Abstract
Integrins are cell adhesion receptors that mediate cell-to-cell, or cell-to-extracellular matrix adhesion. They represent an attractive target for treatment of multiple diseases. Two classes of small molecule integrin inhibitors have been developed. Competitive antagonists bind directly to the integrin ligand binding pocket and thus disrupt the ligand-receptor interaction. Allosteric antagonists have been developed primarily for αLβ2- integrin (LFA-1, lymphocyte function-associated antigen-1). Here we present the results of screening the Prestwick Chemical Library using a recently developed assay for the detection of α4β1-integrin allosteric antagonists. Secondary assays confirmed that the compounds identified: 1) do not behave like competitive (direct) antagonists; 2) decrease ligand binding affinity for VLA-4 ∼2 orders of magnitude; 3) exhibit antagonistic properties at low temperature. In a cell based adhesion assay in vitro, the compounds rapidly disrupted cellular aggregates. In accord with reports that VLA-4 antagonists in vivo induce mobilization of hematopoietic progenitors into the peripheral blood, we found that administration of one of the compounds significantly increased the number of colony-forming units in mice. This effect was comparable to AMD3100, a well known progenitor mobilizing agent. Because all the identified compounds are structurally related, previously used, or currently marketed drugs, this result opens a range of therapeutic possibilities for VLA-4-related pathologies.
Keywords: Adhesion, Allosteric Regulation, Antibodies, Drug Action, Integrin
Introduction
VLA-4 (very late antigen 4,3 α4β1-integrin, CD49d/CD29) plays a major role in the regulation of immune cell recruitment to inflamed endothelia and sites of inflammation. It participates in antigen presenting cell-lymphocyte interactions, retention and mobilization of immature progenitors in the bone marrow (1, 2), cancer cell trafficking, metastasis, and other events (3, 4). Integrins represent an attractive target for several existing drugs for treatment of inflammatory diseases, anti-angiogenic therapy, and anti-thrombotic therapy (5–7). Integrin ligands can also be used as imaging tools, as well as probes for studies of integrin functional activity and molecular conformation (8, 9).
Multiple small molecules have been developed in an attempt to regulate integrin dependent adhesion (6). Competitive antagonists can bind to the natural ligand binding pocket, and block interaction between integrins and natural integrin ligands (10, 11). Because the binding pocket is located between the α-subunit and β-subunit I-like domain they are also termed α/β I-like competitive antagonists (see Fig. 9C in Ref. 10). Multiple compounds of this type were developed for αIIbβ3, αvβ3, and α4β1 integrins. Several integrins have an additional domain that is inserted within the α-subunit β-propeller (A domain or I domain), which is evolutionarily related to the β I-like domain. The I domain serves as a ligand binding site for these integrins (see Fig. 9E in Ref. 10). Two types of allosteric antagonists for these integrins have been described: α/β I-like allosteric antagonists and α I allosteric antagonists (10). No allosteric antagonists have yet been identified for non-I domain containing integrins (such as VLA-4).
One of the features of competitive integrin antagonists is to occupy the ligand binding pocket and induce a conformational change that is similar to the conformational change induced by a natural ligand. Novel antibody epitopes termed ligand-induced binding site (LIBS) epitopes are exposed as a result of this conformational change (12–15). Recently, we showed that this feature can be used for the identification of unknown integrin antagonists, and determination of the ligand binding affinity for unlabeled small integrin ligands (15, 16). We have modified this assay to specifically detect VLA-4 allosteric antagonists, and we performed a high-throughput flow cytometry-based screen of the Prestwick Chemical Library (PCL), which represents one of “smart screening libraries” designed to decrease the number of “low quality” hits.
Here we report the identification of several structurally related compounds that were able to prevent exposure of ligand-induced binding site (LIBS) epitope after the addition of VLA-4-specific ligand, decrease binding affinity of VLA-4-specific ligand, and block VLA-4/VCAM-1-dependent cell adhesion. Because these compounds are previously used or currently marketed drugs (17–19), which are known to possess immunosuppressive properties (20), this effect on VLA-4 ligand binding provides a plausible explanation for the mechanism of immunosuppression (21).
EXPERIMENTAL PROCEDURES
Materials
The VLA-4-specific ligand (22–24) 4-((N′-2-methylphenyl)ureido)-phenylacetyl-l-leucyl-l-aspartyl-l-valyl-l-prolyl-l-alanyl-l-alanyl-l-lysine (LDV), and its FITC-conjugated analog (LDV-FITC probe) were synthesized at Commonwealth Biotechnologies. Mouse anti-human CD29, HUTS-21(PE), isotype control (mouse IgG2a κPE) clone G155–178 were purchased from BD Biosciences and used according to instructions of the manufacturer. Thioridazine hydrochloride and AMD3100 octahydrochloride (Plerixafor) were purchased from Tocris Bioscience, Ellisville, MO. All other reagents were from Sigma-Aldrich. Small molecule stock solutions were prepared in DMSO, at concentrations ∼1000-fold higher than the final concentration. Typically, 1 μl of stock solution was added to 1 ml of cell suspension yielding a 0.1% final DMSO concentration. Control samples were treated with an equal amount of pure DMSO (vehicle).
Cells
The mouse melanoma cell line B78H1 and the human histiocytic lymphoma cell line U937 were purchased from ATCC. Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 and 95% air in RPMI 1640 (supplemented with 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 10 mm HEPES, pH 7.4, and 10% heat-inactivated fetal bovine serum). Cells were then harvested and resuspended in 1 ml of HEPES buffer (110 mm NaCl, 10 mm KCl, 10 mm glucose, 1 mm MgCl2, 1.5 mm CaCl2, and 30 mm HEPES, pH 7.4) containing 0.1% HSA and stored on ice. Cells were counted using the Coulter Multisizer/Z2 analyzer (Beckman Coulter). For experiments, cells were suspended in the same HEPES buffer at 106 cells/ml and warmed to 37 °C for 10 min prior to binding experiments (see below).
For transfection of B78H1 cells, full-length human VCAM-1 cDNA was a kind gift from Dr. Roy Lobb of Biogen Inc. The original construct (25) was subcloned into the pTRACER vector (Invitrogen). Transfection was done using the Lipofectamine Reagent (Invitrogen). High expressors were selected using the MoFlo Flow Cytometer (Cytomation, Inc., Fort Collins, CO).
Detection of VLA-4 Allosteric Antagonists
HUTS-21 antibody binding has been thoroughly studied and described in detail (15, 16). Here we present the assay as adapted for small volumes (384 plate format) at the University of New Mexico Center for Molecular Discovery (UNMCMD). The assay description was uploaded to PubChem (AIDs: 2617, 2674, 2813, 2557). 5 μl RPMI was added to columns 2–24. Column 1 was left empty for flow cytometry data binning purposes. Test compounds in DMSO (Prestwick Chemical Library, PCL) were added and mixed (0.1 μl of 1 mm stock solution). This resulted in 20 μm test compounds (6.7 μm final in 15 μl). Compounds were added to 320 wells (columns 3–22). Column 2 was used for the negative control (no VLA-4 ligand added). Columns 23, 24 were used for the positive control (no test compounds added). Next, 5-μl aliquots of U937 cells were suspended in RPMI 1640 at 3 × 106 cells/ml, loaded in 384 well plates, and mixed (1 × 106 cells/ml final). Cells were incubated for 10 min at room temperature. This time is sufficient for the binding of a small molecule (∼1 kDa) at 10 μm at room temperature. Next, a 5 μl mixture of a non-fluorescent VLA-4 specific ligand (12 nm final) with HUTS-21 PE antibody (25 μl/ml final) was added to columns 3–24) and mixed. This concentration (12 nm) is equal to the dissociation constant of LDV (Kd). A 5-μl aliquot of HUTS-21 (PE) antibody without LDV (25 μl/ml final) were added to a column 2, which was used as a negative control. Addition of LDV resulted in the HUTS-21 LIBS epitope exposure, and therefore initiated antibody binding. Plates were incubated for 1 h at 37 °C. According to the real-time kinetic studies this time was sufficient to reach equilibrium. Next, wells were sampled using the high throughput flow cytometry platform (HyperCyt) at UNMCMD.
After data acquisition by the flow cytometer (CyAnTM ADP), proprietary software was used to analyze the data files (IDLQuery, at UNMCMD, developed by Bruce Edwards). The data were exported into a Microsoft Excel spreadsheet template, and the value for antibody binding inhibition (%) was calculated for each well as follows: % Inhibition = 100(1 − ((MCFtest − MCFneg.control)/(MCFpos.control − MCFneg.control))), where, MCF is mean channel fluorescence of cells with test compounds, and positive or negative control wells. A compound was considered a “hit” if the % inhibition was greater than 50%.
Real-time Binding and Dissociation of VLA-4-specific Ligand (LDV-FITC Probe)
Kinetic analysis of the binding and dissociation of the LDV-FITC probe was described previously (22). Briefly, cells (106 cells/ml) were preincubated in HEPES buffer containing 0.1% HSA at different conditions for 10–20 min at 37 °C. Alternatively, experiments were performed directly in RPMI that was used for growing the cells. Flow cytometric data were acquired for up to 1024 s at 37 °C while the samples were stirred continuously at 300 rpm with a 5 × 2 mm magnetic stir bar (Bel-Art Products). Samples were analyzed for 30–120 s to establish a baseline. The fluorescent ligand was added and acquisition was re-established, creating a 5–10 s gap in the time course.
For real-time inside-out integrin activation experiments, 4 nm LDV-FITC was added after establishing a baseline for unstained cells. Then, data were acquired for 2–3 min, and cells were activated with 100 nm fMLFF (high affinity FPR ligand). Acquisition was re-established, and data were acquired continuously for up to 1024 s. The concentration of the LDV-FITC probe used in the experiments (4 nm) was below the dissociation constant (Kd) for its binding to resting VLA-4 (low affinity state, Kd ∼12 nm), and above the Kd for physiologically activated VLA-4 (high affinity state, Kd ∼1–2 nm) (22). Therefore, the transition from the low affinity to the high affinity receptor state led to increased binding of the probe (from ∼25% to ∼70–80% of receptor occupancy, as calculated based on the one site binding equation), which was detected as an increase in the mean channel fluorescence (MCF). Next, cells were treated with an excess unlabeled LDV containing small molecule (1 μm), or compounds of interest (10–30 μm), and the dissociation of the fluorescent molecule was followed.
For kinetic dissociation measurements without inside-out activation, cell samples were preincubated with the fluorescent probe (25 nm, ∼2 x Kd (12 nm) for the resting state of VLA-4, 68% of receptor occupancy (22)), treated with excess unlabeled LDV containing small molecule (1 μm) or compounds of interest (10–30 μm) and the dissociation of the fluorescent molecule was followed. The resulting data were converted to MCF versus time using FCSQuery software developed by Dr. Bruce Edwards (University of New Mexico).
Real-time Binding of HUTS-21 Antibodies
The ability of a flow cytometer to discriminate between free and bound fluorescent ligand in a homogeneous assay was used to determine the binding kinetics of mAbs in real-time (15, 26). Cells (106 cells/ml) were removed from ice and warmed in HEPES buffer containing 0.1% HSA for 10 min at 37 °C. Flow cytometric data were acquired continuously for up to 2048 s at 37 °C while the samples were stirred continuously at 300 rpm with a 5 × 2 mm magnetic stir bar (Bel-Art Products). First, samples were analyzed for 30–120 s to establish a baseline. Next, the tube was removed and HUTS-21 mAbs (20 μl/1 ml of cells) were added and acquisition was re-established, creating a 5–10 s gap in the time course. In the absence of the LDV ligand no binding of HUTS-21 mAb were observed (15). Screening hits at saturating concentration (10–30 μm final) or DMSO (vehicle) were added at point 0. Next, different concentrations of LDV ligand were added after 60–120 s. Then, acquisition was re-established, and data were acquired continuously for up to 2048 s. The resulting data were converted to MCF versus time using FCSQuery software developed by Dr. Bruce Edwards (University of New Mexico).
Cell Adhesion Assay
The cell suspension adhesion assay has been described previously (23, 27, 28). Briefly, U937 cells stably transfected with FPR were labeled with red fluorescent PKH26GL dye, and B78H1/VCAM-1 transfectants were stained with green fluorescent PKH67GL dye (Sigma-Aldrich). Labeled cells were washed, resuspended in HEPES buffer supplemented with 0.1% HSA or RPMI and stored on ice until used in assays. Control U937 cells were preincubated with the LDV-containing small molecule as a blocking agent. Prior to data acquisition, cells were warmed to 37 °C for 10 min separately and then mixed. During data acquisition, the samples were stirred with a 5 × 2-mm magnetic stir bar (Bel-Art Products, Pequannock, NJ) at 300 rpm and kept at 37 °C. Next, cells were treated with different compounds (screening hits) or LDV (block). The number of cell aggregates containing U937 adherent to B78H1/VCAM-1 (red and green co-fluorescent particles) was followed in real-time. The percentage of aggregates was calculated as follows: % Agg = (number of aggregates/(number of aggregates + number of singlets)) × 100. Experiments were performed using a FACScan flow cytometer and Cell Quest software (Becton Dickinson, San Jose, CA). The data were converted to the number of aggregates versus time using FCSQuery software developed by Dr. Bruce Edwards (University of New Mexico).
Mice
Male C57Bl6 mice (9–13 weeks) were purchased from Jackson Laboratories, Bar Harbor, ME. Mice were acclimated to the facility for at least 1 week on a 12 h light/dark cycle and standard diet. Experiments were conducted between 10:00 and 12:00 AM (lights on at 7:00 AM). Procedures used in this study were conducted by authorized personnel and approved by the Institutional Animal Care and Use Committee of University of New Mexico School of Medicine.
One hour prior to blood collection mice were injected intraperitoneally with vehicle, thioridazine hydrochloride (1.25 mg/kg), or AMD3100 octahydrochloride (Plerixafor, 5 mg/kg). Prior to blood collection mice were anesthetized using isoflurane and monitored for sensitivity. Blood was collected by heart puncture and continued to exsanguination (1–1.4 ml). The blood was collected in a syringe containing heparin and immediately mixed into a heparin-containing tube to prevent clotting.
Hematopoietic Stem and Progenitor Cell (HSPC) Analysis
Collected blood was processed according to the protocol recommended by Stemcell Technologies Inc. Blood was lysed using ammonium chloride lysis buffer (StemCell Technologies Inc.). The cells were then washed with PBS, Iscove's modified Dulbecco medium supplemented with 2% FBS (IMDM), and re-suspended in IMDM supplemented with 2% FBS. An aliquot from each sample was resuspended in PBS and used for nucleated cell enumeration using a Vi-Cell automated cell counter (Becton Dickinson, San Jose, CA). To determine the white blood count (WBC, cell/ml), the total cell count was adjusted for the volume collected. Next, the samples were centrifuged, aspirated and resuspended in IMDM-2% FCS to achieve 3 × 106 cells/ml. The cells were counted again, and these cell counts were utilized as the established load count to determine colony-forming unit (CFU) values, CFU/ml. 300 μl of each load sample was added to a tube containing 3 ml of MethoCult media (MethoCult3534; StemCell Technologies), and mixed. 1.1 ml of the MethoCult media-cell mix was plated in pre-tested 35 mm culture dishes (two per sample) and incubated at 37 °C, 5% CO2. CFU values were counted 14 days later at 10× magnification.
Statistical Analysis
Curve fits, statistics, and EC50 calculations were performed using GraphPad Prism version 4.00 for Windows, GraphPad Software, San Diego CA. Each experiment was repeated at least two times. The experimental curves represent the mean of two or more independent determinations. The standard error of the mean was calculated using GraphPad Prism.
RESULTS
Assay for the Detection of VLA-4 Allosteric Antagonists
Recently, we studied the binding of the conformationally sensitive anti-CD29 antibody (HUTS-21). Our data indicate that intracellular signaling through G-protein-coupled receptors (inside-out signal) does not affect the exposure of the HUTS-21 epitope mapped to the hybrid domain of the β-1 integrin subunit. Exposure of this epitope is solely regulated by the occupancy of the ligand binding pocket, and is independent of integrin affinity state (14, 15). Moreover, exposure of the HUTS-21 epitope can be used to determine the affinity of unlabeled VLA-4 ligands. This was verified using competition between the LDV-FITC ligand and a number of VLA-4-specific ligands identified by us and others with affinities that differ by more than one order of magnitude (16).
To date, all of the competitive VLA-4 antagonists induced exposure of the HUTS-21 mAb epitope (16). Thus, the binding of anti-LIBS antibodies represents an ideal tool to study occupancy of the integrin-binding pocket, and it provides a unique way to discriminate between competitive and allosteric antagonists. Allosteric antagonists, which bind to the allosteric sites of the molecule (by definition), would not induce ligand-induced conformational changes. However they would still block binding of the ligand. As a result, a direct competitor that induces the LIBS epitope would dissociate, and binding of anti-LIBS antibody would decrease. On the contrary, novel competitive ligands, in addition to blocking binding of the control ligand, have the potential to induce the LIBS epitope, and therefore would not be detected. Thus, this assay would specifically detect allosteric antagonists or non-canonical ligands (small molecules that bind to the ligand-binding pocket without inducing LIBS epitope exposure).
HTS Screen Results
The screen of the PCL was performed using the HyperCyt platform in 384-well plate format. The assay was configured to discriminate the nonspecific binding of HUTS-21 as well as its specific binding to VLA-4 in the presence of LDV ligand. HUTS-21 binding in the presence of the compound of interest was calculated as % inhibition where the positive control is 100% and nonspecific binding is 0.
Several chemical libraries have been screened using this specific screening assay. The results of primary and confirmatory assays were uploaded to the PubChem data base (PubChem AIDs: 2557, 2617, 2674, 2813, 449766). Some of the identified compounds are found to be structurally similar to the compounds reported below (for example see “active” compounds in AID: 2674).
The screen identified 36 active molecules. About 31% of them belong to the same structural family, which consists of three different groups (phenothiazines, thioxanthenes, and structurally related 3-ring heterocyclic compounds). All of these compounds exhibit significant structural homology, and represent a single class of drugs that include serotonin-dopamine full and partial antagonists. These hits represent a series of compounds with inhibition ranging from 90 to 51%, which provide nascent SAR data. Here we present the data from secondary and tertiary assays for this dominant group of compounds.
Binding of the LDV-FITC Ligand
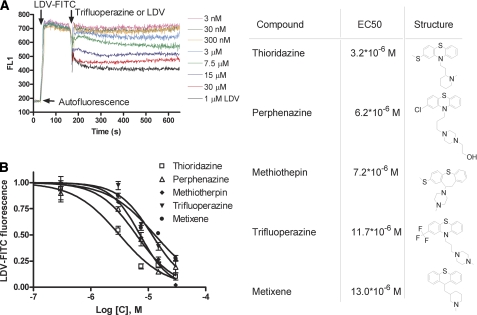
The VLA-4-specific ligand (LDV-FITC) has been used extensively (22, 23, 29) as a tool for studying VLA-4 affinity and conformation. Here we probed whether hits from screening would interfere with the binding of this ligand. Cells were incubated with 25 nm LDV-FITC (Kd ∼12 nm) (22). Next, different concentrations of compound were added. As shown in Fig. 1A, addition of the compound resulted in the rapid dissociation of the LDV-FITC probe. The steady state value of LDV-FITC fluorescence (achieved about ∼300 s after addition) plotted versus compound concentration is shown in Fig. 1B. EC50 values ranged from 3–13 μm. Thus, all five compounds interfered with the binding of the VLA-4-specific ligand to U937 cells at rest (without integrin activation).
FIGURE 1.
Binding and dissociation of the LDV-FITC probe in response to the addition of screening hits. A, LDV-FITC probe binding and dissociation on U937 cells plotted as mean channel fluorescence (FL1) versus time. The experiment involved sequential additions of fluorescent LDV-FITC probe (25 nm), LDV (1 μm, control, excess of unlabeled competitor), or different concentrations of compounds tested. One representative experiment out of two experiments for each compound is shown. B, steady state value of the LDV-FITC fluorescence obtained in experiments analogous to the one shown in panel A, 300–400 s after compound addition, plotted versus compound concentration. LDV-FITC fluorescence was normalized assuming that the value of fluorescence after LDV addition is equal to 0, and after DMSO addition (vehicle) is equal to 1. The data represent means + S.E. (n = 2) for two independent experiments. Curves represent a fit to a sigmoidal dose response equation (variable slope) performed using GraphPad Prism software. EC50 values and compound structures are shown on the right.
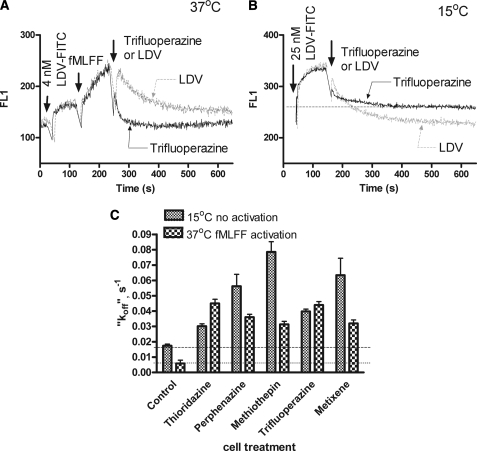
VLA-4 can also be activated by cellular signaling. The “inside-out” signaling pathway can be triggered by G-protein-coupled receptors. This results in a higher affinity of the ligand-binding pocket. To study the effect of compound upon activated cells we used U937 cells stably transfected with a non-desensitizing mutant of the formyl peptide receptor (FPR). The major advantage of this mutant is that in the absence of receptor desensitization, the high affinity state of the VLA-4-binding pocket is preserved for several minutes after addition of its ligand (fMLFF) (24). Addition of compounds after cell activation by fMLFF resulted in the rapid dissociation of the LVD-FITC probe (Fig. 2A). In fact, the LDV-FITC “dissociation rate” was faster than after the addition of excess unlabeled competitor. This result suggests that none of the compounds competed directly with LDV-FITC, where the “dissociation rate” should equal the rate induced by a competitor. In this case the dissociation rate of the probe is determined by the lifetime of the probe-receptor interaction and independent of the nature or affinity of a competitor (added in large excess). This suggests a non-competitive mechanism for the compounds.
FIGURE 2.
Binding and dissociation of the LDV-FITC probe in response to the addition of screening hits. A, LDV-FITC probe binding and dissociation on U937 cells stably transfected with the non-desensitizing mutant of FPR plotted as mean channel fluorescence (FL1) versus time. The experiment involved sequential additions of the fluorescent LDV-FITC probe (4 nm), fMLFF (100 nm), LDV (control, excess unlabeled competitor), or saturating concentrations of compounds tested. One representative experiment (for trifluoperazine) out of two experiments for each compound is shown. B, LDV-FITC probe binding and dissociation on U937 cells plotted as mean channel fluorescence (FL1) versus time at low temperature (15 °C). The experiment involved sequential additions of the fluorescent LDV-FITC probe (25 nm), LDV (control, excess unlabeled competitor), or saturating concentration of compounds tested. One representative experiment (for trifluoperazine) of two experiments for each compound is shown. C, LDV-FITC “dissociation rates” (koff) obtained in kinetic experiments analogous to the experiments shown in panels A and B. The dissociation components of the curves were fitted to a single exponential equation using GraphPad Prism software and plotted for different compounds. Control represents the actual dissociation rates obtained using excess unlabeled competitor (LDV). Notice that for all treatment conditions koff values were larger than in the control sample, representing faster dissociation of the probe.
Recently, we described a signaling pathway that can actively down-regulate the affinity state of the ligand-binding pocket (30). The PCL compounds could also be ligands for GPCRs that provide a signal for integrin de-activation. To study this question, we performed LDV-FITC binding experiments at low temperature (Fig. 2B). This allowed us to compare LDV-FITC dissociation rates without cell activation. At 15 °C without inside-out activation, the resting LDV-FITC dissociation rate was about 4-fold slower than at 37 °C. Nonetheless, the rate of probe dissociation induced by all compounds was faster than for the LDV competitor (Fig. 2C). That the effect of compounds on LDV-FITC binding at 15 °C was as fast as at 37 °C, suggests the lack of involvement of cellular signaling. The ability to down-regulate LDV-FITC binding at rest (without “inside-out” activation) further supports the absence of this type of signaling. The intracellular signaling, which triggers VLA-4 de-activation though the Gαs-coupled pathway was only able to reduce VLA-4 affinity to the resting state, and not below that (30). Thus, all five studied compounds were able to decrease the binding of the LDV-FITC probe at low temperature, and without direct competition with the VLA-4-specific ligand.
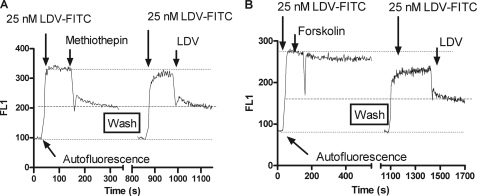
Reversibility of Compound Binding
To test the reversibility of compound binding, cells were consecutively treated with LDV-FITC and compounds of interest. Next, cells were washed three times with medium, and LDV-FITC was added again. Excess LDV competitor was used to determine the nonspecific binding of the compound (Fig. 3A). For all five studied compounds (see Fig. 1) the impact on LDV-FITC binding was fully reversible, indicating that no intracellular signaling was involved. As a control for the effect of the Gαs-coupled signaling pathway, we used forskolin, which activates adenylyl cyclase, increases intacellular cAMP concentration, and is routinely used to mimic Gαs-GPCR activation (30). As expected the impact of forskolin upon LDV-FITC binding was slow and irreversible (Fig. 3B). This further supports our notion that hits from screening do not cause Gαs-GPCR activation.
FIGURE 3.
Reversibility of compound binding assessed using binding and dissociation of the LDV-FITC probe in response to the addition of screening hits. A, LDV-FITC probe binding and dissociation on U937 cells plotted as mean channel fluorescence (FL1) versus time. The experiment involved sequential additions of fluorescent LDV-FITC probe (25 nm), and the compounds tested (30 μm). After dissociation of the LDV-FITC probe the cells were washed three times in RPMI media to remove all traces of the compound. Next, the LDV-FITC probe was replenished, followed by addition of the competitor (LDV). Notice that after the wash step, the binding of LDV-FITC was identical to the binding after the first addition. This indicates that the compound blocking LDV-FITC binding after the first addition was completely removed by the wash. Thus, the binding of the compound was reversible. Analogous data were obtained for all five compounds tested (see Fig. 1 for the list). B, the same experiment as described in panel A was performed using forskolin, an activator of adenylyl cyclase. The addition of forskolin was insufficient to induce the dissociation of LDV-FITC probe to the baseline (indicated by dashed line). A small decrease in the probe binding is attributed to a small number of constitutively active VLA-4 present on the cell surface (a fraction of VLA-4 with slow LDV-FITC dissociation, see panel A after LDV addition). Forskolin treatment reduced the binding affinity for these active receptors to the resting state. Note the slower effect after forskolin addition, and the irreversible inhibition of LDV-FITC that involve intracellular signaling.
Affinity of LDV Ligand Binding
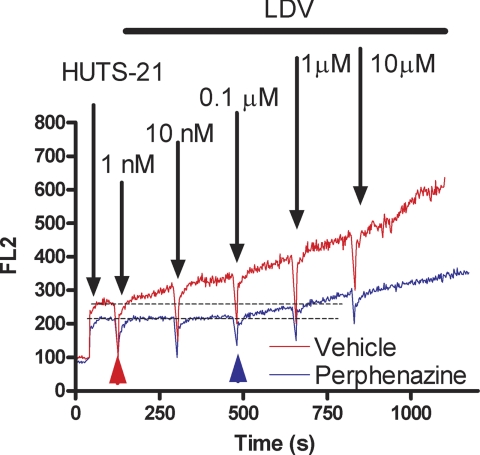
Binding of the LDV-FITC probe can be studied in a homogeneous assay by flow cytometry, for concentrations up to 100 nm or more. At higher concentrations background fluorescence from the fluorescent probe in solution can dominate the analysis. To overcome this problem we developed an assay that relies on a real-time binding analysis of HUTS-21 mAbs in response to the addition of known amounts of unlabeled LDV probe (see Fig. 4A in Ref. 15). Because the HUTS-21 epitope is exposed as a result of ligand binding and the subsequent conformational change, this assay can be used to evaluate how the presence of a hit compound affects the ligand binding affinity at high ligand concentrations (Fig. 4). HUTS-21 binding was detected following the addition of 1 nm LDV in the absence of the compound (red line/arrowhead). In the presence of the compound HUTS-21 binding was detected at a concentration ∼two orders of magnitude higher (blue arrowhead, 0.1 μm). This suggests that the ligand binding affinity was lowered by a factor of 100. These data support previous real-time LDV-FITC binding results. Lowering ligand affinity by about 100 fold produces the dramatic dissociation of the LDV-FITC probe shown in Figs. 2 and 3. This represents a transition at 4 nm (Kd ∼2 nm: FPR activated state) from ∼67% receptor occupancy to ∼2% occupancy (Fig. 2), or a transition at 25 nm (Kd ∼12 nm: resting state) from ∼68% receptor occupancy to ∼2% occupancy (Fig. 3A), respectively.
FIGURE 4.
Kinetics of real-time binding of HUTS-21 (PE) antibodies to U937 cells. Real-time binding of HUTS-21 (LIBS) antibodies plotted as mean channel fluorescence (FL2) versus time. The addition of HUTS-21 antibodies (first arrow) resulted in rapid nonspecific binding of antibodies. The addition of increasing amounts of LDV ligand (arrows) resulted in increased rates of antibody binding in the absence (red), or in the presence of the compound (blue). Compound was added at 0 time point. Notice that binding of HUTS-21 in the absence of the compound starts at 1 nm LDV (red arrowhead). To induce similar binding of HUTS-21 in the presence of the compound 0.1 μm of LDV was required (blue arrowhead). One representative experiment (for perphenazine) is shown. Analogous data were obtained for all five compounds tested (see Fig. 1 for the list).
Cell Aggregation
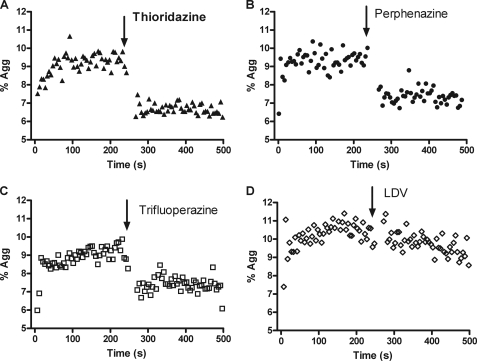
To study the effect of screening hits on VLA-4-dependent cell adhesion, we used a well-characterized cell suspension adhesion assay (24, 27, 28). U937 cells expressing VLA-4 and B78H1 mouse melanoma cells stably transfected with human VCAM-1 (stained with red and green dyes) form VLA-4/VCAM-1-dependent cellular aggregates after mixing. Cell aggregation was followed in real-time. Addition of saturating amounts of screening hits resulted in rapid cellular disaggregation (Fig. 5). Moreover, this disaggregation was much faster than disaggregation induced by LDV (Fig. 5D). The rate of cell disaggregation depends upon the lifetime of the VLA-4/VCAM-1 “bond”, which is determined by the affinity state of the VLA-4-binding pocket, and the number of “bonds” (23, 27). Rapid disruption of cell aggregates is consistent with reduction of the bond life-time, caused by lowering the ligand receptor affinity (as detected in the LDV probe binding assays, see above). The fact that a direct competitor (LDV) induces cellular disaggregation at a slower rate further supports this idea.
FIGURE 5.
Changes in cell adhesion between VLA-4-expressing U937 cells and VCAM-1-transfected B78H1 cells in response to compound addition. U937 cells were stained using a red fluorescent dye (PKH26), and B78H1/VCAM-1 cells were stained using a green fluorescent dye (PKH67). Cells were mixed at 37 °C and sampled continuously using a flow cytometer. Double positive (red and green) cell aggregates were followed as described under “Experimental Procedures,” and plotted as % of aggregates (% Agg) versus time. 30 μm individual compounds (A, B, and C) or 1 μm of LDV competitor (D) were added. Notice the rapid decrease in the number of cell aggregates after the addition of allosteric antagonists. A representative experiment of two experiments is shown.
Mobilization of Hematopoietic Stem and Progenitor Cells (HSPCs) into the Peripheral Blood
VLA-4 plays a specific role in the retention, homing, and engraftment of HSPCs (1, 2). It is expressed on human CD34+ cells, and murine HSPCs (31–33). Blocking the interaction between VLA-4 and its ligands using anti-VLA-4-specific antibodies, or small molecule inhibitors induces mobilization of HSPCs in humans (34, 35), primates (36, 37), and mice (38). Moreover, VLA-4 blockade alone, without additional cytokine treatment, is sufficient to induce HSPC mobilization (see Ref. 37 and references therein). Blockade of other leukocyte integrins, such as β2-integrins using anti-CD18 antibodies, has no effect on progenitor mobilization (36). Thus, the effect of different VLA-4 antagonists on hematopoietic progenitor mobilization is highly VLA-4-specific, and these molecules alone can be used to induce HSPC mobilization.
As compounds of interest (Fig. 1) exhibit properties of VLA-4 antagonists in vitro, we hypothesized that they will also act in vivo in a manner similar to other VLA-4 antagonists, which induce HSPC mobilization. To test this hypothesis, mice were injected with thioridazine, the most potent compound in the series (Fig. 1). As a control, we used the highly selective CXCR4 chemokine receptor antagonist AMD3100 (Plerixafor), which is known to stimulate a rapid increase in the number of circulating HSPC in mice and man (39–41).
We found that administration of thioridazine resulted in a significant increase in the number of CFUs in the peripheral blood (Table 1). The cell mobilizing ability of thioridazine was comparable to AMD3100. However, we also found a significant difference between the two treatments in the ability to modulate WBC. Thirodazine had no effect on WBCs, while AMD3100 significantly increased the WBC count. These data are in agreement with previously published reports. Blockade of VLA-4 using anti-VLA-4 antibodies mobilized hematopoietic progenitors without a significant increase in circulating white cells (36). AMD3100 is shown to induce an increase in the WBC count ranging from 1.5 to 3.1 times the baseline (42). In our experiments we observed ∼2.1-fold WBC count increase. Thus, administration of thioridazine induced mobilization of HSPCs into the peripheral blood in mice. These data support the idea that the identified compounds possess the properties of VLA-4 antagonists.
TABLE 1.
Effect of intraperitoneal administration of thioridazine and AMD3100 upon CFU and WBC in mouse peripheral blood
Mean ± S.E. from three independent experiments performed on different days are shown (4 mice per treatment).
| Treatment | CFU/ml in peripheral blood | WBC count/ml × 106 |
|---|---|---|
| Vehicle | 68.1 ± 9.1 | 3.7 ± 0.6 |
| Thioridazine | 216.8 ± 42.1 | 3.0 ± 0.3 |
| AMD3100 | 511.8 ± 78.8a | 7.8 ± 0.7b |
a Means are significantly different (p < 0.05, according to one-way ANOVA).
b Statistically significant difference was found between vehicle and AMD3100 treatment (p < 0.05, according to unpaired t test). No significant difference was found between vehicle and thioridazine treatment for WBC count.
Taken together our experimental data suggest that all five compounds of interest (thioridazine, perphenazine, methiothepin, trifluoperazine, metixene) exhibit properties of VLA-4 specific allosteric antagonists. Because all these compounds are structurally related FDA-approved drugs, we envision the possibility to reposition these drugs toward VLA-4-dependent diseases.
DISCUSSION
A number of neuroleptic compounds are known to down-modulate the immune response. One of the earliest reports showed that neuroleptic drugs, structurally similar to the compounds tested in the present study, were able to protect mice receiving fatal doses of a bacterial endotoxin (43). Recent reports showed that the use of typical antipsychotic drugs was associated with a dose-dependent increase in the risk for pneumonia in elderly patients (44). However, the most remarkable finding is that the immunosuppressive effect of these drugs is not related to their dopamine antagonistic properties. More specific dopamine antagonists, which are based on an entirely different structural scaffold (such as haloperidol, metoclopramide, or sulpiride), do not possess any immunosuppressive properties (20). These drugs were also present in the Prestwick Chemical Library, and they did not show any antagonistic activity in the HUTS-21-based screen to detect VLA-4 allosteric antagonists. Moreover, the structure-activity relationship of phenothiazines for inhibiting lymphocyte motility, is reported to be different from those for their neuroleptic effects (45). Thus, it is possible that some of the structural features of these compounds are specific for VLA-4 antagonistic properties. This indicates the possibility to develop VLA-4 allosteric antagonists that lack unwanted activity (such as dopamine receptor antagonism or others).
The immunological mechanism of neuroleptic drug-induced immunosuppression is not fully understood, and the modulation of cytokine production or cytokine networks could be an underlying mechanism (46). Our finding that these drugs exhibit properties of VLA-4 allosteric antagonists provides an excellent explanation for such activity. Other VLA-4 specific competitive antagonists in some cases can cause severe immune suppression (47), and increase the risk of opportunistic infections (48). Blocking VLA-4-dependent immune cell adhesion could also explain why these types of compounds selectively affect cell-mediated component of the immune function (20).
One report linked the use of phenothiazines and the appearance of “atypical lymphocytes” in the peripheral blood of schizophrenic and nonschizophrenic patients. Some of these cells morphologically resembled early hematopoietic progenitors (49). This may account for our observation that thioridazine, a widely used phenothiazine, mobilized hematopoietic progenitors into the peripheral blood.
Finally, allosteric antagonists that modulate the binding affinity of natural ligands are envisioned as a potent novel generation of antagonists, with somewhat greater therapeutic potential than competitive antagonists. In some cases this modulation can simply mimic the effects of competitive antagonists (50) (at least for G-protein-coupled receptors). For integrins, which can propagate signals in both directions (inside-out and outside-in) (51), allosteric antagonists would lack the ability to induce an outside-in signal, or at least block natural ligand binding without inducing the ligand-induced binding site (LIBS) epitope.
Acknowledgment
We thank Eric R. Prossnitz for providing U937 cells and plasmids and Bruce S. Edwards for providing FCSQuery and IDLQuery software.
This work was supported, in whole or in part, by National Institutes of Health Grants U54 MH084690 and HL081062 and the Leukemia and Lymphoma Society Grant 7388-06.
- VLA-4
- very late antigen 4
- CFU
- colony-forming unit
- fMLFF
- N-formyl-l-methionyl-l-leucyl-l-phenylalanyl-l-phenylalanine, formyl peptide
- FPR
- formyl peptide receptor 1
- GPCR
- guanine nucleotide-binding protein-coupled receptor
- HSA
- human serum albumin
- HTS
- high throughput screen
- HSPC
- hematopoietic stem and progenitor cell
- LDV-containing small molecule
- 4-((N′-2-methylphenyl)ureido)-phenylacetyl-l-leucyl-l-aspartyl-l-valyl-l-prolyl-l-alanyl-l-alanyl-l-lysine
- LDV-FITC-containing small molecule
- 4-((N′-2-methylphenyl)ureido)-phenylacetyl-l-leucyl-l-aspartyl-l-valyl-l-prolyl-l-alanyl-l-alanyl-l-lysine-FITC
- LFA-1
- lymphocyte function-associated antigen-1
- LIBS
- ligand-induced binding sites
- mAb
- monoclonal antibody
- MCF
- mean channel fluorescence
- PCL
- Prestwick Chemical Library
- VCAM-1
- vascular cell adhesion molecule 1.
REFERENCES
- 1. Lapidot T., Petit I. (2002) Exp. Hematol. 30, 973–981 [DOI] [PubMed] [Google Scholar]
- 2. Lapidot T., Dar A., Kollet O. (2005) Blood 106, 1901–1910 [DOI] [PubMed] [Google Scholar]
- 3. Johnson J. P. (1999) Cancer Metastasis Rev. 18, 345–357 [DOI] [PubMed] [Google Scholar]
- 4. Yoneda T. (2000) J. Orthop. Sci. 5, 75–81 [DOI] [PubMed] [Google Scholar]
- 5. Yusuf-Makagiansar H., Anderson M. E., Yakovleva T. V., Murray J. S., Siahaan T. J. (2002) Med. Res. Rev. 22, 146–167 [DOI] [PubMed] [Google Scholar]
- 6. Jackson D. Y. (2002) Curr. Pharm. Des. 8, 1229–1253 [DOI] [PubMed] [Google Scholar]
- 7. Schmidmaier R., Baumann P. (2008) Curr. Med. Chem. 15, 978–990 [DOI] [PubMed] [Google Scholar]
- 8. Dijkgraaf I., Beer A. J., Wester H. J. (2009) Front Biosci. 14, 887–899 [DOI] [PubMed] [Google Scholar]
- 9. Humphries M. J. (2004) Biochem. Soc. Trans. 32, 407–411 [DOI] [PubMed] [Google Scholar]
- 10. Shimaoka M., Springer T. A. (2003) Nat. Rev. Drug Discov. 2, 703–716 [DOI] [PubMed] [Google Scholar]
- 11. Woodside D. G., Vanderslice P. (2008) Biol. Drugs 22, 85–100 [DOI] [PubMed] [Google Scholar]
- 12. Frelinger A. L., 3rd, Du X. P., Plow E. F., Ginsberg M. H. (1991) J. Biol. Chem. 266, 17106–17111 [PubMed] [Google Scholar]
- 13. Frelinger A. L., 3rd, Cohen I., Plow E. F., Smith M. A., Roberts J., Lam S. C., Ginsberg M. H. (1990) J. Biol. Chem. 265, 6346–6352 [PubMed] [Google Scholar]
- 14. Mould A. P., Barton S. J., Askari J. A., McEwan P. A., Buckley P. A., Craig S. E., Humphries M. J. (2003) J. Biol. Chem. 278, 17028–17035 [DOI] [PubMed] [Google Scholar]
- 15. Chigaev A., Waller A., Amit O., Halip L., Bologa C. G., Sklar L. A. (2009) J. Biol. Chem. 284, 14337–14346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Njus B. H., Chigaev A., Waller A., Wlodek D., Ostopovici-Halip L., Ursu O., Wang W., Oprea T. I., Bologa C. G., Sklar L. A. (2009) Assay. Drug Dev. Technol. 7, 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sudeshna G., Parimal K. (2010) Eur. J. Pharmacol. 648, 6–14 [DOI] [PubMed] [Google Scholar]
- 18. Shen W. W. (1999) Compr. Psychiatry 40, 407–414 [DOI] [PubMed] [Google Scholar]
- 19. Thanacoody H. K. (2007) Br. J. Clin. Pharmacol. 64, 566–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roudebush R. E., Berry P. L., Layman N. K., Butler L. D., Bryant H. U. (1991) Int. J. Immunopharmacol. 13, 961–968 [DOI] [PubMed] [Google Scholar]
- 21. Surman O. S. (1993) Psychosomatics 34, 139–143 [DOI] [PubMed] [Google Scholar]
- 22. Chigaev A., Blenc A. M., Braaten J. V., Kumaraswamy N., Kepley C. L., Andrews R. P., Oliver J. M., Edwards B. S., Prossnitz E. R., Larson R. S., Sklar L. A. (2001) J. Biol. Chem. 276, 48670–48678 [DOI] [PubMed] [Google Scholar]
- 23. Chigaev A., Zwartz G., Graves S. W., Dwyer D. C., Tsuji H., Foutz T. D., Edwards B. S., Prossnitz E. R., Larson R. S., Sklar L. A. (2003) J. Biol. Chem. 278, 38174–38182 [DOI] [PubMed] [Google Scholar]
- 24. Chigaev A., Buranda T., Dwyer D. C., Prossnitz E. R., Sklar L. A. (2003) Biophys. J. 85, 3951–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Osborn L., Hession C., Tizard R., Vassallo C., Luhowskyj S., Chi-Rosso G., Lobb R. (1989) Cell 59, 1203–1211 [DOI] [PubMed] [Google Scholar]
- 26. Sklar L. A., Edwards B. S., Graves S. W., Nolan J. P., Prossnitz E. R. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 97–119 [DOI] [PubMed] [Google Scholar]
- 27. Zwartz G., Chigaev A., Foutz T., Larson R. S., Posner R., Sklar L. A. (2004) Biophys. J. 86, 1243–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Edwards B. S., Curry M. S., Tsuji H., Brown D., Larson R. S., Sklar L. A. (2000) J. Immunol. 165, 404–410 [DOI] [PubMed] [Google Scholar]
- 29. Chigaev A., Waller A., Zwartz G. J., Buranda T., Sklar L. A. (2007) J. Immunol. 178, 6828–6839 [DOI] [PubMed] [Google Scholar]
- 30. Chigaev A., Waller A., Amit O., Sklar L. A. (2008) BMC. Immunol. 9, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gazitt Y. (2004) Leukemia 18, 1–10 [DOI] [PubMed] [Google Scholar]
- 32. Oostendorp R. A., Dörmer P. (1997) Leuk. Lymphoma 24, 423–435 [DOI] [PubMed] [Google Scholar]
- 33. Coulombel L., Auffray I., Gaugler M. H., Rosemblatt M. (1997) Acta Haematol. 97, 13–21 [DOI] [PubMed] [Google Scholar]
- 34. Bonig H., Wundes A., Chang K. H., Lucas S., Papayannopoulou T. (2008) Blood 111, 3439–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zohren F., Toutzaris D., Klärner V., Hartung H. P., Kieseier B., Haas R. (2008) Blood 111, 3893–3895 [DOI] [PubMed] [Google Scholar]
- 36. Papayannopoulou T., Nakamoto B. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9374–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bonig H., Watts K. L., Chang K. H., Kiem H. P., Papayannopoulou T. (2009) Stem Cells 27, 836–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramirez P., Rettig M. P., Uy G. L., Deych E., Holt M. S., Ritchey J. K., DiPersio J. F. (2009) Blood 114, 1340–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Broxmeyer H. E., Orschell C. M., Clapp D. W., Hangoc G., Cooper S., Plett P. A., Liles W. C., Li X., Graham-Evans B., Campbell T. B., Calandra G., Bridger G., Dale D. C., Srour E. F. (2005) J. Exp. Med. 201, 1307–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hatse S., Princen K., Bridger G., De Clercq E., Schols D. (2002) FEBS Lett. 527, 255–262 [DOI] [PubMed] [Google Scholar]
- 41. Liles W. C., Broxmeyer H. E., Rodger E., Wood B., Hübel K., Cooper S., Hangoc G., Bridger G. J., Henson G. W., Calandra G., Dale D. C. (2003) Blood 102, 2728–2730 [DOI] [PubMed] [Google Scholar]
- 42. Hendrix C. W., Flexner C., MacFarland R. T., Giandomenico C., Fuchs E. J., Redpath E., Bridger G., Henson G. W. (2000) Antimicrob. Agents Chemother. 44, 1667–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chedid L. (1954) C.R. Seances Soc. Biol. Fil. 148, 1039–1043 [PubMed] [Google Scholar]
- 44. Trifirò G., Gambassi G., Sen E. F., Caputi A. P., Bagnardi V., Brea J., Sturkenboom M. C. (2010) Ann. Intern. Med. 152, 418–425 [DOI] [PubMed] [Google Scholar]
- 45. Matthews N., Franklin R. J., Kendrick D. A. (1995) Biochem. Pharmacol. 50, 1053–1061 [DOI] [PubMed] [Google Scholar]
- 46. Pollmächer T., Haack M., Schuld A., Kraus T., Hinze-Selch D. (2000) J. Psychiatr. Res. 34, 369–382 [DOI] [PubMed] [Google Scholar]
- 47. Berger J. R., Houff S. (2006) Neurol. Res. 28, 299–305 [DOI] [PubMed] [Google Scholar]
- 48. Rigal E., Gateault P., Lebranchu Y., Hoarau C. (2009) Med. Sci. 25, 1135–1140 [DOI] [PubMed] [Google Scholar]
- 49. Fieve R. R., Blumenthal B., Little B. (1966) Arch. Gen. Psychiatry 15, 529–534 [DOI] [PubMed] [Google Scholar]
- 50. Kenakin T. (2004) Mol. Interv. 4, 222–229 [DOI] [PubMed] [Google Scholar]
- 51. Arnaout M. A., Mahalingam B., Xiong J. P. (2005) Annu. Rev. Cell Dev. Biol. 21, 381–410 [DOI] [PubMed] [Google Scholar]