Abstract
TopBP1 plays important roles in chromosome replication, DNA damage response, and other cellular regulatory functions in vertebrates. Although the roles of TopBP1 have been studied mostly in cancer cell lines, its physiological function remains unclear in mice and untransformed cells. We generated conditional knock-out mice in which exons 5 and 6 of the TopBP1 gene are flanked by loxP sequences. Although TopBP1-deficient embryos developed to the blastocyst stage, no homozygous mutant embryos were recovered at E8.5 or beyond, and completely resorbed embryos were frequent at E7.5, indicating that mutant embryos tend to die at the peri-implantation stage. This finding indicated that TopBP1 is essential for cell proliferation during early embryogenesis. Ablation of TopBP1 in TopBP1flox/flox mouse embryonic fibroblasts and 3T3 cells using Cre recombinase-expressing retrovirus arrests cell cycle progression at the G1, S, and G2/M phases. The TopBP1-ablated mouse cells exhibit phosphorylation of H2AX and Chk2, indicating that the cells contain DNA breaks. The TopBP1-ablated mouse cells enter cellular senescence. Although RNA interference-mediated knockdown of TopBP1 induced cellular senescence in human primary cells, it induced apoptosis in cancer cells. Therefore, TopBP1 deficiency in untransformed mouse and human primary cells induces cellular senescence rather than apoptosis. These results indicate that TopBP1 is essential for cell proliferation and maintenance of chromosomal integrity.
Keywords: Cell Cycle, DNA Damage, DNA Replication, Embryo, Gene Knockout, DNA Damage Response, TopBP1, Cellular Senescence, Embryonic Lethality
Introduction
TopBP15 is conserved in eukaryotes and contains repeats of BRCA1 carboxyl-terminal motifs, which are found in proteins involved in DNA repair and regulation of cell cycle checkpoints (1–6). Recent findings indicate that TopBP1 participates in the loading of Cdc45 for the assembly of the preinitiation complex that is necessary for chromosomal DNA replication (7–11). Knockdown of TopBP1 in human cancer cells showed that TopBP1 is necessary for the activation of cyclin E/CDK2 and Cdc7/Dbf4 kinases as well as the loading of DNA polymerase onto chromatin (12). In response to replication fork stalling or DNA damage, TopBP1 functions as an activator of the ATR·ATRIP complex by binding to Rad9 of the Rad9·Hus1·Rad1 complex (13, 14). The activated ATR phosphorylates Chk1, Nbs1, Smc1, and H2AX (15–18). TopBP1 also mediates ATR-mediated Chk1 activation by facilitating the interaction of claspin and Chk1 (19, 20). Furthermore, association of TopBP1 with NBS1 is involved in double-stranded DNA break-induced homologous recombination repair (21). Therefore, TopBP1 plays crucial roles in the maintenance of genomic integrity.
TopBP1 regulates gene expression. Binding of TopBP1 to E2F1 represses E2F1 proapoptotic activity by recruiting the Brg1·Brm chromatin remodeling complex (22). TopBP1 also binds to the DNA binding domain of p53 and inhibits the promoter binding activity of this protein (23). Therefore, TopBP1 depletion derepresses the proapoptotic activity of p53 and E2F1 and induces cellular apoptosis. In addition, TopBP1 represses Miz-1 expression to inhibit p21 expression (24). The binding and repression of E2F1 and Miz1 require Akt-dependent oligomerization of TopBP1 (25).
Most of the previous studies on the role of TopBP1 in the cell cycle and the DNA damage response have been carried out using human cancer cells. In this study we generated TopBP1 conditional knock-out mice to study the physiological functions of the TopBP1 protein. Inactivation of both TopBP1 alleles led to embryonic lethality at the peri-implantation stage. From TopBP1flox/flox embryos, we prepared mouse embryonic fibroblasts and their immortalized 3T3 cells. Ablation of TopBP1 in these cells using a Cre recombinase-expressing retrovirus produced DNA breakage. This process activated DNA damage response signaling and arrested cell cycle progression, thereby inducing cellular senescence. We also showed that knockdown of TopBP1 in human primary cells by RNA interference caused cellular senescence.
EXPERIMENTAL PROCEDURES
Murine TopBP1 Gene-targeting
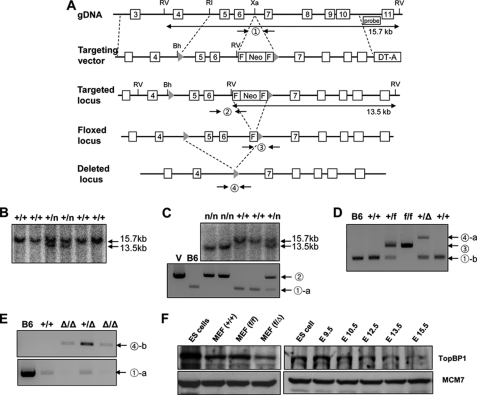
A 16.3-kb DNA fragment containing exon 3 and exon 10 of the murine TopBP1 gene was retrieved from BAC clones (bMQ-304N19, Geneservice) into a pBluescript phagemid system according to a previously reported procedure (26, 27). A 50-bp DNA fragment of the loxP sequence was inserted at the EcoRI restriction site located between exon 4 and 5, and a 1.9-kb neomycin resistance cassette from pL451 (provided by N. Copeland) was inserted at the XbaI restriction site located between exon 6 and 7. The diphtheria toxin A chain cassette was employed as a negative selection marker. The targeting vector was linearized and used for gene targeting in the E14Tg2A ES cell line (129/OlaHsd-derived, Baygenomics). Mouse ES cell culture and electroporation were performed as previously described with the exception of the procedure for neomycin selection (28). Correctly targeted ES clones were injected into C57BL/6 blastocysts for chimera generation. Chimeric males were bred with C57BL/6 females, and germ line transmission of the TopBP1neo allele was verified by PCR and Southern blots (Fig. 1C).
FIGURE 1.
Targeting of the TopBP1 locus. A, schematic representation of the TopBP1 locus, the targeting vector, and the targeted, floxed, and deleted loci is shown. Exons 3–11, Southern blot probe, and restriction sites (RV, EcoRV; RI, EcoRI; Xa, XbaI; Bh, BamHI) are shown. The targeting vector contained a diphtheria toxin A chain (DT-A) gene and a neomycin resistance gene (Neo) flanked by FRT (squares) and loxP (triangles) sites. Mice carrying the TopBP1neo allele were crossed to a Flp and Cre recombinase strain to generate the TopBPflox (conditional) and TopBP1Δ alleles, respectively. B, a Southern blot analysis of a representative TopBP1+/neo ES cell clone is shown. Homologous recombination was verified using external digests and 3′ external probes. For EcoRV digestion, the bands representing wild-type and mutant alleles are 15.7 and 13.5 kb, respectively. The symbol n represents the neomycin allele. C, Southern blot (upper) and PCR (lower) analyses with genomic DNA extracted from mouse tails are shown. In general, mice were genotyped by PCR using the specified primers (supplemental Table S1). Separate reactions were performed to amplify the wild-type and the targeted allele. The circled number represents a PCR reaction with the primers described in supplemental Table S1. D, PCR analyses with genomic DNA extracted from TopBP1flox/flox MEFs and TopBP1+/Δ mice used in this study are shown. TopBP1flox and TopBP1Δ alleles were generated by Flp-FRT and Cre-loxP recombination, respectively, as described under “Experimental Procedures.” E, genotyping of embryos for pre-implantation by PCR analysis is shown. Recovered or in vitro-cultured embryos were genotyped. F, topBP1 expression in mouse ES cells and MEFs and during early embryogenesis is shown. MCM7 protein was used as an internal control.
The TopBP1flox (“floxed”) and TopBP1Δ alleles were generated by crossing the TopBP1neo allele with a Flp deleter strain (FLPeR mice, Jackson laboratory strain 003946) and a Cre deleter strain (zp3-Cre, Jackson laboratory strain 003651) (Fig. 1D), respectively. The PCR primers used and genotyping information for the TopBP1 wild-type, floxed, and mutant alleles are provided in supplemental Table S1.
In Vitro Culture and Genotyping of Pre-implantation Embryos
All blastocysts were generated by natural mating of TopBP1 heterozygote mice. The morning of the day on which a vaginal plug was detected was designated as day E0.5. Blastocysts were collected on E3.5 by flushing the uterus of each mouse with M2 medium (Sigma) followed by culturing in conditioned ES media (knock-out Dulbecco's modified Eagle's medium (DMEM, Invitrogen), 15% FBS (Hyclone), 2 mm l-glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, 55 μm 2-mercaptoethanol (Invitrogen), and 1000 units/ml leukemia inhibitory factor (Chemicon) for 0 or 24 h. Overgrowths were photographed on each day and harvested for genotyping (Fig. 1E). Genotyping of embryonic cells was performed as previously described (29). The sequences of the PCR primers used and genotype information for the TopBP1 wild-type, floxed, and mutant alleles are provided in supplemental Table S1.
Immunostaining of Mouse Embryos
For BrdU-treated embryos, blastocysts were rinsed in PBS and fixed in fresh 4% formaldehyde in PBS for 30 min at room temperature. DNA was denatured after permeabilization with 2 n HCl and 0.5% Triton X-100 for 20 min at room temperature and washed extensively in PBS with 1.0% BSA. Blastocysts were incubated with mouse anti-BrdU (Developmental Studies Hybridoma Bank) overnight at 4 °C. Incubation with Alex Fluor 488 (Invitrogen) secondary antibodies was performed for 1 h at 37 °C followed by staining with 4′,6-diamino-2-phenylindole (DAPI).
Preparation of Mouse Embryonic Fibroblast (MEF) and Immortalized 3T3 Cells
MEFs were generated from embryonic day 13.5 (E13.5) embryos according to standard protocols (30). MEFs were grown in DMEM with 4500 mg/liter d-glucose and supplemented with 10% fetal bovine serum (FBS, Hyclone), 2 mm l-glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, and 55 μm 2-mercaptoethanol in a humidified incubator with 5% CO2 and passaged every 2–3 days. All experiments with MEFs were performed with cells that had been subjected to 3–5 passages. The primary MEF cells were immortalized using the 3T3 protocol (31).
Cell Culture
MEF and 3T3 cells thus obtained were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, 2 mm glutamine (Welgene), and 2β-mercaptoethanol (Invitrogen). U2OS, HeLa, HS68, IMR90, and PT67 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
Cre-retrovirus Infection
PT67 packaging cells containing the retrovirus construct expressing Cre recombinase were generated and cultured with puromycin (3 μg/ml) until confluent. The cells were then incubated in fresh medium without puromycin for 24 h. Supernatant medium was collected and filtered using a 0.45-μm filter. This filtered medium was used to infect MEF and its 3T3 cells. After retroviral infection with Polybrene (6 μg/ml) for 24 h, the virus-infected cells were selected with puromycin (3 μg/ml) for another 24 h.
Synchronization and Fluorescence-activated Cell Sorting (FACS) Analysis
The 3T3 cells were blocked at the G1/S boundary by treatment with 2 mm thymidine for 12 h. After release into fresh medium for 8 h, cells were treated with 2 mm thymidine for an additional 12 h. Nocodazole (100 ng/ml) was added to the cells for 12 h to arrest the cells at the G2/M phase. Cells were trypsinized and then fixed in 70% ethanol at 4 °C. After fixation, the cells were incubated with RNase A (10 μg/ml), Nonidet P-40 (0.05%), and propidium iodide (50 μg/ml) for 1 h. For the BrdU FACS analysis, cells were incubated for 10 min in the presence of BrdU (10 μm). Harvested cells were fixed in 70% ethanol at 4 °C and denatured in 2 n HCl, 0.5% Triton X-100 for 1 h. The cells were then neutralized with 0.1 m Na2B4O7 (pH 8.5) and incubated with BrdU antibody and FITC-conjugated secondary antibody in PBS containing 1% BSA and 0.5% Tween 20 for 1 h each. Cells were stained with propidium iodide solution and then analyzed using the FACSCalibur instrument (BD Biosciences).
Immunostaining and Senescence-associated β-Galactosidase (SA-β-Gal) Staining
Anti-TopBP1 polyclonal antibody was raised against the TopBP1 fragment corresponding to amino acids 1019–1167 (12). The antibody to BrdU was obtained from Abcam. The antibodies to phospho-H2AX (Ser139), phospho-Chk1 (Ser137), and phospho-Chk2 (Thr68) were obtained from Cell Signaling. The antibody to β-actin was obtained from Sigma. Cells grown on coverslips were fixed with 4% paraformaldehyde for 20 min at room temperature. Cells were permeabilized with 0.5% Triton X-100 in phosphate-buffered saline for an additional 20 min. After treatment with each primary and secondary antibody, coverslips were mounted on glass slides with mounting media containing 1 μg/ml DAPI (Vectashield). SA-β-Gal staining was performed using a SA-β-Gal staining kit (#9860; Cell Signaling) according to the manufacturer's instructions.
siRNA Transfection
The sequences of the synthesized siRNA oligonucleotides (Samchully Pharm Co.) were previously described (12): TopBP1 #2, ACCGAGUACGCCACUCUCA, and control siRNA (GL3), CUUACGCUGAGUACUUCGATT. Cells were transfected with siRNAs using Oligofectamine (Invitrogen) according to the manufacturer's instructions.
RESULTS
Murine TopBP1 Gene Targeting
To explore the physiological functions of TopBP1 in embryo development and adult tissues, we generated a conditional TopBP1 allele by gene targeting in mouse ES cells (Fig. 1). The murine TopBP1 gene, located on chromosome 9F1, is distributed over about 45 kb and is composed of 28 exons with a potential translational start codon in exon 2 (as determined from NCBI annotation of NCBI Build 37). In our gene targeting strategy, exons 5 and 6 were flanked by two loxP sequences that are recognized and removed by Cre recombinase. Consequently, only the first 121 of the entire 1515 amino acids of TopBP1 are correctly translated after the frameshift mutation and generation of a null allele.
The TopBP1-targeting construct was electroporated into 129P2/OlaHsd-derived E14K ES cells, and neomycin-resistant clones were screened for homologous recombination by Southern blotting with 5′ external probes and EcoRV digestion (Fig. 1, A and B). Of 196 neomycin-resistant cell lines, 18 (9.1%) displayed the desired target. Correctly targeted ES cell clones were used to derive chimeric mice that transmitted the TopBP1neo allele through the germ line (Fig. 1C). TopBP1+/flox mice were generated as described under “Experimental Procedures” (Fig. 1D).
TopBP1flox/flox mice were viable, fertile, and did not show any abnormalities. The expression levels of TopBP1 in TopBP1flox/flox MEFs were found to be similar to the expression levels observed in the wild-type mouse (Fig. 1F), implying that the TopBP1flox allele does not exert a significant effect on the expression of the protein. Furthermore, the level of TopBP1 protein was found to be higher during early embryogenesis (e.g. ES cell and E9.5) than during late embryogenesis. Furthermore, the level of TopBP1 protein was found to be higher in progenitor cells than in differentiated cells (MEF) (Fig. 1F). These expression patterns of TopBP1 suggest the possibility that the protein is involved in cell proliferation or that it functions in the undifferentiated state.
TopBP1 Is Essential for Development of Peri-implanted Embryos
The expression level of TopBP1 in TopBP1+/Δ MEFs suggests that the TopBP1Δ allele is defective in TopBP1 expression (Fig. 1F). However, heterozygous TopBP1+/Δ mice were apparently normal, healthy, and fertile. There were no developmental abnormalities detected during a 15-month observation period. In contrast, homozygous TopBP1Δ/Δ mice were not produced of 168 live births from TopBP1+/Δ intercrosses (supplemental Table S2), indicating that inactivation of both alleles leads to embryonic lethality.
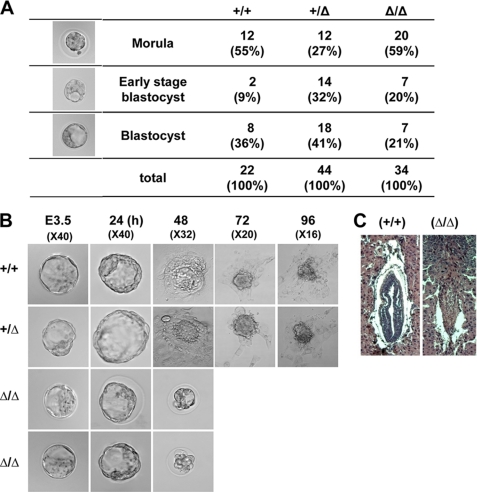
To assess the specific period of TopBP1 knock-out-induced developmental failure, timed heterozygous mating was performed. Embryos or fetuses were collected after various gestation periods, and their genotypes were determined by PCR (Fig. 2A and supplemental Table S2). No homozygous mutant embryos/fetuses were recovered at 8.5 or beyond, and embryos tended to be completely resorbed at E7.5. When blastocysts were recovered at E3.5 from heterozygote intercrosses, 34.9% (72/206) of the total embryos were found to be homozygous TopBP1Δ/Δ. The mutant embryos followed Mendel's law and could not be morphologically distinguished from wild-type and heterozygous embryos. These data indicate that TopBP1-deficient embryos normally develop to the blastocyst stage but die at the peri-implantation stage.
FIGURE 2.
TopBP1 disruption results in early embryonic lethality. A, images of embryos at E3. 5 were obtained under bright-field conditions. The embryos were classified according to their appearance: morula, early stage of blastocyst, and mature blastocyst. B, outgrowth of wild-type, heterozygote, and mutant blastocysts (see “Experimental Procedures”) is shown. Blastocyst stage embryos recovered at E3.5 were cultured in vitro for several days and subsequently genotyped by PCR. Embryo outgrowths are composed of an inner cell mass surrounded by a single layer of trophoblast giant cells. C, hematoxylin and eosin staining of histologic sections were performed to analyze the appearance of E7.5 embryos developing in the uterus. Typical images of wild-type and fully resorbed mutant embryos are displayed in the left and right panels, respectively.
To characterize the growth defects of TopBP1-deficient embryos, blastocysts recovered at E3.5 were cultured in vitro for several days (Fig. 2B). After culturing for 3–4 days, spherical blastocysts became flattened on the culture dish and formed a multicomponent structure in which the inner cell mass grew as a round shape on top of the extra-embryonic trophoblast giant cells (32). In these cultures heterozygous TopBP1+/Δ and wild-type embryonic cells normally developed into inner cell mass and trophoblast giant cells. Although homozygous TopBP1Δ/Δ embryos apparently showed normal growth for 24 h, the embryos did not grow any further after the point when normal embryos hatched. The mutant embryos did not hatch and exhibited morphological shrinkage. Furthermore, we frequently observed complete resorption of the embryos into the uterus at E7.5 (Fig. 2C). These results collectively suggest that homozygous TopBP1Δ/Δ embryos exhibit growth defects after E4.5 and then die at the peri-implantation stage.
Defect in Cell Proliferation and Death of TopBP1Δ/Δ Embryonic Cells
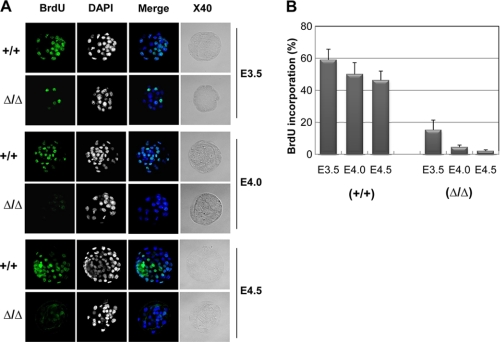
We employed the in vitro cultures of blastocysts to examine whether the developmental failure of mutant embryos at the peri-implantation stage resulted from cell proliferation defects. The proliferation of embryonic cells was measured by the BrdU incorporation assay, which detects proliferative activity of the cells by measuring DNA synthesis (Fig. 3). In this assay, 45–60% of the BrdU-positive cells were detected in wild-type embryos at E3.5, 4.0, and E4.5. On the other hand, less than 15% of the BrdU-positive cells were detected in TopBP1-null embryos at E3.5, and most of the mutant embryos did not exhibit BrdU-positive cells at E4.5. At this point, the TopBP1-null embryos did not develop further and appeared to shrink or die.
FIGURE 3.
Defect in cell proliferation of TopBP1Δ/Δ embryonic cells. A, cell proliferation was gauged by determining the extent of BrdU incorporation into blastocyst culture for 0 (E3.5), 12 (E4.0), and 24 h (E4.5). Embryos were pulsed for 15 min with BrdU and processed for BrdU immunostaining and DAPI counterstaining as described under “Experimental Procedures.” The magnification is ×40. B, a series of z-plane images was stacked and analyzed by confocal microscopy (Zeiss) to quantify the percentage of BrdU-positive cells. Error bars indicate S.D.
The presence of fragmented DNA, which is an indication of apoptotic cells, was detected by staining of TopBP1-null embryos cultured in vitro with DAPI (supplemental Fig. S1). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick-end labeling (TUNEL) staining confirmed that cell death occurred in TopBPΔ/Δ embryos at E4.0 (supplemental Fig. S1). In contrast, the TUNEL-positive cells were rarely detected in wild-type littermates. These results suggest that the dramatic reduction of proliferating cells accompanied by cell death at least in part induces the developmental failure of TopBP1-null embryos.
TopBP1-ablated Mouse Embryonic Fibroblasts Are Arrested during Progression of the Cell Cycle
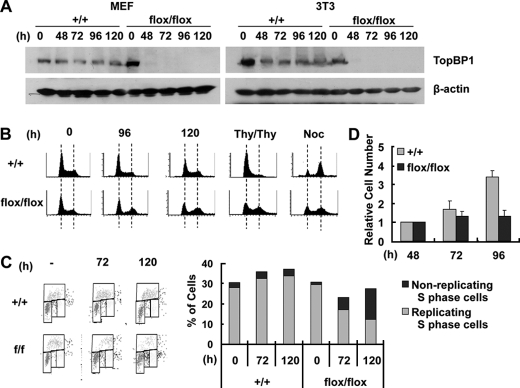
The function of TopBP1 in the cell cycle has been addressed using TopBP1-depleted cancer cells such as HeLa and U2OS, with RNA interference (RNAi) (12, 33, 34). As an alternative, we analyzed the loss of TopBP1 function using TopBP1flox/flox mouse embryonic fibroblast cells prepared from TopBP1flox/flox mice and their immortalized 3T3 cells. Infection of these cells with a retrovirus, which expresses Cre recombinase (Cre-retrovirus), was found to ablate the expression of TopBP1 protein in TopBP1flox/flox MEF and 3T3 cells (Fig. 4). At the indicated time after the infection, TopBP1 protein levels were examined by Western blot analysis (Fig. 4A). In contrast to the wild-type cells, most of the TopBP1 protein was depleted in TopBP1flox/flox cells 48 h after the viral infection. Consistently, as TopBP1 was being depleted, TopBP1 disappeared from the nucleus of TopBP1flox/flox cells (supplemental Fig. S2), although TopBP1 protein is located in the nucleus of wild-type cells during interphase (35, 36).
FIGURE 4.
TopBP1-ablated mouse cells are arrested during the progression of the cell cycle. A, TopBP1+/+ and TopBP1flox/flox MEF and 3T3 cells (left panel, MEF; right panel, 3T3) were infected with Cre-retrovirus for 24 h, selected with puromycin (3 μg/ml) for an additional 24 h, and then harvested at the indicated time after the viral infection. Each protein was detected by immunoblotting with the corresponding antibody. B, the 3T3 cells in the panel A were stained with propidium iodide and analyzed by FACS analysis. At a point 60–80 h after Cre-retrovirus infection, the cells were subjected to a double thymidine block (Thy/Thy) or treatment with nocodazole (Noc). C, the 3T3 cells were pulsed with BrdU (10 μm) for 10 min and then analyzed by FACS with BrdU units/PI staining. The ratios of BrdU-incorporated cells over S-phase cells are indicated in the right panel. D, each cell quantity was normalized with respect to the cell quantity determined 48 h after viral infection.
FACS analysis of TopBP1-ablated cells indicates a reduction of G1-phase cells containing 2N DNA and a reduction of S-phase cells as well as an increase of G2/M-phase cells containing 4N DNA content (Fig. 4B), relative to wild-type cells. To differentiate the cells that undergo normal cell cycle progression from the cells that are arrested in each cell cycle phase, Cre-retrovirus-infected cells were subjected to a double thymidine block or treatment with nocodazole, which arrests cells at the G1/S boundary with 2N DNA content or at prometaphase of mitosis with 4N DNA content, respectively. Whereas the double thymidine block arrested the cell cycle progression of wild-type cells at the G1/S boundary with 2N DNA content, the FACS analysis indicated that the TopBP1-ablated cells yield a significant population of cells containing 4N DNA. The 4N DNA-containing cells undergo arrest of cell cycle progression in the G2 or M phase. Anti-phosphohistone H3 (Ser10) antibody detects both the late G2 and M phase cells, which can be discriminated by their chromosome and nuclear morphologies (37, 38). Most of TopBP1-ablated 4N cells appeared to be arrested in the late G2 phase (supplemental Fig. S3).
Nocodazole treatment of TopBP1-ablated cells produced a higher population of cells containing 2N DNA than the wild-type cells. This implies that the 2N DNA-containing cells were arrested in the G1 phase. As TopBP1 is depleted, BrdU incorporation, which provides an indication of DNA synthesis of proliferating cells, became decreased in the TopBP1-ablated cells (Fig. 4C). Although after 120 h, 27% of the TopBP1-ablated cells were in the S-phase, only 12% of the cells were positive with respect to BrdU incorporation. On the other hand, most of the control S-phase cells had incorporated BrdU. The lower level of BrdU incorporation in the S-phase cells suggests that the cells were not replicating and had undergone cell cycle arrest in the S phase. Inconsistent with the defect of TopBP1-ablated cells in cell cycle progression, the TopBP1-ablated cells did not proliferate (Fig. 4D). These results suggest that the TopBP1-ablated cells undergo cell cycle arrest in G1, S, G2, or M phase. It is likely that the phase in which arrest occurs depends upon the time required for TopBP1 level to fall below the threshold level necessary for progression through each stage of the cell cycle.
TopBP1 Ablation Induces the DNA Damage Response
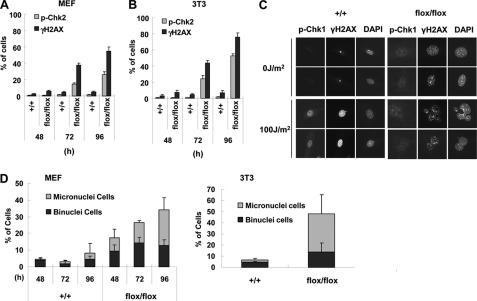
When TopBP1 was depleted by TopBP1-specific siRNAs in cancer cells, the depletion was found to induce phosphorylation of H2AX and activation of the ATM-Chk2 DNA damage pathway even in the absence of external genotoxic stress (34). Because DNA breaks induce H2AX phosphorylation (denoted as γH2AX), which forms foci at the region of the DNA break, the formation of the γH2AX foci together with the activation of the ATM-Chk2 pathway indicates that the TopBP1 knockdown induces the breakage of DNA. These observations suggest that TopBP1 is necessary for maintaining genome integrity. By detecting phosphorylated H2AX and Chk2 using corresponding phospho-specific antibodies, we examined the activation of DNA damage responses of TopBP1-ablated MEFs and 3T3 cells. At a point 72 h after infection with the Cre-retrovirus, the ablation induced increases in the rate of phosphorylation and formation of foci of γH2AX and Chk2 in TopBP1flox/flox cells (Fig. 5, A and B, and supplemental Fig. S4).
FIGURE 5.
TopBP1 ablation activates DNA damage signaling. Cre-retrovirus infected MEF (A) and 3T3 cells (B) were immunostained with anti-p-Chk2 (Thr68) or anti-γH2AX (Ser139) antibody at the indicated times after viral infection (supplemental Fig. S4). Cells found to be positive for p-Chk2, and γH2AX signaling was counted and graphed. Three independent experiments were performed with 300 cells counted at least for each experiment. C, the indicated 3T3 cells were treated with UV (100 J/m2) 96 h after viral infection and then immunostained 3 h after the irradiation. D, at the indicated times (MEF) or 96 h (3T3) after viral infection, the nuclear morphologies were visualized by DAPI staining. The percentages of cells containing abnormal nuclear morphology (binuclear and micronuclear cells) are indicated. Three independent experiments using at least 100 cells for each experiment were averaged.
In response to genotoxic stress, TopBP1 activates Chk1 not only by directly interacting with ATR·ATRIP but also by facilitating activation of claspin and potentiating its ability to bind Chk1 (19, 39). Upon UV radiation, the Chk1 phosphorylation of Cre-retrovirus-infected cells was examined (Fig. 5C). Although the radiation increased the rate of phosphorylation of γH2AX and Chk1 in wild-type cells, Chk1 phosphorylation maintained background levels in the TopBP1-depleted cells. The lack of activation of Chk1 confirmed the previous findings that TopBP1 function is necessary for the activation of Chk1 in response to DNA damage (19, 34, 39). As reported previously (34), TopBP1 depletion increased the number of abnormal nuclei in cells containing binuclei or micronuclei (Fig. 5D and supplemental Fig. S5).
TopBP1 Deficiency Induces Cellular Senescence in Mouse Cells and Human Primary Cells
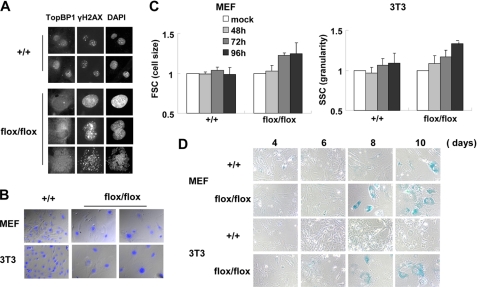
Even in the absence of external genotoxic stress, TopBP1-deficient cells were found to exhibit DNA breaks and activation of Chk2. The DNA breaks detected by γH2AX in the cells persisted for at least 10 days after infection with Cre-retrovirus. This suggests that DNA damage could not be repaired regardless of whether the DNA damage repair mechanism was operating properly (Fig. 6A). Although TopBP1 knockdown by RNA interference induced apoptosis in cancer cells (12, 22, 23, 25, 33), we were not able to detect significant cell death of TopBP1-ablated MEF or 3T3 cells. Instead, TopBP1-ablated cells underwent morphological changes including enlargement and flattening of the cells on the culture dishes (Fig. 6B). At a point 10 days after infection with Cre-retrovirus, the nuclear size of the TopBP1-ablated cells increased by as much as 3–4-fold relative to the wild-type cells. The TopBP1 ablation also increased the cellular granularity of the harvested cells (Fig. 6C). These morphological changes are the typical phenotypes of senescent cells (40, 41). Consistent with these morphological changes, TopBP1-ablated cells were positive with respect to SA-β-gal staining, which detects senescent cells (Fig. 6D). Taken together, the results indicate that TopBP1 ablation induces cellular senescence in mouse MEF and 3T3 cells.
FIGURE 6.
TopBP1-ablated cells induce cellular senescence. A, wild-type and TopBP1flox/flox 3T3 cells were immunostained 10 days after viral infection. B, the nuclei of the cells in panel A were stained with DAPI (blue) and merged with differential interference contrast images. C, cell size and cellular granularity of the cells were determined by forward scattering (FSC) and side scattering (SSC) with FACS. Each value was normalized with control cells that were not infected with the virus. Three independent experiments were performed. D, the cells were stained with SA-β-gal stain and viewed under a phase contrast microscope.
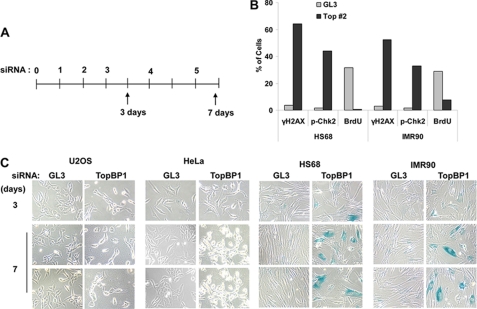
We examined whether TopBP1 deficiency causes cellular senescence in human cells. Using TopBP1-specific siRNA (12), TopBP1 knockdown was carried out in human IMR90 and HS68 primary cells and human HeLa and U2OS cancer cells (Fig. 7). Three days after the siRNA transfections, the human primary cells exhibited generation of DNA breaks detected by phosphorylation of H2AX and Chk2 with the reduction of BrdU-incorporated cells (Fig. 7B and supplemental Fig. S6). At 7 days after the continuous knockdown, most of the human primary cells became senescent. On the other hand, the cancer cell lines were negative for SA-β-gal staining and were observed to undergo apoptosis (Fig. 7C). These results indicate that TopBP1 deficiency also causes cellular senescence in human primary cells in a manner similar to the senescence observed for mouse cells.
FIGURE 7.
TopBP1 knockdown induces cellular senescence in human primary cells. A, the indicated human cell lines were transfected five times with control siRNA (GL3) or TopBP1-specific siRNA as previously described (12). B, 3 days after first siRNA transfection, HS68 and IMR90 cells were immunostained with anti-TopBP1, anti-γH2AX, and anti-p-Chk2 antibodies (supplemental Fig. S6), then the immunostained cells were scored. To detect BrdU incorporation, the cells were incubated with 10 μm BrdU for 30 min followed by detection with anti-BrdU antibody. C, 3–7 days after the first transfection, the cells were stained with SA-β-gal stain and viewed under a phase contrast microscope.
DISCUSSION
In this study we generated and characterized a conditional knock-out mutation in the mouse TopBP1 gene. The TopBP1flox allele was found to be comparable with that of the wild-type allele, and deletion of TopBP1 exons 5 and 6 produced a null allele suitable for determining the roles of TopBP1 in vivo and in primary cells (Fig. 1F). TopBP1-deficient embryos indicate a critical and non-redundant function for TopBP1 during murine peri-implantation development. This conclusion is supported by the following observations; (a) TopBP1 is highly expressed in ES and early embryonic cells (Fig. 1F), (b) about 20% of embryos at E7.5 were fully resorbed (Fig. 2C), and (c) whereas TopBP1Δ/Δ blastocysts recovered at E3.5 were detected at the expected Mendelian ratio (supplemental Table S2) and were morphologically indistinguishable from wild-type, inner cell mass and trophoblast giant cells did not grow upon outgrowth of blastocysts (Fig. 2B). The dramatic reduction of BrdU incorporation in the mutant embryo followed by the observation of cell death suggests that a defect in cell proliferation causes the developmental failure.
It has been reported that TopBP1 is present in mammalian oocytes and is involved in a meiotic recombination checkpoint (28, 42, 43). Also, TopBP1 is highly expressed during early embryogenesis (Fig. 1F). Therefore, the survival of TopBP1-deficient cells to the blastocyst stage suggests that maternal TopBP1 mRNA and/or protein are sufficient to sustain the TopBP1 level through several cell divisions in mutant embryos during early embryogenesis. However, it appears that TopBP1 is depleted at the blastocyst stage, leading to embryonic cell death, as supported by the result shown in Fig. 4C, which indicates that comparable BrdU incorporation in mutant MEFs is sustained until 72 h after TopBP1 ablation.
The functions of TopBP1 in the initiation of chromosome replication, replication fork checkpoint, and DNA damage checkpoint support our proposal that TopBP1 ablation in the mouse MEF cells causes G1 arrest with 2N DNA or DNA replication failure in S phase cells. The failure to complete chromosome replication and DNA damage can lead to arrest of the TopBP1-deficient cells in the G2 phase with 4N DNA content. Also, the DNA breakage taken placed by TopBP1 deficiency can contribute to such defects in the cell cycle progression. TopBP1-depleted cancer cells using siRNA accumulated the cells containing DNA breaks and 4N DNA (34) or G1 arrest cells without DNA damage (12). The functions of TopBP1 support our proposal that TopBP1-deficient cells arrest the cell cycle at each cell cycle phase when TopBP1 is lowered below the threshold level necessary for the progression of each cell phase.
TopBP1 knockdown using siRNAs in cancer cells induces apoptosis (12, 22, 23, 33, 34). TopBP1 recruits Brg1·Brm to E2F1-responsive promoters and inhibits E2F1 transcriptional activity, which represses E2F1-mediated apoptosis during the G1/S transition of the cell cycle (22). Similarly, depletion of TopBP1 up-regulates p53 target genes involved in cell cycle arrest and apoptosis, thereby inducing DNA damage-induced apoptosis because interaction of TopBP1 with the p53 DNA binding domain inhibits the binding of p53 to the promoters (23). These results explain why TopBP1 knockdown induces apoptosis in cancer cells. Although TopBP1 depleted mouse MEFs and 3T3 cells were found to exhibit phenotypes similar to the phenotypes of cancer cells subjected to TopBP1-knockdown, such as generation of DNA breaks, activation of the Chk2 pathway, and inactivation of the Chk1 pathway under UV radiation, the TopBP1-depleted MEFs were found to induce cellular senescence rather than apoptosis. Furthermore, prolonged knockdown of TopBP1 by the addition of siRNA against TopBP1 also triggers cellular senescence in human primary cells. When DNA damage is severe, cells either undergo apoptosis to remove damaged cells from a cell population or initiate cellular senescence that leads to irreversible cell cycle arrest (44). The cell type and the intensity, duration, and nature of the damage may decide whether apoptosis or cellular senescence occurs.
Supplementary Material
Acknowledgments
We thank N. Copeland for providing the pL451 and bacterial strains used in BAC recombineering and YY Kong for providing the Cre-expressing PT-67 cell line.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- TopBP1
- DNA topoisomerase II-binding protein 1
- MEF
- mouse embryonic fibroblast
- Cre-retrovirus
- retrovirus expressing Cre recombinase
- SA-β-Gal
- senescence-associated β galactosidase.
REFERENCES
- 1. Williams J. S., Williams R. S., Dovey C. L., Guenther G., Tainer J. A., Russell P. (2010) EMBO J. 29, 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Taylor R. M., Moore D. J., Whitehouse J., Johnson P., Caldecott K. W. (2000) Mol. Cell. Biol. 20, 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hammet A., Magill C., Heierhorst J., Jackson S. P. (2007) EMBO Rep. 8, 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamane K., Tsuruo T. (1999) Oncogene 18, 5194–5203 [DOI] [PubMed] [Google Scholar]
- 5. Lee M. S., Edwards R. A., Thede G. L., Glover J. N. (2005) J. Biol. Chem. 280, 32053–32056 [DOI] [PubMed] [Google Scholar]
- 6. Xu C., Wu L., Cui G., Botuyan M. V., Chen J., Mer G. (2008) J. Mol. Biol. 381, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balestrini A., Cosentino C., Errico A., Garner E., Costanzo V. (2010) Nat. Cell Biol. 12, 484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. (2010) Cell 140, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sansam C. L., Cruz N. M., Danielian P. S., Amsterdam A., Lau M. L., Hopkins N., Lees J. A. (2010) Genes Dev. 24, 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. (2007) Nature 445, 328–332 [DOI] [PubMed] [Google Scholar]
- 11. Zegerman P., Diffley J. F. (2007) Nature 445, 281–285 [DOI] [PubMed] [Google Scholar]
- 12. Jeon Y., Lee K. Y., Ko M. J., Lee Y. S., Kang S., Hwang D. S. (2007) J. Biol. Chem. 282, 14882–14890 [DOI] [PubMed] [Google Scholar]
- 13. Delacroix S., Wagner J. M., Kobayashi M., Yamamoto K., Karnitz L. M. (2007) Genes Dev. 21, 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J., Kumagai A., Dunphy W. G. (2007) J. Biol. Chem. 282, 28036–28044 [DOI] [PubMed] [Google Scholar]
- 15. Zhao H., Piwnica-Worms H. (2001) Mol. Cell. Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Olson E., Nievera C. J., Lee A. Y., Chen L., Wu X. (2007) J. Biol. Chem. 282, 22939–22952 [DOI] [PubMed] [Google Scholar]
- 17. Wakeman T. P., Xu B. (2006) Mutat. Res. 610, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ward I. M., Chen J. (2001) J. Biol. Chem. 276, 47759–47762 [DOI] [PubMed] [Google Scholar]
- 19. Liu S., Bekker-Jensen S., Mailand N., Lukas C., Bartek J., Lukas J. (2006) Mol. Cell. Biol. 26, 6056–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan S., Lindsay H. D., Michael W. M. (2006) J. Cell Biol. 173, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morishima K., Sakamoto S., Kobayashi J., Izumi H., Suda T., Matsumoto Y., Tauchi H., Ide H., Komatsu K., Matsuura S. (2007) Biochem. Biophys. Res. Commun. 362, 872–879 [DOI] [PubMed] [Google Scholar]
- 22. Liu K., Luo Y., Lin F. T., Lin W. C. (2004) Genes Dev. 18, 673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu K., Bellam N., Lin H. Y., Wang B., Stockard C. R., Grizzle W. E., Lin W. C. (2009) Mol. Cell. Biol. 29, 2673–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herold S., Wanzel M., Beuger V., Frohme C., Beul D., Hillukkala T., Syvaoja J., Saluz H. P., Haenel F., Eilers M. (2002) Mol. Cell 10, 509–521 [DOI] [PubMed] [Google Scholar]
- 25. Liu K., Paik J. C., Wang B., Lin F. T., Lin W. C. (2006) EMBO J. 25, 4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu P., Jenkins N. A., Copeland N. G. (2003) Genome Res. 13, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G. (2005) Nucleic Acids Res. 33, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng P., Schramm R. D., Latham K. E. (2005) Biol. Reprod. 72, 1359–1369 [DOI] [PubMed] [Google Scholar]
- 29. Lee H., Lee D. J., Oh S. P., Park H. D., Nam H. H., Kim J. M., Lim D. S. (2006) Mol. Cell. Biol. 26, 5373–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nash R. A., Caldecott K. W., Barnes D. E., Lindahl T. (1997) Biochemistry 36, 5207–5211 [DOI] [PubMed] [Google Scholar]
- 31. Todaro G. J., Green H. (1963) J. Cell Biol. 17, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nowak R. A., Haimovici F., Biggers J. D., Erbach G. T. (1999) Biol. Reprod. 60, 85–93 [DOI] [PubMed] [Google Scholar]
- 33. Yamane K., Wu X., Chen J. (2002) Mol. Cell. Biol. 22, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim J. E., McAvoy S. A., Smith D. I., Chen J. (2005) Mol. Cell. Biol. 25, 10907–10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mäkiniemi M., Hillukkala T., Tuusa J., Reini K., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Mäkelä T. P., Syväoja J. E. (2001) J. Biol. Chem. 276, 30399–30406 [DOI] [PubMed] [Google Scholar]
- 36. Liu K., Lin F. T., Ruppert J. M., Lin W. C. (2003) Mol. Cell. Biol. 23, 3287–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dunn K. L., Davie J. R. (2005) Oncogene 24, 3492–3502 [DOI] [PubMed] [Google Scholar]
- 38. Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett-Jones D. P., Allis C. D. (1997) Chromosoma 106, 348–360 [DOI] [PubMed] [Google Scholar]
- 39. Garcia V., Furuya K., Carr A. M. (2005) DNA Repair 4, 1227–1239 [DOI] [PubMed] [Google Scholar]
- 40. Chen Q. M., Tu V. C., Catania J., Burton M., Toussaint O., Dilley T. (2000) J. Cell Sci. 113, 4087–4097 [DOI] [PubMed] [Google Scholar]
- 41. Hwang E. S., Yoon G., Kang H. T. (2009) Cell. Mol. Life Sci. 66, 2503–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perera D., Perez-Hidalgo L., Moens P. B., Reini K., Lakin N., Syväoja J. E., San-Segundo P. A., Freire R. (2004) Mol. Biol. Cell 15, 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoo H. Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. (2009) Mol. Biol. Cell 20, 2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. d'Adda di Fagagna F. (2008) Nat. Rev Cancer 8, 512–522 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.