Abstract
PCSK9 (proprotein convertase subtilisin-like/kexin type 9) is an emerging target for pharmaceutical intervention. This multidomain protein interacts with the LDL receptor (LDLR), promoting receptor degradation. Insofar as PCSK9 inhibition induces a decrease in plasma cholesterol levels, understanding the nature of the binding interaction between PCSK9 and the LDLR is of critical importance. In this study, the ability of PCSK9 to compete with apoE3 N-terminal domain-containing reconstituted HDL for receptor binding was examined. Whereas full-length PCSK9 was an effective competitor, the N-terminal domain (composed of the prodomain and catalytic domain) was not. Surprisingly, the C-terminal domain (CT domain) of PCSK9 was able to compete. Using a direct binding interaction assay, we show that the PCSK9 CT domain bound to the LDLR in a calcium-dependent manner and that co-incubation with the prodomain and catalytic domain had no effect on this binding. To further characterize this interaction, two LDLR fragments, the classical ligand-binding domain (LBD) and the EGF precursor homology domain, were expressed in stably transfected HEK 293 cells and isolated. Binding assays showed that the PCSK9 CT domain bound to the LBD at pH 5.4. Thus, CT domain interaction with the LBD of the LDLR at endosomal pH constitutes a second step in the PCSK9-mediated LDLR binding that leads to receptor degradation.
Keywords: Endosomal pH Function, Fluorescence Resonance Energy Transfer (FRET), Lipoprotein Metabolism, Lipoprotein Receptor, Low Density Lipoprotein (LDL)
Introduction
PCSK9 (proprotein convertase subtilisin-like/kexin type 9) is a unique modulator of plasma cholesterol levels (1). Several studies have shown that secreted PCSK9 can reduce LDL receptor (LDLR)2 protein levels by inducing a redistribution of the LDLR from the plasma membrane to lysosomes, where it is subject to degradation (2, 3). This results in decreased LDLR levels at the cell surface and, consequently, increased plasma cholesterol. Insofar as plasma cholesterol is a key determinant of cardiovascular disease risk, an intensive effort is underway to find inhibitors that disrupt or block PCSK9 binding to the LDLR (4).
Naturally occurring “gain-of-function” PCSK9 mutations cause hypercholesterolemia and premature atherosclerosis, whereas the corresponding “loss-of-function” mutations lead to hypocholesterolemia and protection from atherosclerosis (1). Biochemical studies using recombinant proteins have revealed that PCSK9 binds to a specific site on the LDLR, the A repeat in the EGF precursor homology domain (5, 6). Furthermore, PCSK9 binding to EGF-A is significantly enhanced at endosomal pH (5, 7).
PCSK9 is a multidomain protein composed of a prodomain (residues 31–152), a central catalytic domain (residues 153–451) domain, and a C-terminal domain (CT domain; residues 452–692). The structure of PCSK9 has been determined (7–9). The prodomain and catalytic domain (Pro-Cat domain) bind to EGF-A of the LDLR (6, 10, 11). Binding is calcium-dependent and increases dramatically upon reduction in pH from 7 to 5.2. PCSK9 binding to the LDLR interferes with physiological processes related to acid-dependent lipoprotein ligand release and subsequent receptor recycling (10).
The CT domain of PCSK9 consists of three β-domain modules related to one another by a pseudo 3-fold axis (7). The surface of the CT domain is enriched in histidine residues that are located along a surface-exposed region of the second module. Interest in the CT domain has increased in light of the following observations: (i) naturally occurring mutations in the CT domain affect PCSK9-dependent LDLR degradation (12); (ii) a Fab fragment directed against the CT domain interferes with PCSK9-dependent inhibition of LDL uptake (13); and (iii) a truncated PCSK9 variant composed of the Pro-Cat domain binds the LDLR but fails to stimulate receptor degradation (10).
Zhang et al. (10) identified regions in the LDLR and PCSK9 that are required for receptor degradation. These authors found that LDLR variants lacking the classical ligand-binding domain (LBD) or the β-propeller segment fail to be degraded, although they internalize bound PCSK9. Thus, domains in both the LDLR and PCSK9 that are not directly involved in Pro-Cat domain binding to EGF-A are necessary for PCSK9-mediated degradation of the LDLR. In this study, we examined the ability of PCSK9 and truncated variants to compete with apoE-containing reconstituted HDL (rHDL) for binding to an isolated soluble LDLR (sLDLR). The finding that the CT domain of PCSK9 binds to the LBD of the LDLR in a pH-dependent manner provides direct evidence for a second binding step in the pathway whereby PCSK9 mediates receptor degradation.
EXPERIMENTAL PROCEDURES
Recombinant sLDLR Expression, Isolation, and Characterization
Wild-type sLDLR (N-terminal residues 1–699) was isolated from conditioned medium of stably transfected HEK 293 cells as described (14). The truncated variants generated were verified by dideoxy automated DNA sequencing. sLDLR protein was analyzed by SDS-PAGE under reducing and nonreducing conditions as a measure of native protein folding and disulfide bond formation (15).
PCSK9 Isolation and Characterization
A cDNA clone encoding human PCSK9 was a kind gift from Dr. Jay Horton (University of Texas Southwestern Medical School). Stably transfected HEK 293 cells expressing full-length PCSK9, the Pro-Cat domain, and the CT domain were prepared. Each of the constructs generated possessed a C-terminal FLAG tag. SDS-PAGE and immunoblotting confirmed the identity, size, and relative purity of the recombinant protein products.
ApoE3 N-terminal Domain Isolation and rHDL Formation
Recombinant Trp-null apoE3 N-terminal domain (apoE3-NT) was produced and isolated from Escherichia coli culture supernatant as described previously (16). ApoE3-NT rHDL were prepared with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids). The phospholipid was dissolved in chloroform/methanol (3:1, v/v) and dried into a thin film in a glass tube. Following dispersion of the lipid in 50 mm sodium phosphate (pH 7.0) and 150 mm NaCl, apoE3-NT was added. This mixture was bath-sonicated at 24 °C until clear (17). The complexes were stable at 4 °C and were dialyzed into a specified buffer prior to use.
Fluorescence Binding Assay
Four μg of sLDLR (in 50 mm Tris base, 2 mm CaCl2, and 100 mm NaCl (pH 7.3)) was incubated with 1 μg of Trp-null apoE3-NT previously labeled at Cys-112 with the fluorescent probe N-(iodoacetyl)-N′-(5-sulfo-1-naphthyl)ethylenediamine (AEDANS) and complexed with phospholipid. Interaction between AEDANS-labeled Trp-null apoE3-NT rHDL and sLDLR was detected by FRET between excited Trp residues in sLDLR and the AEDANS moiety covalently attached to Trp-null apoE3-NT (14). Following incubation, samples were excited at 280 nm, and fluorescence emission intensity at 470 nm was determined (slit width of 5.0 nm) on a PerkinElmer Model LS 50B luminescence spectrometer.
Direct Binding Assay
sLDLR variants possessing a C-terminal His tag were applied to a HiTrap Ni2+ chelation affinity column in binding buffer (20 mm HEPES (pH 7.3), 100 mm NaCl, and 2 mm CaCl2,). Where specified, after washing, the column buffer was adjusted to pH 5.4 (20 mm Tris succinate, 100 mm NaCl, and 2 mm CaCl2). Subsequently, full-length PCSK9 or a given PCSK9 variant was applied to the column. After washing with buffer containing 50 mm imidazole, bound proteins were eluted with buffer containing 500 mm imidazole, dialyzed, and subjected to Western blot analysis.
Analytical Methods
Protein concentrations were determined by the bicinchoninic acid assay using bovine serum albumin as a standard. SDS-PAGE was performed on 4–20% acrylamide gradient slabs at 35-mA constant current under reducing or nonreducing conditions and stained with Coomassie Blue.
Immunoblotting
For immunoblotting, size-separated proteins were transferred to a 0.2-μm PVDF membrane (Bio-Rad). Nonspecific binding sites on the membrane were blocked with 0.1% buffer containing 0.1% Tween 20, 20 mm Tris, and 150 mm NaCl (pH 7.2). Anti-FLAG tag monoclonal antibody (Sigma) or anti-His tag polyclonal antibody (Abcam) was used as the primary antibody. Alkaline phosphatase-conjugated anti-mouse or anti-rabbit IgG (Cell Signaling Technology) was used as the secondary antibody. Subsequently, the membrane was incubated with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (Thermo Scientific) until an optimal signal was detected.
RESULTS
ApoE3-NT rHDL-PCSK9 Competition Binding Experiments
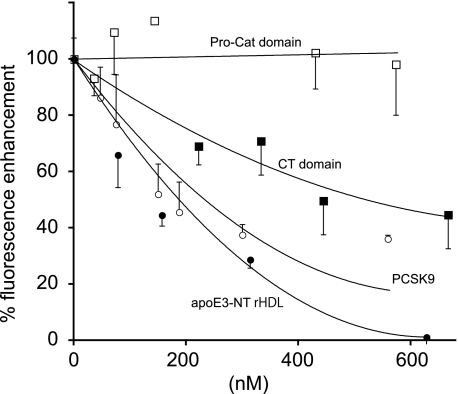
Despite the fact that PCSK9 binds to EGF-A in the EGF precursor homology domain of the LDLR and apoE binds to the LBD (18), the possibility exists that binding of one ligand may interfere with binding of another. Initial studies were conducted to determine whether full-length PCSK9 or truncated variants are able to compete with apoE3-NT rHDL for binding to sLDLR. ApoE3-NT rHDL binding was detected in solution using a fluorescence-based competition assay. When Trp-null apoE3-NT was labeled with an extrinsic fluorophore and complexed with phospholipid to form rHDL, binding to sLDLR was accompanied by an enhancement in FRET from excited Trp residues in sLDLR to an AEDANS fluorophore covalently attached to apoE3-NT. Unlabeled apoE3-NT rHDL competed for binding to sLDLR, resulting in a concentration-dependent decrease in AEDANS fluorescence intensity (Fig. 1). When unlabeled full-length PCSK9 was introduced as a competitor, a similar decrease in apoE3-NT AEDANS fluorescence intensity was observed. Thus, full-length PCSK9 competes for apoE3-NT rHDL binding to sLDLR. These data complement and extend the work of Fisher et al. (11), who reported that preincubation of the LDLR with PCSK9 reduces LDL binding. Given that apoE and PCSK9 are known ligands for the LDLR, this result was not unexpected. However, insofar as apoE3-NT and PCSK9 bind to distinct sites on the LDLR, the apparent similarity in concentration-dependent competition observed between unlabeled apoE3-NT rHDL and PCSK9 was surprising. To investigate this further, the ability of truncated PCSK9 variants to compete with AEDANS-labeled apoE3-NT rHDL for binding to the LDLR was investigated. When the isolated Pro-Cat domain was studied, no competition was observed. In this case, the lack of competition may be explained if the CT domain of PCSK9 exerts a steric effect, hindering access of apoE3-NT rHDL to the LBD. Thus, when the CT domain is absent, the Pro-Cat domain alone is unable to interfere with apoE3-NT rHDL access to the receptor. This interpretation is not consistent, however, with the finding that a PCSK9 variant corresponding to the CT domain effectively competed for apoE3-NT rHDL binding to the LDLR. Indeed, this observation implies that the CT domain alone can serve as an LDLR ligand.
FIGURE 1.
Effect of full-length PCSK9, the Pro-Cat domain, and the CT domain on AEDANS-labeled apoE3-NT rHDL binding to sLDLR. One μg of AEDANS-labeled Trp-null apoE3-NT rHDL and 4 μg of sLDLR were incubated in the presence of increasing concentrations of unlabeled apoE3-NT rHDL, full-length PCSK9, the Pro-Cat domain, or the CT domain. Samples (300-μl final volume) were excited at 280 nm, and fluorescence emission intensity at 470 nm was determined. ●, unlabeled apoE3-NT rHDL; ○, full-length PCSK9; □, Pro-Cat domain; ■, CT domain. Values are the mean ± S.D. (n = 3).
Direct Binding of PCSK9 to sLDLR
To further investigate the apparent, albeit unexpected, binding of the isolated CT domain of PCSK9 to sLDLR, a direct binding assay was developed to complement the competition binding assay employed with fluorescent-labeled apoE3-NT rHDL. The first choice was surface plasmon resonance spectroscopy. After extensive evaluation and despite the fact that this method could potentially provide key information about relative binding affinities, reproducible detection of binding by surface plasmon resonance was not successful. Instead, an on-column binding assay was developed. In this assay, sLDLR was bound to a HiTrap Ni2+ chelation affinity chromatography column via an engineered C-terminal His tag. Following interaction of sLDLR with the column matrix, PCSK9 or variants thereof were passed over the column. Binding was detected after elution of sLDLR and Western blot analysis. If PCSK9 or a given variant binds to sLDLR, a corresponding band will be detected upon analysis of the column eluate.
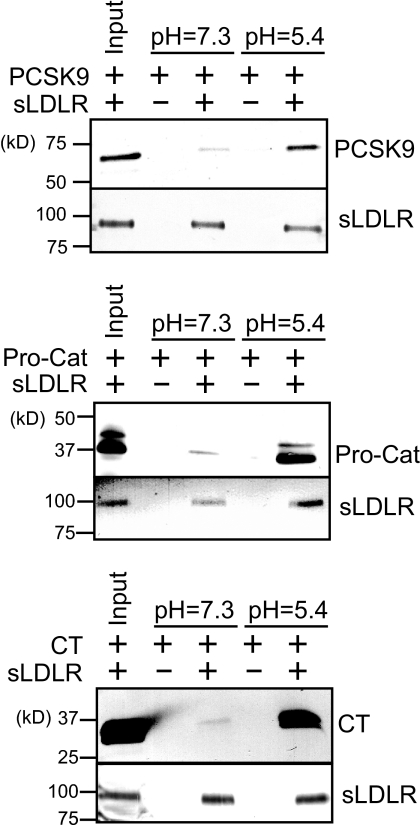
In the case of full-length PCSK9, at pH 7.3, a small amount of PCSK9 was recovered with sLDLR (Fig. 2). By contrast, significant binding was observed at pH 5.4, consistent with results reported by others (5, 7). Under both pH conditions, similar amounts of sLDLR were recovered, indicating there was no pH-dependent loss of the LDLR from the column matrix. In a manner similar to full-length PCSK9, when the Pro-Cat domain was passed over the sLDLR affinity column matrix, little binding was detected at pH 7.3, with enhanced binding observed at pH 5.4. Insofar as the Pro-Cat domain contains structural elements required for interaction with EGF-A (6, 10), this result is expected. To investigate CT domain binding to sLDLR, the same binding assay design was employed. When the CT domain was passed over the sLDLR affinity column, little binding occurred at pH 7.3. On the other hand, at pH 5.4, CT domain binding increased, indicating that, at low pH, the PCSK9 CT domain binds sLDLR.
FIGURE 2.
PCSK9 binding to an sLDLR affinity column. sLDLR was bound to a HiTrap Ni2+ chelation affinity column via an engineered C-terminal His tag. Following column equilibration at pH 7.3 (20 mm HEPES, 100 mm NaCl, and 2 mm CaCl2,) or pH 5.4 (20 mm Tris succinate, 100 mm NaCl, and 2 mm CaCl2), full-length PCSK9 (upper panel), the Pro-Cat domain (middle panel), or the CT domain (lower panel) was applied to the column. After washing, sLDLR was eluted with buffer containing 500 mm imidazole and subjected to SDS-PAGE and immunoblot detection of sLDLR and PCSK9.
Characterization of PCSK9 CT Domain Binding to sLDLR
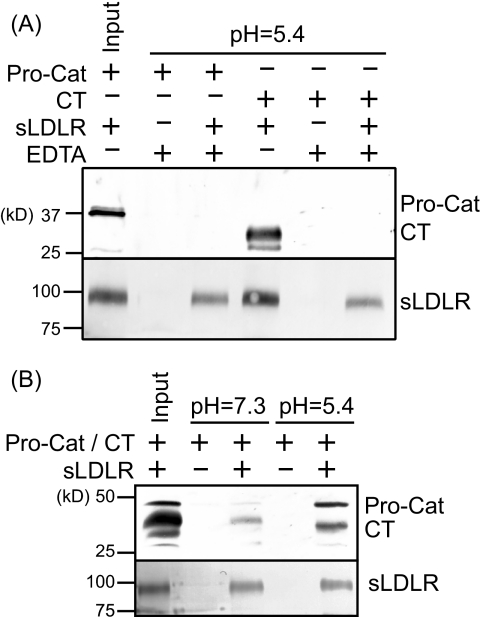
It is recognized that lipoprotein ligand binding to the LDLR, as well as Pro-Cat domain binding to EGF-A, requires calcium. To further characterize CT domain interaction with sLDLR, the effect of EDTA (4 mm) on CT domain binding was examined at pH 5.4 (Fig. 3). Both Pro-Cat domain and CT domain binding to sLDLR required Ca2+, indicating that these interactions are specific. Next, we examined whether the Pro-Cat domain can compete with the CT domain for binding to sLDLR. At pH 7.3, low Pro-Cat domain binding was observed, whereas more CT domain was bound. In co-incubation experiments at pH 5.4, both ligands bound to sLDLR, suggesting that they bind to different regions of the receptor.
FIGURE 3.
Factors affecting the interaction between PCSK9 domains and sLDLR. A, the Pro-Cat domain or CT domain was applied to an sLDLR affinity column in the presence or absence of 4 mm EDTA. B, the Pro-Cat domain and CT domain were applied to the sLDLR affinity column simultaneously. After washing, sLDLR was eluted with buffer containing 500 mm imidazole and subjected to SDS-PAGE and immunoblot detection of sLDLR and PCSK9.
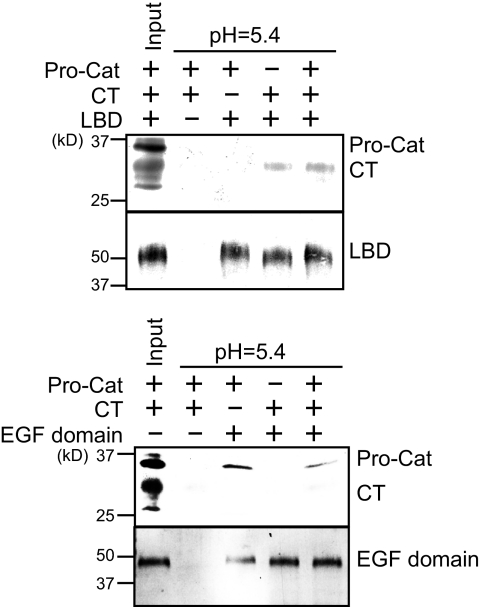
PCSK9 Variant Binding to LDLR Fragments
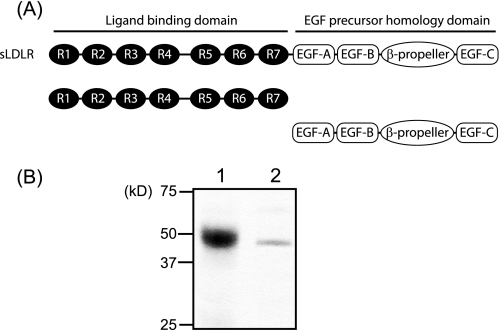
Based on its ability to compete with apoE3-NT rHDL for binding to sLDLR, it is plausible that the PCSK9 CT domain binds the LBD of the LDLR. To better define the interaction between specific PCSK9 domains and the LDLR, two receptor fragments were expressed in stably transfected HEK 293 cells and isolated. One fragment corresponded to the classical LBD (encompassing repeats 1–7), whereas the second corresponded to the EGF precursor homology domain, including EGF-A, EGF-B, the β-propeller, and EGF-C (Fig. 4). The two fragments were bound to a HiTrap Ni2+ chelation column, and specific PCSK9 variants were applied to the respective affinity columns at pH 5.4. When the Pro-Cat domain was passed over the LBD affinity column, no binding was observed (Fig. 5). On the other hand, the CT domain bound to this region of the LDLR. In addition, the presence of the Pro-Cat domain had no effect on CT domain binding under these conditions. In the case of the EGF precursor homology domain affinity column, Pro-Cat domain binding was observed, whereas CT domain binding was not. Furthermore, the CT domain had no effect on Pro-Cat domain binding to the EGF precursor homology domain affinity column.
FIGURE 4.
Characterization of the LBD and EGF precursor homology domain of LDLR. A, schematic diagram of the LBD and EGF precursor homology domain of the LDLR. R, repeat. B, following expression and purification, the isolated domains were separated on a 4–20% SDS-polyacrylamide gel and stained with Coomassie Blue. Lane 1, LBD; lane 2, EGF domain.
FIGURE 5.
Pro-Cat domain and CT domain binding to LBD and EGF domain affinity columns. The LBD or EGF domain was bound to a HiTrap Ni2+ chelation affinity column and equilibrated in 20 mm Tris succinate (pH 5.4), 100 mm NaCl, and 1 mm CaCl2. After washing, the PCSK9 Pro-Cat and/or CT domain was applied to the LBD (upper panel) and EGF domain (lower panel) columns. After washing, bound proteins were eluted with buffer containing 500 mm imidazole and subjected to SDS-PAGE and immunoblot detection of sLDLR and PCSK9.
DISCUSSION
PCSK9 is a prime target for therapeutic intervention as a means to improve plasma cholesterol levels. Various approaches include humanized monoclonal antibodies, antisense oligonucleotides, RNA interference, and small molecule inhibitors. Recent success in studies with antibodies directed against PCSK9 has further increased interest in this target (19, 20). As progress toward therapeutic intervention of PCSK9-LDLR interactions advances, it is important to understand the molecular basis of the biological events involved. A priori, it is rather surprising that a protein with no apparent connection to lipoprotein metabolism exerts such profound effects on plasma cholesterol levels. Thus far, no compelling connection between these seemingly “strange bedfellows” has emerged.
Elegant studies by Hobbs and co-workers (5, 10) documented the nature of the binding interaction between the Pro-Cat domain of PCSK9 and EGF-A of the LDLR. Structure determination of a co-crystal of PCSK9 and EGF-A/B provides a clear view of this interaction (6). In this structure, EGF-A binds a surface of PCSK9 encompassing residues 367–381. An antiparallel β-sheet is formed between residues 377–379 of PCSK9 and residues 308–310 of EGF-A. As may be expected, this interaction has been the target of approaches designed to interfere with PCSK9-LDLR interactions (21). At the same time, unexplained results have emerged indicating that Pro-Cat domain binding to EGF-A is not sufficient for manifestation of LDLR degradation activity (10). Furthermore, the fact that mutations in the CT domain or antibodies directed against this domain interfere with PCSK9-mediated degradation of the LDLR is curious. How does the CT domain contribute to PCSK9-mediated degradation of the LDLR? As suggested by Hobbs and co-workers (10), this domain may interfere with an endosomal pH-induced conformational change in the LDLR that is necessary for ligand release/receptor recycling. This process was originally proposed on the basis of deletion experiments with the LDLR (22). Subsequent determination of the three-dimensional structure of sLDLR at pH 5.3 has provided a new understanding of this conformational change. Rudenko et al. (23) showed that the receptor adopts a “closed” conformation wherein the β-propeller segment contacts residues in ligand-binding repeats 4 and 5. The nature of this interaction is ionic and, importantly, involves key histidine residues. The concept emerging from these studies is that the transition from neutral pH at the cell surface to low pH in the endosomal compartment activates a “histidine switch” that promotes the aforementioned intramolecular interaction between receptor domains (24). A critical aspect of this conformational change is that it promotes ligand release, thereby facilitating receptor recycling to the cell surface, where it is available for another round of endocytosis. PCSK9-mediated interference with this process causes the LDLR to traffic to lysosomes, where it is degraded.
Physiologically, the process whereby endocytosed LDLR recycles to the cell surface is closely associated with ligand release. One question that has yet to be answered is whether PCSK9-mediated LDLR degradation requires co-internalization of a lipoprotein ligand. The finding that LDLR variants possessing a minimum of three ligand-binding repeats retain susceptibility to PCSK9-mediated degradation suggests that lipoprotein ligand binding is not required. Because larger deletions in the LDLR LBD abrogate PCSK9-mediated receptor degradation, however, it is clear that elements within the LBD participate in this process. Given the combination of deletion mutants examined by Zhang et al. (10), it is apparent that no specific ligand-binding repeat is essential (10). Furthermore, the observation that PCSK9-mediated receptor degradation activity is compromised in the case of ΔLBD repeat 1–4 and ΔLBD repeat 4–7 LDLR variants indicates that at least four LBD repeats are needed for manifestation of optimal PCSK9-dependent LDLR degradation activity.
Zhang et al. (10) also examined PCSK9 fragment binding to the LDLR using an immunoprecipitation assay. Their conclusion that the CT domain does not bind the LDLR was based on the results of immunoprecipitation experiments conducted under conditions in which low binding is expected (i.e. PBS). Likewise, in the case of cell-based binding studies, failure to detect CT domain binding to the LDLR may be related to their use of an LDLR variant that lacks EGF-A (10). Indeed, in intact PCSK9, Pro-Cat domain binding to EGF-A may facilitate CT domain interaction with the LBD by effectively localizing the CT domain in the vicinity of the LBD. Our results indicate that CT domain binding to ligand-binding repeats in the LDLR is strongly pH-dependent, with much greater binding at pH 5.4. Thus, it is conceivable that PCSK9 contacts the LDLR at neutral pH primarily via its Pro-Cat domain, and following internalization and localization to the low pH environment of the endosome, increased positive charge density in the CT domain (owing to side chain ionization of its numerous surface-exposed histidine residues (7–9)) promotes interaction with elements in the LBD. Such a model would explain the observation that whereas the Pro-Cat domain of PCSK9 binds the LDLR, it does not support receptor degradation. Whether the PCSK9 CT domain can bind to the same site in the LBD as the β-propeller segment does in full-length LDLR is not known. Based on the work of Zhang et al. (10), no specific ligand-binding repeat is required for PCSK9-mediated degradation of the LDLR. Under normal physiological conditions, the CT domain may bind to a specific site on the LDLR, although it appears that other ligand-binding repeats can substitute in the case of certain LDLR deletion mutants.
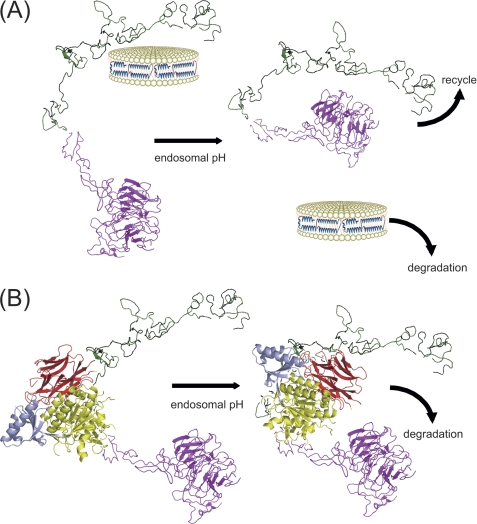
Cunningham et al. (7) also studied PCSK9 binding to the LDLR. Using surface plasmon resonance, these authors found a strong pH dependence on binding and, interestingly, observed two populations of binding sites at pH 5.4, a high affinity binding site (Kd ∼ 1 nm) and a lower affinity binding site (Kd ∼ 50 nm). Whether these may correspond to Pro-Cat domain binding to EGF-A and CT domain binding to the LBD, respectively, remains to be determined. Taken together, our data permit a two-step model to be invoked wherein the Pro-Cat domain initiates contact with EGF-A of the receptor, followed by interaction of the CT domain with the LBD, driving the LDLR toward degradation (Fig. 6). This binding may be affected by lipoprotein ligands (11). In any event following internalization of the LDLR, with or without low pH discharge of bound lipoprotein, it is conceivable that the CT domain of PCSK9 interferes with “normal” conformational adaptations in the LDLR required for receptor recycling. An obvious candidate is disruption of the interaction between the β-propeller segment and ligand-binding repeats 4 and 5. Although Beglova et al. (25) reported that the interface between ligand-binding repeat 7 and EGF-A adopts a pH-insensitive rigid conformation, it is conceivable that flexible linkers between ligand-binding repeats expand the conformational sampling boundary of the LBD such that access to the CT domain of EGF-A-bound PCSK9 can be achieved. Moreover, the inherent flexibility of the CT domain with respect to the remainder of PCSK9 (8) would facilitate such an interaction. Once achieved, CT domain binding to the LBD could interfere with conformational changes in the LDLR that are critical to receptor recycling.
FIGURE 6.
Two-step model of PCSK9 binding to the LDLR. A, in the absence of PCSK9, lipoprotein binding (depicted as apoE-NT rHDL) (27) to the LDLR (left) leads to receptor-mediated endocytosis. The low pH environment of the endosome (right) induces a conformational change in the LDLR (with the LBD shown in green and the EGF domain in magenta), resulting in discharge of bound lipoprotein ligand and interaction between the β-propeller segment and ligand-binding repeats 4 and 5 (23). This event permits the segregation and separate trafficking of the LDLR to the cell surface and the lipoprotein ligand to the lysosome, respectively. B, in the first step (left), PCSK9 binds EGF-A of the LDLR at the cell surface via its Pro-Cat domain (with the prodomain shown in cyan and the catalytic domain in yellow) (6). Following internalization and exposure to the low pH environment of the endosome, the CT domain (shown in red) of PCSK9 binds the LBD (second step; right), disrupting the ability of the receptor to adopt a recycling-competent conformation and promoting trafficking of the PCSK9-LDLR complex to the lysosome.
In conclusion, the data presented herein document a pH-dependent binding interaction between the CT domain of PCSK9 and the LBD of the LDLR. The results provide a biochemical explanation for unexplained findings, including (a) the isolated CT domain associates with LDLR on the surface of transfected CHO cells (26); (b) the CT domain is required for PCSK9-mediated LDLR degradation; and (c) antibodies directed against the CT domain inhibit PCSK9 effects on the LDLR. Given the apparent importance of this interaction, future studies designed to characterize the specific nature of this binding, perhaps via co-crystal structure determination, may provide new strategies for intervention.
Acknowledgment
We thank Alison Zheng for skilled technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant HL 64159. This work was also supported by California Tobacco-Related Disease Research Program New Investigator Award 17KT-0026 (to T. Y.).
- LDLR
- LDL receptor
- CT domain
- C-terminal domain
- Pro-Cat domain
- prodomain and catalytic domain
- LBD
- ligand-binding domain
- rHDL
- reconstituted HDL
- sLDLR
- soluble LDLR
- apoE3-NT
- apoE3 N-terminal domain
- AEDANS
- N-(iodoacetyl)-N′-(5-sulfo-1-naphthyl)ethylenediamine.
REFERENCES
- 1. Horton J. D., Cohen J. C., Hobbs H. H. (2009) J. Lipid Res. 50, S172–S177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lagace T. A., Curtis D. E., Garuti R., McNutt M. C., Park S. W., Prather H. B., Anderson N. N., Ho Y. K., Hammer R. E., Horton J. D. (2006) J. Clin. Invest. 116, 2995–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McNutt M. C., Kwon H. J., Chen C., Chen J. R., Horton J. D., Lagace T. A. (2009) J. Biol. Chem. 284, 10561–10570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davignon J., Dubuc G., Seidah N. G. (2010) Curr. Atheroscler. Rep. 12, 308–315 [DOI] [PubMed] [Google Scholar]
- 5. Zhang D. W., Lagace T. A., Garuti R., Zhao Z., McDonald M., Horton J. D., Cohen J. C., Hobbs H. H. (2007) J. Biol. Chem. 282, 18602–18612 [DOI] [PubMed] [Google Scholar]
- 6. Kwon H. J., Lagace T. A., McNutt M. C., Horton J. D., Deisenhofer J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cunningham D., Danley D. E., Geoghegan K. F., Griffor M. C., Hawkins J. L., Subashi T. A., Varghese A. H., Ammirati M. J., Culp J. S., Hoth L. R., Mansour M. N., McGrath K. M., Seddon A. P., Shenolikar S., Stutzman-Engwall K. J., Warren L. C., Xia D., Qiu X. (2007) Nat. Struct. Mol. Biol. 14, 413–419 [DOI] [PubMed] [Google Scholar]
- 8. Hampton E. N., Knuth M. W., Li J., Harris J. L., Lesley S. A., Spraggon G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14604–14609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Piper D. E., Jackson S., Liu Q., Romanow W. G., Shetterly S., Thibault S. T., Shan B., Walker N. P. (2007) Structure 15, 545–552 [DOI] [PubMed] [Google Scholar]
- 10. Zhang D. W., Garuti R., Tang W. J., Cohen J. C., Hobbs H. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13045–13050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fisher T. S., Lo Surdo P., Pandit S., Mattu M., Santoro J. C., Wisniewski D., Cummings R. T., Calzetta A., Cubbon R. M., Fischer P. A., Tarachandani A., De Francesco R., Wright S. D., Sparrow C. P., Carfi A., Sitlani A. (2007) J. Biol. Chem. 282, 20502–20512 [DOI] [PubMed] [Google Scholar]
- 12. Kotowski I. K., Pertsemlidis A., Luke A., Cooper R. S., Vega G. L., Cohen J. C., Hobbs H. H. (2006) Am. J. Hum. Genet. 78, 410–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ni Y. G., Condra J. H., Orsatti L., Shen X., Di Marco S., Pandit S., Bottomley M. J., Ruggeri L., Cummings R. T., Cubbon R. M., Santoro J. C., Ehrhardt A., Lewis D., Fisher T. S., Ha S., Njimoluh L., Wood D. D., Hammond H. A., Wisniewski D., Volpari C., Noto A., Lo Surdo P., Hubbard B., Carfí A., Sitlani A. (2010) J. Biol. Chem. 285, 12882–12891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamamoto T., Lamoureux J., Ryan R. O. (2006) J. Lipid Res. 47, 1091–1096 [DOI] [PubMed] [Google Scholar]
- 15. Bajari T. M., Strasser V., Nimpf J., Schneider W. J. (2005) Methods 36, 109–116 [DOI] [PubMed] [Google Scholar]
- 16. Fisher C. A., Wang J., Francis G. A., Sykes B. D., Kay C. M., Ryan R. O. (1997) Biochem. Cell Biol. 75, 45–53 [PubMed] [Google Scholar]
- 17. Weers P. M., Narayanaswami V., Ryan R. O. (2001) Eur. J. Biochem. 268, 3728–3735 [DOI] [PubMed] [Google Scholar]
- 18. Russell D. W., Brown M. S., Goldstein J. L. (1989) J. Biol. Chem. 264, 21682–21688 [PubMed] [Google Scholar]
- 19. Chan J. C., Piper D. E., Cao Q., Liu D., King C., Wang W., Tang J., Liu Q., Higbee J., Xia Z., Di Y., Shetterly S., Arimura Z., Salomonis H., Romanow W. G., Thibault S. T., Zhang R., Cao P., Yang X. P., Yu T., Lu M., Retter M. W., Kwon G., Henne K., Pan O., Tsai M. M., Fuchslocher B., Yang E., Zhou L., Lee K. J., Daris M., Sheng J., Wang Y., Shen W. D., Yeh W. C., Emery M., Walker N. P., Shan B., Schwarz M., Jackson S. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9820–9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bottomley M. J., Cirillo A., Orsatti L., Ruggeri L., Fisher T. S., Santoro J. C., Cummings R. T., Cubbon R. M., Lo Surdo P., Calzetta A., Noto A., Baysarowich J., Mattu M., Talamo F., De Francesco R., Sparrow C. P., Sitlani A., Carfí A. (2009) J. Biol. Chem. 284, 1313–1323 [DOI] [PubMed] [Google Scholar]
- 21. Ni Y. G., Di Marco S., Condra J. H., Peterson L. B., Wang W., Wang F., Pandit S., Hammond H. A., Rosa R., Cummings R. T., Wood D. D., Liu X., Bottomley M. J., Shen X., Cubbon R. M., Wang S. P., Johns D. G., Volpari C., Hamuro L., Chin J., Huang L., Zhao J. Z., Vitelli S., Haytko P., Wisniewski D., Mitnaul L. J., Sparrow C. P., Hubbard B., Carfi A., Sitlani A. (2011) J. Lipid Res. 52, 78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davis C. G., Goldstein J. L., Südhof T. C., Anderson R. G., Russell D. W., Brown M. S. (1987) Nature 326, 760–765 [DOI] [PubMed] [Google Scholar]
- 23. Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J. (2002) Science 298, 2353–2358 [DOI] [PubMed] [Google Scholar]
- 24. Yamamoto T., Chen H. C., Guigard E., Kay C. M., Ryan R. O. (2008) Biochemistry 47, 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beglova N., Jeon H., Fisher C., Blacklow S. C. (2004) Mol. Cell 16, 281–292 [DOI] [PubMed] [Google Scholar]
- 26. Nassoury N., Blasiole D. A., Tebon Oler A., Benjannet S., Hamelin J., Poupon V., McPherson P. S., Attie A. D., Prat A., Seidah N. G. (2007) Traffic 8, 718–732 [DOI] [PubMed] [Google Scholar]
- 27. Raussens V., Fisher C. A., Goormaghtigh E., Ryan R. O., Ruysschaert J. M. (1998) J. Biol. Chem. 273, 25825–25830 [DOI] [PubMed] [Google Scholar]