite encouraging results from in vitro experimental screenings and preclinical animal trials, their therapeutic efficiency is limited by a short serum half-life, ranging from minutes to a few hours (1–4). The main reasons for this rapid elimination are their small molecular size, below the renal clearance threshold, as well as susceptibility to degradation by serum and intracellular proteases.
However, a number of strategies have been developed to improve the pharmacokinetic properties of therapeutics. These include increasing the molecular size by chemical modifications such as conjugation with polyethylene glycol (5, 6) or genetic fusion to human serum albumin (HSA)2 (7–9) or the Fc portion of human IgG (hIgG) (10). In addition, noncovalent association with albumin or IgG has been explored as an alternative to direct fusion. Pioneering approaches included fusion to naturally occurring albumin binding domains derived from SpG, and an increased in vivo half-life was demonstrated in mice, rats, and primates (11, 12). Since then, a minimal three-helical albumin-binding module within SpG has been widely used as a fusion partner for Fab fragments (13, 14), single chain diabodies (15, 16), and Affibody molecules (17). Other prominent albumin targeting molecules, selected by phage display technology, include the albumin-binding peptide developed by Dennis and co-workers (18–20) and the AlbudAbs, domain antibodies with specificity for albumin, developed by Holt et al. (21) and Walker et al. (22).
The incentives for targeting albumin and IgG are that they constitute the most abundant serum proteins in blood, and they both have an extraordinary long half-life of ∼2–3 weeks in humans (23, 24). In addition to having a molecular size above the renal clearance threshold, the long half-lives are attributed to the efficient receptor-mediated recycling pathway involving the neonatal Fc receptor (FcRn) (25–27). FcRn is a major histocompatibility class I-related protein that resides predominantly within acidified endosomes of endothelial and hematopoietic cells (28–31). It interacts with IgG and albumin in a strictly pH-dependent manner, i.e. binding at acidic pH and no binding or release at physiological pH. Pinocytosed IgG and albumin bound by the receptor within acidified endosomes are transported back to the cell surface where the physiological pH of the blood triggers release of the ligands into the blood circulation. The intracellular nonbound fractions are targeted for lysosomal degradation (30, 32, 33).
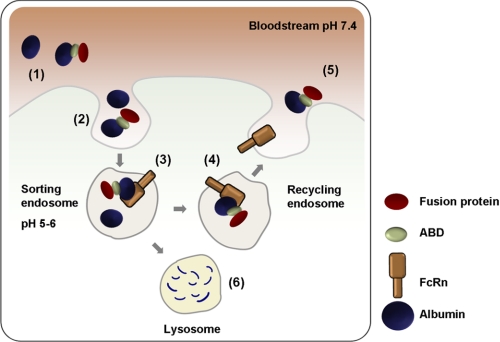
The strategy of indirect targeting of FcRn is schematically illustrated in Fig. 1. Several basic criteria must be met to achieve successful co-recycling of ABD fusion proteins. First, the binding sites for ABD and FcRn on albumin must be nonoverlapping. Second, albumin must not undergo any conformational changes upon binding of ABD or FcRn that prevents or disrupts binding of the other molecule. Third, the pH-dependent interaction between albumin and FcRn must be preserved, and fourth, the ABD fusion protein must remain bound to albumin at the acidic pH of the endosome. As for the first criterion, the binding sites for ABD and FcRn on HSA are known to be distally located. The 67-kDa HSA consists of three independently folding domains denoted I, II, and III. Structural data of a complex between HSA and an ABD homologue (the GA domain of protein PAB) derived from Finegoldia magna revealed that helices two and three of ABD interact with domain II of HSA (34). The interaction site between HSA and FcRn has not been mapped in detail, but deletion studies have located the FcRn-binding site to domain III of HSA (35, 36).
FIGURE 1.
Schematic illustration of ABD-based indirect targeting of FcRn. Schematic represents a hematopoietic or endothelial cell surrounded by blood containing large amounts of albumin (∼40 mg/ml) and a minor fraction of albumin that is associated with an exogenously given ABD fusion protein (1). Both albumin and ABD-associated albumin are continually taken up by fluid phase endocytosis (2). FcRn resides predominantly within acidified intracellular compartments where the pH triggers binding of albumin and ABD-albumin complexes to the FcRn heavy chain. The acidic pH herein does not affect the interaction between ABD and albumin (3). The complex is then recycled back to the cell surface through a pH gradient until it is exposed to the physiological pH of the blood that subsequently disrupts the binding affinity for FcRn (4) followed by release of albumin and ABD-albumin complexes back into the bloodstream (5). Albumin and ABD fusions that escape binding to FcRn within in the acidified recycling compartments will go to lysosomal degradation (6).
In this study, we have explored the prerequisites for using the ABD as a general carrier molecule for half-life extension of short lived proteins. We have performed interaction studies by enzyme-linked immunosorbent assay (ELISA) and surface plasmon resonance (SPR) to ensure that ABD in complex with albumin does not interfere with pH-dependent binding of FcRn to either albumin or IgG. The interaction studies were repeated with the ABD amino- or carboxyl-terminally fused to a mono- or divalent Affibody molecule (37) targeting HER2 (38), and its ligand binding properties were investigated. Finally, the biodistribution and blood clearance rate of isotope-labeled ABD fusion protein were compared with that of rat serum albumin (RSA) injected simultaneously into rats, confirming a similar half-life.
EXPERIMENTAL PROCEDURES
Construction and Production of Soluble FcRn Variants
The construction of a truncated human FcRn mutant variant (C48S/C251S) cDNA encoding the three ectodomains (α1–α3) genetically fused to a cDNA encoding the Schistosoma japonicum glutathione S-transferase (GST) has been described previously (39, 40). A mutant hFcRn heavy chain variant, denoted H166A, was constructed using the template described in Ref. 40 followed by site-directed mutagenesis (41). A vector containing a cDNA segment encoding a truncated mouse FcRn heavy chain (pcDNA3-mFcRn-GST-hβ2m-oriP) was constructed as described previously (42). A rat liver cDNA library (Zyagen) was used to PCR-amplify a cDNA encoding a truncated version of the rat FcRn heavy chain (including the endogenous native leader sequence, α1, α2, and α3 domains; 293 amino acids) using the primers rFcRnForw (5-att gaa ttc acc atg ggg atg tcc cag ccc ggg-3) and rFcRnRev (5-ata tac tcg agt agg tcc aca gtg aga ggc tg-3). Primers were designed to allow in-frame ligation of the fragment upstream of a cDNA encoding a GST tag into the pcDNA3-GST-hβ2m-oriP vector. The final vector was sequenced and denoted pcDNA3-rFcRn-GST-hβ2m-oriP. All vectors described above also contain a cDNA encoding human β2-microglobulin and the Epstein-Barr virus origin of replication (oriP). The soluble FcRn variants were produced in HEK 293E cells, and secreted receptors were purified using a GSTrap column as described previously (40).
Construction and Production of Isolated and Affibody-fused ABD Variants
ABD was cloned and produced essentially as described previously (43). cDNA fragments of ZHER2:342 (44) were PCR-amplified with AccI overhangs and inserted into AccI-digested vectors with a sequence encoding ABD either upstream (pAY00540) or downstream (pAY01138) of the restriction site. The resulting vectors encoding ZHER2:342-ABD and ABD-ZHER2:342, respectively, were transformed into Escherichia coli BL21(DE3)-competent cells, and proteins were produced by fermentation. Pelleted cells were solved in 25 mm Tris-HCl, 200 mm NaCl, 1 mm EDTA, pH 8.0, and disrupted by sonication on ice. The clarified supernatants were affinity-purified on HSA (Sigma) in-house coupled to CNBr-activated Sepharose (GE Healthcare). Washing was performed with 1× TST (25 mm Tris-HCl, 200 mm NaCl, 1 mm EDTA, 0.05% Tween 20, pH 8.0) followed by 5 mm NH4Ac, pH 5.5. Bound proteins were eluted with 0.5 m HAc, pH 2.8. ABD-ZHER2:342 was buffer-exchanged to 0.5× PBS (1.34 mm KCl, 0.74 mm KH2PO4, 68.5 mm NaCl, 4.05 mm Na2HPO4, pH 7.4) on a column packed with Sephadex G-25 medium (GE Healthcare), and ZHER2:342-ABD was further purified by reverse phase chromatography on a 3-ml SourceTM 15 RPC column (GE Healthcare). 2% acetonitrile, 0.065% TFA in water was used as running buffer, and protein was eluted using a linear gradient of 0–50% of 80% acetonitrile, 0.05% TFA in water over 30 min. Buffer exchange to 5 mm sodium phosphate, 150 mm NaCl, pH 7.2, was performed using a NAP-5 column (GE Healthcare).
Cloning and production of ABD-(ZHER2:342)2 have been described previously (17), and (ZHER2:342)2-ABD was produced likewise.
Competitive ELISA Analyses
Wells of MaxiSorp ELISA plates (NUNC) were coated with 100 μl of 3-iodo-4-hydroxy-5-nitrophenacetyl (NIP)-conjugated bovine serum albumin (BSA) at 1 μg/ml (a kind gift from T. E. Michaelsen, National Institute of Public Health, Norway) and incubated overnight at 4 °C. The wells were then blocked with 4% skimmed milk (Skm; Acumedia) diluted in PBS for 1 h at room temperature (RT) and washed four times with 1× PBS with 0.005% Tween 20 (PBS/T; Sigma). Portions of hIgG1 anti-NIP (0.25 μg/ml, also a generous gift from T. E. Michaelsen) diluted in 4% Skm/PBS/T were added, incubated for 1 h at RT, and then washed four times with PBS/T, pH 6.0. Purified shFcRn-GST at 0.25 μg/ml was added alone or together with titrated amounts of 1000 to 4 nm ABD, HSA, or hIgG1 with irrelevant specificity (hIgGIr; palivizumab, MedImmune, Inc., Gaithersburg, MD). The reactions were incubated for 1 h at RT and washed four times with PBS/T, pH 6.0. An HRP-conjugated anti-GST IgG (from goat, polyclonal; GE Healthcare), diluted 1:5000 in 4% Skm/PBS/T, pH 6.0, was added to each well. The plates were incubated for 1 h at RT followed by four washes with PBS/T, pH 6.0. Bound shFcRn-GST was detected by adding 100 μl of the substrate 3,3′,5,5′-tetramethylbenzidine (Calbiochem) to each well. The absorbance was measured at 620 nm using Sunrise TECAN spectrophotometer (TECAN). The same ELISA was performed with monomeric HSA (Sigma) directly coated in wells followed by detection of bound shFcRn-GST in the absence or presence of ABD, hIgGIr, or monomeric HSA.
ELISA Analyses
Wells of MaxiSorp ELISA plates (Nunc) were coated with serial dilutions (2 to 0.027 μg/ml) of ABD or the fusion variants described above and incubated overnight at 4 °C. The wells were then blocked with 4% Skm diluted in PBS for 1 h at RT and washed four times with PBS/T, pH 6.0. Monomeric HSA, RSA, or MSA (50 μg/ml) diluted in 4% Skm/PBS/T, pH 6.0 was added to each well, incubated for 1 h at RT, and washed four times with PBS/T, pH 6.0. Purified shFcRn-GST (0.5 μg/ml), shFcRn H166A-GST (2 μg/ml), smFcRn-GST (1.0 μg/ml), or srFcRn-GST (1.0 μg/ml) was preincubated with an HRP-conjugated anti-GST IgG (GE Healthcare; diluted 1:5000) in 4% Skm/PBS/T, pH 6.0, and added to each well. After incubation for 1 h at RT the wells were washed four times with PBS/T, pH 6.0, and bound receptors were detected by adding 100 μl of the substrate 3,3′,5,5′-tetramethylbenzidine (Calbiochem) to each well. The absorbance was measured at 620 or 450 nm using Sunrise spectrophotometer (TECAN). The same ELISA was performed in the presence of 1 μm of palivizumab and also using PBS buffer at pH 7.4 in all steps.
Binding Studies Using Surface Plasmon Resonance
For the shFcRn binding studies a Biacore 3000 instrument (GE Healthcare) was used. CM5 sensor chips were coupled with shFcRn-GST (∼600–1000 RU) or smFcRn-GST (∼600–1000 RU) using amine coupling chemistry as described by the manufacturer. The coupling was performed by injecting 10–12 μg/ml of each protein into 10 mm sodium acetate, pH 4.5 (GE Healthcare). All experiments were performed in phosphate buffer (67 mm phosphate buffer, 0.15 m NaCl, 0.005% Tween 20) at pH 6.0.
Portions of 1 μm of size exclusion chromatography-isolated monomeric HSA (Sigma) or MSA (Calbiochem) were injected, alone or together, with the ABD or ABD fusion variants (1–2 μm) over immobilized receptor with a flow rate of 40 μl/min at 25 °C. The surfaces were gently regenerated by dissociation of bound molecules using buffer, pH 6.0. As controls, the ABD and ABD fusions were injected over immobilized receptors.
HER2 binding studies were performed using a Biacore 2000 instrument (GE Healthcare). CM5 sensor chips were coupled with HER2-Fc (R&D Systems; 2430 RU) using amine coupling chemistry, and 400 and 80 nm (ZHER2:342)2-ABD, ABD-(ZHER2:342)2, and nonfused (ZHER2:342)2 preincubated with 4 μm HSA were injected in duplicates at a flow rate of 50 μl/min. The experiments were performed in HBS-EP buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% Surfactant P20) including 4 μm HSA, and the surfaces were regenerated using 25 mm HCl. The same experiments were performed in the absence of HSA.
In all experiments, data were zero adjusted, and the reference cell value was subtracted. Binding analyses were performed using the BIAevaluation 4.1 Wizard.
Radiolabeling of RSA and ABD-(ZHER2:342)2 for Dual Label Pharmacokinetic Study
Metal impurities were removed from buffers using a Chelex-100 resin (sodium form, Bio-Rad). The chelator CHX-A″/DTPA (Macrocyclics, Dallas, TX) was coupled to RSA (Sigma) or ABD-(ZHER2:342)2 at a 1:1 molar ratio to avoid overmodification; 200 μl of RSA (5 mg/ml in 0.07 m borate buffer, pH 9.2) was mixed with 10 μl of a freshly prepared solution of CHX-A″/DTPA (1 mg/ml in 0.07 m borate buffer, pH 9.2), and 290 μl of 0.07 m borate buffer, pH 9.2, was added to the mixture. Likewise, 330 μl of ABD-(ZHER2:342)2 (1.66 mg/ml in PBS) was mixed with 20 μl of CHX-A″/DTPA, and 150 μl of borate buffer was added. The mixtures were incubated for 18 h at 37 °C. Buffer exchange and removal of the unreacted chelator were performed using disposable NAP-5 columns (GE Healthcare) pre-equilibrated and eluted with 1.0 m ammonium acetate buffer, pH 5.5, according to the manufacturer's instructions.
900 μl (900 μg) of CHX-A″/DTPA/RSA solution was mixed with 5 μl of (3.5 MBq) 111InCl3 in 0.05 m HCl (Tyco Mallinckrodt, The Netherlands) The mixture was incubated at RT for 30 min, and the labeling efficiency was checked using ITLC SG (Pall Corp., East Hills, NY) eluted with 0.2 m citric acid. With a labeling efficiency of 99.9%, additional purification was not necessary. The solution was diluted to 1 ml with PBS.
500 μl (45 μg) of CHX-A″/DTPA/ABD-(ZHER2:342)2 solution was mixed with 5.3 μl (3 MBq) of 177LuCl3 solution in 0.05 m HCl (IDB Holland, The Netherlands). The mixture was incubated at RT for 30 min resulting in a labeling efficiency of 99.4%, as determined above. The solution was diluted to 1 ml with PBS.
An injection formulation was prepared by mixing 55 μl of the 177Lu-CHX-A″/DTPA/ABD-(ZHER2:342)2 solution with 115 μl of the 111In-CHX-A″/DTPA/RSA solution, followed by dilution with 8.83 ml of sterile PBS.
Biodistribution of 111In-Labeled RSA and 177Lu-Labeled ABD Fusion Protein
The animal study was approved by the Uppsala Committee of Animal Research Ethics. Female Sprague-Dawley rats (Taconic, Lille Skensved, Denmark, 6 weeks old at arrival) were acclimatized for 1 week. Animals (mean 240 g, range 227–250 g) were anesthetized using isoflurane, and an intravenous catheter was introduced into the tail vein for subsequent injection of premixed 177Lu-CHX-A″/DTPA/ABD-(ZHER2:342)2 (3.6 kBq, 0.06 μg) and 111In-CHX-A″/DTPA/RSA (9 kBq, 2.3 μg) in a total volume of 0.2 ml. Animals (n = 3 at each time point) were sacrificed at 0.25, 4, 20, 48, 72, 168, and 240 h post-injection, by means of intravenous pentobarbital. Samples of blood, hind leg muscle, and skin, as well as whole liver and kidneys, were excised and weighed, and radioactivity was measured using an automatic γ-spectrometer (1480 Wizard, Wallac Oy, Turku, Finland). Whole spectra of each sample, as well as a standard of injected solution and authentic samples of 177Lu and 111In, were recorded. Radioactivity of 177Lu was measured as total counts in the energy region 60–130 keV and radioactivity of 111In in the region 380–460 keV. Data were corrected for background, dead time, and spillover. Uptake was calculated and expressed as percent of injected radioactivity/g.
Volumes of distribution and distributional clearance were assessed by an i.v. bolus two-compartment model (WinNonLin 4.0, Pharsight). Data are expressed as regression mean ± S.D.
The space accessible for plasma proteins in the interstitium and blood is limited due to tissue exclusion and blood cells, respectively. Thus, the actual concentration in the accessible space is higher than that calculated from tissue weight. Concentration in muscle, skin, and plasma was therefore adjusted for this based on data from albumin distribution (45) and blood plasma fraction (46). In hind limb skin, the fractional interstitial volume and fractional interstitial exclusion of albumin are 43 and 45%, respectively, whereas corresponding values for hind limb muscle are 6.1 and 27%, respectively (45). Hence, an applied volume correction factor for skin is 1/0.43/(1–0.45) = 4.2 and for muscle 1/0.061/(1–0.27) = 22.5. The plasma fraction in blood is 54% and the plasma correction factor is thus 1/0.54 = 1.85 (46).
RESULTS
ABD in Complex with Albumin Does Not Interfere with shFcRn Binding
The impact of ABD on the pH-dependent ligand binding of shFcRn was analyzed by SPR and by a set of ELISA experiments. First, the binding of shFcRn to HSA in the presence of ABD was investigated. SPR was performed at pH 6.0 with shFcRn covalently immobilized on a CM5 biosensor chip. Injection of monomeric HSA preincubated with ABD resulted in additive and reversible binding of the complex to the receptor (Fig. 2A). Importantly, the binding kinetics of HSA in complex with ABD completely resembled that of HSA injected alone, except from a higher response reflecting the increase in molecular weight of the complex. At pH 7.4, binding of neither the complex nor HSA alone was detected (data not shown).
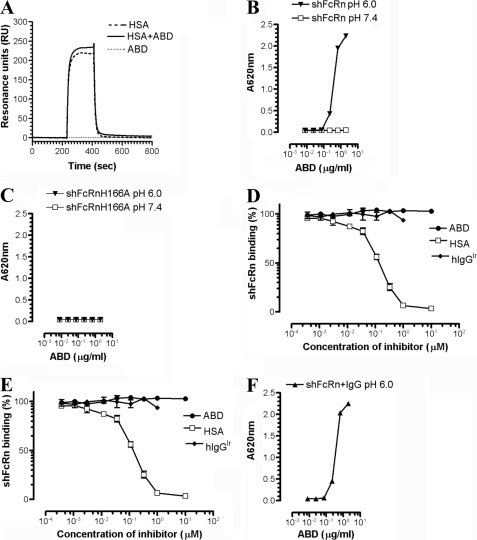
FIGURE 2.
ABD in complex with HSA does not affect the pH-dependent binding of shFcRn to HSA or IgG. A, representative SPR sensorgrams showing the binding responses of 1 μm of monomeric HSA, 2 μm of ABD, and HSA in complex with ABD when injected over immobilized shFcRn (∼1500 RU) at pH 6.0. Injections were performed at a flow rate of 40 μl/min at 25 °C. B, ELISA measurements showing pH-dependent binding of shFcRn to HSA in complex with ABD. C, ELISA measurements showing no binding of shFcRnH166A to HSA in complex with ABD. D, competitive ELISAs show the inhibitory responses of ABD, HSA, and IgGIr on HSA. E, hIgG1NIP binds to shFcRn at pH 6.0. F, ELISA measurements showing pH-dependent binding of shFcRn to HSA in complex with ABD in the presence of hIgG1. The numbers given represent the mean of triplicates.
In the ELISA, titrated amounts of HSA were captured on ABD coated directly in MaxiSorp wells. HSA in complex with ABD was shown to bind shFcRn in a strictly pH-dependent manner, binding at pH 6.0 but no detectable binding at pH 7.4 (Fig. 2B). An shFcRn mutant, H166A, which previously was shown to completely eliminate HSA binding while retaining the hIgG binding property (41), did not bind HSA when captured on ABD (Fig. 2C). Furthermore, a competitive ELISA in which wells were coated with monomeric HSA followed by preincubation of shFcRn with titrated amounts of ABD, HSA, or hIgG1 with irrelevant specificity (hIgGIr) at pH 6.0 showed that high amounts of ABD did not influence the binding of shFcRn to HSA (Fig. 2D).
Because FcRn is a bifunctional receptor that binds IgG and albumin simultaneously and rescues both proteins from intracellular degradation (25–27), the impact of ABD on shFcRn binding to IgG was also investigated. Human-mouse chimeric IgG1 with specificity for the hapten NIP (hIgG1NIP) was captured on NIP-conjugated BSA coated in MaxiSorp wells. Importantly, the NIP-conjugated BSA did not bind shFcRn under the conditions used (data not shown). Titrated amounts of ABD, HSA, and hIgGIr preincubated with shFcRn at pH 6.0 showed that neither ABD nor HSA affected receptor binding to hIgG1NIP (Fig. 2E). Inversely, a mixture of preformed shFcRn-hIgG1 complexes was shown to bind HSA captured on ABD as efficiently as in the absence of hIgG1 (Fig. 2F).
ABD Fusion Proteins and Impact on pH-dependent FcRn Binding
For therapeutic or diagnostic purposes, it was of interest to couple an effector molecule to the ABD molecule. Therefore, we next extended the approach and accordingly investigated whether a protein genetically fused to ABD would interfere with shFcRn binding of HSA. The Affibody molecule ZHER2:342 (38, 44), which selectively binds the HER2 oncogene protein, was chosen. HER2 is a highly relevant target because it is overexpressed in a number of malignant phenotypes such as carcinomas of breast and ovary (38, 47). Notably, the ZHER2:342 molecule binds to a different epitope than the anti-HER2 monoclonal IgG antibody trastuzumab (Herceptin®) registered for cancer treatments (38).
The ZHER2:342 molecule was genetically fused to ABD in a divalent fashion at either the amino or carboxyl terminus of ABD, as illustrated in Fig. 3A. The bacterially produced fusion proteins were purified with expected molecular weights (supplemental Fig. 1) and used in ELISA and SPR analyses in the same way as described above. Neither binding of (ZHER2:342)2-ABD nor ABD-(ZHER2:342)2 to HSA interfered with pH-dependent binding of shFcRn to HSA. As before, the presence of hIgG1 had no impact on the interaction (Fig. 3, B and C). Furthermore, SPR showed that the binding kinetics was essentially unaffected by fusion to ABD, and additive, reversible, and pH-dependent binding was seen when complexes of ABD fusion protein and HSA were injected over immobilized shFcRn (Fig. 3, D and E).
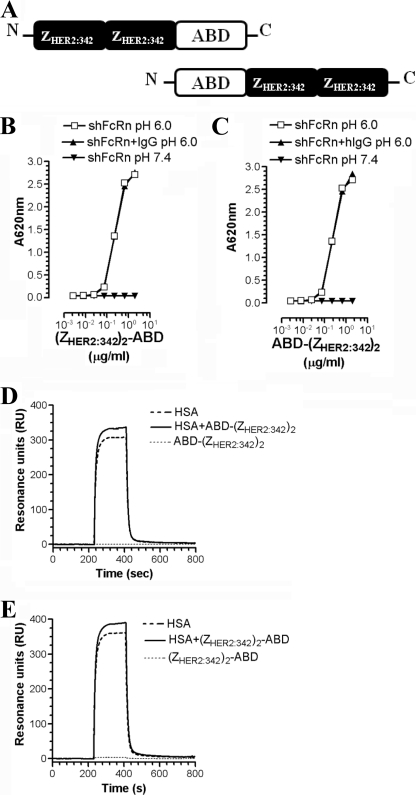
FIGURE 3.
Genetic fusion of bivalent (ZHER2:342)2 to ABD does not interfere with the pH-dependent binding of HSA to shFcRn. A, illustration of the divalent (ZHER2:342)2 fused to the amino- or carboxyl-terminal end of ABD. B, ELISA measurements showing pH-dependent binding of shFcRn to HSA in complex with (ZHER2:342)2-ABD, and C, ABD-(ZHER2:342)2 in the absence or presence of hIgG1 at pH 6.0 and 7.4. The numbers given represent the mean of triplicates. Representative SPR sensorgrams showing the binding responses of 1 μm of monomeric HSA in complex with 2 μm (ZHER2:342)2-ABD (D) and 2 μm ABD-(ZHER2:342)2 (E) when injected over immobilized shFcRn (∼1500 RU) at pH 6.0, flow rate 40 μl/min at 25 °C.
Thus, genetic fusion of a divalent anti-HER2 Affibody molecule to either side of the minimal ABD scaffold has no impact on the binding kinetics of HSA to shFcRn. Similar results were obtained when monomeric ZHER2:342 was fused to the amino- or the carboxyl-terminal end of ABD (supplemental Fig. 2).
Retained Target Binding Capacity of an ABD Fusion Protein in Complex with Albumin
It was investigated whether the HER2 binding properties of (ZHER2:342)2 fused to ABD, either amino- or carboxyl-terminally, had any impact on target binding. Soluble recombinant HER2 was immobilized on a CM5 biosensor chip and 400 and 80 nm of purified ABD-(ZHER2:342)2, (ZHER2:342)2-ABD, or nonfused (ZHER2:342)2 premixed with 4 μm HSA were injected over the surface. Both fusion variants showed retained HER2 binding capacity, although the ABD-(ZHER2:342)2 bound HER2 less efficiently than the (ZHER2:342)2-ABD (Fig. 4 and supplemental Fig. 3). Thus, fusion of ABD to the carboxyl-terminal end of (ZHER2:342)2 is preferable for optimal HER2 targeting.
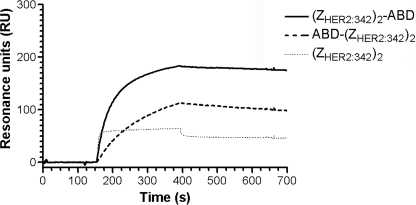
FIGURE 4.
Impact of ABD fusion on HER2 binding. Representative SPR sensorgram showing injections of 80 nm of purified (ZHER2:342)2-ABD, ABD-(ZHER2:342)2, and nonfused (ZHER2:342)2 over immobilized recombinant HER2 (2430 RU) and in the presence of 4 μm HSA. The samples were injected with a flow rate of 50 μl/min at 25 °C.
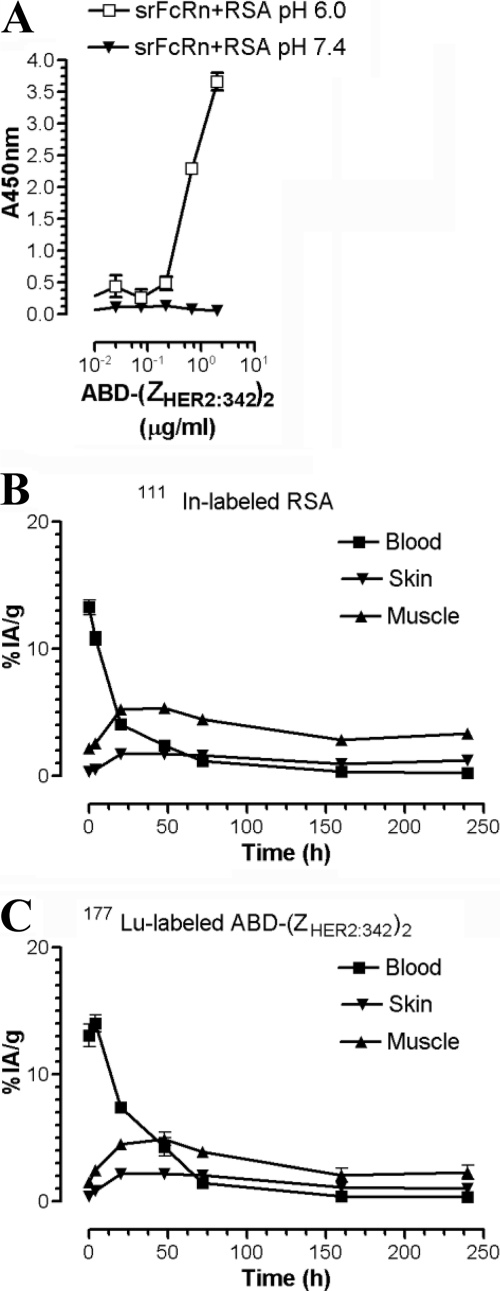
In Vivo Evaluation Shows Similar Biodistribution for the ABD-(ZHER2:342)2 Fusion Protein and RSA
Half-life extension of ABD fusion proteins in vivo requires pH-independent association with endogenous albumin, followed by pH-dependent binding of the complex to FcRn expressed intracellularly within endothelial and hematopoietic cells. A successful co-recycling of the ABD fusion protein according to the illustration in Fig. 1 would result in an extended half-life similar to that of endogenous albumin. To address this issue, a dual label biodistribution study was performed in rat by simultaneous injection of 111In-labeled RSA and 177Lu-labeled ABD-(ZHER2:342)2 and subsequent radioactivity measurement of selected organs up to 250 h post-injection. Importantly, it has previously been shown that rat albumin binds ABD (43), and here we show that srFcRn binds pH-dependently to the RSA in complex with ABD-(ZHER2:342)2 (Fig. 5A). Measured concentrations (percent of injected radioactivity/g) in different organs are listed in supplemental Table 1. The obtained data show similar blood clearance and tissue deposition in skin and muscle for 111In-labeled RSA and 177Lu-labeled ABD-(ZHER2:342)2 (Fig. 5, B and C). Furthermore, the central distribution volumes (13.1 ± 0.8 ml for ABD-(ZHER2:342)2 and 13.5 ± 0.6 ml for RSA) are in accordance with rat blood volume (58 ml/kg) (46).
FIGURE 5.
In vivo biodistribution of ABD-fused (ZHER2:342)2 in a preclinical rat model. A, ELISA measurements showing pH-dependent binding of srFcRn to RSA in complex with ABD-(ZHER2:342)2. The numbers given represent the mean of triplicates. The blood, skin, and muscle biodistribution of 111In-labeled-RSA (B) and 177Lu-labeled-ABD-(ZHER2:342)2 (C) in Sprague-Dawley rats (three rats/group and time point). Organ uptake is expressed as percent of injected radioactivity/g (%IA/g), and error bars indicate the S.E. The concentrations are based on the biodistribution data presented in supplemental Table 1 and corrected for interstitial volume as described under “Experimental Procedures.”
During the first 48 h, the blood concentration of ABD-(ZHER2:342)2 exceeds that of RSA. The distribution clearance was slightly more rapid for RSA than for ABD-(ZHER2:342)2 (data not shown). Corrected tissue concentration in muscle and skin exceeds that of plasma after 72 h for both ABD-(ZHER2:342)2 and RSA.
Taken together, the data indicate that the biodistribution profiles for the two molecules are similar and that ABD-(ZHER2:342)2 displays the wide distribution typical for albumin, as well as extended serum half-life, in contrast to a non-ABD-fused Affibody molecule (44, 48).
DISCUSSION
The use of albumin fusion or albumin binding for the purpose of extending the serum half-life of short lived molecules has been extensively described previously (25, 49, 50). However, no reports have so far addressed the impact of such fusion or targeting in relation to pH-dependent binding to FcRn. To fully benefit from the long half-life of serum albumin, its recycling mechanism mediated by FcRn must not be disrupted.
In this study, we have shown that the ABD in complex with HSA does not affect the strictly pH-dependent binding of shFcRn to HSA or IgG. This indicates that binding of ABD and FcRn to albumin are noncompetitive and with no allosteric effects interfering with binding of one or the other molecule. The same result was obtained with ABD genetically fused to a divalent HER2 targeting Affibody molecule (ZHER2:342)2, either at the amino- or carboxyl-terminal end. Furthermore, the ability to bind albumin was preserved for both fusion variants. This offers flexibility in the design of the fusion protein, which is of key importance because different fusion partners may require either a free amino or carboxyl terminus to retain full functionality when incorporated into a fusion protein with ABD.
For optimal target binding capacity, the (ZHER2:342)2 used in this study should preferably be fused to ABD via its carboxyl-terminal end. This can be explained as follows: the HER2-binding site is located to helix one and two within the three-helical Affibody molecule, and ABD fused to the amino-terminal end of ZHER2:342 causes sterical interference. In the presence of HSA, the ABD fusion proteins showed a slower on-rate and a slightly faster off-rate compared with the nonfused (ZHER2–342)2, which does not associate with HSA. However, when the same experiment was performed in the absence of HSA, the (ZHER2:342)2-ABD fusion protein bound with almost identical kinetics as the (ZHER2–342)2 variant (supplemental Fig. 5). Thus, fusion to ABD as such does not alter the HER2 binding capacity of (ZHER2–342)2. Incorporating a linker between the ABD molecule and the Affibody molecule may improve HER2 binding, in particular for the ABD-(ZHER2–342)2 fusion variant.
The in vitro data demonstrating that ABD or the ABD fusion proteins used in this study do not interfere with the albumin-FcRn interaction is supported by the in vivo dual labeling experiment performed in rat. 177Lu-Labeled ABD-(ZHER2:342)2 exhibited a similar biodistribution profile and blood terminal half-life as 111In-labeled-RSA, except possibly for a slightly slower distribution rate. The latter may be due to the about 30% higher mass of a complex between this molecule and RSA compared with RSA alone. No difference was observed in the elimination rate of ABD-(ZHER2:342)2 compared with RSA. However, using radioactively labeled substrates, it cannot be excluded that unlabeled albumin may have a lower elimination rate. The ABD used in the animal study interacts with rodent albumin with a KD of about 0.1 nm,3 but using an affinity-maturated variant of ABD, such as ABD035, for which the KD of the albumin interaction is about 2 orders of magnitude stronger (43), may cause the elimination rate of the ABD-fused molecule to become identical to elimination of endogenous serum albumin.
A therapeutic molecule fused to ABD in a complex with albumin benefits from an extended half-life, as demonstrated in Tolmachev et al. (17), and it shows a rapid and wide distribution. The latter has an advantage over therapeutic antibodies, for instance, showing slow tissue penetration and more restricted distribution. This study showed that the corrected tissue concentration in muscle and skin exceeds that of plasma after 72 h for both ABD-(ZHER2:342)2 and RSA, equal to the time to reach steady state using continuous infusion of RSA (72 h) (51). This is faster than what has been determined with infusion of IgG (120 h) (52). Thus, if IgG and ABD-(ZHER2:342)2 were labeled with the same isotope, ABD-(ZHER2:342)2 would reach steady state earlier, suggesting a more rapid accumulation of radioactivity in tissues and metastases.
By not interfering with the pH-dependent binding of HSA to shFcRn, ABD meets the fundamental criterion for a carrier molecule to be used for half-life extension based on the FcRn recycling mechanism. Noncovalent association with serum albumin offers several advantages over conjugation to polyethylene glycol and direct fusion to a carrier protein. First, the production may be more cost efficient because ABD is readily produced in bacteria. Second, the small ABD is less likely to interfere with the function of the fused protein. Third, the clearance rate might be modulated by using engineered ABD variants with different affinity. Finally, a major advantage for an ABD fusion is that the ABD binds serum albumin from several species, including albumin from the two important toxicology species, rat and cynomolgus macaques (43). Thus, both toxicology and preclinical efficacy studies are readily performed, in contrast to an HSA fusion protein where a rodent model is inappropriate for preclinical evaluation because mouse and human FcRn show dramatic differences in cross-species ligand binding. Specifically, smFcRn binds very weakly to HSA, whereas human FcRn binds strongly to MSA (42).
Taken together, we have presented in vitro and in vivo data suggesting that ABD can be used as a carrier molecule for half-life extension based on indirect targeting of FcRn. Further support for the proposed mechanism comes from recent data showing that bi-specific single chain diabodies fused to ABD had a 2-fold reduced serum half-life in FcRn heavy chain knock-out mice compared with wild-type mice (14). In line with this is the fact that ABD in complex with MSA binds smFcRn pH dependently (supplemental Fig. 4). We conclude that the ABD fusion technology is a widely applicable strategy for extending the half-life and improving the biodistribution of therapeutics via indirect targeting of FcRn.
Supplementary Material
Acknowledgments
We thank Sathiyaruby Manikam Vadivelu for excellent technical assistance; Dr. Per Jonasson and Dr. Andreas Jonsson for protein production; Anna Sjöberg for help with the Biacore experiments; and Assistant Prof. Anna Orlova for help with the in vivo experiments.
This work was supported by grants from the Steering Board for Research in Molecular Biology, Biotechnology, and Bioinformatics at the University of Oslo (to J. T. A.) and The Norwegian Research Council (to J. T. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Table 1.
A. Jonsson, unpublished data.
- HSA
- human serum albumin
- FcRn
- neonatal Fc receptor
- ABD
- albumin binding domain
- SpG
- streptococcal protein G
- hIgG
- human IgG
- SPR
- surface plasmon resonance
- RSA
- rat serum albumin
- NIP
- 3-iodo-4-hydroxy-5-nitrophenacetyl
- hIgGIr
- hIgG1 with irrelevant specificity
- RT
- room temperature
- Skm
- skimmed milk
- DTPA
- diethylenetriaminepentaacetic acid
- MSA
- mouse serum albumin
- RU
- resonance unit.
REFERENCES
- 1. Binz H. K., Amstutz P., Plückthun A. (2005) Nat. Biotechnol. 23, 1257–1268 [DOI] [PubMed] [Google Scholar]
- 2. Brekke O. H., Sandlie I. (2003) Nat. Rev. Drug Discov. 2, 52–62 [DOI] [PubMed] [Google Scholar]
- 3. McGregor D. P. (2008) Curr. Opin. Pharmacol. 8, 616–619 [DOI] [PubMed] [Google Scholar]
- 4. Nelson A. L., Reichert J. M. (2009) Nat. Biotechnol. 27, 331–337 [DOI] [PubMed] [Google Scholar]
- 5. Harris J. M., Chess R. B. (2003) Nat. Rev. Drug. Discov. 2, 214–221 [DOI] [PubMed] [Google Scholar]
- 6. Yamamoto Y., Tsutsumi Y., Yoshioka Y., Nishibata T., Kobayashi K., Okamoto T., Mukai Y., Shimizu T., Nakagawa S., Nagata S., Mayumi T. (2003) Nat. Biotechnol. 21, 546–552 [DOI] [PubMed] [Google Scholar]
- 7. Chuang V. T., Kragh-Hansen U., Otagiri M. (2002) Pharm. Res. 19, 569–577 [DOI] [PubMed] [Google Scholar]
- 8. Halpern W., Riccobene T. A., Agostini H., Baker K., Stolow D., Gu M. L., Hirsch J., Mahoney A., Carrell J., Boyd E., Grzegorzewski K. J. (2002) Pharm. Res. 19, 1720–1729 [DOI] [PubMed] [Google Scholar]
- 9. Subramanian G. M., Fiscella M., Lamousé-Smith A., Zeuzem S., McHutchison J. G. (2007) Nat. Biotechnol. 25, 1411–1419 [DOI] [PubMed] [Google Scholar]
- 10. Jazayeri J. A., Carroll G. J. (2008) BioDrugs 22, 11–26 [DOI] [PubMed] [Google Scholar]
- 11. Makrides S. C., Nygren P. A., Andrews B., Ford P. J., Evans K. S., Hayman E. G., Adari H., Uhlén M., Toth C. A. (1996) J. Pharmacol. Exp. Ther. 277, 534–542 [PubMed] [Google Scholar]
- 12. Nygren P. A., Flodberg P., Andersson R., Wigzell H., Uhlen M. (1991) Vaccines 91, pp. 363–368, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13. Smith B. J., Popplewell A., Athwal D., Chapman A. P., Heywood S., West S. M., Carrington B., Nesbitt A., Lawson A. D., Antoniw P., Eddelston A., Suitters A. (2001) Bioconjug. Chem. 12, 750–756 [DOI] [PubMed] [Google Scholar]
- 14. König T., Skerra A. (1998) J. Immunol. Methods 218, 73–83 [DOI] [PubMed] [Google Scholar]
- 15. Stork R., Campigna E., Robert B., Müller D., Kontermann R. E. (2009) J. Biol. Chem. 284, 25612–25619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stork R., Müller D., Kontermann R. E. (2007) Protein Eng. Des. Sel. 20, 569–576 [DOI] [PubMed] [Google Scholar]
- 17. Tolmachev V., Orlova A., Pehrson R., Galli J., Baastrup B., Andersson K., Sandström M., Rosik D., Carlsson J., Lundqvist H., Wennborg A., Nilsson F. Y. (2007) Cancer Res. 67, 2773–2782 [DOI] [PubMed] [Google Scholar]
- 18. Dennis M. S., Jin H., Dugger D., Yang R., McFarland L., Ogasawara A., Williams S., Cole M. J., Ross S., Schwall R. (2007) Cancer Res. 67, 254–261 [DOI] [PubMed] [Google Scholar]
- 19. Dennis M. S., Zhang M., Meng Y. G., Kadkhodayan M., Kirchhofer D., Combs D., Damico L. A. (2002) J. Biol. Chem. 277, 35035–35043 [DOI] [PubMed] [Google Scholar]
- 20. Nguyen A., Reyes A. E., 2nd., Zhang M., McDonald P., Wong W. L., Damico L. A., Dennis M. S. (2006) Protein Eng. Des. Sel. 19, 291–297 [DOI] [PubMed] [Google Scholar]
- 21. Holt L. J., Basran A., Jones K., Chorlton J., Jespers L. S., Brewis N. D., Tomlinson I. M. (2008) Protein Eng. Des. Sel. 21, 283–288 [DOI] [PubMed] [Google Scholar]
- 22. Walker A., Dunlevy G., Rycroft D., Topley P., Holt L. J., Herbert T., Davies M., Cook F., Holmes S., Jespers L., Herring C. (2010) Protein Eng. Des. Sel. 23, 271–278 [DOI] [PubMed] [Google Scholar]
- 23. Waldmann T. A., Strober W. (1969) Prog. Allergy 13, 1–110 [DOI] [PubMed] [Google Scholar]
- 24. Peters T., Jr. (1985) Adv. Protein Chem. 37, 161–245 [DOI] [PubMed] [Google Scholar]
- 25. Andersen J. T., Sandlie I. (2009) Drug Metab. Pharmacokinet 24, 318–332 [DOI] [PubMed] [Google Scholar]
- 26. Anderson C. L., Chaudhury C., Kim J., Bronson C. L., Wani M. A., Mohanty S. (2006) Trends Immunol. 27, 343–348 [DOI] [PubMed] [Google Scholar]
- 27. Roopenian D. C., Akilesh S. (2007) Nat. Rev. Immunol. 7, 715–725 [DOI] [PubMed] [Google Scholar]
- 28. Qiao S. W., Kobayashi K., Johansen F. E., Sollid L. M., Andersen J. T., Milford E., Roopenian D. C., Lencer W. I., Blumberg R. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9337–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akilesh S., Christianson G. J., Roopenian D. C., Shaw A. S. (2007) J. Immunol. 179, 4580–4588 [DOI] [PubMed] [Google Scholar]
- 30. Montoyo H. P., Vaccaro C., Hafner M., Ober R. J., Mueller W., Ward E. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ward E. S., Zhou J., Ghetie V., Ober R. J. (2003) Int. Immunol. 15, 187–195 [DOI] [PubMed] [Google Scholar]
- 32. Chaudhury C., Mehnaz S., Robinson J. M., Hayton W. L., Pearl D. K., Roopenian D. C., Anderson C. L. (2003) J. Exp. Med. 197, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ober R. J., Martinez C., Vaccaro C., Zhou J., Ward E. S. (2004) J. Immunol. 172, 2021–2029 [DOI] [PubMed] [Google Scholar]
- 34. Lejon S., Frick I. M., Björck L., Wikström M., Svensson S. (2004) J. Biol. Chem. 279, 42924–42928 [DOI] [PubMed] [Google Scholar]
- 35. Andersen J. T., Daba M. B., Sandlie I. (2010) Clin. Biochem. 43, 367–372 [DOI] [PubMed] [Google Scholar]
- 36. Chaudhury C., Brooks C. L., Carter D. C., Robinson J. M., Anderson C. L. (2006) Biochemistry 45, 4983–4990 [DOI] [PubMed] [Google Scholar]
- 37. Nord K., Gunneriusson E., Ringdahl J., Ståhl S., Uhlén M., Nygren P. A. (1997) Nat. Biotechnol. 15, 772–777 [DOI] [PubMed] [Google Scholar]
- 38. Wikman M., Steffen A. C., Gunneriusson E., Tolmachev V., Adams G. P., Carlsson J., Ståhl S. (2004) Protein Eng. Des. Sel. 17, 455–462 [DOI] [PubMed] [Google Scholar]
- 39. Andersen J. T., Justesen S., Fleckenstein B., Michaelsen T. E., Berntzen G., Kenanova V. E., Daba M. B., Lauvrak V., Buus S., Sandlie I. (2008) FEBS J. 275, 4097–4110 [DOI] [PubMed] [Google Scholar]
- 40. Berntzen G., Lunde E., Flobakk M., Andersen J. T., Lauvrak V., Sandlie I. (2005) J. Immunol. Methods 298, 93–104 [DOI] [PubMed] [Google Scholar]
- 41. Andersen J. T., Dee Qian J., Sandlie I. (2006) Eur. J. Immunol. 36, 3044–3051 [DOI] [PubMed] [Google Scholar]
- 42. Andersen J. T., Daba M. B., Berntzen G., Michaelsen T. E., Sandlie I. (2010) J. Biol. Chem. 285, 4826–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jonsson A., Dogan J., Herne N., Abrahmsén L., Nygren P. A. (2008) Protein Eng. Des. Sel. 21, 515–527 [DOI] [PubMed] [Google Scholar]
- 44. Orlova A., Magnusson M., Eriksson T. L., Nilsson M., Larsson B., Höidén-Guthenberg I., Widström C., Carlsson J., Tolmachev V., Ståhl S., Nilsson F. Y. (2006) Cancer Res. 66, 4339–4348 [DOI] [PubMed] [Google Scholar]
- 45. Wiig H., Reed R. K., Tenstad O. (2000) Am. J. Physiol. Heart Circ. Physiol. 278, H1627–H1639 [DOI] [PubMed] [Google Scholar]
- 46. Kwon Y. (2001) Handbook of Essential Pharmacokinetics, Pharmacodynamics and Drug Metabolism for Industrial Scientists, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 47. Aunoble B., Sanches R., Didier E., Bignon Y. J. (2000) Int. J. Oncol. 16, 567–576 [DOI] [PubMed] [Google Scholar]
- 48. Orlova A., Nilsson F. Y., Wikman M., Widström C., Ståhl S., Carlsson J., Tolmachev V. (2006) J. Nucl. Med. 47, 512–519 [PubMed] [Google Scholar]
- 49. Kratz F. (2008) J. Control Release 132, 171–183 [DOI] [PubMed] [Google Scholar]
- 50. Andersen J. T., Sandlie I. (2009) in Recombinant Antibodies for Immunotherapy (Little M. ed) pp. 293–310, Cambridge University Press, New York [Google Scholar]
- 51. Wiig H., DeCarlo M., Sibley L., Renkin E. M. (1992) Am. J. Physiol. 263, H1222–H1233 [DOI] [PubMed] [Google Scholar]
- 52. Wiig H., Kaysen G. A., al-Bander H. A., De Carlo M., Sibley L., Renkin E. M. (1994) Am. J. Physiol. 266, H212–H219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.