Abstract
All members of the kinesin superfamily of molecular motors contain an unusual structural motif consisting of an α-helix that is interrupted by a flexible loop, referred to as L5. We have examined the function of L5 in the mitotic kinesin Eg5 by combining site-directed mutagenesis of L5 with transient state kinetics, molecular dynamics simulations, and docking using cryo electron microscopy density. We find that mutation of a proline residue located at a turn within this loop profoundly slows nucleotide-induced structural changes both at the catalytic site as well as at the microtubule binding domain and the neck linker. Molecular dynamics simulations reveal that this mutation affects the dynamics not only of L5 itself but also of the switch I structural elements that sense ATP binding to the catalytic site. Our results lead us to propose that L5 regulates the rate of conformational change in key elements of the nucleotide binding site through its interactions with α3 and in so doing controls the speed of movement and force generation in kinesin motors.
Keywords: Electron Microscopy (EM), Fluorescence, Fluorescence Resonance Energy Transfer (FRET), Kinesin, Kinetics, Molecular Dynamics, Molecular Motors
Introduction
The kinesin superfamily of molecular motors share several evolutionarily conserved structural features with myosins and G-proteins. These include domains that coordinate the β phosphate of bound nucleotide (P loop), that sense the γ phosphate (switch I), and that induce conformational changes in response to phosphate release (switch II) (1–3). However, kinesins also contain an unusual structural element not found in these other motors and switches. This consists of an α-helix (α2) on the carboxyl terminal end of the P loop, which is interrupted by a stem and loop motif referred to as L5 (Ref. 4 and Fig. 1). The length of L5 varies considerably between different kinesin superfamily members, from short in kinesin 1 and CENP-E to longest in the mitotic kinesin Eg5 (Fig. 2).
FIGURE 1.
Crystallographic structures of L5 in ADP, ADP + monastrol, and AMPPNP. Orthogonal views of Eg5 crystallized in the presence of ADP (PDB 1II6), AMPPNP (PDB 3HQD), and ADP + monastrol (PDB 1X88). Major differences between the ADP and AMPPNP-bound structures include a shortening and bending of α3, associated with a ring stacking interaction between Trp-127 in L5 and Tyr-211, and an unfolding of the switch I helix. The ADP + monastrol structure represents a hybrid, with an ADP-like α3 and an “ATP-like” L5. Several key residues in L5 are depicted with their side chains, and prolines 121 and 131 are also colored blue.
FIGURE 2.
Sequence alignments of the α2a-L5-α2b domains from various kinesin family members. The conserved proline residues are highlighted in red. The proline at position 121 of human Eg5 aligns with a proline in position 102 in Nod. The proline at position 131 of human Eg5 is in a more structured region of L5.
The function of L5 remains unclear, although three lines of evidence suggest that it may play important roles in regulating nucleotide and microtubule binding. First, several small molecules induce L5 in Eg5 to fold over, generating a hydrophobic pocket bounded by L5, α2, and α3 (5, 6). In this conformation both ADP release and microtubule binding are prevented (7, 8). The analogous site in CENP-E is also the binding site for a novel inhibitor stabilizing the microtubule-motor complex, which appears to stabilize this motor in a strong microtubule binding conformation (9). Second, labeling of L5 with an EPR spin probe demonstrates that its mobility is affected by nucleotide binding, and in turn, deletion of L5 affects nucleotide binding (10). Third, cryo-EM reconstructions of Eg5-decorated microtubules in the presence of AMPPNP suggests that L5 can interact with α3, when the Eg5 motor is bound to the microtubule (11). Because the α3 helix is on the amino-terminal side of switch I, changes in the L5-α3 interaction might have downstream effects on nucleotide affinity and, secondarily, on microtubule binding. However, this study generated a docking model using crystallographic structures of Eg5 in the presence of ADP, and the authors noted unoccupied density in their cryo EM reconstructions that could not be explained by the crystallographic structures of L5 or α3. In addition, an L5-α3 interaction is also supported by crystallographic models of the kinesin 10 family member Nod, which show that the L5 interacts directly with α3 (12). This interaction is mediated by two proline residues (Pro-101 and Pro-102) in L5 that make hydrophobic interactions with α3 (12). In this regard we note that L5 in Eg5 contains two prolines (Fig. 2). One of these, Pro-121, corresponds to Pro-102 of Nod, whereas the second, Pro-131, is located in a more ordered portion of this loop and appears to induce a turn in this structure (Fig. 1).
In this study we have investigated the role of L5 in Eg5 by examining the effects of mutating these two proline residues on the presteady state and steady state kinetics of its ATPase cycle, and we have correlated our findings with results of molecular dynamics simulations and the current highest resolution cryoEM reconstruction. Our results lead us to propose a model in which the L5 domain of Eg5 plays a key role in modulating the kinetics of specific structural transitions in this mitotic kinesin.
EXPERIMENTAL PROCEDURES
Materials
PCR primers, TaqDNA polymerase, paclitaxol, QSY7, AEDANS,2 FlAsH, and MDCC-labeled phosphate-binding protein were purchased from Invitrogen. Restriction enzymes and ligase were purchased from New England Biolabs, Inc. (Ipswitch, MA). Bacterial culture media was bought from EMD Chemicals, Inc. and Fisher. Buffer reagents were purchased from Sigma and Fisher except for imidazole, which was purchased from EMD Chemicals, Inc. Protein assay reagents and gels were purchased from Bio-Rad. 2′deoxy-3′mant ATP and ADP were synthesized as previously described (26).
Mutagenesis
The cysteine-light Eg5 monomers with a single cysteine on the neck-linker (V365C) were generated as previously described (13). The proline mutations were generated using the following primers and their reverse compliment: the 5′ primer for the P121A sequence, AATGGAAGGTGAAAGGTCAGCTAATGAAGAGTATACCTGG, and the 5′ primer for the P131A sequence, GTATACCTGGGAAGAGGATGCCTTGGCTGGTATAATTCCA. Amino-terminal tetracysteine (CCPGCC) variants of the two proline mutants and the cysteine-light Eg5 were generated by using the 5′ primer sequence, CGGAATTCGAAATGTGTTGTCCTGGTTGTTGTGCGTCGCAGCCAAATTCGTCTGC, and the 3′ primer sequence, CTCGAGACA TTCAGGCTTATTCAATATGTTCTTTGCTC. The PCR products were digested with XhoI and EcoRI and ligated into pET21a plasmids for expression in BL21 cells. All DNA sequences were confirmed by sequencing.
Bacterial Cell Culture and Protein Expression
Expression plasmids were transformed into Bl21(DE3) RIL cells (Stratagene, Inc., La Jolla, CA) in an enriched medium (2% Tryptone, 1% yeast extract, 0.5% NaCl, 0.2% glycerol, 50 mm Na2HPO4, 50 mm K2HPO4, 50 mg/liter ampicillin, 50 mg/liter chloramphenicol). Five liters of culture were grown to an absorbance at 595 nm of 0.6–1.0 and induced with 0.5 mm isopropylthio-β-galactoside at 18 °C for 48 h. Typical yields were 5 mg of protein/liter of cells. After thawing, proteins were filtered through a prepacked PD-10 column containing Sephadex G-25 M (GE Healthcare) using the desired buffer.
Steady State ATPase Assays
ATPase assays were performed using EnzChek phosphate assay kit (Invitrogen) with motor concentrations in the 5–20 nm range and 25–2000 nm polymerized tubulin. ATPase assays were performed in 20 mm KCl, 25 mm HEPES, 2 mm MgCl2, and 1 mm DTT, pH 7.50. kcat for P131A was calculated from fitting the Michaelis-Menten equation to ATPase data taken at an MT concentration 5-fold above the K0.5,MT, whereas kcat for wild type and P121A were calculated from the MT-dependent ATPase data while holding ATP constant at a value 8–10-fold higher than the K0.5,ATP.
Fluorescence Methodologies
Labeling of the amino-terminal CCPGCC domain of an Eg5 construct with FlAsH was accomplished by incubating a 1:1 ratio of dye to motor overnight at 4 °C in 100 mm KCl, 25 mm Hepes, pH 7.5, 2 mm MgCl2, 0.2 mm Tris(2-carboxyethyl)phosphine, 0.2 mm ADP. This resulted in an ∼90% labeling efficiency. QSY7 was then added in 5-fold excess for 3 h on ice, and the reaction product was then desalted over a PD-10 column to remove excess dye. Labeling with QSY7 was >90% efficient. Labeling with a 2-fold excess of FlAsH resulted in 1:1 molar stoichiometries.
Kinetic Methodologies
Presteady state kinetics of 2′-deoxy-3′mant ATP (2′dmT) and 2′-deoxy-3′mantADP (2′dmD) binding, nucleotide-dependent MT release, and Pi release were performed on a KinTex SF-2004 stopped-flow as previously described (13, 27, 28). For experiments using the FlAsH-QSY7 donor acceptor pair, FlAsH was excited at 520 nm, and a 550-nm-long pass filter was used to monitor changes in FlAsH fluorescence emission. Motor-MT complexes were formed before stopped-flow experiments by removing unbound nucleotide through gel filtration (PD10) followed by the addition of 0.2 units/ml of apyrase.
Molecular Dynamics Simulations
Simulations were based on the crystal structure of the human Eg5 in complexed with ADP and 1Mg2+ (PDB code 1II6).
Structure of P131A Mutant
The side chain of residue proline 131 was changed to alanine, and a local energy minimization was carried out so as to generate a starting conformation for molecular dynamics (MD) simulations. The local minimization consisted in two rounds of 350 minimization steps (50 steepest descent steps followed by 300 conjugate gradient steps) where only the atoms within 4 or 6 Å from residue 131 were, respectively, left without constraint in the first and second round.
Structure of the ΔL5 Mutant
Residues 102–118 were removed from the Eg5 structure, and a local energy minimization was performed so as to generate a starting conformation for MD simulations in the presence of ADP. The local minimization consisted in two rounds of 400 minimization steps (50 steepest descent steps followed by 350 conjugate gradient steps) where only the atoms within 6 or 8 Å from residues 101 or 119 were left without constraint in, respectively, the first and the second round.
Structures of the Nucleotide-free States
The structures of ADP-bound Eg5 motor domain, P131A, and the ΔL5 mutant were used to generate the respective apo structures. In each case the ADP and the Mg2+ were removed, and a local energy minimization was performed. Three rounds of 400 minimization steps (50 steepest descent steps followed by 350 conjugate gradient steps) were performed. Only atoms within 4, 6, or 8 Å from ADP or Mg2+ were, respectively, left without constraints during the first, second, and third minimization rounds.
Molecular Dynamics Simulations
All molecular dynamics simulations were performed with Gromacs-3.2.1 (29) using the GROMOS force field. In all cases proteins were embedded in a cubic box filled with water molecules so that no atom of the protein is closer than 10 Å from the box boundaries. In the 12 simulations (Eg5, kinesin Eg5, P131A mutant, and ΔL5 mutant in the presence or in the absence of ADP), the simulation box size was ∼90 × 90 × 90 Å, and each system contained slightly more than 71,000 atoms. We ensured global charge neutrality of the different systems by adding counterions to the box as needed. Periodic boundary conditions were assumed in all cases. A uniform integration step of 2 fs was used for all types of interactions, and all bonds were constrained using the LINCS algorithm throughout all simulations. A cutoff of 12 Å was used for van der Waals interactions, and electrostatic interactions were calculated with the particle mesh technique for Ewald sums with a cutoff of 12 Å. A Nose-Hoover thermostat was used to maintain a constant temperature of 300 K, and constant pressure of 1 atm was maintained using a Parrinello-Rahman barostat.
All simulations started with an equilibration of the system consisting of four heating steps of 250 ps each, with the water molecules free to move and protein heavy atoms constrained. The production phase was then run, and frames were recorded at 0.1-ps intervals. The production phase lasted 18 ns for all simulations. Root mean square fluctuations of the backbone atoms (i.e. C, Cα and N atoms) were computed using the g_rmsf module of Gromacs. In a first step each frame of the trajectory was superposed with the first frame via a structural alignment of the protein backbone.
Cryo-EM Docking Models
The cryo-EM reconstruction of the kinesin-5 KLP61F motor domain bound to AMPPNP and paclitaxel-stabilized microtubules was calculated as previously described (11) and is deposited in the Electron Microscopy Data Bank, reference EMD-1604. The fitted coordinates of tubulin and the KLP61F homology model are deposited in the PDB, code 2WBE. UCSF Chimera (30) was used for visualization and rigid-body docking of kinesin crystal structures (Eg5-AMPPNP, PDB code 3HQD (24); Eg5-ADP PDB code 1II6 (4); Nod-ADP PDB code 3DC4 (12)) into the cryo-EM reconstruction. Fits were first performed manually and were refined computationally.
RESULTS
The Eg5 ATPase Cycle
We had previously described a model for the ATPase cycle of the Eg5 motor that is summarized in Fig. 3 (13). In this earlier study, we found that the neck linker in Eg5 assumes three different orientations during the ATPase cycle. These are indicated in Fig. 3 with the subscripts R (for rigor), T (for ATP), and D (for ADP). The R state cannot bind ATP, and binding of this nucleotide is, therefore, rate-limited by a conformational change that alters neck linker orientation (R → T). ATP hydrolysis generates a weak microtubule binding state that leads to dissociation of the motor from the microtubule and phosphate release. This is associated with a second reorientation of the neck linker (T → D). Rebinding of Eg5·ADP to the microtubule leads to resumption of the rigor conformation and ADP release, regenerating the cycle.
FIGURE 3.
The mechanochemical cycle of Eg5. We had previously shown that the neck linker in Eg5 undergoes two structural transitions during the ATPase cycle. These are indicated by the subscripts R (for rigor), T (for ATP), and D (for ADP). Beginning in the rigor state (far left), structural transitions that induce movement of the neck linker into the T orientation occur with rate constants k1 and k−1 and gate the maximum rate ATP can bind (k2). Hydrolysis and phosphate release lead to dissociation from the microtubule (k3). This leads to an isomerization of the neck linker from the T to the D state, occurring with forward and reverse rate constants k4 and k−4. Rebinding to the MT (k5) is followed by MT-stimulated ADP release (k6), returning the catalytic domain to the rigor state. At low ionic strength, the chemical processivity of P121A is significantly greater than for Eg5 and implies that phosphate can be released without dissociation and is indicated by the dashed arrow in the figure.
Steady State ATPase of P121A and P131A (Table 2)
TABLE 2.
Effect of ionic strength on steady state ATPase parameters
| Constant | Condition | Eg5 | P121A | P131A |
|---|---|---|---|---|
| kcat (s−1) | 100 mm KCl | 8.6 ± 0.7 | 5.7 ± 0.6 | |
| 20 mm KCl | 8.3 ± 0.5 | 15.2 ± 1.0 | 11.8 ± 2.8 | |
| K0.5,MT (μm) | 100 mm KCl | 6.3 ± 1.2 | 0.7 ± 0.2 | |
| 20 mm KCl | 1.1 ± 0.2 | 0.02 ± 0.01 | 0.3 ± 0.04 | |
| K0.5,ATP (μm) | 100 mm KCl | 51 ± 11 | 25 ± 7 | 791 ± 343 |
| 20 mm KCl | 68 ± 28 | 96 ± 41 | 1285 ± 570 | |
| kcat/K0.5,MT (μm−1 s−1) | 20 mm KCl | 7.5 | 760.1 | 39.3 |
We have used the scheme depicted in Fig. 3 as a framework for studying the effect of mutating Pro-121 and Pro-131 within the L5 domain of human Eg5. We were interested in determining how mutating each of these prolines to alanine would affect nucleotide binding, microtubule binding, and the kinetics of the steps that constitute the Eg5 ATPase cycle. We started our analysis by measuring steady state ATPase parameters for these two monomeric mutants and comparing them to an Eg5 motor domain monomer containing the amino-terminal 367 amino acids (Table 2). As Table 2 shows, the major effect of the P131A mutation on the steady state ATPase parameters is to increase K0.5,ATP by nearly 15–20-fold in a manner that shows little ionic strength dependence. By contrast, the major effect of the P121A mutation is to reduce K0.5,MT by nearly 10-fold in 100 mm KCl and >50-fold at 20 mm KCl. To identify which steps in the Eg5 ATPase cycle are perturbed by these mutations, we next examined the kinetics of the individual steps in the cycle.
ATP Binding and Release (K1k+2 and k−2 in Table 1)
TABLE 1.
Summary of rate and equilibrium constants for Eg5, P121A, and P131A
PBP, phosphate binding protein.
| Constanta | Description | Methodb | Eg5 | P121A | P131A |
|---|---|---|---|---|---|
| (k+1) (s−1) | Isomerization of catalytic site | 2′dmT | 62–79c | 19.6 ± 9.4 | |
| (K1k+2) (μm−1s−1) | ATP binding | 2′dmT | 1.1 ± 0.1c | 1.0 ± 0.1 | 0.02 ± 0.01 |
| (k−2) (s−1) | ATP dissociation | 2′dmT | 19 ± 9.6c | 21.5 ± 13.9 | 1.7 ± 0.4 |
| Phosphate release (s−1) | MDCC-PBP | 7.9 ± 0.9 | 7.8 ± 0.6 | 5.8 ± 0.9 | |
| (k3) (s−1) | ATP-induced MT dissociation | Turbidity | 8.7 ± 1.9 | 5.1 ± 0.2 | 3.4 ± 0.2 |
| ATP-induced neck linker isomerization (s−1) | FlAsH → QSY7 | 10.4 ± 0.8 | 4.6 ± 0.2 | 1.8 ± 0.1 | |
| ADP-induced neck linker isomerization (s−1) | FlAsH → QSY7 | 16.5 ± 1.0 | 7.8 ± 0.4 | 2.4 ± 0.1 | |
| (k+5) (μm−1s−1) | Eg5·ADP binding to MT (20 mm KCl) | AEDANS | 5.9 ± 0.8 | 2.3 ± 0.4 | 3.4 ± 1.2 |
| (k−5) (s−1) | Dissociation of Eg5·ADP (20 mm KCl) | AEDANS | 44.1 ± 6.5 | 3.2 ± 1.8 | 11.5 ± 5.5 |
| (k−5) (s−1) | Dissociation of Eg5·ADP (100 mm KCl) | AEDANS | 14.2 ± 0.4 | 14.2 ± 0.6 | 3.9 ± 0.2 |
| Turbidity | 9.6 ± 0.4 | 11.0 ± 0.7 | 4.2 ± 0.3 | ||
| (k−6) (μm−1s−1) | ADP binding | 2′dmD | 0.3 ± 0.1c | 0.07 ± 0.03 | 0.09 ± 0.01 |
| (k+6) (s−1) | ADP dissociation | 2′dmD | 76 ± 15c | 14.9 ± 2.5 | 9.9 ± 0.9 |
a Rate and equilibrium constants as depicted in Fig. 2.
b Methods include the following: 1) 2′dmT and 2′dmD, FRET from motor + microtubule tryptophans to 2′dmT or 2′dmD fluorophor; 2) MDCC-PBP, fluorescence enhancement of MDCC-labeled phosphate-binding protein; 3) turbidity, measured in the stopped flow spectrometer at 350 nm; 4) FlAs → SY7, FRET from FlAsH on the amino terminus of Eg5 or mutants to QSY7 on the carboxyl terminus; 5) AEDANS, FRET from microtubule tryptophans to AEDANS on the Eg5 or mutant neck linker.
c From Rosenfeld et al. (13).
We measured binding of the fluorescent ATP analog 2′dmT to complexes of microtubule with P121A or with P131A in 100 mm KCl buffer by monitoring FRET from tryptophan 127 in L5 to the 2′dmT in the active site. For both mutants ∼80% of the resulting transient consists of a phase of increasing FRET whose rate constant, reflecting binding to the empty catalytic site, varies with [2′dmT] (Fig. 4). For the P121A mutant, this variation is hyperbolic, defining values for apparent second order rate constant, dissociation rate constant, maximum forward rate constant, and apparent dissociation constant that are all within a factor of 2–3 of the corresponding values for Eg5 (Fig. 4C, Table 1). As with Eg5, 2′dmT binding to the P121A mutant appears to saturate with a rate constant nearly 50-fold less than that for kinesin 1, suggesting that ATP binding is gated by a rate-limiting conformational change (13). Binding of 2′dmT to P131A also produces a transient (Fig. 4B). However, the rate constant in this case varies linearly with 2′dmT (Fig. 4C), and a maximum rate could not be observed. The apparent second order rate constant for this process is nearly 50-fold lower than the corresponding value for Eg5 (Table 1).
FIGURE 4.
Kinetics of binding of 2′dmT to the P121A and P131A mutants of human Eg5. A, shown is a schematic of the experimental design. Complexes of either mutant with an excess of microtubules were mixed with 2′dmT in the stopped flow, and the resulting fluorescence increase was monitored by FRET from vicinal tryptophans to the mant fluorophor. B, fluorescence transients produced by mixing 5 μm P121A (gray) or 5 μm P131A (black) + 10 μm microtubules with 40 μm 2′dmT are shown. For both mutants ∼80% of the resulting transient consists of a phase of increasing FRET. C, the rate constant for this process varies hyperbolically with 2′dmT for P121A (gray), defining a maximum forward rate constant and dissociation rate constant listed in Tables 1 and 2. By contrast, the rate corresponding rate constant for P131A (black) varies linearly with 2′dmT, defining an apparent second order rate constant and dissociation rate constant listed in Tables 1 and 2.
ATP-induced Dissociation of Motor from Microtubule (k3 in Table 1)
We measured motor-microtubule dissociation in 100 mm KCl buffer by mixing complexes of microtubules + Eg5, P121A, and P131A with ATP or ADP in the stopped flow and monitoring turbidity. In each case the resulting decrease in turbidity fit a single exponential decay with a linear tail, as has been previously described for kinesin 1 (14). The rate constant for these two constructs varies hyperbolically with nucleotide concentration (supplemental Fig. 1), defining maximum rates of dissociation by ATP and ADP that are summarized in Table 1. The rate constant for dissociation of P131A by ATP is ∼3-fold slower than for Eg5, whereas that for P121A is ∼2-fold slower (Table 1).
Phosphate Release
Mixing complexes of microtubules + Eg5, P121A, or P131A with ATP in the stopped flow in the presence of MDCC-labeled phosphate-binding protein in 100 mm KCl buffer produces a fluorescence increase in two phases, the first of which could be described by a single exponential term and the second by a linear term (supplemental Fig. 2). The exponential term corresponds to the process of phosphate release in the first turnover, whereas the linear term reflects subsequent ATP turnover in the steady state. The rate constant for the exponential phase varies hyperbolically with [ATP] (supplemental Fig. 3), defining maximum rates and dissociation constants that are summarized in Table 1. Neither mutation appreciably changes the rate of phosphate release.
Microtubule Binding and Release by Motor·ADP (k+5 and k−5 in Table 1)
Association of Eg5·ADP to the microtubule leads to formation of a strong binding state that is followed by ADP release. We measured the kinetics of this process at 20 mm KCl buffer for the P121A and P131A mutants by using a technique we previously described (13). Both mutant constructs contain a single reactive cysteine at position 365, in the neck linker. We labeled the Eg5 constructs at this site with the fluorescent probe AEDANS and monitored binding to microtubules by FRET from microtubule tryptophans to the AEDANS probe. Mixing AEDANS-labeled motor·ADP in the stopped flow with a 3–4-fold excess of microtubules produces a fluorescence increase that fits a double exponential process. The rate constant for the faster phase, reflecting microtubule binding, varies linearly with microtubule concentration, defining apparent second order rate constants that are summarized in supplemental Fig. 4 and Table 1. Extrapolation of the line to the ordinate provides a measure of the rate constant for dissociation of motor·ADP from the microtubule at 20 mm KCl. As Table 1 demonstrates, this rate constant for the P121A mutant is >10-fold slower than that for Eg5, which partially accounts for the lower K0.5,MT for this mutant. We also measured the kinetics of P121A·microtubule and P131A·microtubule dissociation directly by mixing with ADP in 100 mm KCl buffer and monitoring both the AEDANS FRET signal and turbidity at 350 nm, and as Table 1 demonstrates, both methods give similar results. In addition, the effect of altering ionic strength on the value of k−5 (Fig. 3, Table 1) also suggests the effect of the P121A mutation on MT release depends on electrostatic interactions.
ADP Binding and Release (k+6 and k−6 in Table 1)
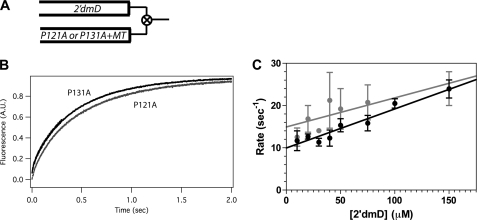
We measured binding of 2′dmD to complexes of nucleotide-free P121A and P131A with microtubules by mixing with this ADP analog in the stopped flow. The resulting fluorescence transients were qualitatively similar to those for described for 2′dmT above (Fig. 5A). However, as Fig. 5B demonstrates, the dependence of rate constant versus 2′dmD varies linearly for both P121A and P131A. The slopes of these lines, defining an apparent second order rate constant for binding of 2′dmD, are similar to each other (Table 1). In contrast to the case of 2′dmT, the kinetics of binding of 2′dmD to the two proline-to-alanine mutants are very similar to each other.
FIGURE 5.
Kinetics of binding of 2′dmD to the P121A and P131A mutants of human Eg5. A, a schematic of the experimental design is shown. Complexes of either mutant with an excess of microtubules were mixed with 2′dmD in the stopped flow, and the resulting fluorescence increase was monitored by FRET from vicinal tryptophans to the mant fluorophor. B, fluorescence transients produced by mixing 5 μm P121A (gray) or 5 μm P131A (black) + 10 μm microtubules with 40 μm 2′dmD are shown. Fluorescence transients are qualitatively similar to those for 2′dmT. C, the rate constant for this process varies linearly with 2′dmD for both P121A (gray) and P131A (black), defining apparent second order rate constants and dissociation rate constants listed in Tables 1 and 2.
Neck Linker Reorientation
We had previously identified three steps in the Eg5 mechanochemical cycle where the neck linker moves relative to the microtubule (13). The first occurs in concert with a conformational change that allows ATP binding, the second occurs after the motor dissociates from the microtubule, and the third occurs with microtubule rebinding and ADP release. Although the approach used in that study involving FRET pairs on the microtubule and the Eg5 neck linker had the sensitivity to monitor the kinetics of these steps, the potential for multiple fluorophors on the microtubule to excite a single acceptor on the neck linker limited our ability to make structurally interpretable conclusions. We have, therefore, utilized a different approach in this current study to monitor the kinetics of neck linker movement in Eg5 as well as in the P121A and P131A mutants. We generated versions of these three constructs that contain the sequence CCPGCC at the amino terminus as well as a single reactive cysteine at position 365 in the neck linker. The former sequence binds the bis-arsenical derivative of fluorescein, FlAsH, specifically and with high affinity (15). We labeled these constructs with FlAsH at the amino terminus and the non-fluorescent acceptor QSY7 at position 365 with ∼1:1 stoichiometries. These modifications did not affect the ATPase activity or the kinetics of nucleotide binding or microtubule detachment (data not shown). The rationale for this labeling strategy is that changes in neck linker orientation might be measurable as changes in FRET between the amino-terminal FlAsH and the neck linker QSY7 (illustrated in Fig. 6A).
FIGURE 6.
Kinetics of ATP and ADP induced neck linker reorientation in Eg5, measured by FRET from FlAsH to QSY7 in Eg5 and in the P121A and P131A mutants. A, shown is a schematic of the experimental design. The amino terminus of these constructs containing the sequence CCPGCC is labeled with the fluorescence donor FlAsH (small open circle), whereas the neck linker is labeled with the non-fluorescent acceptor QSY7 (small closed circle). Separation of the donor and acceptor as would occur with neck linker reorientation increases donor fluorescence (depicted by the star). B, fluorescence transient resulting from mixing a complex of FlAsH-QSY7-labeled Eg5 + microtubules with ATP (gray) or ADP (black) is shown. The resulting fluorescence enhancement fits a single exponential rise following a lag for ATP, as evidenced by an inflection in the transient. The lag phase was absent when the experiment was repeated with ADP. C, the rate constant for the rising phase in fluorescence in B varies in a hyperbolic manner with nucleotide concentration, defining maximum rate constants listed in Tables 1 and 2. D, the corresponding experiment with the P121A mutant is qualitatively similar to Eg5. E, rate constants for neck linker reorientation show similar hyperbolic dependence on nucleotide concentration, although the extrapolated rates for both ATP and ADP are approximately half those for Eg5 (Tables 1 and 2). F, the corresponding experiment with the P131A mutant demonstrates that both ATP and ADP produce qualitatively very similar transients, neither of which is associated with a lag phase. G, rate constants for neck linker reorientation also show hyperbolic dependence on nucleotide concentration, although the extrapolated rates for both ATP and ADP are approximately 20% of those for Eg5 (Tables 1 and 2).
Mixing a complex of microtubules + Eg5 labeled with FlAsH and QSY7 with ATP in the stopped flow produces a biphasic fluorescence transient (Fig. 6B). The first phase consists of a lag, associated with a rate constant of 40–60 s−1, which does not vary with [ATP] (data not shown). We had previously shown that neck linker docking in Eg5 occurs with a similar rate constant and rate limits ATP binding (13). The second phase consists of an increase in fluorescence, whose rate constant varies hyperbolically with [ATP] (Fig. 6, B and C), with a maximum rate that is similar to that for ATP-induced dissociation (Tables 1 and 2). Repeating this experiment with ADP produces a monophasic increase in fluorescence without an initial lag, with a rate constant that also varies hyperbolically with [ADP] and is similar to that for ADP-induced dissociation. Repeating this experiment in the absence of either microtubules or of the QSY7 quencher results in no significant change in the fluorescence of the FlAsH donor (data not shown).
Repeating this experiment with the P121A mutant produces results that are very similar to those of Eg5 (Fig. 6D and Table 1). By contrast, the corresponding experiment with the P131A mutant + microtubules produces a fluorescence increase with no lag phase for both ATP and ADP (Fig. 6E and Tables 1 and 2). In both cases, the maximum rate constants are similar to the corresponding rates of nucleotide-induced dissociation from the microtubule.
Structural Insights from Molecular Dynamics
Our results demonstrate that point mutations in L5 can affect nucleotide affinity, microtubule affinity, and the kinetics of coupling of nucleotide binding to neck linker orientation. However, these studies do not provide any structural insight into how L5 affects these steps in the ATPase cycle. To investigate this issue, we performed equilibrium MD simulations on three sets of structures: monomeric Eg5, the P131A mutant, and a mutant where the L5 has been deleted (ΔL5). Each simulation was performed under 2 conditions, in the presence or the absence of ADP and Mg2+, and for each of these 6 systems, two independent simulations were performed and carried out to 18 ns.
The comparison of the root mean square fluctuations (r.m.s.f., a measure of the average atomic mobility) of backbone atoms along the motor domain during the different simulations is depicted in Fig. 7 and supplemental Figs. 4 and 5. The P131A mutation increases the local r.m.s.f. around this mutation (arrow and asterisk in Fig. 7A). This is consistent with a role of prolines in reducing the flexibility of polypeptide loops by limiting accessible Ramachandran space (16). Deletion of L5 as well as the P131A mutation affects the dynamics of both the α3 helix and switch I, which is on the immediate carboxyl terminal side of this helix (Fig. 2). In the ADP-bound state, the removal of L5 results in an increased mobility of the entire α3 helix. Remarkably, we also found that the introduction of the P131A mutation has a similar, albeit less profound effect (Fig. 7). By contrast, the r.m.s.f. of switch I, which directly interacts with bound nucleotide, is not affected by either of these two mutations over the timescale of the simulations.
FIGURE 7.
r.m.s.f. of the backbone atoms in the region of the α1, α2, and α3 helices and surrounding switch I. A, the color code for the different curves is indicated in the top right panel on the graph, and secondary structure elements are represented at the bottom. The P-loop, switch I, and proline 131 are represented as indicated in the legend. The asterisk and arrow denote the location of the P131A mutation. B, shown is the effect of ADP removal on the r.m.s.f. of backbone atoms in the region of the α3 helix and switch I for Eg5 (left) and the P131A mutant (right). The color code for the different curves is the same in the three graphs and is indicated in the top left panel on the left graph. Secondary structure elements are indicated at the bottom of the graphs.
In Eg5, removal of ADP markedly enhances the r.m.s.f. of the P loop and switch I as well as of both the α2 and α3 helices (Fig. 7, supplemental Figs. 3 and 4). By contrast, this enhancement in r.m.s.f. is markedly blunted in the P131A mutant. These results suggest that the conformational flexibility of L5 can affect the structure of not only its immediately neighboring domain (α2) but also of more distant domains that are involved in nucleotide binding and sensing (P loop and switch I) through a pathway that involves α3.
DISCUSSION
L5 Is Found in All Kinesins, but Its Function Remains Unclear
The presence of this unusual structural motif in all kinesins argues that it plays an important role in the ATPase cycle of this superfamily of molecular motors. There has, however, been little information to explain what it does. As Fig. 2 illustrates, L5 is in close proximity to the P loop, which coordinates the β-phosphate of bound ATP or ADP, and to switch I, which senses the γ-phosphate of bound ATP (2, 17, 18). Furthermore, cryoEM reconstructions indicate that L5 is in close contact to α3, which is on the amino-terminal side of switch I (Figs. 1 and 7 and Ref. 11). Considering the proximity of L5 to these functionally important nucleotide binding domains, it is not surprising that two lines of evidence already suggest that L5 is somehow involved in regulating the structure of the nucleotide binding site. First, a number of small molecules induce L5 to fold over, generating a hydrophobic cleft that is bounded by this loop, along with α2 and α3. In this state, ADP binding is enhanced, and microtubule binding is weakened (7, 8, 19, 20). Second, binding of nucleotide to the catalytic site of the mitotic kinesin Eg5 affects the conformation of L5, as measured by the fluorescence emission of a tryptophan residue (127) located within this loop (21). However, neither experimental approach provides a mechanistic explanation for how the state of L5 could affect the structure of the P loop and switch I.
The length of L5 shows a high degree of variability among different kinesins with different physiologies and ATPase activities. However, several features of the L5 sequence appear to be shared among kinesin family members. First, the loop is delimited from the α2a and α2b helices with glycine residues. Second, there is a proline residue (Pro-131 in human Eg5) toward the carboxyl terminal end of L5. In human Eg5, Pro-131 appears to induce a bend in the structure of this loop, consistent with one of the roles proline residues play in protein structure (16). Finally, in Nod as well as in several members of the Eg5 subfamily, a second proline (Pro-121 in human Eg5) appears to be conserved. Consequently, we elected to mutate Pro-121 and Pro-131 in human Eg5 to determine whether the resulting perturbations in enzymatic activity could reveal insights into the role of L5 in this kinesin family member.
The Proline-to-alanine Mutations Define Roles for L5 in Regulating Nucleotide and Microtubule Binding
The most striking effect of the P131A mutation is on the kinetics of nucleotide binding and release. This is reflected in a ∼15–20-fold increase in K0.5,ATP compared with Eg5 and correlates in turn with a >50-fold slowing in the rate of ATP binding and a nearly 10-fold slowing in ATP release (Tables 1 and 2; K1k2, k−2). ADP binding and release are also slower, but to a lesser degree, and in contrast to the case for ATP, the apparent ADP affinity is slightly increased. This mutation also slows the dissociation of P131A from the microtubule by both ATP and ADP by ∼5-fold, although it has little effect on the kinetics of phosphate release or on the second order rate constant for binding of P131A·ADP to the microtubule. These results suggest that the P131A mutation has two major effects. First, it alters the conformation of key structural elements of the catalytic site and thereby markedly slows ATP binding. Second, it also slows the rates of the conformational changes that occur subsequent to nucleotide binding and that lead to microtubule dissociation. This latter effect would be expected to prolong the lifetime of strongly bound states, enhance the duty cycle, and increase motor processivity. As previously described (22), the average time a kinesin motor remains attached to the microtubule can be approximated by the ratio 1/(k5·K0.5,MT). At 20 mm KCl this “persistence time” is nearly 23-fold longer for the P131A mutant (Tables 1 and 2), implying that at this ionic strength P131A can undergo multiple cycles of ATP hydrolysis and phosphate release without dissociating from the microtubule (illustrated by the dashed line in Fig. 3). Although a prior study of this mutant demonstrated an ∼3-fold higher value of K0.5,MT, we note that the ATPase assay used in this study was performed under different buffer conditions (5), and the affinity of Eg5 for the microtubule shows a strong ionic strength dependence (Tables 1 and 2).
By contrast, the P121A mutation has only modest effects on the affinities of ATP and ADP and on the kinetics of nucleotide binding and release. Rather, the major effect of this mutation is to enhance apparent microtubule affinity at both low and physiologic ionic strength (Tables 1 and 2). At 20 mm KCl, this corresponds to a 145-fold increase in persistence time and a 10-fold reduction in the rate constant for ADP-induced microtubule dissociation (Tables 1 and 2, k−5) compared with wild type Eg5. These findings imply that like the P131A mutation, the P121A mutant is capable of undergoing multiple rounds of ATP hydrolysis and phosphate release without dissociating from the microtubule (Fig. 3, dashed line). These results suggest that L5 can modulate microtubule affinity both through its effects on the catalytic site (as illustrated by the P131A mutation) as well as through separate, nucleotide-independent effects on the Eg5 microtubule binding domains (as illustrated in the P121A mutation). This latter point is also underscored by a recent study of GSK923295, a small molecule inhibitor of another mitotic kinesin, CENP-E (9). GSK923295 binds to the region in CENP-E bounded by L5, α2, and α3, like monastrol does for Eg5. However, unlike monastrol, binding of this inhibitor to CENP-E induces the motor to assume a strong binding conformation and inhibits microtubule dissociation, as the P121A mutation appears to do for Eg5.
The Neck Linker in Eg5 Has Discrete Orientations in Rigor, ATP, and ADP, and the P131A Mutation Defines a Role for L5 in Regulating Neck Linker Movement
In our previous study (13), we had used a variety of FRET pairs to examine the timing of neck linker orientation changes during the Eg5 ATPase cycle, and we found that there were three points when this occurs; with ATP binding (R → T transition in Fig. 3), after ATP hydrolysis and microtubule dissociation (T → D) and after rebinding of the Eg5·ADP complex to the microtubule (D → R). For this study, we have re-examined the kinetics of neck linker conformational changes in Eg5 and in the P121A and P131A mutants with a new FRET pair-FlAsH and QSY7. Changes in donor (FlAsH) fluorescence in this assay can be due to one of three possibilities; 1) a change in FlAsH quantum yield, 2) a change in κ2, a term reflecting donor orientation relative to acceptor, or 3) a change in distance between the donor and acceptor. We see no significant fluorescence change when we mix Eg5 labeled with donor only + microtubules with nucleotide, implying that donor quantum yield does not change with nucleotide binding and microtubule dissociation. Furthermore, the amino terminus, where the donor is attached, is unstructured in crystallographic models of Eg5 (4), and the FlAsH donor is attached to an additional sequence (CCPGCC) that should not be involved in cover strand interactions with the neck linker. These features imply that the donor fluorophore is isotropic and makes it highly unlikely that alterations in κ2 could account for any observed changes in FRET efficiency. We, therefore, conclude that the observed fluorescence enhancement in this assay mirrors an increase in distance between the neck linker and the amino terminus in response to nucleotide binding.
We first utilized the FlAsH-QSY7 FRET pair with an Eg5 monomeric construct. The fluorescence transient resulting from mixing an Eg5·microtubule complex with ATP is consistent with our prior model (13) if we make three assumptions; 1) the R → T transition, which we had previously shown gates ATP binding and occurs at a rate of 40–60 s−1, is not detected by this FRET pair, and it accounts for the initial lag phase, 2) the T → D transition produces a fluorescence increase, reflecting a separation between the amino terminus and the neck linker (Fig. 6B), and 3) the neck linker isomerization step (k4 + k−4 in Fig. 3) is faster than the rates of ATP or ADP-induced dissociation, so the observed rate constant for the change in FlAsH → QSY7 FRET is rate-limited by the rate of microtubule dissociation. This latter assumption is consistent with our previous study, which calculated a value of 28–31 s−1 for k4 + k−4 (13).
Our finding that ADP-induced dissociation produces a change in FlAsH → QSY7 energy transfer also implies that the position of the neck linker in Eg5 in rigor is different from that in the presence of ADP. Although this is in marked contrast to currently accepted models of kinesin 1, where the neck linker is presumed to be disordered in both states (23), it is consistent with EPR studies, which detect differences in the mobility of a spin probe on the neck linker between a rigor and an ADP-bound Eg5·microtubule complex (10). However, our previous study disagrees with Larson et al. (10) as it demonstrated that the rigor orientation of the neck linker is also distinct from that in ATP (Ref. 13); summarized in Fig. 3). Our model also predicts that mixing a labeled Eg5·microtubule complex with ADP will generate a fluorescence increase without a lag phase and with a rate that is faster than for ATP, as this reaction would not proceed through the R → T transition or through subsequent ATP hydrolysis and phosphate release. As Fig. 6B demonstrates, this is in fact the case.
We find that the kinetics of ATP and ADP-induced neck linker movement for the P121A mutant are qualitatively and quantitatively similar to those for Eg5 (Fig. 6C). By contrast, the P131A mutation markedly slows both ATP- and ADP-induced separation of the neck linker and amino terminus. Furthermore, we can no longer detect a lag phase with ATP (Fig. 6D, Tables 1 and 2). We can explain these results through either of two possibilities, 1) the P131A mutation markedly slows the R → T and R → D conformational changes (Fig. 3) that occur in response to ATP and ADP binding, respectively, or 2) this mutation induces neck linker docking without any added nucleotide and eliminates the R → T transition altogether. The latter explanation is consistent with a recent study, which observed that deletion of L5 reduces the mobility of the Eg5 neck linker in a nucleotide-independent manner and which interpreted this finding to suggest that deletion of L5 induces neck linker docking (10). As we have shown above our molecular dynamics simulations demonstrate that the P131A mutation has effects on the dynamics of α3 and switch I that are qualitatively similar to those produced by deletion of L5 (Fig. 7A).
Molecular Dynamics Simulations Suggest That L5 Regulates the Structure and Responsiveness of the Catalytic Site in Eg5
Our kinetic and steady state studies demonstrate that the P131A mutation has profound effects on nucleotide binding and subsequent downstream conformation responses, including neck linker movements. The relative importance of this residue is underscored by our finding that mutation of a second proline (Pro-121) has by contrast only modest effects on the enzymology of the Eg5 mechanochemical cycle. L5 is strategically located near switch I, and our molecular dynamics simulations provide insight into how this loop might regulate the structure of this nucleotide-coordinating domain. As Fig. 7 illustrates, removal of ADP from the active site of wild type Eg5 produces a large increase in the r.m.s.f. of α3 and switch I, implying a correspondingly large increase in flexibility in these regions. Switch I senses the γ phosphate of ATP, and we propose that this resulting increase in flexibility is necessary for switch I to alter its conformation to accommodate ATP in the catalytic site (as illustrated in Fig. 1). As the figure also shows, the major effect of the P131A mutation is to blunt this increase in r.m.s.f.. We propose that the result of this effect would be to slow the conformational change in switch I needed to accommodate ATP, which would thereby make the process of ATP binding rate-limiting, as we observe experimentally.
Our simulations also provide insight into how L5 can regulate the process of nucleotide binding. Deleting L5 increases the r.m.s.f. of the entire α3 helix (Fig. 7A). As Fig. 2 demonstrates, the ADP and AMPPNP structures of Eg5 differ not only in the conformation of switch I but also in the structure of α3 as well. With AMPPNP, α3 comes closer to α2 and assumes a shorter length. This conformation is stabilized by specific interactions between L5 and the α3 helix. In particular there is a salt bridge between lysine 207, at the amino terminus of α3, and glutamate 128 of L5 and a hydrophobic contact between tyrosine 211, in the middle of α3, and tryptophan 127 of L5 interactions that are absent in the ADP structure. These crystal structures as well as our simulations suggest that L5 functions by interacting with the α3 helix. Changes in this interaction that might occur in transitions from rigor to ATP to ADP states could then be transmitted to switch I and, ultimately, to other downstream elements involved in microtubule binding and neck linker orientation.
Cryo-EM Reconstructions Suggest That Motor Binding to the Microtubule Stabilizes an L5-α3 Interaction
Previous cryo-EM reconstructions of Eg5-decorated microtubules in the presence of AMPPNP demonstrated an extra density not accounted for by docking of available crystal structures (11). This was attributed to a new conformation of L5, which the authors proposed was closely apposed to α3 when the motor was strongly bound to the microtubule. This is illustrated in the orthogonal projections in Fig. 8, panels A and C, which show that docking of the Eg5·AMPPNP crystal structure in the cryo-EM reconstructions (yellow) leaves L5 protruding from the surface density, away from the density that is presumed to reflect the microtubule-bound L5 orientation (dotted outline, (24)). This is also true for the Eg5·ADP crystal structure (Fig. 8, B and D, green and Ref. 4), and consistent with our experimental data and MD simulations, we interpret this as providing evidence that L5 undergoes conformational changes during Eg5 MT binding. The MT is an active, if allosteric, participant in these conformational changes, explaining why crystal structures, necessarily calculated in the absence of MTs, do not capture this conformation. However, the kinesin motor Nod does demonstrate a clear interaction between L5 and α3 (12), and we have also docked this crystallographic model into the cryo-EM density. This is depicted as the lilac structure in Fig. 8, B and D, and reveals that Pro-102 in this motor (equivalent to Pro-121 in Eg5) makes close contact with α3 and approaches the putative L5 density in the Eg5 cryo-EM reconstruction. We propose that a similarly close contact forms between Eg5 Pro-121 and α3. In contrast, the invariant position of Pro-131 in all the Eg5 crystal structures suggests that its side chain is essential in guiding the conformation of L5 through the Eg5 ATPase cycle and explains why mutating it has a profound effect on motor function.
FIGURE 8.
Docking of kinesin crystal structures into the cryo-electron microscopy reconstruction of KLP61F (Drosophila Eg5)·MT complex. A and C, shown are orthogonal views of the docked Eg5-AMPPNP crystal structure which closely matches the cryo-EM density (11) for the length of α3 (red) and length and orientation of α4 (not shown). However, L5 protrudes from the cryo-EM structure, and the putative density for the L5 in the MT-bound motor (dotted oval) remains unfilled. B and D, shown are orthogonal views of docked Eg5-AMPPNP crystal structure compared with the Eg5-ADP crystal structure (green (4)) and the Nod-ADP crystal structure (lilac (12)). Note that the position of Pro-131, corresponding to Pro-107 in Nod, is very similar in all of the structures. Pro-121 is in a similar position for Eg5-AMPPNP and Eg5-ADP, whereas the corresponding residue in Nod (Pro-102, indicated by a red asterisk) is positioned toward α3, although it also still protrudes from the putative L5 cryo-EM density.
L5 Can Act as a Conformational Latch
We propose that L5 can reversibly contact α3 and when it does so, induces a conformation in switch I that can accommodate ATP. If this conformational change were required for ATP binding, it could be rate-limiting for this process and might explain the relatively slow kinetics of ATP binding that we previously reported for this motor (13). This model could also explain the effect of the P131A mutation. This proline appears to induce a bend in this loop (Fig. 1), consistent with a role prolines play in protein structure (16). This proline-induced bend would be expected to affect the global orientation of L5, which in turn could affect how L5 interacts with α3. Any disruption in this bend might destabilize the L5-α3 interaction and markedly slow the conformational change in switch I needed for ATP binding and for subsequent conformational changes, including neck linker movement and microtubule dissociation. Proline 121 is in the middle of the loop portion of L5 (Fig. 1), and its mutation to alanine might, therefore, be expected to have less of an effect on the structure of nucleotide binding site. However, as noted above, this mutation produces an ionic strength-dependent slowing of microtubule dissociation. We note that in the strong microtubule binding AMPPNP crystallographic structure (Fig. 1), an ionic bond (Lys-207—Glu-128) stabilizes an interaction between α3 and L5, and this bond appears to be broken in the weak binding, ADP structure. We postulate that the P121A mutation stabilizes this bond, perhaps by altering the flexibility of L5 and allowing a closer approach of these residues to each other. This might be expected to make the transition from a strong microtubule binding, “AMPPNP” structure to a weak microtubule binding, “ADP” structure slower at low ionic strength and thereby enhance apparent microtubule affinity and chemical processivity.
Taken together, our results suggest that L5 can function as a “conformational latch” that by reversibly interacting with α3 regulates how quickly ATP can bind to the catalytic site and induce downstream changes in microtubule binding and neck linker orientation. Consistent with this are prior studies, which have shown that nucleotide and microtubule binding can change the mobility and orientation of L5 (10, 21). ATP binding induces movement in kinesins, so we might a priori expect that kinesins with different physiologic roles would differ in the kinetics of ATP binding. This variability in ATP binding kinetics might, therefore, be a reflection in a corresponding variability in the length of L5. This role of L5 in modulating the kinetics of strong ATP binding is also consistent with the effect of small molecule inhibitors, such as monastrol, which by binding to this loop, interfere with its interaction with α3 and induce this helix to remain elongated, in an “ADP-like” conformation (Fig. 1 and Ref. 25). These conclusions in turn suggest that L5 is a dynamic structure whose conformation changes through the course of the ATPase cycle, and the timing and nature of these changes will be investigated in future studies.
Supplementary Material
Acknowledgment
We thank Mr. Christopher Beadle for his excellent technical support.
This work was supported, in whole or in part, by National Institutes of Health Grant AR048565 (NIAMS; to S. S. R.). This work was also supported by the Biotechnology and Biological Sciences Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- AEDANS
- 5-({2-[(iodoacetyl)amino]ethyl}amino)naphthalene-1-sulfonic acid
- mant
- N-methyl anthraniloyl
- 2′dmD
- 2′-deoxy,3′-mant ADP
- 2′dmT
- 2′-deoxy,3′-mant ATP
- FlAsH
- 4′,5′-bis(1,3,2-dithiarsolan-2-yl) fluorescein
- FRET
- fluorescence resonance energy transfer
- MDCC
- N-[2-(1-maleimidyl)ethyl]-7 (diethylamino)coumarin-3-carboxamide
- r.m.s.f.
- root mean square fluctuation
- AMPPNP
- adenosine 5′-(β,γ-imino)triphosphate
- MT
- microtubule.
REFERENCES
- 1. Marx A., Müller J., Mandelkow E. (2005) Adv. Protein Chem. 71, 299–344 [DOI] [PubMed] [Google Scholar]
- 2. Kull F. J., Endow S. A. (2002) J. Cell Sci. 115, 15–23 [DOI] [PubMed] [Google Scholar]
- 3. Kikkawa M., Sablin E. P., Okada Y., Yajima H., Fletterick R. J., Hirokawa N. (2001) Nature 411, 439–445 [DOI] [PubMed] [Google Scholar]
- 4. Turner J., Anderson R., Guo J., Beraud C., Fletterick R., Sakowicz R. (2001) J. Biol. Chem. 276, 25496–25502 [DOI] [PubMed] [Google Scholar]
- 5. Brier S., Lemaire D., DeBonis S., Forest E., Kozielski F. (2006) J. Mol. Biol. 360, 360–376 [DOI] [PubMed] [Google Scholar]
- 6. Kim E. D., Buckley R., Learman S., Richard J., Parke C., Worthylake D. K., Wojcik E. J., Walker R. A., Kim S. (2010) J. Biol. Chem. 285, 18650–18661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DeBonis S., Simorre J. P., Crevel I., Lebeau L., Skoufias D. A., Blangy A., Ebel C., Gans P., Cross R., Hackney D. D., Wade R. H., Kozielski F. (2003) Biochemistry 42, 338–349 [DOI] [PubMed] [Google Scholar]
- 8. Luo L., Carson J. D., Dhanak D., Jackson J. R., Huang P. S., Lee Y., Sakowicz R., Copeland R. A. (2004) Biochemistry 43, 15258–15266 [DOI] [PubMed] [Google Scholar]
- 9. Wood K. W., Lad L., Luo L., Qian X., Knight S. D., Nevins N., Brejc K., Sutton D., Gilmartin A. G., Chua P. R., Desai R., Schauer S. P., McNulty D. E., Annan R. S., Belmont L. D., Garcia C., Lee Y., Diamond M. A., Faucette L. F., Giardiniere M., Zhang S., Sun C. M., Vidal J. D., Lichtsteiner S., Cornwell W. D., Greshock J. D., Wooster R. F., Finer J. T., Copeland R. A., Huang P. S., Morgans D. J., Jr., Dhanak D., Bergnes G., Sakowicz R., Jackson J. R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5839–5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Larson A. G., Naber N., Cooke R., Pate E., Rice S. E. (2010) Biophys. J. 98, 2619–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bodey A. J., Kikkawa M., Moores C. A. (2009) J. Mol. Biol. 388, 218–224 [DOI] [PubMed] [Google Scholar]
- 12. Cochran J. C., Sindelar C. V., Mulko N. K., Collins K. A., Kong S. E., Hawley R. S., Kull F. J. (2009) Cell 136, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosenfeld S. S., Xing J., Jefferson G. M., King P. H. (2005) J. Biol. Chem. 280, 35684–35695 [DOI] [PubMed] [Google Scholar]
- 14. Moyer M. L., Gilbert S. P., Johnson K. A. (1998) Biochemistry 37, 800–813 [DOI] [PubMed] [Google Scholar]
- 15. Adams S. R., Campbell R. E., Gross L. A., Martin B. R., Walkup G. K., Yao Y., Llopis J., Tsien R. Y. (2002) J. Am. Chem. Soc. 124, 6063–6076 [DOI] [PubMed] [Google Scholar]
- 16. Creighton T. E. (1983) Proteins: Structures and Molecular Principles, W. H. Freeman and Co., New York [Google Scholar]
- 17. Klumpp L. M., Mackey A. T., Farrell C. M., Rosenberg J. M., Gilbert S. P. (2003) J. Biol. Chem. 278, 39059–39067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nitta R., Kikkawa M., Okada Y., Hirokawa N. (2004) Science 305, 678–683 [DOI] [PubMed] [Google Scholar]
- 19. Lad L., Luo L., Carson J. D., Wood K. W., Hartman J. J., Copeland R. A., Sakowicz R. (2008) Biochemistry 47, 3576–3585 [DOI] [PubMed] [Google Scholar]
- 20. Maliga Z., Kapoor T. M., Mitchison T. J. (2002) Chem. Biol. 9, 989–996 [DOI] [PubMed] [Google Scholar]
- 21. Cochran J. C., Gilbert S. P. (2005) Biochemistry 44, 16633–16648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hackney D. D. (1995) Nature 377, 448–450 [DOI] [PubMed] [Google Scholar]
- 23. Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., Vale R. D. (1999) Nature 402, 778–784 [DOI] [PubMed] [Google Scholar]
- 24. Parke C. L., Wojcik E. J., Kim S., Worthylake D. K. (2010) J. Biol. Chem. 285, 5859–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maliga Z., Mitchison T. J. (2006) BMC Chem. Biol. 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hiratsuka T. (1983) Biochim. Biophys. Acta 742, 496–508 [DOI] [PubMed] [Google Scholar]
- 27. Maliga Z., Xing J., Cheung H., Juszczak L. J., Friedman J. M., Rosenfeld S. S. (2006) J. Biol. Chem. 281, 7977–7982 [DOI] [PubMed] [Google Scholar]
- 28. Rosenfeld S. S., Jefferson G. M., King P. H. (2001) J. Biol. Chem. 276, 40167–40174 [DOI] [PubMed] [Google Scholar]
- 29. Lindahl E., Hess B., Van Der Spoel D. (2001) J. Mol. Mod. 7, 306–317 [Google Scholar]
- 30. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.