Abstract
Eukaryotic organisms have developed diverse mechanisms for the acquisition of iron, which is required for their survival. Graminaceous plants use a chelation strategy. They secrete phytosiderophore compounds, which solubilize iron in the soil, and then take up the resulting iron-phytosiderophore complexes. Bacteria and mammals also secrete siderophores to acquire iron. Although phytosiderophore secretion is crucial for plant growth, its molecular mechanism remains unknown. Here, we show that the efflux of deoxymugineic acid, the primary phytosiderophore from rice and barley, involves the TOM1 and HvTOM1 genes, respectively. Xenopus laevis oocytes expressing TOM1 or HvTOM1 released 14C-labeled deoxymugineic acid but not 14C-labeled nicotianamine, a structural analog and biosynthetic precursor of deoxymugineic acid, indicating that the TOM1 and HvTOM1 proteins are the phytosiderophore efflux transporters. Under conditions of iron deficiency, rice and barley roots express high levels of TOM1 and HvTOM1, respectively, and the overexpression of these genes increased tolerance to iron deficiency. In rice roots, the efficiency of deoxymugineic acid secretion was enhanced by overexpression of TOM1 and decreased by its repression, providing further evidence that TOM1 encodes the efflux transporter of deoxymugineic acid. We have also identified two genes encoding efflux transporters of nicotianamine, ENA1 and ENA2. Our identification of phytosiderophore efflux transporters has revealed the final piece in the molecular machinery of iron acquisition in graminaceous plants.
Keywords: Amino Acid Transport, Iron, Metals, Oocyte, Plant, Phytosiderophores
Introduction
Iron (Fe) is essential for plant growth, being required for various cellular activities ranging from photosynthesis to respiration. It is also required for human survival, and Fe deficiency is one of the most widespread human nutritional problems in the world (1). Increasing the level of bioavailable Fe in food plants would therefore have a dramatic positive impact on human health.
Under aerobic conditions, Fe is oxidized and poorly soluble in water. Thus, although mineral soils contain 6% Fe by weight, most of this Fe is not available to plants. This problem is exacerbated in soils of high pH, including calcareous soils, constituting a major problem for crop production. Fe deficiency leads to leaf chlorosis, poor yield, and decreased nutritional quality. As a Fe acquisition strategy, graminaceous plants, including important staple crops such as rice, wheat, and barley, secrete phytosiderophore from their roots (2). Phytosiderophores are natural Fe chelators; in the soil, the secreted phytosiderophores solubilize Fe, forming Fe-phytosiderophore complexes that are absorbed into root cells through the Fe-phytosiderophore transporter YS1 (3). The secretion of phytosiderophores in barley follows a distinct diurnal rhythm with a peak just after sunrise or initial illumination (4). The diurnal rhythm of the secretion of phytosiderophores in rice has not been fully characterized. Terrestrial and marine microorganisms also release low molecular weight compounds, siderophores, to chelate and acquire Fe for their survival (5). In addition, siderophores in mammal were identified recently (6, 7). 2,3-Dihydroxybenzoate and 2,5-dihydroxybenzoate from mammalian cells were identified as intracellular ligands for Fe. Their Fe complexes were bound to the siderocalin, the secreted protein (Scn-Ngal) (6). In addition, a subset of catechols chelates Fe and binds to the Scn-Ngal, with subnanomolar affinity (7). The Scn-Ngal-catechol-Fe complexes could deliver Fe to cells and might contribute to constant Fe supply in some organs, such as kidney. Catechols are excreted in the urine; however, the transporters involved in the excretion have not been identified.
Phytosiderophores are synthesized from l-methionine through a nicotianamine (NA)3 intermediate (8–10). Phytosiderophores (11–13) and NA (14, 15) are also essential for the translocation of metal nutrients within plants. All of the genes encoding the phytosiderophore biosynthetic enzymes and the Fe-phytosiderophore uptake transporters have been isolated from barley, maize, and rice (16–19). The genes involved in the transcriptional regulation of cellular Fe homeostasis in rice have been identified, and their regulatory network has been delineated (20, 21). However, until the present study, the genes responsible for phytosiderophore efflux had not been identified, and the molecular mechanism of phytosiderophore secretion was largely unknown.
In this report, we show that TOM1 in rice and HvTOM1 in barley are directly involved in the efflux of deoxymugineic acid (DMA), the primary member of the phytosiderophore family. Xenopus laevis oocytes expressing TOM1 or HvTOM1 released 14C-labeled DMA but not 14C-labeled NA, indicating that TOM1 and HvTOM1 are the phytosiderophore efflux transporters. In addition, we found that ENA1 and ENA2 encode NA efflux transporters; X. laevis oocytes expressing ENA1 or ENA2 released 14C-labeled NA but not 14C-labeled DMA. Expression of TOM1 or HvTOM1 was strongly induced in Fe-deficient roots, with a strongly diurnal pattern in their expression. In rice, the level of TOM1 expression directly correlates with the level of phytosiderophore secretion from the roots. Moreover, overexpression of TOM1 or HvTOM1 resulted in increased tolerance of Fe deficiency. This study, therefore, has succeeded in identifying the efflux transporter of DMA, the missing piece in the mechanics of the Fe acquisition system of grain plants.
EXPERIMENTAL PROCEDURES
Gene Cloning and Plasmid Construction
The TOM1 gene was amplified from a rice Oryza sativa L. cv. Nipponbare cDNA library template using the primers 5′-TCTAG AATGA GTGAG GAGGC ACCGC CATCG-3′ and 5′-TCTAG ATTAA TTTGG TATTG CCAGG AACGG-3′. The amplified DNA fragment was subsequently cloned into the pCR4Blunt-TOPO vector (Invitrogen) to create plasmid pTOM1. The HvTOM1 gene was isolated using the colony hybridization method (22) with the full-length ORF of TOM1 as a probe and cloned into the pCR4Blunt-TOPO vector to create plasmid pHvTOM1.
Construction of Oocyte Expression Vectors
The TOM1, HvTOM1, ENA1, ENA2, and AK068840 open reading frames (ORFs) were amplified from a rice O. sativa L. cv. Nipponbare cDNA library template using the following primer pairs: TOM1, 5′-aagct tATGA GTGAG GAGGC ACCGC C-3′ and 5′-ctcga gTTAA TTTGG TATTG CCAGG AAC-3′; HvTOM1, 5′-gaatt cATGG GTGAC ACTAC CAGGA A-3′ and 5′-ctcga gTCAT AATGT TTTGG AACTG CTAG-3′; ENA1, 5′-aagct tATGC ACCTG CTGCT TGGAC T-3′ and 5′-ctcga gTCAT GGTTG AGCTA ATAGT G-3′; ENA2, 5′-aagct tATGC GCATT GCCCG GATTC C-3′ and 5′-cccgg gTCAG CTGAG CTCGA GGAAG C-3′; AK068840; 5′-aagct tATGG CCAGC CACGC CGTCA C-3′ and 5′-ctcga gCTAC ATGAC ATCAC AACCG T-3′. The amplified TOM1, HvTOM1, ENA1, ENA2, and AK068840 genes were ligated into pENTR/D-TOPO (Invitrogen) and then subcloned into a previously constructed plasmid (23). Capped complementary RNA (cRNA) derived from these plasmids was then injected into X. laevis oocytes (23).
Construction of the GFP Fusion Gene and Observation of TOM1-sGFP Localization
The TOM1 ORF was subcloned into pENTR/D-TOPO to create plasmid pENTR-TOM1. A subsequent attL/attR substrate recombination reaction between pDEST35S-sGFP (24) and pENTR-TOM1 generated an expression vector containing the cauliflower mosaic virus (CaMV) 35S promoter-TOM1-sGFP gene fusion sequence (35S::TOM1-GFP). Onion epidermal cells were transformed with 35S::TOM1-GFP using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories), and sGFP fluorescence was observed.
Construction of Plant Expression Vectors and Transgenic Plants
The TOM1 coding sequence was excised from pTOM1 and cloned into the pIG121Hm vector (25) to construct the CaMV 35S promoter–TOM1 cassette 35S::TOM1. Partial fragments of the TOM1 coding sequence (350 bp) or 3′-noncoding region (312 bp) were amplified from pTOM1 or a rice O. sativa L. cv. Nipponbare cDNA library template using the primer pairs 5′-CACCA GTTGC AGATC GTATA GGGAG GAA-3′ and 5′-TCGGA AAATA CATTT GGATA TTGCT-3′ or 5′-CACCA TCAGT TGAAG TAGAA GCT-3′ and 5′-GAGAG AGACA GGCTT GCGTA CA-3′, respectively, and cloned into the pENTR/D-TOPO vector (Invitrogen). The resulting intermediate entry vectors were recombined with the pIG121-RNAi-DEST binary vector (26) using LR Clonase (Invitrogen), according to the manufacturer's instructions, to construct the RNAi cassettes TOM1RNAi (line 1 or line 17 and 20). The HvTOM1 coding region was amplified from pHvTOM1 using primers 5′-TCTAG AATGA GTGAG GAGGC ACCGC CATCG-3′ and 5′-TCTAG ATTAA TTTGG TATTG CCAGG AACGG-3′ and cloned into plasmid pCR4Blunt-TOPO. The HvTOM1 ORF was subsequently cloned into the pIG121Hm vector (25) to construct the CaMV 35S promoter–HvTOM1 cassette 35S::HvTOM1. The pIG121 binary vector (25) was used as a vector control. To construct β-glucuronidase (GUS) reporter fusion genes, 1.5-kb fragments of the 5′ upstream regions of TOM1, TOM2, or TOM3 gene were amplified from rice genomic DNA (cv. Nipponbare) using the primer pairs 5′-AAGCT TGTCC ACCTG CATGG CAGCC CGCTC-3′ and 5′-TCTAG AAGCC ATCCA GTACT TCACA CTTAA-3′, 5′-CTCGA GCGGT GTATT TTACT CATGA ACAAA-3′ and 5′-TCTAG ACAAC GGCGA CGCTC TCAAT GGAGT-3′, or 5′-CTCGA GTGCT ACTGA TGCTC AACCA GGGCT C-3′ and 5′-TCTAG AAGGT CGCCC ATCAA TTCAC CAATC T-3′, respectively, and ligated into the pCR4Blunt-TOPO vector. The gene fragments were then excised and subcloned into the pIG121Hm vector (25) upstream of the GUS ORF to create the TOM1 promoter::GUS, TOM2 promoter::GUS, and TOM3 promoter::GUS constructs. To confirm cellular localization of TOM1 in rice plant, TOM1-sGFP fusion gene was expressed under the control of CaMV 35S promoter. The GUS gene in the pIG121Hm vector was replaced by sGFP with XbaI and SacI. The TOM1 ORF was subsequently cloned into the modified pIG121Hm vector to construct the CaMV 35S promoter–TOM1–sGFP cassette 35S::TOM1–sGFP. Agrobacterium tumefaciens strain C58 carrying these constructs was then used to transform rice (O. sativa L. cv. Tsukinohikari) (25).
Production of 14C-labeled NA and DMA
S-Adenosyl-l-[14C]methionine (2.1 GBq/mmol; Amersham Biosciences) was converted into 14C-labeled NA by barley NA synthase 1 enzyme (HvNAS1) as described previously (27), with some modifications. HvNAS1 was expressed in and purified from Escherichia coli strain XL1-Blue. The 14C-labeled NA was purified on thin layer chromatography LK6 plates (Whatman, Clifton, NJ), extracted with water, and purified with an equal volume of chloroform. The supernatant fraction was used as 14C-labeled NA. 14C-Labeled NA was converted into 14C-labeled DMA using barley NA aminotransferase 1 enzyme (HvNAAT1) and NaBH4 according to the methods described previously (28), with some modifications. The 14C-labeled DMA was purified on cellulose plates (Avicel SF; Asahi Chem. Industry Co., Tokyo, Japan) according to the described methods (10) and then extracted with water.
Efflux Experiments
X. laevis oocytes were isolated (23), injected with 23 ng of TOM1, HvTOM1, ENA1, ENA2, or AK068840 cRNA or water, and then incubated at 17 °C (23). After incubation for 2 days, five oocytes were selected from each group, injected with either 23 nl of 24.18 μm [14C]DMA or 20.16 μm [14C]NA in sodium barth buffer, and rinsed three times with 750 μl of sodium barth buffer. The oocytes were then incubated at 17 °C with periodic removal of 650-μl aliquots of the buffer for radioactivity measurements. The buffer was replenished with fresh buffer. At the end of each experiment, the oocytes were dissolved in 10% SDS. The radioactivity of the sampled external solutions and that remaining in the oocytes was measured using full spectrum disintegrations/minute counting in a liquid scintillation analyzer. Radioactivity efflux was expressed as a percentage of the total radioactivity injected.
Plant Growth Conditions and Elemental Analysis
Rice and barley plants were grown hydroponically. Seeds were surface-sterilized with a 2.5% sodium hypochlorite solution and then germinated for a week. After germination, the seedlings were transferred to a 20-liter plastic container containing a nutrient solution of the following composition: 0.7 mm K2SO4, 0.1 mm KCl, 0.1 mm KH2PO4, 2.0 mm Ca(NO3)2, 0.5 mm MgSO4, 10 μm H3BO3, 0.5 μm MnSO4, 0.2 μm CuSO4, 0.5 μm ZnSO4, 0.05 μm Na2MoO4 and 0.1 mm Fe-EDTA. The pH of the nutrient solution was adjusted daily to 5.5 with 1 m HCl. Fe deficiency was initiated 4 weeks after germination by transfer of the plants to an Fe(III)-EDTA-free culture medium. For Northern blotting, GUS assay, and microarray analyses, plants were harvested 5–7 days after the transition to Fe-deficient medium. For Northern blot analysis of diurnal expression of TOM1, plants were harvested 2 weeks after the transition (29, 30). For DMA measurement, plants were harvested after 10 days of Fe deficiency. DMA was extracted from harvested rice plants and secreted for 6 h from the start of illumination, and endogenous DMA levels were analyzed using high performance liquid chromatography (HPLC) (31). For SPAD (chlorophyll) analysis, plants were grown hydroponically as described above. At the time of transition to Fe-deficient medium and again 12 days later, the newest and oldest leaves were removed and analyzed for chlorophyll content using a SPAD-5 chlorophyll meter (Konika Minolta). The concentrations of Fe, Zn, and Cu were determined using inductively coupled plasma mass spectroscopy (20). Experiments were all performed in triplicate.
GUS Activity Assay
The GUS activity in roots and shoots of transgenic plants was determined using a histochemical assay (32). GUS activity in reproductive organs and germinating seeds was also determined histochemically according to methods described previously (33).
Northern Blot Analysis
Total RNA (20 μg) was denatured, electrophoresed on 1.2% agarose gels containing 5% (v/v) formaldehyde, and blotted onto nylon membranes (Hybond-N+; Amersham Biosciences). The blotted membranes were hybridized with γ-32P-labeled probes specific for the 3′-noncoding regions of TOM1, TOM2, or TOM3.
Quantitative Real Time PCR of TOM1
Total RNA was treated with RNase-free DNase I (Takara) to remove contaminating genomic DNA. First-strand cDNA was synthesized using ReverTra Ace reverse transcriptase (Toyobo; Tokyo, Japan) by priming with oligo(dT)30. A fragment was amplified by PCR in a StepOnePlus real time PCR system (Applied Biosystems, Foster City, CA) with SYBR Green I and ExTaqTM RealTime-PCR Version (Takara). The primers used for real time PCR were same to the primers used for RNAi cassettes TOM1RNAi. The primers used for internal control in real time PCR were OsActin1 forward, 5′-ACACC GGTGT CATGG TCGG-3′; OsActin1 reverse, 5′-ACACG GAGCT CGTTG TAGAA-3′. The sizes of the amplified fragments were confirmed by agarose gel electrophoresis.
Oligonucleotide DNA Microarray Analysis
We performed a microarray expression analysis using a Rice Gene Expression 44K Microarray (Agilent Technology) with 42,023 unique synthetic 60-mer oligonucleotides that were designed based on sequence data from the Rice Full-length cDNA Project. Total RNA was extracted from shoots and roots using an RNeasy plant kit (Qiagen), following the manufacturer's instructions; the yield and RNA purity were determined spectrophotometrically. The integrity of the RNA was checked using an Agilent 2100 Bioanalyzer. Total RNA (200 ng) was labeled with Cy-3 or Cy-5 using an Agilent Low RNA Input Fluorescent Linear Amplification kit. Fluorescently labeled targets were hybridized to Agilent rice 44K oligonucleotide DNA microarrays. Microarray hybridization, scanning, and data analysis were performed as described previously (33). The biological replicate was one. The reproducibility of the microarray analysis was assessed by a dye swap in each experiment. Genes showing differential expression with significant Student's t test (p < 0.001) were further analyzed. To identify the up-regulated genes under Fe-deficient conditions, the ratio was calculated as (signal values of Fe-deficient (−Fe) sample)/(signal values of Fe-sufficient (+Fe) sample), and these ratios were used for identifying the Fe deficiency induction of the genes. To analyze the transgenic plants, the ratio was calculated as (signal values of OXOs, OXHv, or RNAi sample)/(signal values of nontransformant sample), and these ratios were used for identifying the genes whose expression was changed by modification of TOM1.
RESULTS
Identification of TOM1, ENA1, and ENA2
To identify the genes involved in phytosiderophore secretion in rice, we performed a high resolution microarray analysis to identify genes that were induced under conditions of Fe deficiency. We focused on genes that were identified as candidate efflux transporters of phytosiderophores due to their apparent similarities to genes of a transporter family involved in the efflux of substrates such as small peptides and amino acids. As a result, we identified four genes: AK069533 (Gene ID: Os11g0134900), AK102457 (Os11g0151500), AK064089 (Os06g0695800), and AK068840 (Os01g0871500) (Table 1).
TABLE 1.
Oligonucleotide microarray analysis of the roots and the shoots of rice under Fe-sufficient or Fe-deficient conditions
Ratio was calculated as (signal values of Fe-deficient (−Fe) sample)/(signal values of Fe-sufficient (+Fe) sample).
| Accession number | Roots |
Shoots |
Putative gene description | ||||
|---|---|---|---|---|---|---|---|
| Signal |
Ratio −Fe/+Fe | Signal |
Ratio −Fe/+Fe | ||||
| −Fe | +Fe | −Fe | +Fe | ||||
| AK069533, Os11g0134900 | 11,570 | 1,826 | 6.33 | 597 | 106 | 5.70 | Major facilitator superfamily antiporter |
| AK102457, Os11g0151500 | 1,002 | 264 | 3.80 | 129 | 309 | 4.42 | Major facilitator superfamily antiporter |
| AK064089, Os06g0695800 | 5,520 | 4,635 | 1.19 | 7,390 | 7,756 | 0.95 | ABC transporter-related domain-containing protein |
| AK068840, Os01g0871500 | 38,732 | 8,372 | 4.64 | 41,855 | 3,346 | 12.60 | TGF-β receptor, type I/II extracellular region family protein |
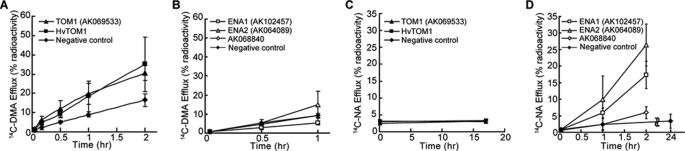
Rice produces and secretes only one member of the phytosiderophore family, DMA. To examine DMA transport activity, 14C-labeled DMA was synthesized from S-adenosyl-l-[14C]methionine using recombinant NAS (27) and NA aminotransferase (28), and the final product was chemically reduced with NaBH4. We injected water (negative control) or capped AK069533, AK102457, AK064089, or AK068840 cRNA into X. laevis oocytes and incubated them for 2 days. The oocytes were then loaded with 14C-labeled DMA and monitored for the release of 14C into the medium (Fig. 1). Oocytes expressing AK069533 exhibited a higher rate of DMA efflux than that of control oocytes or oocytes expressing AK102457, AK064089, or AK068840 (Fig. 1, A and B). The oocytes expressing AK069533 did not show any efflux activity for NA, the structural analog and biosynthetic precursor of DMA (Fig. 1C).
FIGURE 1.
Efflux activity of TOM1-, HvTOM1-, ENA1-, ENA2-, and AK068840-encoded proteins in X. laevis oocytes. Time-dependent efflux of [14C]DMA and [14C]NA is shown. Oocytes expressing TOM1, HvTOM1, ENA1, ENA2, or AK068840 and control oocytes were injected with water or with [14C]DMA (A and B) or [14C]NA (C and D), and the efflux of 14C was measured for 1–24 h. Values are expressed as the percentage of the total radioactivity injected. Data shown represent means ± S.D. (error bars; n = 3).
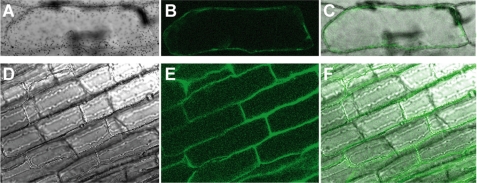
We next investigated the subcellular localization of the TOM1 protein by fusing the coding sequence for GFP to TOM1. In onion epidermal cells transiently expressing the TOM1-GFP fusion protein (under control of the CaMV 35S promoter), fluorescence was localized to the cell membrane (Fig. 2, A–C). In addition, we confirmed the plasma membrane localization of TOM1-GFP fusion protein in rice roots stably expressing TOM1-GFP (Fig. 2, D–F), indicating that AK069533 encodes a DMA efflux transporter capable of transporting DMA out of cells. Therefore, we designated AK069533 as O. sativa L. transporter of mugineic acid family phytosiderophores 1 (TOM1). From barley, we isolated the TOM1 homolog HvTOM1. Like TOM1, HvTOM1 significantly increased the release of DMA, but not NA, when expressed in oocytes (Fig. 1, A, and C).
FIGURE 2.
Subcellular localization of TOM1 in onion epidermal cells (A–C) and in rice roots (D–F). A and D, differential interference contrast image. B and E, fluorescence image. C and F, overlay.
Sequence Analysis of TOM1 and HvTOM1
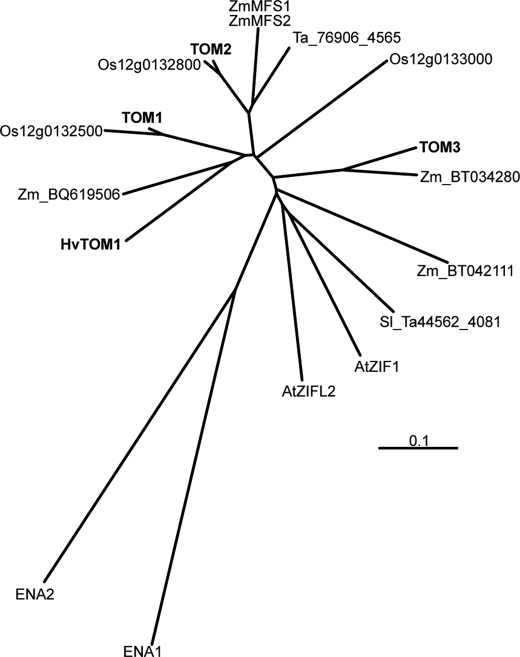
TOM1 and HvTOM1 are members of the major facilitator superfamily (MFS; Ref. 34). The proximal promoter region of TOM1 contains sequences similar to cis-acting elements iron deficiency-responsive element 1 (IDE1) and IDE2, which are recognized by IDEF1 and IDEF2, transcription factors that regulate the response to Fe deficiency (20, 21). The predicted TOM1 and HvTOM1 proteins are ∼80% identical in amino acid sequence (Fig. 3A). TOM1 has two homologs, AK121911 (Os11g0135000; TOM2) and AK064297 (Os11g0135900; TOM3), which are located in tandem with TOM1 on chromosome 11 (Fig. 3B). We found TOM1 orthologs (Fig. 4) in other graminaceous plants (maize, barley, and wheat) and in dicots (Arabidopsis and tomato).
FIGURE 3.
Characters of TOM1 family. A, amino acid sequence of HvTOM, TOM1, TOM2, and TOM3. The 12 putative membrane-spanning domains predicted using the TM-pred program are shown in upperlines. B, location of TOM family on the chromosome 11.
FIGURE 4.
Phylogenetic tree of plant TOM transporters. Os, rice; Hv, barley; Zm, maize; At, Arabidopsis; Sl, tomato; Ta, wheat. Scale bar, 0.1 substitution/site.
Interestingly, oocytes expressing AK102457 and AK064089 displayed NA, but not DMA, efflux activity (Fig. 1, B and D). Therefore, we designated these genes as O. sativa L. efflux transporter of NA 1 (ENA1) and 2 (ENA2), respectively. ENA1 is also a member of the MFS family, with few similarities to the TOM family (Fig. 4).
Expression Analysis of TOM1 and HvTOM1
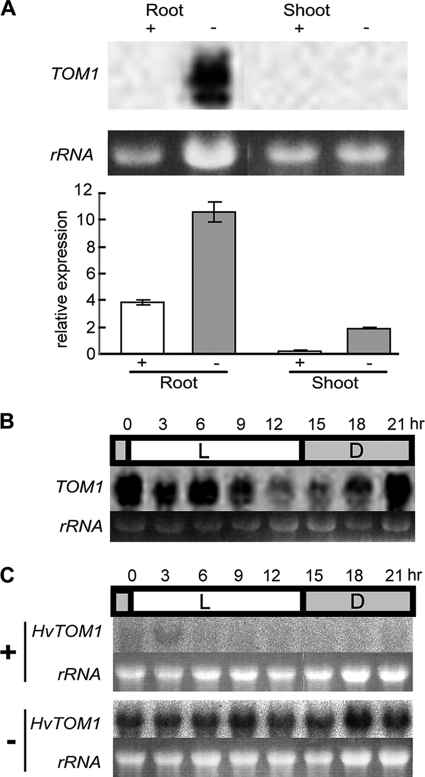
We found that TOM1 was expressed primarily in Fe-deficient rice roots (Fig. 5A). Levels of TOM1 expression were induced both in roots and shoots by Fe deficiency (Fig. 5A). In Fe-deficient roots, the level of TOM1 mRNA accumulation underwent diurnal changes, decreasing with light and increasing after dark (Fig. 5B). Similarly, HvTOM1 expression was highly induced in Fe-deficient barley roots, with a diurnal rhythm similar to that of TOM1 (Fig. 5C).
FIGURE 5.
Expression of TOM1. A, regulation of TOM1 mRNA levels by Fe availability in rice. The upper panel shows the Northern blot analysis, and the lower panel shows the quantitative RT-PCR analysis of TOM1. +, Fe-sufficient; −, Fe-deficient. In the Northern blot analysis, ethidium bromide-stained rRNA is shown as a loading control. In the quantitative real time PCR analysis, the values were normalized with the expression of OsActin1 and represent the mean ± S.D. (error bars) from three reactions. B, diurnal changes in TOM1 expression in Fe-deficient rice roots. L, light; D, dark. C, diurnal changes in HvTOM1 expression in Fe-deficient (−) and Fe-sufficient (+) barley roots. L, light; D, dark.
Promoter-GUS Analysis of TOM1
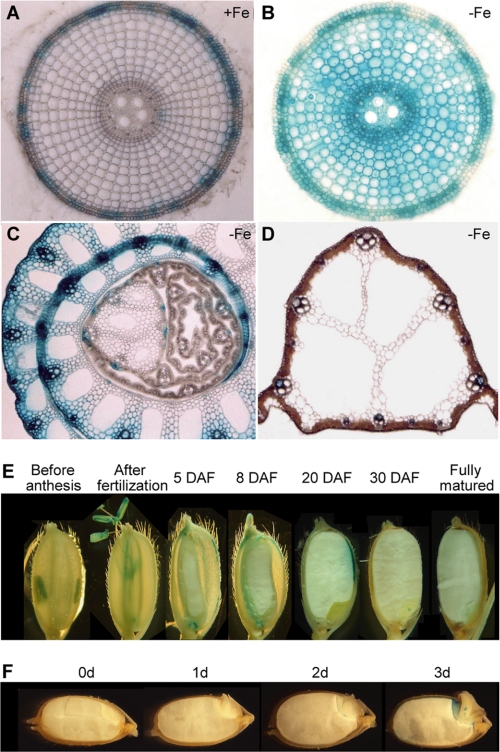
To assess the tissue-specific localization of TOM1, we generated transgenic plants in which the GUS gene uidA was driven by the TOM1 promoter (Fig. 6). Histochemical analysis of GUS activity revealed GUS reporter gene expression in some restricted parts of the exodermis of roots grown under Fe-sufficient conditions (Fig. 6A and supplemental Fig. S1, A and D). In roots grown under Fe-deficient conditions, GUS staining was observed not only in the epidermis, exodermis, cortex, and endodermis, but also in the central cylinder (Fig. 6B; and supplemental Fig. S1, B, C, E, and F). Furthermore, GUS staining was observed in the vascular bundles of leaf sheaths (Fig. 6C and supplemental Fig. S1, G and H) and leaf phloem (Fig. 6D and supplemental Fig. S1I). It was also observed in the pollen and dorsal vascular bundle in developing seeds (Fig. 6E) and in the epithelium of germinating seeds (Fig. 6F).
FIGURE 6.
Tissue distribution of TOM1 protein expression in rice during vegetative stage (A–D), seed development (E), and germination (F) as shown by GUS staining. A, Fe-sufficient root cross-section. B, Fe-deficient root cross-section. C, Fe-deficient leaf sheath cross-section. D, Fe-deficient leaf cross-section. E, seeds before anthesis; after fertilization; 5, 8, 20, and 30 days after flowering (DAF), and immediately before full maturation. F, fully mature seeds (0d) and germinating rice seeds 1–3 days after sowing.
TOM1 Function in Rice
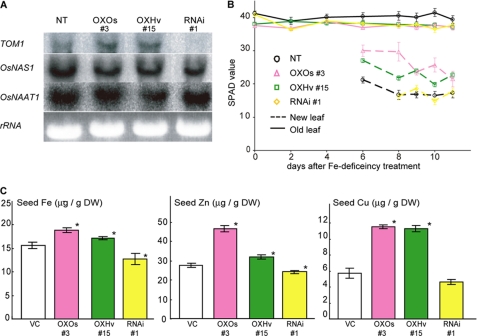
To examine TOM1 function in rice plants, we repressed its expression using RNA interference (RNAi plants) and increased its expression using the CaMV 35S promoter (OXOs plants). We also generated transgenic rice plants overexpressing HvTOM1 (OXHv plants). Northern blot and microarray analysis of the RNAi plant roots confirmed that the expression of TOM1 (and not TOM2 or TOM3) was repressed (Fig. 7A and supplemental Table S1) and that the expression of genes encoding DMA biosynthetic enzymes was not affected (Fig. 7A and supplemental Table S1).
FIGURE 7.
In vivo function of TOM1 in rice. A, Northern blot analysis in Fe-deficient roots of NT plants; plants overexpressing TOM1 (OXOs) or HvTOM1 (OXHv); and plants with RNAi-repressed TOM1 expression (RNAi). B, SPAD values (chlorophyll content) of the newest and oldest leaves of OXOs, OXHv, RNAi, and NT plants during progression of Fe deficiency stress (0 days) and 12 days later. Error bars represent S.E. (n = 3–6). C, Fe, Zn, and Cu contents in OXOs, OXHv, RNAi, and vector control (VC) seeds. Error bars represent S.D. *, significant difference (p < 0.01) by Student's t test.
Under Fe-sufficient conditions, RNAi, OXOs, and OXHv plants showed no significant differences in vegetative growth. However, when the plants were grown in Fe-deficient hydroponic culture medium, the chlorophyll content (in SPAD units) of the youngest leaves of the OXOs and OXHv plants was higher than that of the youngest leaves of the RNAi and nontransformed (NT) plants (Fig. 7B), suggesting that the OXOs and OXHv plants were tolerant of Fe deficiency. We found that not only Fe but also Zn and Cu contents in the OXOs, OXHv seeds were higher than vector control (VC), whereas that of RNAi seeds were lower than VC seeds (Fig. 7C).
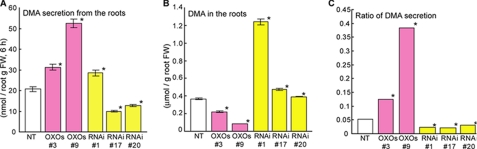
We examined the amounts of DMA secreted from the roots of NT, OXOs, and RNAi plants (Fig. 8A). The germination rate of RNAi plants made using a fragment of the TOM1 ORF was so low that we were able to use only one line (line 1). Surmising that the knock-out of TOM1 expression might result in a lethal phenotype, we made new RNAi plants in which TOM1 expression was repressed by the 3′-noncoding region of TOM1 (supplemental Fig. S2, RNAi, lines 17 and 20). As shown in Fig. 8A, compared with the NT plant roots, the OXOs roots secreted more DMA (lines 3 and 9) and the RNAi roots secreted less DMA (lines 17 and 20). In addition, compared with the NT plant roots, the RNAi roots had a much higher endogenous DMA content, and the OXOs roots had a lower DMA content (Fig. 8B). When we calculated the fraction of secreted DMA amount divided by the DMA amount synthesized in the root (Fig. 8C), we found that the DMA secretion fraction was decreased in RNAi plants and increased in OXOs plants. Fe and Cu contents in the roots were correlated with DMA secretion from the roots; Fe and Cu levels in OXOs tended to be higher than NT, whereas Fe and Cu levels in RNAi tended to be lower than NT (supplemental Fig. S3). On the other hand, the expression levels of the genes encoding the enzymes for phytosiderophore biosynthesis did not differ between the plant types (Fig. 7A and supplemental Table S1). Thus, the amount of DMA produced in RNAi roots appeared to be unaffected by the level of TOM1 expression, leading us to conclude that RNAi plants are defective in DMA secretion and not in DMA synthesis.
FIGURE 8.
Analysis of levels of endogenous DMA in and DMA secretion from Fe-deficient OXOs, OXHv, RNAi, and NT plant roots after a 6-h exposure to light. A, secreted DMA. B, endogenous DMA. C, fraction of the total DMA synthesized in the root that was secreted. Values shown represent the means of three replicates. Error bars represent S.D. *, significant difference (p < 0.01) by Student's t test.
DISCUSSION
In this study, we have identified the efflux transporters of phytosiderophores, TOM1 and HvTOM1, from rice and barely, thereby completing the repertoire of molecular components of the phytosiderophore-based iron acquisition system of graminaceous plants.
TOM1 and HvTOM1 Belong to MFS Family
TOM1 and HvTOM1 belong to the MFS (34), including the EntS gene product of E. coli, involved in siderophore export necessary in Fe acquisition (35). Phytosiderophore and siderophore are both Fe chelators; however, these structures are diverse. It is interesting that same family transporters are involved in Fe acquisition in different organism. ENA1 also belongs to the MFS. ENA1 is similar to AtZIF1 and AtZIFL2 (Fig. 2A). Notably, AtZIF1 is induced by excess Zn and localizes on the vacuolar membrane (36), and an AtZIF1-knock-out mutant is sensitive to excess Zn, suggesting that AtZIF1 is involved in Zn detoxification through sequestration into the vacuole. ENA1 is predicted by the PSORT (Prediction of Protein Sorting Signals and Localization Sites in Amino Acid Sequences) Web server to be localized on the vacuolar membrane. ENA1 might participate in metal detoxification by transporting nicotianamine into the vacuole.
TOM1 and HvTOM1 Showed Diurnal Expression Patterns
The expression level of TOM1 and HvTOM1 was changed diurnally, and their expression levels were the highest in the night. Importantly, these findings are consistent with the previously noted diurnal rhythm of phytosiderophore secretion (4). In addition, this diurnal expression pattern of TOM1 was similar to that of OsNAS2, a gene involved in NA synthesis (29). It was suggested that the diurnal phytosiderophore secretion must be regulated in the molecular level in rice and barley.
TOM1 Was Expressed Not Only in Roots but Also Shoots
TOM1 was expressed mainly in the roots, and their expression levels were induced strongly by Fe deficiency. The promoter region of TOM1 contains sequences similarity to IDE1 and IDE2 (20, 21), suggesting that TOM1 is regulated by IDEF1 and IDEF2. OsIRO2, the transcription factor involved in phytosiderophore biosynthesis (26), is regulated by IDEF1 (20). Indeed, the TOM1 expression level was dramatically enhanced by OsIRO2 overexpression but decreased by OsIRO2 repression.4
Under the Fe-sufficient condition, TOM1 expression was observed in exodermis, and under the Fe-deficient condition, TOM1 expression was observed in not only whole root cell but also in leaf sheaths and leaf. This pattern of TOM1 expression appeared to be similar to that of genes involved in DMA biosynthesis (18, 32, 37). Because DMA is responsible for both Fe acquisition from the soil and internal Fe transport in plants (18, 38), these results strongly suggest that TOM1 is involved not only in DMA secretion from the root but also efflux of DMA to the phloem and xylem for internal Fe transport.
Recently, the enzymes involved in phytosiderophore biosynthesis and Fe transporters were shown to be expressed during reproductive stage and germination (19, 33, 39, 40). TOM1 expression was also observed during reproductive stage and germination. In addition, the RNAi seed exhibited an extremely low rate of seed germination compared with OXOs, OXHv, and NT seeds. Because DMA chelates not only Fe, but also Zn and Cu (41), and is important for metal transport during rice seed germination, TOM1 must have an important role in the mobilization of the metal during reproductive stage and germination.
In conclusion, we identified the phytosiderophore efflux transporters that have been a final piece in the molecular machinery of iron acquisition by graminaceous plants. Previously, we have shown that a transgenic approach to increase the tolerance of rice to low iron availability is a practical way to improve agricultural productivity in adverse soils (17, 31, 42, 43). Our identification of phytosiderophore efflux transporters provides a new powerful strategy for the development of transgenic crops that are tolerant of low iron availability in adverse soils. Furthermore, these crops may prove valuable for increasing the amount of dietary iron available to peoples dependent on plant-based diets.
Supplementary Material
Acknowledgments
We thank A. Sato for technical support in the oocyte experiments, Dr. Y. Nagamura for oligonucleotide microarray analysis, Y. Nozoye, R. N. Itai, and Dr. Pax Blamey for reading and commenting on the manuscript.
Note Added in Proof
ENA2 is the same gene as STAR1 named by Huang et al. (44).
This work was supported by a research grant of CREST from Japan Science and Technology Agency (to N. K. N.) and by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to N. K. N. and N. U.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
The nucleotide sequence data reported in this paper are deposited in the RAP-DB/DDBJ/GenBank/TIGR nucleotide sequence databases under the accession numbers AK069533 (Os11g0134900), AK102457 (Os11g0151500), AK064089 (Os06g0695800), AK068840 (Os01g0871500), AK121911 (Os11g0135000), AK064297 (Os11g0135900), Os12g0132500, Os12g0132800, Os12g0133000, BQ619506, BT034280, BT042111, Ta44562_4081, Ta76906_4565.
Y. Ogo, R. N. Itai, T. Kobayashi, M. Sann Aung, H. Nakanishi, and N. K. Nishizawa, unpublished data.
- NA
- nicotianamine
- CaMV
- cauliflower mosaic virus
- DMA
- deoxymugineic acid
- GUS
- β-glucoronidase
- IDE1, 2
- iron deficiency-responsive element 1, 2
- MFS
- major facilitator superfamily
- NAS
- nicotianamine synthase
- NT
- nontransformed
- TOM1
- transporter of mugineic acid 1
- SPAD
- soil and plant analyzer development.
REFERENCES
- 1. Welch R. M., Graham R. D. (2004) J. Exp. Bot. 55, 353–364 [DOI] [PubMed] [Google Scholar]
- 2. Takagi S. (1976) Soil Sci. Plant Nutr. 45, 993–1002 [Google Scholar]
- 3. Curie C., Panaviene Z., Loulergue C., Dellaporta S. L., Briat J. F., Walker E. L. (2001) Nature 409, 346–349 [DOI] [PubMed] [Google Scholar]
- 4. Takagi S., Kamei S., Takemoto T. (1984) J. Plant Nutr. 7, 469 [Google Scholar]
- 5. Miethke M., Marahiel M. A. (2007) Microbiol. Mol. Biol. Rev. 71, 413–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Devireddy L. R., Hart D. O., Goetz D. H., Green M. R. (2010) Cell 141, 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bao G., Clifton M., Hoette T. M., Mori K., Deng S. X., Qiu A., Viltard M., Williams D., Paragas N., Leete T., Kulkarni R., Li X., Lee B., Kalandadze A., Ratner A. J., Pizarro J. C., Schmidt-Ott K. M., Landry D. W., Raymond K. N., Strong R. K., Barasch J. (2010) Nat. Chem. Biol. 6, 602–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mori S., Nishizawa N. K. (1987) Plant Cell Physiol. 28, 1081–1092 [Google Scholar]
- 9. Shojima S., Nishizawa N. K., Mori S. (1989) Plant Cell Physiol. 30, 673–677 [Google Scholar]
- 10. Shojima S., Nishizawa N. K., Fushiya S., Nozoe S., Irifune T., Mori S. (1990) Plant Physiol. 93, 1497–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mori S., Nishizawa N. K., Hayashi H., Chino M., Yoshimura E., Ishihara J. (1991) Plant and Soil 130, 143–156 [Google Scholar]
- 12. Kawai S., Kamei S., Matsuda Y., Ando R., Kondo S., Ishizawa A., Alam S. (2001) Soil Sci. Plant Nutr. 47, 265–272 [Google Scholar]
- 13. Suzuki M., Takahashi M., Tsukamoto T., Watanabe S., Matsuhashi S., Yazaki J., Kishimoto N., Kikuchi S., Nakanishi H., Mori S., Nishizawa N. K. (2006) Plant J. 48, 85–97 [DOI] [PubMed] [Google Scholar]
- 14. Hell R., Stephan U. W. (2003) Planta 216, 541–551 [DOI] [PubMed] [Google Scholar]
- 15. Takahashi M., Terada Y., Nakai I., Nakanishi H., Yoshimura E., Mori S., Nishizawa N. K. (2003) Plant Cell 15, 1263–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mori S. (1999) Curr. Opin. Plant Biol. 2, 250–253 [DOI] [PubMed] [Google Scholar]
- 17. Kobayashi T., Nakanishi H., Takahashi M., Mori S., Nishizawa N. K. (2008) Rice 1, 144–153 [Google Scholar]
- 18. Inoue H., Takahashi M., Kobayashi T., Suzuki M., Nakanishi H., Mori S., Nishizawa N. K. (2008) Plant Mol. Biol. 66, 193–203 [DOI] [PubMed] [Google Scholar]
- 19. Inoue H., Kobayashi T., Nozoye T., Takahashi M., Kakei Y., Suzuki K., Nakazono M., Nakanishi H., Mori S., Nishizawa N. K. (2009) J. Biol. Chem. 284, 3470–3479 [DOI] [PubMed] [Google Scholar]
- 20. Kobayashi T., Ogo Y., Itai R. N., Nakanishi H., Takahashi M., Mori S., Nishizawa N. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19150–19155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ogo Y., Kobayashi T., Nakanishi Itai R., Nakanishi H., Kakei Y., Takahashi M., Toki S., Mori S., Nishizawa N. K. (2008) J. Biol. Chem. 283, 13407–13417 [DOI] [PubMed] [Google Scholar]
- 22. Nakanishi H., Okumura N., Umehara Y., Nishizawa N. K., Chino M., Mori S. (1993) Plant Cell Physiol. 34, 401–410 [PubMed] [Google Scholar]
- 23. Kato Y., Sakaguchi M., Mori Y., Saito K., Nakamura T., Bakker E. P., Sato Y., Goshima S., Uozumi N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6488–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishimaru Y., Suzuki M., Kobayashi T., Takahashi M., Nakanishi H., Mori S., Nishizawa N. K. (2005) J. Exp. Bot. 56, 3207–3214 [DOI] [PubMed] [Google Scholar]
- 25. Hiei Y., Ohta S., Komari T., Kumashiro T. (1994) Plant J. 6, 271–282 [DOI] [PubMed] [Google Scholar]
- 26. Ogo Y., Itai R. N., Nakanishi H., Kobayashi T., Takahashi M., Mori S., Nishizawa N. K. (2007) Plant J. 51, 366–377 [DOI] [PubMed] [Google Scholar]
- 27. Higuchi K., Suzuki K., Nakanishi H., Yamaguchi H., Nishizawa N. K., Mori S. (1999) Plant Physiol. 119, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Takahashi M., Yamaguchi H., Nakanishi H., Shioiri T., Nishizawa N. K., Mori S. (1999) Plant Physiol. 121, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nozoye T., Itai R. N., Nagasaka S., Takahashi M., Nakanishi H., Mori S., Nishizawa N. K. (2004) Soil Sci. Plant Nutr. 50, 1125–1131 [Google Scholar]
- 30. Nagasaka S., Takahashi M., Itai R. N., Bashir K., Nakanishi H., Mori S., Nishizawa N. K. (2009) Plant Mol. Biol. 69, 621–631 [DOI] [PubMed] [Google Scholar]
- 31. Takahashi M., Nakanishi H., Kawasaki S., Nishizawa N. K., Mori S. (2001) Nat. Biotechnol. 19, 466–469 [DOI] [PubMed] [Google Scholar]
- 32. Inoue H., Higuchi K., Takahashi M., Nakanishi H., Mori S., Nishizawa N. K. (2003) Plant J. 36, 366–381 [DOI] [PubMed] [Google Scholar]
- 33. Nozoye T., Inoue H., Takahashi M., Ishimaru Y., Nakanishi H., Mori S., Nishizawa N. K. (2007) Plant Mol. Biol. 64, 35–47 [DOI] [PubMed] [Google Scholar]
- 34. Pao S. S., Paulsen I. T., Saier M. H., Jr. (1998) Microbiol. Mol. Biol. Rev. 62, 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Furrer J. L., Sanders D. N., Hook-Barnard I. G., McIntosh M. A. (2002) Mol. Microbiol. 44, 1225–1234 [DOI] [PubMed] [Google Scholar]
- 36. Haydon M. J., Cobbett C. S. A. (2007) Plant Physiol. 143, 1705–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bashir K., Inoue H., Nagasaka S., Takahashi M., Nakanishi H., Mori S., Nishizawa N. K. (2006) J. Biol. Chem. 281, 32395–32402 [DOI] [PubMed] [Google Scholar]
- 38. Kakei Y., Yamaguchi I., Kobayashi T., Takahashi M., Nakanishi H., Yamakawa T., Nishizawa N. K. (2009) Plant Cell Physiol. 50, 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takahashi M., Nozoye T., Kitajima N., Fukuda N., Hokura A., Terada Y., Nakai I., Ishimaru Y., Kobayashi T., Nishizawa N. K. (2009) Plant and Soil 325, 39–51 [Google Scholar]
- 40. Aoyama T., Kobayashi T., Takahashi M., Nagasaka S., Usuda K., Kakei Y., Ishimaru Y., Nakanishi H., Mori S., Nishizawa N. K. (2009) Plant Mol. Biol. 70, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nomoto K., Ohfune Y. (1982) J. Syn. Org. Chem. Japan 40, 401–414 [Google Scholar]
- 42. Ishimaru Y., Kim S., Tsukamoto T., Oki H., Kobayashi T., Watanabe S., Matsuhashi S., Takahashi M., Nakanishi H., Mori S., Nishizawa N. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7373–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suzuki M., Morikawa K. C., Nakanishi H., Takahashi M., Saigusa M., Mori S., Nishizawa N. K. (2008) Soil Sci. Plant Nutr. 54, 77–85 [Google Scholar]
- 44. Huang C. F., Yamaji N., Mitani N., Yano M., Nagamura Y., Ma J. F. (2009) Plant Cell 21, 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.