Abstract
Plasmodium IMC1 (inner membrane complex 1) proteins comprise components of the subpellicular network, a lattice of intermediate filaments that form a structural part of the pellicle in the zoite stages of malaria parasites. Family members IMC1a and IMC1b are differentially expressed in sporozoites and ookinetes, respectively, but have functionally equivalent roles affecting cell morphology, strength, motility, and infectivity. Because of the coincident effects of previous imc1 gene disruptions on both zoite shape and locomotion, it has been impossible to ascribe a direct involvement in motility to these proteins. We show here that a third family member, IMC1h, has a distinct differential expression pattern and localizes to the pellicle of both ookinetes and sporozoites. Knock-out of IMC1h mimics the loss-of-function phenotypes of IMC1a and IMC1b in their respective life stages, indicating that IMC1 proteins could be operating co-dependently. By generating double null mutant parasites for IMC1h and IMC1b, we tested this hypothesis: double knock-out exacerbated the phenotypes of the single knock-outs in terms of ookinete strength, motility, and infectivity but did not further affect ookinete morphology. These findings provide the first genetic evidence that IMC1 proteins can function independently of each other and contribute to gliding motility independently of cell shape.
Keywords: Cell Motility, Cytoskeleton, Gene Knock-out, Membrane Function, Parasitology
Introduction
Malaria remains the most harmful parasitic infection in people, causing an estimated half a billion clinical cases and up to one million deaths annually (1). Malaria control efforts are hampered by widespread resistance to antiparasitic drugs and insecticides, emphasizing the urgent need for novel malaria control strategies. The intervention of parasite transmission by mosquitoes is considered a vital component of a successful malaria control program. Malaria parasite transmission begins when male and female gametocytes circulating in the blood of a Plasmodium-infected host are taken up in the blood meal of a feeding vector mosquito. The following drop in temperature and rise in pH that occur in the mosquito midgut initiate the process of gametogenesis. This completes within ∼15 min, with each gametocyte producing either eight male microgametes or a single female macrogamete. After fertilization, zygotes transform over a 16–20-h period into ookinetes, which leave the midgut lumen by invading and crossing the midgut epithelium. Under the basal lamina, ookinetes transform into oocysts, which, in the following 2 weeks, grow and undergo nuclear division and ultimately cytokinesis to generate thousands of progeny sporozoites. The sporozoites leave the oocyst and invade and inhabit the insect's salivary glands, where they await transmission to a new host upon mosquito bite to initiate new malaria infections.
The motile invasive life stages (zoites) of malaria parasites (i.e. merozoites, ookinetes, and sporozoites), as well as zoites of other apicomplexan parasites, are characterized by possessing a unique cortical structure named the pellicle. Apicomplexan parasites move by substrate-mediated gliding motility that is powered by an actin-myosin motor, which, together with its associated molecules (referred to as the glideosome), is situated within the pellicle of the zoite (2). The pellicle is composed of the parasite plasma membrane, a double membrane structure named the inner membrane complex (IMC),2 and a cytoskeletal structure named the subpellicular network (SPN) that underlies the IMC (3–5). In Toxoplasma gondii, the SPN was shown to be composed of different intermediate filaments that form a two-dimensional lattice, which acts as a membrane skeleton supporting the pellicular IMC membranes and providing mechanical strength to the cell (6, 7). TgIMC1, a protein structurally related to articulins (membrane skeleton proteins of free-living protists), was identified in T. gondii as a main component of the SPN. This, in turn, led to the identification of a family of IMC1 proteins structurally related to TgIMC1 in Plasmodium and other apicomplexan parasites (8). Related proteins from dinoflagellate algae and ciliates have recently been added to this family, named alveolins, which now define the protist infrakingdom Alveolata (9).
In the genus Plasmodium, eight conserved IMC1 protein/alveolin family members, named IMC1a–IMC1h, were initially identified by us (8), while an additional family member orthologous to Pfs77 was identified subsequently (9). Two of these, IMC1a and IMC1b, were shown to be differentially expressed in sporozoites and ookinetes, respectively, and to form part of their pellicle structures in the rodent malaria species Plasmodium berghei (8, 10). IMC1a and IMC1b are structurally and functionally homologous and involved in parasite morphology, mechanical strength, gliding motility, and infectivity, in accordance with their roles as membrane skeleton proteins (8, 10). Despite these and other studies (7, 11, 12), membrane skeleton assembly and function in Plasmodium species and related parasites remain poorly understood processes. In this study, we show that a third IMC1 protein family member, IMC1h, is found in the pellicle of both ookinetes and sporozoites and acts in a very similar way to IMC1b and IMC1a, pointing to the possibility that IMC1 proteins could be operating in a mutually dependent fashion. We tested this hypothesis by generating and phenotypically analyzing an IMC1h/IMC1b double null mutant parasite line. The results obtained provide evidence that IMC1 proteins are in fact functionally autonomous and can operate independently of each other.
As shown in previous studies (8, 10), knock-out of IMC1b and IMC1a leads, in both cases, to an abnormal morphology as well as a reduction in gliding motility of the affected zoite stage. We show in this study that the same is true for IMC1h null mutants. However, because it cannot be ruled out that the observed reductions in locomotion in these null mutant parasites are caused by the zoites' irregular shape, the precise participation of IMC1 proteins in gliding motility remains unclear. Using our IMC1h/IMC1b double null mutant parasite line, we were able to address this fundamental question. Our findings provide evidence for a direct involvement of IMC1 proteins in gliding motility uncoupled from cell shape. The biological relevance of this in relation to other biochemical evidence is discussed.
EXPERIMENTAL PROCEDURES
P. berghei ANKA clone 234 parasites were maintained as cryopreserved stabilates or by mechanical blood passage and regular mosquito transmission. To purify parasites for genomic DNA extraction, white blood cells were removed from parasitemic blood by passage through CF11 columns (Whatman). Ookinete cultures were set up from gametocytemic blood as described previously (13). Where desired, ookinetes were purified from 18–20-h-old cultures via ice-cold 0.17 m ammonium chloride lysis and centrifugation at 800 × g for 15 min, followed by PBS washes. Mosquito transmission assays were carried out by direct feeds on anesthetized, parasite-infected mice with comparable parasitemia and gametocytemia as described previously (14–17). Unfed mosquitoes were removed post-feeding. Parasite transfection, pyrimethamine selection, and dilution cloning were performed as described previously (18). Prior to performing transfections, plasmid DNA was digested with KpnI and SacII to remove the vector backbone. Genomic DNA extraction and Southern blotting were performed as described previously (14). For accurate cell shape determination, images of Giemsa-stained ookinetes from neat cultures were captured by microscopy, and the cell length and width of at least 100 ookinetes were measured as described (10). Statistical analyses were carried out using Student's t test (ookinete width/length) or the Mann-Whitney U test (ookinete motility and infectivity). Western blotting and confocal microscopy were carried out as described (10).
Osmotic Shock and Viability Assays
Ookinetes in neat cultures were subjected to hypo-osmotic shock of 0.5× normal osmotic strength by adding an equal volume of water. After 5 min, normal osmotic conditions were restored by adding an appropriate amount of 10× PBS. Cell viability was scored by fluorescence microscopy in the presence of 5 ml/liter propidium iodide and 1% Hoechst 33258. Ookinetes whose nuclei stained positive for both propidium iodide and Hoechst were scored as nonviable, whereas ookinetes whose nuclei stained positive only for Hoechst were scored as viable.
Construction of Transfection Plasmids
The coding sequence of imc1h plus 0.57 kb of the 5′-UTR were PCR-amplified with primers pDNR-IMC1h-F (ACGAAGTTATCAGTCGACGGTACCTAATGGAAAAATTATCTCGTACG) and pDNR-IMC1h-R (ATGAGGGCCCCTAAGCTTATGTTTTTCTGGATAGCTTTAACATC) and introduced into SalI/HindIII-digested pDNR-EGFP (19) via In-Fusion cloning (Takara Biotech) to give plasmid pDNR-IMC1h/EGFP. A 0.71-kb sequence corresponding to the 3′-UTR of imc1h was PCR-amplified with primers pLP-IMC1h-F (AGAGCGGCCGCCCCCTAATCTTATCTTTACAAATATTAAAACAATG) and pLP-IMC1h-R (AGCTGGAGCTCCACCGCGGTTGAATAAGGGGCATGAATG) and introduced into SacII-digested pLP-hDHFR (20) via In-Fusion cloning to give plasmid pLP-DHFR/IMC1h. The IMC1h-specific sequence from pDNR-IMC1h/EGFP was introduced into pLP-DHFR/IMC1h via Cre-loxP site-specific recombination (New England Biolabs) to give the final transfection construct pLP-IMC1h/EGFP, used to introduce a GFP-tagged version of imc1h into its locus.
Plasmid pLP-IMC1h/EGFP served as a template for PCR using primers IMC1hKO-F (ATTTAGCTATGAGTAAAGGAGAAGAACTTTTCAC) and IMC1hKO-R (TTACTCATAGCTAAATTGTCATAATTCAGTTCATC). The amplified plasmid DNA was recircularized via In-Fusion cloning, resulting in the transfection construct pLP-ΔIMC1h/EGFP, in which all but the first 25 amino acids of the imc1h ORF have been removed. This plasmid was used to introduce a GFP reporter gene into the imc1h locus.
Diagnostic PCR of Parasite Genomic DNA
Genomic DNA was extracted from purified parasites using a Wizard genomic DNA purification kit (Promega). Confirmation of correct targeting and integration into the imc1h and imc1b loci was carried out with diagnostic PCR across the integration sites using primer pair hDHFR/ERI-F (ACAAAGAATTCATGGTTGGTTCGCTAAACT) and IMC1h-3′R (TTCTATAATCTTTAATTGTTCAGAAATGTG) for parasite lines IMC1h/GFP and IMC1h-KO (where KO is knock-out) and primer pair TgDHFRcas-3′F (TCGTGGGCTACGTCCCGCAC) and IMC1b-3′R (TGGTTATATTTTCATTTTGAATTAATAATATATG) for parasite line IMC1b-KO. Confirmation of the presence/absence of the WT allele was carried out with primer pair pDNR-IMC1h-F and IMC1h-3′R (for imc1h) and primer pair pDNR-IMC1b-F (10) and IMC1b-3′R (for imc1b).
RESULTS
Sequence and Structure of IMC1h
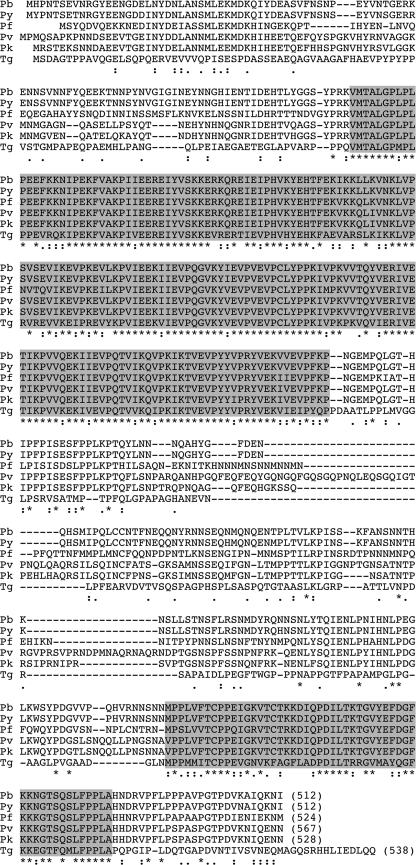
The P. berghei (Pb) imc1h gene was identified from BLAST searches of P. berghei genomic sequences with the IMC1h sequence from Plasmodium yoelii (DDBJ/GenBankTM/EBI accession number EAA20426) (8). The predicted gene product, PbIMC1h, is encoded by a single exon. The full-length protein is composed of 512 amino acids with a calculated Mr of 58,697, and it shares 96.9% amino acid sequence identity with its ortholog in P. yoelii. BLAST searches identified orthologs in all other Plasmodium species examined, including Plasmodium falciparum (AAN36292), Plasmodium vivax (EDL47576), Plasmodium chabaudi (CAH75026), and Plasmodium knowlesi (CAQ42162). An ortholog named TgIMC3 (AAT49041) was also identified in the closely related apicomplexan parasite T. gondii. TgIMC3 was previously demonstrated to be a pellicle-associated protein of tachyzoites (21). As shown in Fig. 1, the Plasmodium IMC1h and TgIMC3 proteins share two conserved domains: a central articulin-like domain related to the IMCp domain superfamily (Pfam 12314) that defines the alveolins and a C-terminal domain that is unique to Plasmodium IMC1h and TgIMC3. None of the Plasmodium IMC1h proteins identified possess the cysteine motifs at N and C termini described in TgIMC1 and PbIMC1a (6, 8).
FIGURE 1.
Sequence and structure of IMC1h. Shown is a multiple amino acid sequence alignment of the predicted IMC1h proteins from P. berghei (Pb), P. yoelii (Py), P. falciparum (Pf), P. vivax (Pv), and P. knowlesi (Pk), as well as the T. gondii ortholog TgIMC3 (Tg). The length in amino acids of each of the proteins is indicated by the numbers in parentheses. The C terminus of TgIMC3 has 38 amino acids not shown. Indicated are two conserved domains (shaded) and gaps introduced to allow optimal alignment (hyphens). Conserved amino acid identities (asterisks) and similarities (colons and periods) are indicated underneath. The alignment was made with ClustalW with default parameters.
Generation of Genetically Modified Parasites
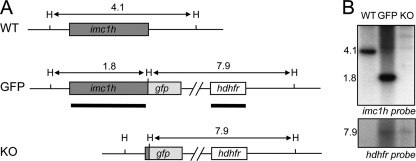
To study the expression and localization of IMC1h, a genetically modified P. berghei parasite line stably expressing full-length IMC1h fused to a C-terminal enhanced GFP tag was generated. This was achieved by replacing, via double crossover homologous recombination, the native imc1h allele with a recombinant allele linked in-frame to the enhanced gfp coding sequence. Concomitantly, a human dihydrofolate reductase (hdhfr) gene cassette was introduced, which confers resistance to the antimalarial drugs pyrimethamine and WR99210. To study the function of IMC1h and its contribution to parasite development, a genetically modified P. berghei parasite line was generated in which all of the imc1h coding sequence was removed except for the first 25 residues. In this parasite line, GFP acts as a reporter gene under the control of the native imc1h promoter. After transfection of purified schizont preparations, pyrimethamine-resistant parasites were selected and cloned. Diagnostic PCR across the predicted integration sites showed correct integration of the hDHFR gene cassette into the imc1h locus as well as the absence of the unmodified imc1h alleles (data not shown). This was confirmed by assessing the integrity of the genetically modified parasite lines (named IMC1h/GFP and IMC1h-KO, respectively) by Southern blot analysis of HindIII-digested genomic DNA. Two DNA probes were used: one specific to the imc1h coding sequence and one specific to the hDHFR gene (Fig. 2A). The imc1h probe gave rise to a 4.1-kb band in the parental WT parasites, a 1.8-kb band in IMC1h/GFP parasites, and no signal in IMC1h-KO parasites, as predicted (Fig. 2B). The hDHFR probe gave rise to a hDHFR-specific signal of ∼7.9 kb in IMC1h/GFP and IMC1h-KO parasites, but not in WT parasites, as expected (Fig. 2B). These combined results confirmed the correct integration of the recombinant imc1h and hDHFR alleles into the imc1h locus.
FIGURE 2.
Southern analysis of genetically modified parasite lines IMC1h/GFP and IMC1h-KO. A, schematic diagram of WT and genetically modified imc1h loci on genomic DNA. Indicated are coding sequences (bars), positions of the HindIII restriction sites (H), and expected HindIII restriction fragments (horizontal arrows) with sizes shown in kb. Sequences corresponding to probes used are indicated by thick lines. B, Southern blot of HindIII-digested parasite genomic DNA using probes specific to imc1h and hDHFR sequences.
Life Stage Expression and Subcellular Localization of IMC1h
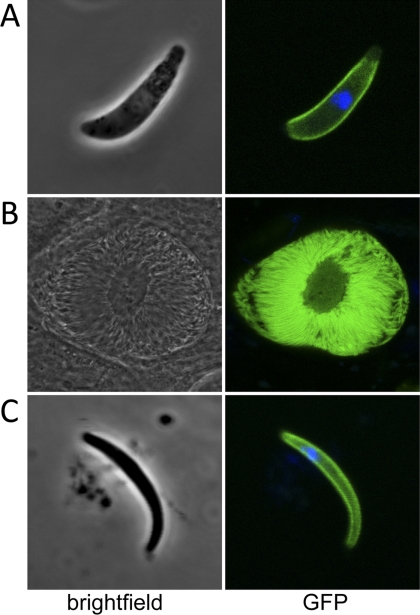
Expression of the imc1h gene product was studied using parasite line IMC1h/GFP, which expresses, from its native promoter, a full-length version of IMC1h that is C-terminally tagged with GFP. Parasites of line IMC1h/GFP developed normally in mice and mosquitoes and were readily transmitted by infected mosquito bites, demonstrating that the addition of the GFP tag to the IMC1h protein did not adversely affect parasite development. Examination of IMC1h/GFP parasites by confocal microscopy revealed strong GFP-based fluorescence in ookinetes, sporulating oocysts, and sporozoites (Fig. 3) but no fluorescence in blood stage parasites (data not shown). In ookinetes as well as in sporozoites, the majority of the GFP-based fluorescence was found at the periphery of the cells (Fig. 3). These observations demonstrate a new differential expression profile and are supportive of a pellicular localization of IMC1h in ookinetes and sporozoites, consistent with its predicted function as a membrane skeleton protein. This subcellular localization is also consistent with that of its T. gondii ortholog TgIMC3 (21).
FIGURE 3.
Expression and subcellular localization of IMC1h. Shown are confocal microscope bright-field and fluorescence images of a typical P. berghei ookinete (A), a sporulating oocyst (B), and a sporozoite (C). Hoechst 33258 stain (blue) was used to mark the position of the nucleus.
IMC1h Loss-of-function Phenotypes
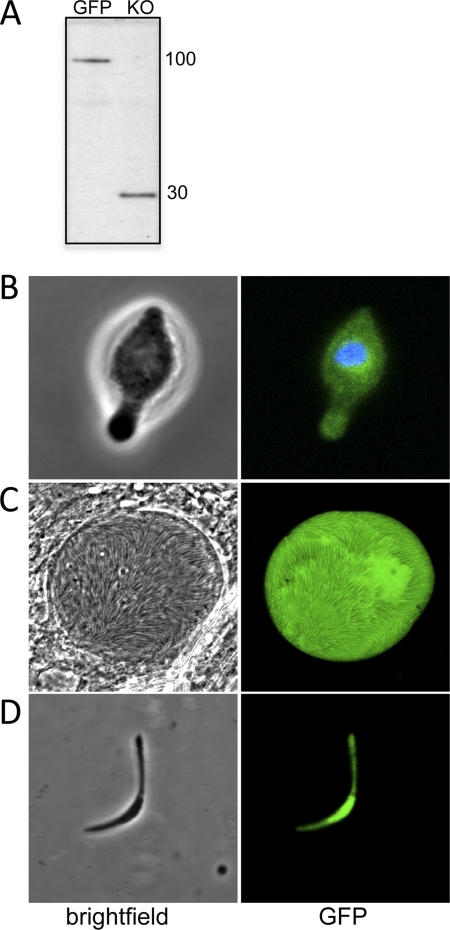
The function of IMC1h and its contribution to parasite development and infectivity were studied using parasite line IMC1h-KO, an IMC1h null mutant. IMC1h-KO blood stage parasites developed normally in mice, gametogenesis occurred normally in vitro, and ookinetes developed in vitro in numbers comparable with WT parasites (data not shown). Western blot analysis of purified ookinete samples using anti-GFP antibody revealed bands with apparent sizes of ∼100 and 30 kDa in parasite lines IMC1h/GFP and IMC1h-KO, respectively (Fig. 4A), corresponding to the IMC1h::GFP fusion protein and to GFP fused to the N-terminal 25 residues of IMC1h. This confirmed the correct expression of the recombinant imc1h allele in parasite line IMC1h/GFP and is in agreement with the absence of IMC1h expression in parasite line IMC1h-KO.
FIGURE 4.
Western blot and morphology of IMC1h-KO parasites. A, Western blot of purified ookinetes (50,000/well) from parasite lines IMC1h/GFP and IMC1h-KO. B, confocal bright-field and fluorescence images of an IMC1h-KO ookinete. C, confocal images of an IMC1h-KO sporulating oocyst. D, confocal images of an IMC1h-KO sporozoite. Hoechst 33258 stain (blue) was used to mark the position of the nucleus.
Closer examination of IMC1h-KO ookinetes revealed an abnormal morphology. Compared with IMC1h/GFP ookinetes, which express fully functional IMC1h, typical IMC1h-KO ookinetes were wider (mean widths of 1.97 ± 0.04 μm for IMC1h/GFP and 2.54 ± 0.06 μm for IMC1h-KO (n = 100); p < 0.0001) and shorter (mean lengths of 11.32 ± 0.15 μm for IMC1h/GFP and 9.36 ± 0.16 μm for IMC1h-KO (n = 100); p < 0.0001), and these ookinetes possessed a bulging area typically in the central part of the cell (Fig. 4B). As expected, GFP expressed in these ookinetes was no longer targeted to the pellicle structure, resulting in a dispersed green fluorescence (Fig. 4B). This phenotype is the same as that observed in ookinetes of IMC1b null mutant parasites (10) and is consistent with a role of IMC1h as a membrane skeleton protein.
To assess the infectivity of IMC1h-KO ookinetes, Anopheles stephensi vector mosquitoes were infected and analyzed for oocyst development at 10 days post-infection in direct comparison with IMC1h/GFP parasites expressing functional IMC1h. Reproducibly, IMC1h-KO parasite-infected mosquitoes had markedly lower oocyst numbers (∼30-fold; p < 0.0001) and a lower prevalence of infection (i.e. the percentage of mosquitoes with at least one oocyst) than IMC1h/GFP parasite-infected mosquitoes (Table 1), indicating that knock-out of IMC1h expression adversely affects ookinete infectivity.
TABLE 1.
Impact of imc1h gene disruption on P. berghei oocyst development in A. stephensi mosquitoes
| Parasite line | Mean no. of oocysts/mosquito (range)a | Prevalence of infection | Reduction |
|---|---|---|---|
| % | -fold | ||
| Exp. I | |||
| IMC1h/GFP | 71 (2–170) | 100 | |
| IMC1h-KO | 2.2 (0–6) | 70 | 32 |
| Exp. II | |||
| IMC1h/GFP | 122 (18–312) | 100 | |
| IMC1h-KO | 4.4 (0–13) | 80 | 28 |
a Based on dissection and analysis of 20 mosquitoes/parasite line.
Oocysts in PbIMC1h-KO parasite-infected mosquitoes appeared to develop normally, forming large numbers of sporozoites (Fig. 4C). However, closer examination of the PbIMC1h-KO sporozoites revealed that they, too, were of abnormal shape, typically possessing a bulging area near the middle of the sporozoite (Fig. 4D) reminiscent of IMC1a null mutant sporozoites (8). We did not observe sporozoites of normal morphology. We failed to detect IMC1h-KO sporozoites in the salivary glands, and consistent with this, we could not transmit this parasite to naive mice by sporozoite-infected mosquito bites (data not shown). These results indicate that IMC1h is necessary for normal sporozoite cell shape and is essential for sporozoite infectivity.
Generation of IMC1h/IMC1b Double Knock-out Parasites
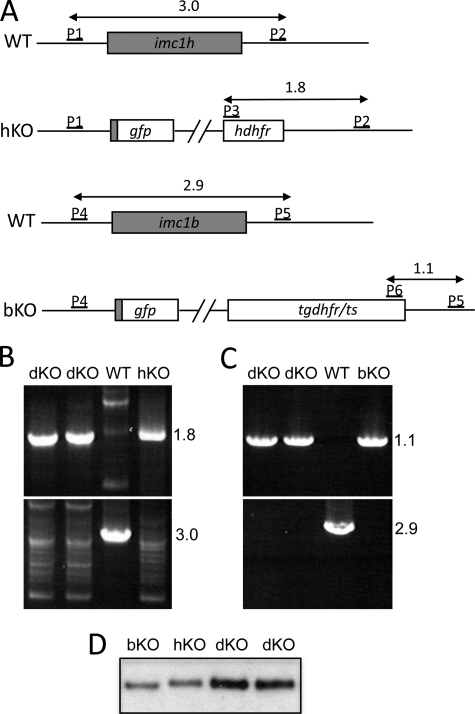
Ookinetes of parasite line IMC1h-KO are morphologically indistinguishable from IMC1b null mutant ookinetes, and both imc1h and imc1b gene disruptions lead to a considerable, albeit sublethal, drop in ookinete infectivity (Table 1) (10). Likewise, sporozoites of parasite line IMC1h-KO display the same abnormal cell shape and reduced infectivity as sporozoites of an IMC1a null mutant parasite (8). These striking similarities in the loss-of-function phenotypes of IMC1a, IMC1b, and IMC1h in the affected life stages were suggestive of a scenario of mutually dependent IMC1 protein function. In this scenario, knock-out of a single IMC1 protein component would prevent assembly of a functional membrane skeleton, and consequently, similar loss-of-function phenotypes for IMC1h, IMC1b, and IMC1a in their respective life stage(s) would be the expected outcome, as indeed observed. To test this hypothesis, we generated an IMC1h/IMC1b double null mutant parasite line. Because both IMC1h and IMC1b are expressed in the ookinete, the double knock-out parasite would allow us to compare the effects of the double knock-out with those of the single knock-outs in the same life stage. Generating a double knock-out parasite line was achieved by a genetic cross of parasite lines IMC1h-KO and IMC1b-KO, a strategy that did not require new gene targeting events. The two parasite lines were crossed in vitro, followed by membrane feeding of the resulting ookinete population to A. stephensi vector mosquitoes. Three weeks later, the resulting sporozoite population was transmitted to a naive mouse by mosquito bites. Clones were obtained by limiting dilution from the ensuing blood stage infection after WR99210 selection. In two selected clones, the presence of both the disrupted imc1h and imc1b alleles in the double knock-out parasite line (named IMC1h/b-dKO), as well as the absence of the respective WT alleles, was confirmed by diagnostic PCR. PCR diagnostic for integration into the imc1h locus produced a specific band of 1.8 kb in the two double knock-out clones as well as in parasite line IMC1h-KO, whereas PCR diagnostic for the presence of the WT imc1h allele gave a specific band of 3.0 kb only in WT parasites (Fig. 5, A and B). Likewise, PCR diagnostic for integration into the imc1b locus produced a specific band of 1.1 kb in the two double knock-out clones as well as in the single IMC1b null mutant, whereas PCR diagnostic for the presence of the WT imc1b allele gave a specific band of 2.9 kb only in WT parasites (Fig. 5, A and C). In full agreement with the established genotype of these parasites, Western blot analysis of IMC1h/b-dKO ookinetes with anti-GFP antibody confirmed the presence of both the IMC1h- and IMC1b-specific GFP reporters (Fig. 5D). These reporters have slightly different electrophoretic motilities in protein gels due to their distinct N-terminal fusions.
FIGURE 5.
Molecular analyses of IMC1h/b-dKO, IMC1h-KO, IMC1b-KO, and WT parasites. A, schematic diagram of WT and disrupted imc1h and imc1b alleles indicating the oligonucleotide primers (P1–P6) used for diagnostic PCR. Indicated are coding sequences (bars) and expected PCR products (horizontal arrows) with sizes shown in kb. For primer sequences, see “Experimental Procedures.” P1, pDNR-IMC1h-F; P2, IMC1h-3′R; P3, hDHFR/ERI-F; P4, pDNR-IMC1b-F; P5, IMC1b-3′R; P6, TgDHFRcas-3′F. B, diagnostic PCR using primers specific to the disrupted (upper panel; P2/P3) and WT (lower panel; P1/P2) imc1h alleles. C, diagnostic PCR using primers specific to the disrupted (upper panel; P5/P6) and WT (lower panel; P4/P5) imc1b alleles. D, Western blot of purified ookinetes (50,000/well).
IMC1h/IMC1b Double Knock-out Ookinetes Have Exacerbated Phenotypes Compared with Parental Single Knock-out Ookinetes
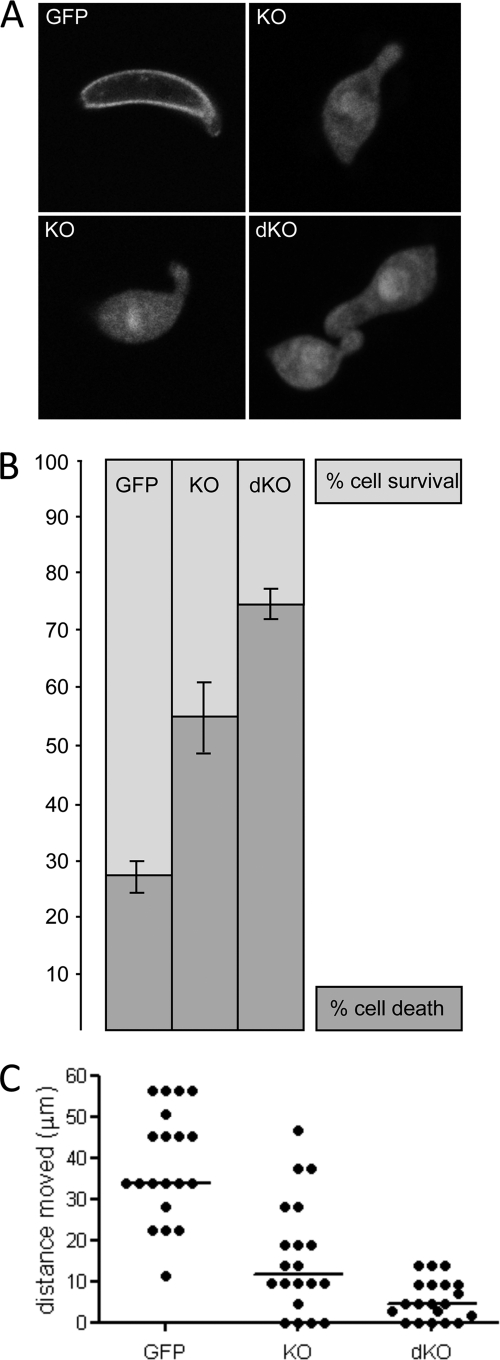
Development of asexual and sexual blood stages of IMC1h/b-dKO parasites in mice appeared normal, gametogenesis occurred normally in vitro, and ookinetes developed in vitro in numbers comparable with WT parasites (data not shown). Morphologically, the shape of IMC1h/b-dKO ookinetes was again abnormal compared with WT ookinetes, with the cells typically being shorter and wider (p < 0.0001) and possessing a bulging area in the center of the cell (Fig. 6A). However, this shape was not significantly different from that of IMC1h-KO ookinetes (mean length of 9.14 ± 0.13 μm and mean width of 2.65 ± 0.05 μm (n = 100); p = 0.6 and 0.1, respectively). Thus, IMC1h-KO ookinete cell shape appears not to be further affected by the additional imc1b gene disruption.
FIGURE 6.
Shape, mechanical strength, and motility of IMC1h mutant ookinetes. A, confocal microscope fluorescence images of IMC1h/GFP, IMC1h-KO, and IMC1h/b-dKO ookinetes. B, percentage ookinete survival/death after hypo-osmotic shock. Values are normalized to 100% viability in untreated cells. Error bars indicate S.D. from two independent experiments. At least 100 ookinetes were scored for each sample. C, scatter plot of ookinete motility expressed as distance traveled (in μm) over a 20-min period. Horizontal bars indicate median values.
We assessed the mechanical strength of our genetically modified parasite lines by subjecting ookinetes to hypo-osmotic shock. These conditions cause cells to draw in water and swell, and the degree of hypo-osmotic stress a cell can tolerate is a measure of its mechanical strength (22). Exposure to hypo-osmotic conditions caused nearly twice as much cell death in IMC1h-KO ookinetes (54%) as it did in ookinetes expressing functional GFP-tagged IMC1h (28%) (Fig. 6B). Cell death in response to osmotic shock was further increased significantly in the IMC1h/b-dKO parasites (74%) (Fig. 6B). These results demonstrate that like IMC1b, IMC1h is involved in the mechanical stability of ookinetes, which is consistent with it being an ookinete membrane skeleton component. They also show that there is a cumulative effect of multiple ookinete-specific imc1 gene disruptions on ookinete mechanical strength.
We also assessed the gliding motility of ookinetes in vitro by assessing the distance moved over a period of 20 min (Fig. 6C). Gliding of IMC1h-KO ookinetes was markedly reduced compared with ookinetes expressing functional GFP-tagged IMC1h (∼2.9-fold; p < 0.0001). These results show that like IMC1b, IMC1h is not essential for ookinete gliding motility in vitro; however, its disruption does adversely affect the ability of ookinetes to glide normally. The gliding motility of IMC1h/b-dKO ookinetes in vitro displayed a further reduction compared with IMC1h single knock-out ookinetes (∼2.6-fold; p < 0.005) (Fig. 6C). This shows that there is a cumulative effect of multiple ookinete-specific imc1 gene disruptions on ookinete locomotion.
To assess the infectivity of the IMC1h/b-dKO ookinetes, we infected vector mosquitoes and assessed oocyst development at 10 days post-infection. Mean oocyst numbers obtained from IMC1h/b-dKO parasite feeds were significantly lower than those obtained from mosquitoes infected with IMC1h-KO parasites (∼6-fold; p < 0.01) (Table 2). Moreover, compared with mosquitoes infected with IMC1h/GFP parasites, which express functional IMC1h, the oocyst numbers of double knock-out parasites were also clearly reduced (∼98-fold; p < 0.0001) (Table 2), and this reduction was greater than that obtained with IMC1h single knock-out parasites (Table 1). These observations again point to a cumulative effect of multiple imc1 gene disruptions, this time on ookinete infectivity. The few oocysts that developed in IMC1h/b-dKO parasite-infected mosquitoes gave rise to sporozoites that displayed the cell shape typical of IMC1h-KO sporozoites (data not shown). These results are fully expected: because IMC1b is not expressed in sporozoites, the IMC1h/b-dKO parasite line reproduces the phenotype of IMC1h-KO sporozoites.
TABLE 2.
Impact of imc1h and imc1b double gene disruptions on P. berghei oocyst development in A. stephensi mosquitoes
| Parasite line | Mean no. of oocysts/mosquito (range)a | Prevalence of infection | Reduction |
|---|---|---|---|
| % | -fold | ||
| Exp. I | |||
| IMC1h/GFP | 39 (10–172) | 100 | |
| IMC1h/b-dKO | 0.4 (0–2) | 25 | 98 |
| Exp. II | |||
| IMC1h-KO | 7.2 (0–27) | 70 | |
| IMC1h/b-dKO | 1.2 (0–8) | 35 | 6 |
a Based on dissection and analysis of 20 mosquitoes/parasite line.
DISCUSSION
In this study, we have shown that IMC1h, a member of the Plasmodium IMC1 protein family of membrane skeleton proteins (also called alveolins), is a component of the pellicle structure and is functionally equivalent to the previously characterized family members IMC1a and IMC1b (Table 3). Structurally, IMC1a and IMC1b are closely related with an identical three-domain composition, whereas homology with IMC1h is limited to a single alveolin domain. Nonetheless, IMC1h displays a similar subcellular localization and loss-of-function phenotype to its family members, suggesting that the alveolin module could be instrumental in defining these functions. In support of this, it was very recently shown that the alveolin domain of TgIMC3 on its own is sufficient to target the protein to the pellicle (12). Whereas IMC1a and IMC1b are differentially expressed in P. berghei sporozoites and ookinetes, respectively, IMC1h is expressed in both these zoite stages. Thus, IMC1h is the first family member to be differentially expressed in multiple, albeit not all, zoite stages of the parasite, adding to the diversity in Plasmodium imc1 gene expression between life stages. There are currently no proteome data available for P. falciparum ookinetes, but the P. falciparum IMC1h ortholog has been identified in the sporozoite and oocyst proteomes (23, 24), pointing to a similar expression profile for IMC1h in these human and rodent malaria species.
TABLE 3.
Summary of the effects of P. berghei imc1 gene disruptions on ookinete and sporozoite morphology, strength, motility, and infectivity
| imc1a (8) | imc1b (10) | imc1h | imc1b + imc1h | |
|---|---|---|---|---|
| Ookinete morphology | Normal | Abnormal | Abnormal | Abnormal |
| Ookinete strength | Not assessed | Reduced ∼2.0-fold | Reduced ∼1.9-fold | Reduced ∼2.6-fold |
| Ookinete motility | Not assessed | Reduced ∼3.9-fold | Reduced ∼2.9-fold | Reduced ∼7.4-fold |
| Ookinete infectivity | Normal | Reduced ∼9-fold | Reduced ∼30-fold | Reduced ∼98-fold |
| Sporozoite morphology | Abnormal | Normal | Abnormal | Abnormal |
| Sporozoite strength | Reduced ∼7.2-fold | Not assessed | Not assessed | Not assessed |
| Sporozoite motility | Reduced ∼5.0-fold | Not assessed | Not assessed | Not assessed |
| Sporozoite infectivity | None detected | Normal | None detected | None detected |
To investigate the impact of multiple imc1 gene disruptions on ookinete behavior, we carried out a genetic cross between IMC1h null mutant and IMC1b null mutant parasites to generate ookinetes that lack both IMC1h and IMC1b. This strategy was possible because the imc1h and imc1b genes are situated on distinct P. berghei chromosomes (25, 26). Because of chromosome pairing prior to meiotic division, which occurs around 2 h after fertilization (27), a proportion of heterokaryotic oocysts derived from the cross are anticipated to produce a mixture of sporozoites that are genotypically either imc1h+imc1b+ or imc1h−imc1b−. The latter sporozoite population is expected to be phenotypically normal because of the expression, in the same oocyst, of IMC1h protein from the WT allele. Hence, the sporozoites can be normally transmitted by mosquito bite. From the ensuing blood stage infection, we were able to efficiently select parasite clones with the double knock-out genotype after a single round or WR99210 selection. The selectable marker hDHFR has already been successfully employed to achieve sequential genetic manipulation of parasite lines already containing other pyrimethamine selectable markers (28). Our observations further indicate that parasites that carry both TgDHFR/thymidylate synthase (TS) and hDHFR resistance alleles at the same time have a selective advantage not only over parasites that carry just a copy of TgDHFR/TS (in our case, parasite line IMC1b-KO) but also over those that carry just a copy of hDHFR (in our case, parasite line IMC1h-KO). Similar results were reported with parasites carrying copies of both PbDHFR/TS and hDHFR resistance alleles, which displayed a 2-fold increased resistance in vitro to WR99210 over parasites carrying only hDHFR (28). This demonstrates that, in certain cases, genetic crosses can be a useful alternative to sequential parasite transfection.
Our results show that the abnormal ookinete shape resulting from the absence of IMC1h is the same as that of IMC1b null mutants. Notably, the absence of both IMC1h and IMC1b does not further affect ookinete shape. This latter observation is interesting because it implies that the shape change in the absence of IMC1h or IMC1b (or both) is somehow constrained. A likely explanation is that there must be additional cytoskeletal elements that maintain this “minimal” shape of the ookinete in the absence of a complete IMC1 protein repertoire. Additional IMC1 proteins expressed in the ookinete could be responsible for this (26). The fact that the same shape is also imposed on sporozoites that are lacking either IMC1h or IMC1a suggests that a similar scenario applies to this zoite stage.
According to current models of gliding motility (5, 29–33), ectodomains of adhesive proteins on the surface of the zoite bind to cell surfaces and other specific substrates. The same adhesins are connected by their cytoplasmic tails to actin filaments, an interaction that is mediated by aldolases and that occurs in the space between plasma membrane and IMC. The class XIV myosin component of the motor, present in the same space, is anchored to the outer membrane of the IMC via the glideosome-associated proteins GAP45 and GAP50 (34, 35) and is thought to power directional movement of the zoite by pulling the actin filaments the opposite way. The data presented here show that knock-out of IMC1h adversely affects ookinete gliding motility in a similar way to IMC1b null mutant ookinetes (10). As it is quite conceivable that a shorter ookinete with a bulge could be less effective at substrate-mediated gliding than its WT shape counterparts, we cannot, from these observations alone, ascertain whether the reduction in gliding is an effect of the interrupted interaction of the IMC1 protein with the molecular locomotion machinery or whether it is merely caused by the abnormal cell shape. The IMC1h/IMC1b double null mutant ookinete sheds light on this conundrum: the gliding locomotion of the double knock-out ookinete is further reduced compared with the IMC1h single knock-out ookinete, but without displaying further change in shape. Hence, the anomalous shape of these ookinetes alone cannot account for the reduction in gliding. From this, we infer that the IMC1 proteins contribute directly to gliding motility, most likely by interaction with the glideosome. These findings are supported by recent biochemical evidence for an interaction of IMC1 proteins with GAPM proteins (36). The GAPM proteins are multipass membrane proteins situated in the inner membrane of the IMC that also interact with components of the actin-myosin motor and hence are postulated to connect this motor with the membrane skeleton (36). Our data provide supporting genetic evidence for this model and point toward the SPN as the ultimate structure onto which the glideosome is anchored.
We have also shown in this work that IMC1h is involved in providing mechanical strength to the ookinete because, in its absence, the ookinete's ability to withstand osmotic stress is markedly reduced. Again, this observation is similar to what was found in IMC1b null mutant ookinetes (10) and seems to be a general feature of membrane skeleton protein mutants. Our experiments also show that additional knock-out of IMC1b expression further reduces mechanical stability of the IMC1h null mutant ookinete. It is no surprise to see that the effects of the IMC1h and IMC1b disruptions on both the gliding motility and mechanical strength of the ookinetes impact on the ability of these cells to invade the midgut epithelium and form oocysts. Gliding motility is essential for ookinete infectivity (14, 37, 38), and hence, a reduction in motility is likely to reduce infectivity. In addition, ookinetes undergo considerable constrictions as they migrate through the peritrophic matrix and through midgut epithelial cells (39, 40), and hence, structurally weakened ookinetes are likely to be less able to successfully invade or be more prone to damage during such events.
With the exception of ookinete morphology, IMC1h/IMC1b double null mutant ookinetes have aggravated phenotypes compared with the parental single knock-out ookinete populations. The most important conclusion that we can draw from these observations is that the function of these IMC1 proteins is not co-dependent. If this were the case, the double knock-out would have had a similar phenotype to the single knock-out parasites. As this clearly is not the case, individual IMC1 proteins must be able to operate autonomously, at least in the case of IMC1h and IMC1b. As is shown in this and other studies (8, 10), removal of IMC1 proteins by targeted gene disruption leaves parasites with a weakened but still partially functional SPN, which is manifested by a reduction in mechanical strength, gliding motility, and infectivity. This study goes on to show that the more IMC1 proteins removed, the greater the impact on the parasite. Although there is as yet no evidence that IMC1 proteins can form filaments, these combined observations fit with a model in which expressed IMC1 protein family members would each assemble into distinct intermediate filaments before being interconnected into a functional network, similar to other intermediate filament systems (41). Intermediate filament-based networks have primary roles in cell architecture and plasticity and as mechanical stress absorbers (42, 43). It is therefore likely that the three zoite stages of Plasmodium have different requirements of their membrane skeletons to operate successfully in their specific microenvironments. Simply by expressing a diverse repertoire of IMC1 proteins, the zoites would acquire membrane skeletons of different intermediate filament composition and consequently with distinct physical properties to suit their specific needs.
Our data presented here further underpin the vital roles of IMC1 proteins in malaria parasite development, infectivity, and transmission. There is evidence that other members of the IMC1 protein family are expressed in the blood stages (6, 9, 26). Thus, IMC1 protein assembly into functional structures could form a prime therapeutic target for parasite intervention that could impact across the parasite life cycle. The functional autonomy of IMC1 proteins shown here makes it more feasible that, as shown for other intermediate filament proteins, an in vitro system for filament assembly could be developed using recombinantly expressed IMC1 proteins, which could allow for the screening and identification of specific inhibitors.
Acknowledgments
We thank E. McCarthy and R. Gregory for assistance with the confocal microscopy.
This work was supported by the Wellcome Trust.
- IMC
- inner membrane complex
- SPN
- subpellicular network
- Tg
- T. gondii
- Pb
- P. berghei
- hDHFR
- human dihydrofolate reductase.
REFERENCES
- 1. Greenwood B. M., Bojang K., Whitty C. J., Targett G. A. (2005) Lancet 365, 1487–1498 [DOI] [PubMed] [Google Scholar]
- 2. Keeley A., Soldati D. (2004) Trends Cell Biol. 14, 528–532 [DOI] [PubMed] [Google Scholar]
- 3. Bannister L. H., Hopkins J. M., Fowler R. E., Krishna S., Mitchell G. H. (2000) Parasitol. Today 16, 427–433 [DOI] [PubMed] [Google Scholar]
- 4. Morrissette N. S., Sibley L. D. (2002) Microbiol. Mol. Biol. Rev. 66, 21–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Santos J. M., Lebrun M., Daher W., Soldati D., Dubremetz J. F. (2009) Int. J. Parasitol. 39, 153–162 [DOI] [PubMed] [Google Scholar]
- 6. Mann T., Beckers C. (2001) Mol. Biochem. Parasitol. 115, 257–268 [DOI] [PubMed] [Google Scholar]
- 7. Mann T., Gaskins E., Beckers C. (2002) J. Biol. Chem. 277, 41240–41246 [DOI] [PubMed] [Google Scholar]
- 8. Khater E. I., Sinden R. E., Dessens J. T. (2004) J. Cell Biol. 167, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gould S. B., Tham W. H., Cowman A. F., McFadden G. I., Waller R. F. (2008) Mol. Biol. Evol. 25, 1219–1230 [DOI] [PubMed] [Google Scholar]
- 10. Tremp A. Z., Khater E. I., Dessens J. T. (2008) J. Biol. Chem. 283, 27604–27611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu K., Mann T., Striepen B., Beckers C. J., Roos D. S., Murray J. M. (2002) Mol. Biol. Cell 13, 593–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson-White B. R., Ivey F. D., Cheng K., Szatanek T., Lorestani A., Beckers C. J., Ferguson D. J., Sahoo N., Gubbels M. J. (2011) Cell. Microbiol. 13, 18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arai M., Billker O., Morris H. R., Panico M., Delcroix M., Dixon D., Ley S. V., Sinden R. E. (2001) Mol. Biochem. Parasitol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 14. Dessens J. T., Beetsma A. L., Dimopoulos G., Wengelnik K., Crisanti A., Kafatos F. C., Sinden R. E. (1999) EMBO J. 18, 6221–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dessens J. T., Mendoza J., Claudianos C., Vinetz J. M., Khater E., Hassard S., Ranawaka G. R., Sinden R. E. (2001) Infect. Immun. 69, 4041–4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dessens J. T., Sidén-Kiamos I., Mendoza J., Mahairaki V., Khater E., Vlachou D., Xu X. J., Kafatos F. C., Louis C., Dimopoulos G., Sinden R. E. (2003) Mol. Microbiol. 49, 319–329 [DOI] [PubMed] [Google Scholar]
- 17. Le Chat L., Sinden R. E., Dessens J. T. (2007) Mol. Biochem. Parasitol. 153, 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waters A. P., Thomas A. W., van Dijk M. R., Janse C. J. (1997) Methods 13, 134–147 [DOI] [PubMed] [Google Scholar]
- 19. Carter V., Shimizu S., Arai M., Dessens J. T. (2008) Mol. Microbiol. 68, 1560–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saeed S., Carter V., Tremp A. Z., Dessens J. T. (2010) Mol. Biochem. Parasitol. 170, 49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gubbels M. J., Wieffer M., Striepen B. (2004) Mol. Biochem. Parasitol. 137, 99–110 [DOI] [PubMed] [Google Scholar]
- 22. Menke A., Jockusch H. (1991) Nature 349, 69–71 [DOI] [PubMed] [Google Scholar]
- 23. Lasonder E., Ishihama Y., Andersen J. S., Vermunt A. M., Pain A., Sauerwein R. W., Eling W. M., Hall N., Waters A. P., Stunnenberg H. G., Mann M. (2002) Nature 419, 537–542 [DOI] [PubMed] [Google Scholar]
- 24. Florens L., Washburn M. P., Raine J. D., Anthony R. M., Grainger M., Haynes J. D., Moch J. K., Muster N., Sacci J. B., Tabb D. L., Witney A. A., Wolters D., Wu Y., Gardner M. J., Holder A. A., Sinden R. E., Yates J. R., Carucci D. J. (2002) Nature 419, 520–526 [DOI] [PubMed] [Google Scholar]
- 25. Kooij T. W., Carlton J. M., Bidwell S. L., Hall N., Ramesar J., Janse C. J., Waters A. P. (2005) PLoS Pathog. 1, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hall N., Karras M., Raine J. D., Carlton J. M., Kooij T. W., Berriman M., Florens L., Janssen C. S., Pain A., Christophides G. K., James K., Rutherford K., Harris B., Harris D., Churcher C., Quail M. A., Ormond D., Doggett J., Trueman H. E., Mendoza J., Bidwell S. L., Rajandream M. A., Carucci D. J., Yates J. R., 3rd, Kafatos F. C., Janse C. J., Barrell B., Turner C. M., Waters A. P., Sinden R. E. (2005) Science 307, 82–86 [DOI] [PubMed] [Google Scholar]
- 27. Sinden R. E., Hartley R. H., Winger L. (1985) Parasitology 91, 227–244 [DOI] [PubMed] [Google Scholar]
- 28. de Koning-Ward T. F., Fidock D. A., Thathy V., Menard R., van Spaendonk R. M., Waters A. P., Janse C. J. (2000) Mol. Biochem. Parasitol. 106, 199–212 [DOI] [PubMed] [Google Scholar]
- 29. Soldati D., Meissner M. (2004) Curr. Opin. Cell Biol. 16, 32–40 [DOI] [PubMed] [Google Scholar]
- 30. Kappe S. H., Buscaglia C. A., Bergman L. W., Coppens I., Nussenzweig V. (2004) Trends Parasitol. 20, 13–16 [DOI] [PubMed] [Google Scholar]
- 31. Sibley L. D. (2004) Science 304, 248–253 [DOI] [PubMed] [Google Scholar]
- 32. Baum J., Papenfuss A. T., Baum B., Speed T. P., Cowman A. F. (2006) Nat. Rev. Microbiol. 4, 621–628 [DOI] [PubMed] [Google Scholar]
- 33. Matuschewski K., Schüler H. (2008) Subcell. Biochem. 47, 110–120 [DOI] [PubMed] [Google Scholar]
- 34. Gaskins E., Gilk S., DeVore N., Mann T., Ward G., Beckers C. (2004) J. Cell Biol. 165, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson T. M., Rajfur Z., Jacobson K., Beckers C. J. (2007) Mol. Biol. Cell 18, 3039–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bullen H. E., Tonkin C. J., O'Donnell R. A., Tham W. H., Papenfuss A. T., Gould S., Cowman A. F., Crabb B. S., Gilson P. R. (2009) J. Biol. Chem. 284, 25353–25363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yuda M., Sakaida H., Chinzei Y. (1999) J. Exp. Med. 190, 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Templeton T. J., Kaslow D. C., Fidock D. A. (2000) Mol. Microbiol. 36, 1–9 [DOI] [PubMed] [Google Scholar]
- 39. Han Y. S., Thompson J., Kafatos F. C., Barillas-Mury C. (2000) EMBO J. 19, 6030–6040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vernick K. D., Fujioka H., Aikawa M. (1999) Exp. Parasitol. 91, 362–366 [DOI] [PubMed] [Google Scholar]
- 41. Herrmann H., Aebi U. (2004) Annu. Rev. Biochem. 73, 749–789 [DOI] [PubMed] [Google Scholar]
- 42. Ausmees N., Kuhn J. R., Jacobs-Wagner C. (2003) Cell 115, 705–713 [DOI] [PubMed] [Google Scholar]
- 43. Herrmann H., Strelkov S. V., Burkhard P., Aebi U. (2009) J. Clin. Invest. 119, 1772–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]