Abstract
The DExH protein RNA helicase A (RHA) plays numerous roles in cell physiology, and post-transcriptional activation of gene expression is a major role among them. RHA selectively activates translation of complex cellular and retroviral mRNAs. Although RHA requires interaction with structural features of the 5′-UTR of these target mRNAs, the molecular basis of their translation activation by RHA is poorly understood. RHA contains a conserved ATPase-dependent helicase core that is flanked by two α-β-β-β-α double-stranded RNA-binding domains at the N terminus and repeated arginine-glycine residues at the C terminus. The individual recombinant N-terminal, central helicase, and C-terminal domains were evaluated for their ability to specifically interact with cognate RNAs by in vitro biochemical measurements and mRNA translation assays in cells. The results demonstrate that N-terminal residues confer selective interaction with retroviral and junD target RNAs. Conserved lysine residues in the distal α-helix of the double-stranded RNA-binding domains are necessary to engage structural features of retroviral and junD 5′-UTRs. Exogenous expression of the N terminus coprecipitates junD mRNA and inhibits the translation activity of endogenous RHA. The results indicate that the molecular basis for the activation of translation by RHA is recognition of target mRNA by the N-terminal domain that tethers the ATP-dependent helicase for rearrangement of the complex 5′-UTR.
Keywords: Gene Expression, RNA-binding Protein, RNA Helicase, RNA Structure, RNA Viruses, Translational Control, Virus
Introduction
RNA helicase A (RHA)2 is a ubiquitous DEIH superfamily 2 helicase that is necessary for translation of retroviral and selected cellular mRNAs (1–3). A unifying feature of RHA-responsive mRNA templates is a structurally complex 5′-UTR that is a platform for alternative RNA-protein interactions that modulate gene expression (8). All retroviruses require a complex 5′-UTR for productive viral replication (8). Alternative 5′-UTR RNA-RNA and RNA-protein interactions generate alternatively spliced mRNAs that are templates for synthesis of viral proteins or unspliced genome-length RNA that is dimerized and assembled into virions. The later serves as a template for reverse transcription upon infection. RHA is necessary for the translation of the junD proto-oncogene and the retroviruses that infect a variety of metazoan host cells (1–3, 21, 22). RHA interacts with structural features of their complex 5′-UTR and facilitates polyribosome loading and productive translation (1–3). JunD is an AP1 transcription factor that is necessary for healthy cell growth and the expression of which is stringently controlled at the transcriptional and post-transcriptional levels (23). The necessary role of RHA in translation of junD and retroviruses posits the hypothesis that conserved features of RHA determine recognition of these target mRNAs.
Site-directed mutagenesis and reporter assays have determined structural features of the complex 5′-UTR that are necessary for RHA interaction and translation activity (1–3, 24). RHA-dependent translation activity is conferred by orientation-dependent activity of the ∼150-nucleotide 5′ terminus, which is designated the post-transcriptional control element (PCE) (13, 14). PCE activity is attributable to two functionally redundant stem-loop structures (A and C) (2, 21). Mutations that disrupt structural features of structures A and C (referred to as mutAC) in spleen necrosis virus (SNV), an avian retrovirus, eliminate translation activity and detectable interaction with epitope-tagged RHA (FLAG-RHA) in cells, whereas compensatory mutations restore translation activity and interaction with FLAG-RHA (1–3, 24). These observations provide a framework to determine the residues of RHA necessary for functionally relevant interaction with PCE.
The architecture of RHA is complex, with a conserved ATPase-dependent helicase domain that is flanked by motifs frequently observed in functionally distinct post-transcriptional regulatory proteins (4–6). The N-terminal residues contain two repeats of a double-stranded RNA-binding domain (dsRBD) that is observed widely among RNA-interactive proteins (5), e.g. the dsRBD in protein kinase R (PKR) and Dicer direct interaction with generic viral double-stranded RNA and small duplex RNAs, respectively (4, 7). By comparison, RHA selectively recognizes the complex 5′-UTR of a subset of cap-dependent mRNA templates (8). The possible role for the dsRBD in the selective recognition of RHA target mRNAs is unknown.
The C terminus of RHA is highly conserved from canine species to Caenorhabditis elegans (NCBI Homolog Database) and is characterized by repeated arginine and glycine (RG) residues that are substrates for methylation by PRMT1 (9). A similar RG-rich domain is observed in nucleolin and heterogeneous nuclear ribonucleoprotein A1 and interacts with protein cofactors that tether target mRNA (10–12). Moreover, the RG-rich domain of fragile X mental retardation protein directly engages target RNA at G quartets (13, 14). These findings suggest the possibility that the RG-rich domain of RHA is important for interaction with target mRNAs.
DExH/D box proteins have been identified among all prokaryotes and eukaryotes examined and affect all steps in RNA metabolism (6, 15). Named for the amino acid residues that are adjacent to the ATP-binding region (16, 17), the helicase domain spans 350–400 amino acids and exhibits RNA-binding capability (18–20). For instance, mutations in the helicase domain of retinoic acid inducible gene-I (RIG-I) disrupt binding of RIG-I to poly(I:C) or in vitro transcribed dsRNA (19). The possibility that the helicase domain of RHA binds target mRNA remains to be determined.
Herein, individual recombinant RHA domains were evaluated for binding activity to PCE in comparison with nonfunctional PCE and generic control double-stranded RNAs (dsRNAs). The results of biochemical and biophysical experiments show that the N-terminal domain exhibits higher binding affinity for PCE than for nonfunctional mutant RNA or control dsRNA. Highly conserved surface-exposed lysine residues were required for selective interaction with PCE RNA. By comparison, the isolated DEIH domain lacked detectable binding to the PCE RNAs tested in our study, and the C-terminal RG-rich domain bound to negative control dsRNAs with greater affinity than to the PCE RNAs tested here. In cells, the N-terminal domain directed interaction with PCE, mRNA and its exogenous expression blocked the translation activity of endogenous RHA. Our results demonstrate that N-terminal residues are necessary for recognition of PCE RNA and that this interaction is necessary for the translation activation of selected cellular and viral mRNAs.
EXPERIMENTAL PROCEDURES
RNA Transcription
In vitro transcripts were generated in the RiboMAXTM large-scale RNA production system (Promega) in the presence of [α-32P]UTP/[α-32P]CTP (PerkinElmer Life Sciences), treated with DNase (Promega), separated on 8% denaturing urea gels, and eluted in passive gel elution buffer (Ambion). RNAs were precipitated in 95% ethanol and 0.3 m NaOAc at −80 °C for 20 min, collected by centrifugation, resuspended in diethyl pyrocarbonate-treated water, and monitored by scintillation counting. The details of template and primer DNA sequences are provided under supplemental “Experimental Procedures.”
Expression and Purification of Recombinant Proteins
The preparation of expression plasmids for the RHA N-terminal (N-term), DEIH, and C-terminal (C-term) domains is described under supplemental “Experimental Procedures.” The proteins were expressed in BL21-CodonPlus optimized cells (Stratagene). After induction with 1 mm isopropyl-β-d-thiogalactopyranoside (United States Biochemical Corp.) for 2.5 h at 33 °C, cells were resuspended in PBS with 10 μl/ml protease inhibitor mixture (Sigma) and 1 μl/ml dithiothreitol (1 m) and lysed with an Aminco French pressure cell at 5000 units of pressure. Soluble proteins were harvested by centrifugation in a Sorvall RC5C SS-34 rotor at 12,000 rpm for 20 min at 4 °C.
Wild-type and mutant N-terminal domains were purified from the soluble protein lysate on glutathione-Sepharose beads (Pierce). Lysate was exposed to the beads overnight, followed by four washes with 50 mm HEPES and 150 mm NaCl (pH 7.0), one wash with 50 mm HEPES and 500 mm NaCl (pH 7.0), and one wash with thrombin cleavage buffer (20 mm HEPES, pH 8, 0.15 m NaCl, and 2.5 m CaCl2). Biotinylated thrombin (10 units) was used to release N-term from the GST tag and was removed from the solution by incubation with streptavidin. For purification of the DEIH domain, soluble protein lysate was incubated with 2 ml of glutathione-Sepharose (GE Healthcare) for 30 min at room temperature. The beads were then washed with 20 ml of buffer containing 50 mm HEPES, pH 7.3, 500 mm NaCl, 5 mm DTT, and 1 mm EDTA. The DEIH domain was eluted from the beads using buffer containing 50 mm HEPES, pH 7.3, 25 mm reduced glutathione, 500 mm NaCl, 5 mm DTT, and 1 mm EDTA. Glutathione from the protein preparation was removed by dialysis of protein fractions in the same buffer without glutathione. C-term was purified from the soluble protein lysate on nickel-Sepharose (GE Healthcare). After a 1-h incubation of the lysate with 2 ml of nickel beads at room temperature, nonspecifically bound proteins were removed by extensive washing of the beads with 20 ml of buffer containing 50 mm HEPES, pH 7.4, 500 mm NaCl, 7.5 mm CHAPS, 20 mm imidazole, and 4 mm β-mercaptoethanol. C-term was then eluted in 50 mm HEPES, pH 7.4, 500 mm NaCl, 7.5 mm CHAPS, 500 mm imidazole, 5 mm EDTA, and 4 mm β-mercaptoethanol and dialyzed against the same buffer without imidazole. Protein preparations were evaluated for purity by PAGE and quantified using the Bio-Rad DC protein assay.
EMSA
EMSAs were performed as described (1) with recombinant protein and 100,000 cpm of in vitro transcribed α-32P-labeled RNA (see supplemental “Experimental Procedures”) in EMSA buffer (2% glycerol, 0.8 mm EGTA, 0.2 mm EDTA, 2 mm Tris, pH 7.6, 14 mm KCl, and 0.2 mm Mg(OAc)2). Control experiments to establish equilibrium conditions varied the incubation time and temperature and RNA and protein concentrations. Electrophoresis was performed at 4 °C in 5% native Tris borate/EDTA-acrylamide gels, which were fixed, dried, and exposed overnight in a PhosphorImager cassette. EMSAs were repeated with at least three independent preparations of protein and RNA. For competition experiments, α-32P-labeled PCE RNA and recombinant protein were combined prior to the addition of unlabeled competitor RNA.
Fluorescence Anisotropy (FA) Measurements
Synthetic RNA oligonucleotides labeled at the 5′-nucleotide with fluorescein were purchased from Dharmacon. To fold RNA, 3 μl of 20 μm RNA was incubated in 5.25 μl of diethyl pyrocarbonate-treated water, 3.75 μl of 100 mm HEPES, pH 7.5, and 1.5 μl of 1 m NaCl at 80 °C for 2 min and at 60 °C for 2 min, followed by the addition of 1.5 μl of 100 mm MgCl2. Diethyl pyrocarbonate-treated water was added to a final volume of 300 μl, and the mixture was incubated on ice for 30 min. FA measurements were performed in triplicate in Corning 3676 low volume 384-well black nonbinding surface polystyrene plates with 20 nm RNA and the indicated amounts of recombinant protein in EMSA buffer. Reactions were incubated for 30 min at room temperature in the dark to allow samples to reach equilibrium. Samples were excited at 485 nm, and the emission intensities at 530 nm from the parallel and perpendicular planes were measured on a SpectraMax M5 plate reader system (Molecular Devices), which measures significant anisotropy changes above a cutoff of 0.04. The equilibrium dissociation constants (Kd) were obtained by fitting the binding curves to a single-binding site model on KaleidaGraph as described previously (25). Weighted averages and S.D. were calculated as described (26).
RNA Coprecipitation
HEK293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic (Invitrogen). For transfection, 1 × 106 HEK293 cells in a 10-cm dish were cultured overnight, and 5 μg of the respective plasmid was transfected in duplicate with FuGENE 6 (Roche Applied Science; 3:1 (w/v) FuGENE 6/DNA) following the manufacturer's protocol at 37 °C in 5% CO2 for 48 h. Cells were harvested in PBS and resuspended in 100 μl of polysome buffer (100 mm KCl, 5 mm MgCl2, 10 mm HEPES, pH 7.0, 0.5% Nonidet P-40, and 1 mm DTT) supplemented with RNaseOUT (Invitrogen) and a protease inhibitor mixture of serine, cysteine, and aspartic proteases and aminopeptidase (Sigma). Immunoblotting with rabbit anti-FLAG antiserum (Sigma) verified protein expression. The soluble lysate was incubated with anti-FLAG monoclonal antibody M2-conjugated agarose beads (Sigma) that were equilibrated in NT2 buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm MgCl2, and 0.05% Nonidet P-40) overnight at 4 °C. The messenger ribonucleoprotein complexes were washed with NT2 buffer. An equivalent aliquot of the input lysates and beads (15 μl) was immunoblotted with anti-FLAG antiserum to verify similar immunoprecipitation efficiency. RNA was extracted from the beads with TRIzol (Invitrogen) following the manufacturer's protocol. As described (1), reverse transcription reactions used random hexamer primer, and PCR utilized gene-specific primers (KB1303/KB1304 for junD and KB750/KB752 for GAPDH (supplemental Table S1).
RHA Translation Reporter Assay
As described (2, 8), 1 × 106 HEK293 cells in duplicate 10-cm dishes were cultured overnight and transfected with 1 μg of pYW100 and the indicated RHA expression plasmid. Cells were harvested in PBS from duplicate wells for protein and RNA analysis. Protein was detected by HIV-1 Gag ELISA (ZeptoMetrix) and immunoblotting with anti-FLAG (Stratagene) and anti-RHA (Vaxron) antisera. RNA was detected by real-time RT-PCR analysis with gene-specific primers (KB1614/KB1615 for gag and KB1252/KB1253 for actin) (supplemental Table S1) (3).
RESULTS
Specific Interaction of N-term with PCE of Viral and Cellular Origin
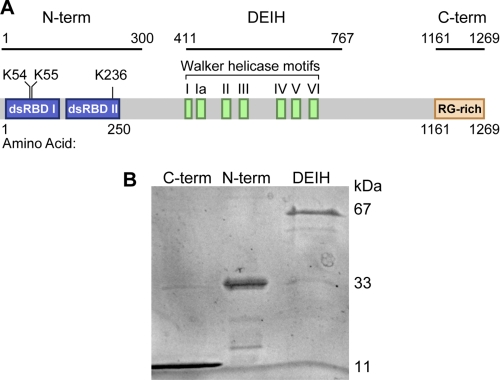
Human RHA is composed of three domains that are observed in functionally distinct post-transcriptional regulatory proteins (Fig. 1A). The helicase domain is necessary for RHA translation activity on PCE target mRNA (3). However the domain(s) necessary for RHA recognition of PCE RNAs is unknown. Therefore, to test whether selective interaction is attributable to the N-terminal, DEIH, or C-terminal domain, epitope-tagged recombinant proteins were expressed in Escherichia coli and purified by affinity chromatography (Fig. 1, A and B). EMSAs were used for initial screening of PCE-binding activity, and then fluorescence anisotropy was employed for quantitative assessment of RNA affinity. EMSAs were carried out with the following 32P-labeled transcripts: 165-nucleotide (nt) SNV PCE and 138-nt junD PCE, nonfunctional 162-nt SNV mutAC, and negative control dsRNAs tRNALys and 7SL RNA. Equilibrium binding conditions were determined by varying the incubation temperature and time. As shown in supplemental Fig. S1A, incubation for 30 or 60 min produces similar binding patterns indicating equilibrium binding by 30 min (27), and subsequent incubations were performed at 30 min.
FIGURE 1.
RHA domain structure and isolation of recombinant N-term, C-term, and DEIH polypeptides. A, depiction of the 1269-amino acid RHA and amino acid coordinates of recombinant domains evaluated in this study; B, SDS-PAGE analysis of recombinant protein preparations.
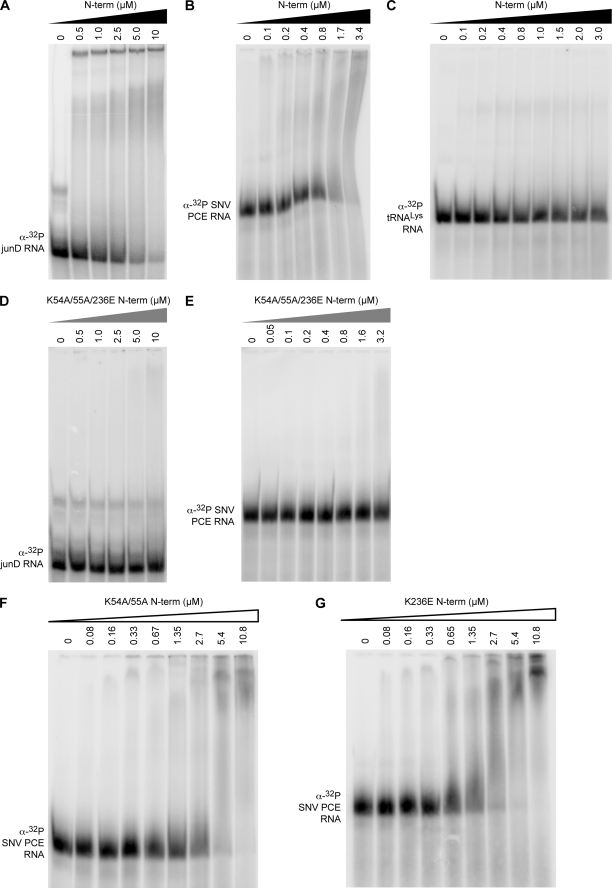
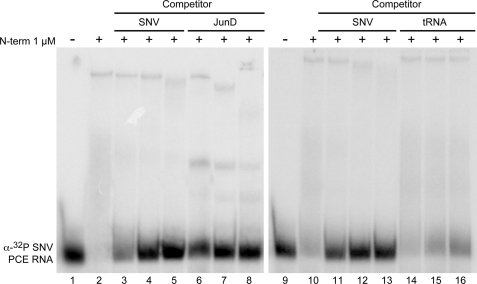
RNA EMSA detected interaction of N-term with 32P-labeled junD PCE and SNV PCE as indicated by the shift in probe mobility and diminished unbound probe (Fig. 2, A and B). By contrast, N-term did not interact significantly with 76-nt 32P-labeled tRNALys (Fig. 2C). The specificity of the N-term-PCE interaction was evaluated by competition with unlabeled SNV PCE and junD PCE. As shown in Fig. 3, robust interaction of N-term and 32P-labeled SNV PCE (compare lanes 1 and 2 or 9 and 10) was eliminated by the addition of SNV PCE RNA in 10-, 50-, and 100-fold excess (lanes 3–5 and 11–13). Moreover, unlabeled junD PCE also competed for N-term interaction with 32P-labeled SNV PCE (lanes 6–8), indicating that N-term has a similar binding affinity for SNV PCE and junD PCE. By contrast, N-term interaction with 32P-labeled SNV PCE was not competed by tRNALys in 10-, 50-, or 100-fold excess (lanes 14–16). The results indicate that N-term exhibits specific interaction with viral and cellular PCEs.
FIGURE 2.
Conserved lysines in dsRBD1 and dsRBD2 are essential for effective binding of N-term to PCE RNA. Shown are the results from EMSAs of the indicated recombinant protein and 32P-labeled RNA. A, wild-type N-term and 32P-labeled junD PCE; B, wild-type N-term and 32P-labeled SNV PCE; C, wild-type N-term and 32P-labeled tRNALys; D, K54A/K55A/K236E N-term and 32P-labeled junD PCE; E, K54A/K55A/K236E N-term and 32P-labeled SNV PCE; F, K54A/K55A N-term and 32P-labeled SNV PCE; G, K236E N-term and 32P-labeled SNV PCE.
FIGURE 3.
N-term exhibits specific interaction with PCE RNA of viral and cellular origin. Shown are the results from EMSA of N-term and 32P-labeled SNV PCE RNA (0.02 μm) (lanes 1, 2, and 9–10) upon the addition of unlabeled SNV PCE (lanes 3–5 and 11–13), junD PCE (lanes 6–8), or tRNALys (lanes 14–16). The concentrations of competitor RNA were 0.2, 1, and 2 μm, respectively. The protein concentration was 1 μm and shifted all 32P-labeled SNV PCE RNA.
Similar experiments assessed DEIH and C-terminal domains for RNA-binding activity. The DEIH domain lacked detectable binding to 32P-labeled junD PCE or SNV PCE in our assay (supplemental Fig. S2, A and B). C-term exhibited weak interaction with 32P-labeled junD PCE or SNV PCE that required a high protein concentration (10–20 μm) (supplemental Fig. S2, C and D). Moreover, nonfunctional mutant PCE exhibited slightly higher affinity for C-term (supplemental Fig. S2E). These results indicate that isolated DEIH and C-term RNA-binding activity is not selective for PCE RNA.
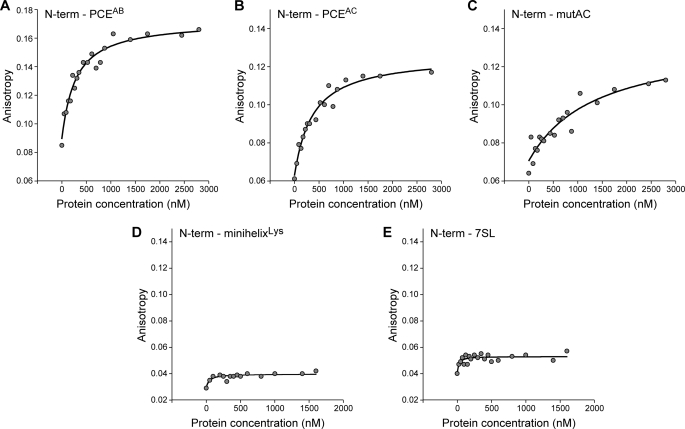
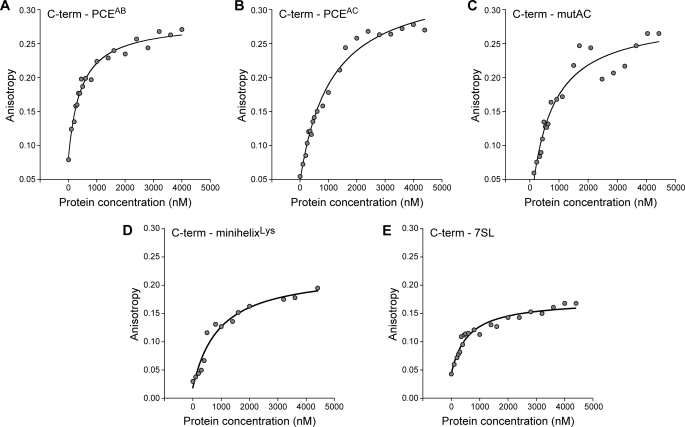
FA assays using synthetic 5′-fluorescein-tagged SNV PCE RNAs were carried out to verify the RNA-binding trends and to measure the RNA affinity. Because of the length constraints of chemical RNA synthesis (<100 nt), shorter RNAs were used for these studies. The 96-and 98-nt functionally redundant stem-loop structures PCEAC and PCEAB, which are necessary for RHA translation activity (24) and for precipitation of epitope-tagged RHA in cells (1), were chosen for this work. Control dsRNAs were again used to compare RNA binding. The mutAC structural mutant of PCEAC, which lacks translation activity and does not coprecipitate RHA in cells, was used to measure the expected reduction in binding affinity. The minihelixLys 35-nt RNA derived from the acceptor-TΨC stem of human tRNA3Lys (28) and the human 7SL 27-nt hairpin RNA provided generic control dsRNAs that lack PCE activity. PAGE confirmed the integrity of the synthetic RNAs (data not shown), and the FA of each RNA (20 nm) was measured with titration of each recombinant domain (25–3000 nm). Consistent with the EMSA, N-term bound PCEAB and PCEAC with comparable affinity (Fig. 4, A and B, and Table 1). In contrast, N-term exhibited lower affinity for mutAC by a factor of 3, indicating that the loss of functional activity by mutAC is accompanied by weakened but not eliminated binding ability for N-term. Moreover, N-term and negative control dsRNAs minihelixLys RNA and mini7SL RNA exhibited anisotropy changes of <0.04, which is below the threshold value for a statistically significant interaction (Fig. 4, C–E, and Table 1). By contrast, C-term exhibited binding to all RNAs examined (Fig. 5). As expected from the EMSA, C-term exhibited weak affinity for PCEAC and PCEAB (Table 1). However, slightly increased affinity was observed for mutAC. Moreover, C-term exhibited 4–5-fold higher affinity for the minihelixLys and 7SL negative control dsRNAs (Table 1). The results indicate that C-term exhibits greater affinity for control dsRNAs than for PCE RNA. These results argue against C-term conferring selective recognition of PCE RNA. Similar analysis of the DEIH domain determined less than detectable affinity for PCEAB, PCEAC, and mutAC by this assay (Table 1 and supplemental Fig. S3). In sum, the results of EMSA and FA measurements indicate that N-term, but not DEIH or C-term, is essential for selective recognition of PCE RNA.
FIGURE 4.
N-term binding to PCE is recapitulated and quantified by FA measurements. Shown are the results from representative FA assays to measure the binding affinity of N-term for the indicated RNA. A, PCEAB; B, PCEAC; C, mutAC; D, minihelixLys; E, 7SL RNA. Anisotropy changes >0.04 are statistically significant.
TABLE 1.
Apparent equilibrium dissociation constants for binding of recombinant RHA domains to 5′-fluorescein-labeled RNAs
FA measurements were carried out as described under “Experimental Procedures.” Values represent the mean ± S.D. of three or more independent experiments.
| RNA | Apparent Kd for indicated RHA domain |
|||||
|---|---|---|---|---|---|---|
| N-term | C-term | DEIH | K54A/K55A/K236E N-term | K54A/5K5A N-term | K236E N-term | |
| nm | ||||||
| PCEAB | 284 ± 33 | 1170 ± 92 | <MDa | <MD | 414 ± 47 | 437 ± 53 |
| PCEAC | 348 ± 50 | 1590 ± 170 | <MD | <MD | 515 ± 85 | 721 ± 120 |
| mutAC | 829 ± 160 | 993 ± 190 | <MD | ND | ND | ND |
| MinihelixLys | <MD | 222 ± 37 | NDa | ND | ND | ND |
| 7SL | <MD | 305 ± 48 | ND | ND | ND | ND |
a <MD, less than minimum detectable; ND, not determined.
FIGURE 5.
C-term lacks specificity for PCE RNA. Shown are the results from representative FA assays to measure the binding affinity of C-term for the indicated RNA. A, PCEAB; B, PCEAC; C, mutAC; D, minihelixLys; E, 7SL RNA.
N-term Is Necessary for RNA Binding
All dsRBDs exhibit the α-β-β-β-α topology and conserved lysine residues (4). The crystal structure of the African clawed frog Xlrbpa dsRBD, a homolog of human PKR, in complex with a 10-nt dsRNA revealed that these basic residues are surface-exposed (29). In RHA, these residues correspond to Lys-54 and Lys-55 in dsRBD1 and to Lys-236 in dsRBD2. To assess their potential roles in PCE interaction, the following mutant proteins were prepared: (i) the triple mutant containing K54A, K55A, and K236E substitutions (referred to hereafter as K54A/K55A/K236E N-term); (ii) the double mutant containing K54A and K55A (designated K54A/K55A N-term); and (iii) the single K236E mutant (termed K236E N-term). The RNA-binding affinity of these proteins was assessed by EMSA and FA measurement.
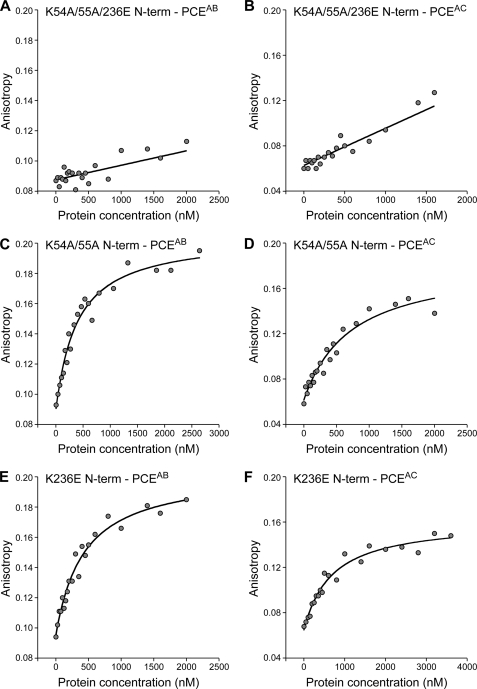
The EMSA results determined that K54A/K55A/K236E N-term produced a severe reduction in interaction with junD PCE or SNV PCE compared with N-term (Fig. 2, compare D and E with A and B). By contrast, K54/K55A N-term and K236E N-term exhibited relatively minor reduction in interaction with SNV PCE (Fig. 2, compare F, G, and B). Similar trends were observed in the FA measurements. K54A/K55A/K236E N-term severely reduced the affinity for PCEAB or PCEAC (Fig. 6, A and B, and Table 1), whereas K54A/K55A N-term or K236E N-term sustained weak interaction (Fig. 6, C–F, and Table 1). The FA measurements (Table 1) determined that K54A/K55A N-term in dsRBD1 or K236E N-term in dsRBD2 produced a minor (factor of <2) reduction in binding affinity for PCEAB RNA or PCEAC RNA relative to N-term. However, K54A/K55A/K236E N-term eliminated detectable interaction with PCEAB or PCEAC. In sum, the results indicate that conserved lysine residues of both dsRBD1 and dsRBD2 contribute to PCE RNA interaction. Mutation of the conserved lysine residues in both dsRBD1 and dsRBD2 is necessary to eliminate interaction with PCE RNA.
FIGURE 6.
Mutation of conserved lysine residues eliminates affinity of N-term for PCE RNA. Shown are the results from representative FA assays to measure the binding affinity of the indicated N-term mutant protein and RNA. A, K54A/K55A/K236E N-term and PCEAB; B, K54A/K55A/K236E N-term and PCEAC; C, K54A/K55A N-term and PCEAB; D, K54A/K55A N-term and PCEAC; E, K236E N-term and PCEAB; F, K236E N-term and PCEAC.
N-term Is Sufficient for Interaction with PCE RNA in Cells
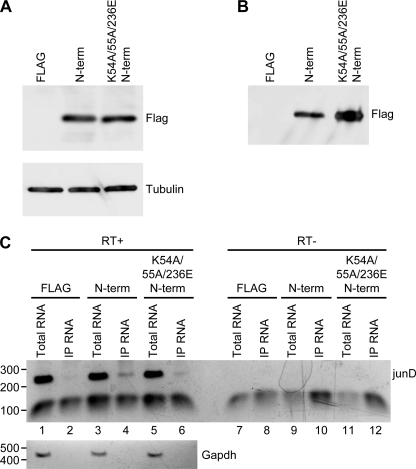
RNA coprecipitation was used to determine whether or not the PCE RNA-binding activity of N-term is recapitulated in cells. HEK293 cells were transfected with plasmids that express FLAG-tagged N-term or K54A/K55A/K236E N-term in four independent experiments. Western blotting verified equivalent expression of FLAG-tagged N-term and K54A/K55A/K236E N-term and also that the proteins were immunoprecipitated (Fig. 7, A and B). Total RNA and RNA isolated from the immunoprecipitations were harvested and evaluated by RT-PCR with primers specific for junD and GAPDH (Fig. 7C). As expected, total RNA preparations displayed junD and GAPDH transcripts (Fig. 7C, lanes 1, 3, and 5). N-term immunoprecipitated RNA exhibited junD but lacked detectable GAPDH transcript (lane 4), as observed previously for FLAG-RHA (1). K54A/K55A/K236E N-term immunoprecipitated RNA exhibited a severe reduction in junD RNA (lane 6). The negative control FLAG immunoprecipitate lacked detectable junD RNA (lane 2). No amplification products were detected in reactions that lacked reverse transcriptase (lanes 7–12). The results demonstrate that N-term is sufficient for interaction with PCE RNA in cells and that the interaction is severely reduced by the lysine mutations.
FIGURE 7.
N-term interacts with PCE RNA in cells. Shown are the results from a representative immunoprecipitation (IP) assay to compare PCE RNA coprecipitation with N-term proteins. Empty FLAG plasmid, FLAG-tagged N-term, or FLAG-tagged K54A/K55A/K236E N-term was expressed in transfected HEK293 cells, and FLAG-tagged proteins were subjected to immunoprecipitation with anti-FLAG antibody (mouse) conjugated to agarose beads. Immunoblotting of equivalent aliquots of input lysate or immunoprecipitate on agarose beads determined that the immunoprecipitation efficiency was similar for each N-term protein. RNA was harvested from the remaining beads and subjected to RT-PCR with junD or GAPDH primer. A, Western blotting with FLAG-specific antiserum (rabbit) detected equivalent expression of N-term and K54A/K55A/K236E N-term. The 9-amino acid FLAG peptide was not resolved. B, Western blotting with FLAG-specific antiserum (rabbit) detected that immunoprecipitation was similar for N-term and K54A/K55A/K236E N-term. C, PCR amplification products from the indicated RNA preparation incubated with or without reverse transcriptase (RT+ or RT−, respectively). Representative samples of total RNA and RNA isolated from immunoprecipitated samples were evaluated with junD- or GAPDH-specific primers. The junD amplification product is 232 bp, and the lower band is the primer dimer.
N-term Dominantly Interferes with Translation of PCE Reporter RNA
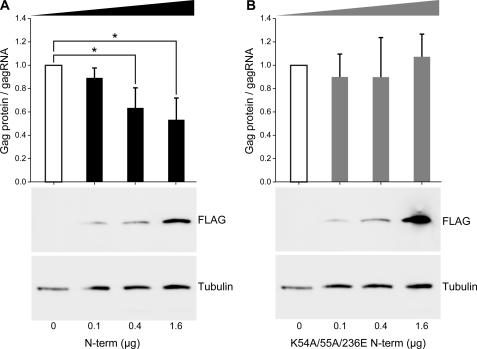
Because N-term binds to PCE RNA in 293 cells (Fig. 7), we hypothesized that it could inhibit translation of PCE target RNA by endogenous RHA. To test this, HEK293 cells were transfected with PCE reporter plasmid and increasing amounts of plasmids that express FLAG-tagged N-term or K54A/K55A/K236E N-term. Western blotting with anti-FLAG antiserum verified comparable expression of N-term and K54A/K55A/K236E N-term; control anti-tubulin antiserum verified equivalent protein loading (Fig. 8, A and B). To determine PCE activity, HIV-1 Gag protein production was measured by ELISA, and gag RNA levels were quantified by quantitative RT-PCR (2). The results of three independent transfection experiments indicated that PCE activity was decreased in response to N-term (p < 0.03) (Fig. 8A). By contrast, PCE activity was not affected by K54A/K55A/K236E N-term (Fig. 8B). In sum, the RNA immunoprecipitation and translation assay results demonstrate that the PCE-binding activity of N-term interferes with the translation activity of endogenous RHA, presumably by blocking productive interaction with endogenous RHA.
FIGURE 8.
Exogenously expressed N-term dominantly inhibits translation of PCE reporter RNA. Shown are the results from a representative transfection assay to determine the translation activity of endogenous RHA upon exogenous expression of N-term. HEK293 cells were transfected with pYW100 SNV PCE reporter plasmid and increasing amounts of the indicated N-term expression plasmid. Total cell protein was subjected to Gag ELISA and Western blotting with anti-FLAG and anti-tubulin antisera. Total cellular RNA was subjected to real-time RT-PCR, and control reactions lacked reverse transcriptase. PCE reporter activity was determined in three independent assays summarized graphically as Gag protein relative to gag RNA standardized to actin. Error bars indicate S.D. *, statistically significant difference (p = < 0.03). A, PCE activity is inhibited by FLAG-tagged N-term. B, PCE activity is not changed by FLAG-tagged K54A/K55A/K236E N-term.
DISCUSSION
We have demonstrated that the N-terminal domain of RHA contributes to selective recognition of viral and cellular PCE RNAs. Exogenously expressed N-term interacts with PCE target RNA and requires the conserved lysine residues within RHA dsRBD1 and dsRBD2. We observed that PCE interaction with exogenously expressed N-term blocked RHA translation activity. Conversely, mutations of its conserved lysine residues eliminated interaction with PCE RNA in cells and did not block translation of PCE RNA. Our results indicate that N-term basic residues of RHA are necessary to engage the RNA structure and to deliver the ATP-dependent helicase activity that is necessary for translation of PCE mRNA (3). This N-term interaction is selective for functional PCE because the binding affinity for nonfunctional PCE (mutAC) is reduced by a factor of 2–3, and binding to negative control dsRNAs is not detectable.
The conserved lysine residues are positioned in the second conserved α-helix of the dsRBD α-β-β-β-α topology. The crystal structure of the Xlrbpa dsRBD-RNA complex determined that the second highly conserved α-helix positions the lysine residues for interaction with the RNA backbone (29). In RHA dsRBD1, PKR, and TAR RNA-binding protein, the amino acid conservation of this α-helix is 65%, whereas conservation of the β-sheets is <20%. We postulate that the divergent residues of the β-sheets provide conformational or electrostatic interactions that are unique among the dsRBDs and facilitate their affinity for a particular category of target RNA. The position of the β-sheets on top of the α-helices directs solvent-exposed side chains away from the RNA. These residues are strong candidates for interaction with important protein cofactors.
Our results document that, despite the wide conservation in RNA-binding proteins, the dsRBDs are necessary for interaction with functionally distinct populations of substrate RNAs. Our results are similar in principle to the selective interaction of the budding yeast Rnt1p dsRBD with an AGNN tetraloop, which is directed by the first α-helix of the dsRBD (30, 31). Drosophila Staufen dsRBDIII contacts the stem-loop of dsRNA at the first α-helix (32). Thus, the conserved α-β-β-β-α topology is essential for contacting the target mRNA but is malleable to accommodate variable residues that generate the sequence or structure-specific recognition.
In this study, we observed that mutations of the conserved lysine residues introduced separately in dsRBD1 or dsRBD2 reduced but did not eliminate affinity for PCE RNA. By contrast, mutation of either dsRBD was sufficient to eliminate RHA translation activity on retroviral PCE RNA (data not shown). Possible explanations are as follows. 1) PCE RNA binding of the lysine residues of both dsRBD1 and dsRBD2 is necessary to reshape the conformation of the β-sheets to facilitate interaction with protein cofactor(s). This scenario is similar in principle to augmentation of eIF4A activity by interaction with eIF4B during cap-dependent translation initiation (33). 2) Alternatively, an initial interaction with protein cofactor(s) is necessary to rearrange the lysine residues for productive interaction with PCE RNA. Notably, the results of this and previous studies (1, 3) determined direct binding of RHA to PCE. However, the possibility remains that cofactor interaction refines intramolecular RNA structure to enable productive ribosome scanning of PCE mRNA.
We have shown that the dsRBDs play a predominant role in the selectivity of the RHA-PCE interaction. Our EMSA and FA measurements failed to detect PCE RNA-binding activity for the DEIH domain and revealed a low nonspecific RNA-binding affinity for C-term. Additionally, co-incubation with C-term did not enhance or inhibit binding of N-term to the PCE RNA (data not shown). An important future direction is to assess the possible role of these domains in the context of full-length RHA. In the cellular environment, similar or increased RNA affinity may be conferred by domain interactions that produce conformational changes or cofactor interaction. In the case of heterogeneous nuclear ribonucleoprotein A1, the C-terminal RGG-rich domain interacts intramolecularly and with other heterogeneous nuclear ribonucleoproteins to influence activity (11). Likewise, the RGG-rich domain of nucleolin is required for interaction with ribosomal protein L3 (10, 12). Similar to RHA, PKR possesses two N-terminal dsRBDs, and they interact with the distal kinase domain (34). For PKR, the engagement of generic dsRNA by dsRBD reveals the kinase domain in an active conformation (34). By analogy, our results indicate that selective engagement of PCE RNA is necessary for RHA translation activity. The possibility remains that the N-terminal dsRBD(s) interact with distal RHA domains; future experiments to test this model are warranted. The results will contribute to understanding the essential role of RHA in normal cells and the apparent contribution of RHA dysfunction to neoplastic growth (35–37). The outcomes will fuel fundamental understanding of RNA helicases across cell biology and at the virus-host interface.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. J. Marcela Hernandez for discussion, Dr. Michael Ignatov for assistance with FA calculations, and Tim Vojt for illustration.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1CA108882 (to K. B.-L.), P30CA100730 (to K. B.-L. and M. K.), RO1GM065056 (to K. M.-F.), and P01CA16058 (to the Ohio State University Comprehensive Cancer Center). This work was also supported by a Glenn Barber predoctoral fellowship from the Ohio State University College of Veterinary Medicine (to A. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S3, and Table 1.
- RHA
- RNA helicase A
- PCE
- post-transcriptional control element
- SNV
- spleen necrosis virus
- dsRBD
- double-stranded RNA-binding domain
- PKR
- protein kinase R
- dsRNA
- double-stranded RNA
- N-term
- N-terminal domain
- C-term
- C-terminal domain
- FA
- fluorescence anisotropy
- nt
- nucleotide(s).
REFERENCES
- 1. Hartman T. R., Qian S., Bolinger C., Fernandez S., Schoenberg D. R., Boris-Lawrie K. (2006) Nat. Struct. Mol. Biol. 13, 509–516 [DOI] [PubMed] [Google Scholar]
- 2. Bolinger C., Yilmaz A., Hartman T. R., Kovacic M. B., Fernandez S., Ye J., Forget M., Green P. L., Boris-Lawrie K. (2007) Nucleic Acids Res. 35, 2629–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bolinger C., Sharma A., Singh D., Yu L., Boris-Lawrie K. (2010) Nucleic Acids Res. 38, 1686–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fierro-Monti I., Mathews M. B. (2000) Trends Biochem. Sci. 25, 241–246 [DOI] [PubMed] [Google Scholar]
- 5. Burd C. G., Dreyfuss G. (1994) Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 6. Fuller-Pace F. V. (2006) Nucleic Acids Res. 34, 4206–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saunders L. R., Barber G. N. (2003) FASEB J. 17, 961–983 [DOI] [PubMed] [Google Scholar]
- 8. Bolinger C., Boris-Lawrie K. (2009) Retrovirology 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith W. A., Schurter B. T., Wong-Staal F., David M. (2004) J. Biol. Chem. 279, 22795–22798 [DOI] [PubMed] [Google Scholar]
- 10. Ghisolfi L., Kharrat A., Joseph G., Amalric F., Erard M. (1992) Eur. J. Biochem. 209, 541–548 [DOI] [PubMed] [Google Scholar]
- 11. Cartegni L., Maconi M., Morandi E., Cobianchi F., Riva S., Biamonti G. (1996) J. Mol. Biol. 259, 337–348 [DOI] [PubMed] [Google Scholar]
- 12. Bouvet P., Diaz J. J., Kindbeiter K., Madjar J. J., Amalric F. (1998) J. Biol. Chem. 273, 19025–19029 [DOI] [PubMed] [Google Scholar]
- 13. Darnell J. C., Jensen K. B., Jin P., Brown V., Warren S. T., Darnell R. B. (2001) Cell 107, 489–499 [DOI] [PubMed] [Google Scholar]
- 14. Zanotti K. J., Lackey P. E., Evans G. L., Mihailescu M. R. (2006) Biochemistry 45, 8319–8330 [DOI] [PubMed] [Google Scholar]
- 15. Rocak S., Linder P. (2004) Nat. Rev. Mol. Cell Biol. 5, 232–241 [DOI] [PubMed] [Google Scholar]
- 16. Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. (1989) Nature 337, 121–122 [DOI] [PubMed] [Google Scholar]
- 17. Tanner N. K., Linder P. (2001) Mol. Cell 8, 251–262 [DOI] [PubMed] [Google Scholar]
- 18. Linder P. (2006) Nucleic Acids Res. 34, 4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bamming D., Horvath C. M. (2009) J. Biol. Chem. 284, 9700–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cordin O., Banroques J., Tanner N. K., Linder P. (2006) Gene 367, 17–37 [DOI] [PubMed] [Google Scholar]
- 21. Butsch M., Hull S., Wang Y., Roberts T. M., Boris-Lawrie K. (1999) J. Virol. 73, 4847–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hull S., Boris-Lawrie K. (2002) J. Virol. 76, 10211–10218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hernandez J. M., Floyd D. H., Weilbaecher K. N., Green P. L., Boris-Lawrie K. (2008) Oncogene 27, 4757–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roberts T. M., Boris-Lawrie K. (2003) J. Virol. 77, 11973–11984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stewart-Maynard K. M., Cruceanu M., Wang F., Vo M. N., Gorelick R. J., Williams M. C., Rouzina I., Musier-Forsyth K. (2008) J. Virol. 82, 10129–10142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taylor J. R. (1997) An Introduction to Error Analysis: The Study of Uncertainties in Physical Measurements, University Science Books, Sausalito, CA [Google Scholar]
- 27. Hellman L. M., Fried M. G. (2007) Nat. Protoc. 2, 1849–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stello T., Hong M., Musier-Forsyth K. (1999) Nucleic Acids Res. 27, 4823–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ryter J. M., Schultz S. C. (1998) EMBO J. 17, 7505–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nagel R., Ares M., Jr. (2000) RNA 6, 1142–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lebars I., Lamontagne B., Yoshizawa S., Aboul-Elela S., Fourmy D. (2001) EMBO J. 20, 7250–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramos A., Grünert S., Adams J., Micklem D. R., Proctor M. R., Freund S., Bycroft M., St Johnston D., Varani G. (2000) EMBO J. 19, 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rogers G. W., Jr., Richter N. J., Lima W. F., Merrick W. C. (2001) J. Biol. Chem. 276, 30914–30922 [DOI] [PubMed] [Google Scholar]
- 34. Nanduri S., Rahman F., Williams B. R., Qin J. (2000) EMBO J. 19, 5567–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schlegel B. P., Starita L. M., Parvin J. D. (2003) Oncogene 22, 983–991 [DOI] [PubMed] [Google Scholar]
- 36. Erkizan H. V., Kong Y., Merchant M., Schlottmann S., Barber-Rotenberg J. S., Yuan L., Abaan O. D., Chou T. H., Dakshanamurthy S., Brown M. L., Uren A., Toretsky J. A. (2009) Nat. Med. 15, 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guénard F., Labrie Y., Ouellette G., Beauparlant C. J., Durocher F. (2009) J. Hum. Genet. 54, 152–161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.