Abstract
Lipid droplets (LDs), the major intracellular storage sites for neutral lipids, consist of a neutral lipid core surrounded by a phospholipid monolayer membrane. In addition to their function in lipid storage, LDs participate in lipid biosynthesis and recently were implicated in proteasomal protein degradation and autophagy. To identify components of the protein degradation machinery on LDs, we studied several candidates identified in previous LD proteome analyses. Here, we demonstrate that the highly conserved and broadly expressed ancient ubiquitous protein 1 (AUP1) localizes to LDs, where it integrates into the LD surface in a monotopic fashion with both termini facing the cytosol. AUP1 contains a C-terminal domain with strong homology to a domain known as G2BR, which binds E2 ubiquitin conjugases. We show that AUP1, by means of its G2BR domain, binds to Ube2g2. This binding is abolished by deletion or mutation of the G2BR domain, although the LD localization of AUP1 is not affected. The presence of the AUP1-Ube2g2 complex at LDs provides a direct molecular link between LDs and the cellular ubiquitination machinery.
Keywords: Lipid Droplet, Microscopic Imaging, Protein Degradation, Protein Domains, Ubiquitin-conjugating Enzyme (Ubc), AUP1, ERAD, G2BR Domain, Ube2g2
Introduction
Specialized organelles called lipid droplets (LD)2 (1) play a key role in intracellular turnover and storage of neutral lipids. In LDs, a hydrophobic core of neutral lipids, usually triacylglycerides and esterified sterols, is surrounded by a monolayer of phospholipids with several embedded or associated proteins, most prominently proteins of the PAT family, containing perilipin, adipophilin, and the tail-interacting protein of 47 kDa (TIP-47) (2, 3). The compartmentalization via a monolayer of phospholipids, instead of the conventional bilayer, is the most characteristic difference of LDs to other organelles (4, 5). LDs were originally regarded as immobile storage sites for triacylglyceride in adipocytes. LDs perform additional functions in many cell types, e.g. in signaling and transport events and as a general reservoir for hydrophobic and otherwise toxic substances (6, 7). LDs are ubiquitous, motile, and highly dynamic organelles (reviewed in Refs. 8, 9), which interact with many other organelles, e.g. the ER, mitochondria, endosomes, and peroxisomes (10–14). Progress in our knowledge about the cell biology of LDs was the subject of several recent reviews (9, 15–20), but essential questions are still open, like the mechanism of targeting or degradation of LD proteins or the machinery for regulation of LD size.
Recent studies from several laboratories have provided comprehensive insight into the proteome of LDs of various cell types (21–26). Comparative analysis of these LD proteomes revealed the repeated identification of AUP1 as a component of LDs (22, 23, 26). Originally, the AUP1 gene was identified as a part of the mnd2 (motor neuron degeneration 2) locus in mouse, mutation of which results in a lethal neuromuscular disorder (27). Independently, AUP1 was found as a binding partner of adenoviral proteins (28, 29). AUP1 was also reported as a cytosolic protein that binds to integrin α subunits and supports inside-out signaling in platelets (30, 31). Very recently, AUP1 was identified as a component of the Sel1l complex at the ER (32, 33), indicating an involvement of AUP1 in protein degradation processes.
Here, we present evidence that a major fraction of AUP1 resides on lipid droplets. We show that AUP1 is a monotopic membrane protein with both termini facing the cytosol. We demonstrate that AUP1 binds to the E2 conjugase Ube2g2 and recruits it to LDs. Thereby AUP1 provides a molecular link between LDs and ubiquitination.
EXPERIMENTAL PROCEDURES
Antibodies
Polyclonal rabbit antisera against recombinant His6-AUP1(221–410), His6-NSDHL(1–211), and His6-TIP47(1–168) were raised by Eurogentec and were affinity-purified against the antigens. Additionally, we used the following antibodies: anti-HA (clone F-7, Santa Cruz Biotechnology), anti-protein-disulfide isomerase (StressGen), Alexa555- and Alexa488-conjugated secondary antibodies (Invitrogen), and HRP-coupled secondary antibodies (Jackson ImmunoResearch).
Cell Culture
A431 and COS7 cells were maintained in DMEM (Invitrogen 31966), supplemented with 10% FCS. Huh7 cells were cultured in RPMI (Invitrogen 31870) with 10% FCS, 0.1 mm nonessential amino acids, 2 mm l-glutamine, and 10 mm HEPES. All cells were kept at 37 °C and 5% CO2.
DNA Constructs
DNA sequences were PCR-amplified from expressed sequence tags and cloned into 3HA, EGFP, GST, MBP, or His6 expression vectors. For details see supplemental Table 1. All constructs were verified by sequencing.
Sequence Alignment
Members of the AUP1 family were identified by reciprocal BLAST searches against the nonredundant protein data base at the NCBI (release of January 2007). The multiple sequence alignment and a distance-based neighbor tree were generated using Clustal.
Bacterial Expression and Purification of Recombinant Proteins
Plasmids as described in supplemental Table 1 were transformed in Escherichia coli BL21/DE3 or ER2566 strains. Bacteria were grown in LB supplemented with ampicillin and chloramphenicol (BL21/DE3) or ampicillin (ER2566), induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and shaken at 18–32 °C for 4–16 h. Bacteria were collected by centrifugation and pellets resuspended in 30–50 ml of Lysis Buffer (as recommended by the manufacturer's protocols for the different fusion tags, i.e. His6, GST, MBP, always including Complete inhibitor tablets without EDTA; Roche Applied Science). All the following steps were performed at 4 °C. Cells were lysed in the Emulsiflex (Avestin), and the lysate was centrifuged at 50,000 × g for 15 min. The supernatant was incubated with 2–6 ml of the respective affinity matrix. Beads were collected and washed, and the fusion proteins eluted with imidazole, reduced glutathione, or maltose according to standard protocols.
GST Pulldown
GST fusion constructs of Ube2g1 and Ube2g2 were bound to GSH-Sepharose beads, and the beads were incubated with a stoichiometric amount of His6-AUP1(221–410) in PBS, 1% Triton X-100, 1 mm DTT. GST fusion constructs of AUP1(361–410) and AUP1(361–410)-M1 were mixed with MBP-Ube2g2 at a stoichiometric ratio of 2:1 in 20 mm HEPES/KOH, pH 7.5, 100 mm KCl, 1 mm EDTA, 1 mm DTT, and the complex was bound to GSH-Sepharose beads. For all experiments, beads were washed several times with PBS supplemented with 1% Triton X-100, 250 mm NaCl and eluted with Elution Buffer (20 mm GSH, 50 mm Tris, pH 8.0). Eluates were analyzed via SDS-PAGE/Coomassie Brilliant Blue staining.
Anti-AUP1 Pulldown
Twenty 15-cm dishes of A431 cells were grown in medium supplemented with 20 μm oleate. Cells were washed with PBS and scraped into 16 ml of Binding Buffer (50 mm HEPES, pH 7.4, 70 mm sodium acetate, 1 mm NaF, 20 mm β-glycerol phosphate, 5 mm magnesium acetate, 0.5 mm DTT, 2× Complete protease inhibitor mixture). The suspension was adjusted to 1% Triton X-100 and cleared by two sequential centrifugations (4500 × g for 10 min at 4 °C) and (50,000 × g for 10 min at 4 °C). Supernatant was incubated with Affi-Gel 10 beads with or without coupled anti-AUP1 antibody for 24 h. Beads were washed, and bound proteins were eluted with 3 ml of Elution Buffer (0.1 m glycine, pH 2.4, 10% acetonitrile, and 0.5% Triton X-100). Proteins were precipitated with chloroform/methanol, redissolved in 50 μl of Sample Buffer, separated with SDS-PAGE/Coomassie Brilliant Blue staining, and analyzed by mass spectrometry.
Purification of LDs
Cells were grown in four 10-cm dishes in medium supplemented with 100 μm oleate for the last 24 h. Cells were washed with cold PBS and Buffer A (50 mm Tris-HCl, pH 7.4, 20 mm sucrose, supplemented with Complete Protease inhibitor mixture; Roche Applied Science) and scraped into a total of 1 ml of Buffer A. Cells were cracked by passing through a 22-gauge needle (0.7 × 40 mm) four times and further disrupted by 13 strokes in a ball bearing cell cracker (isobiotec, Heidelberg, Germany, inner diameter 8.020 mm, ball diameter 8.004 mm). The cell homogenate was centrifuged (1500 × g/10 min/4 °C) to pellet nuclei and intact cells. The supernatant was mixed 1:1 with buffer containing 50 mm Tris-HCl, pH 7.4, 2 m sucrose, supplemented with Complete Protease inhibitor mixture (Roche Applied Science), transferred to an SW60Ti ultracentrifugation tube, and overlaid with Buffer A. LDs were floated to the top of the gradient by centrifugation (100,000 × g for 3 h at 4 °C).
Fluorescence Microscopy of Fixed Samples
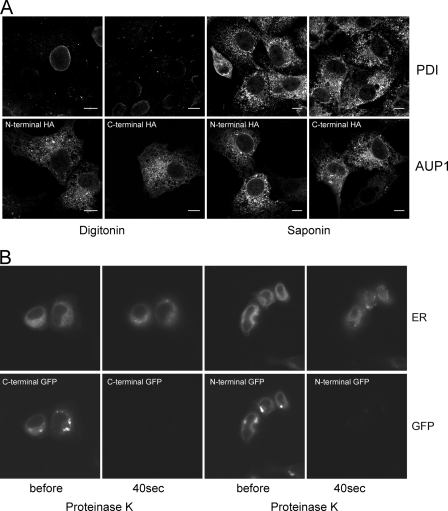
Cells were grown on coverslips, washed with PBS, and fixed with 3.7% (w/v) paraformaldehyde in PBS for 30 min. After washing with PBS, cells were blocked and permeabilized with Blocking Buffer (0.5% BSA and 0.1% saponin in PBS). Primary and secondary antibodies were diluted in Blocking Buffer and incubated for 1 h. Nuclei and LDs were stained with DAPI and Bodipy 493/503, respectively. Finally, cells were washed with PBS, rinsed with water, and mounted in Mowiol 4-88 containing 2.5% 1,4-diazabicyclo[2.2.2]octane. Images were acquired at a Zeiss LSM510 confocal microscope equipped with a 100× NA1.3 oil objective or at a Zeiss Axio Observer.Z1 with a 63× NA1.4 objective. For selective permeabilization of the plasma membrane (see Fig. 5), saponin in the Blocking Buffer was replaced by 0.001% digitonin (Applichem A1905,0100).
FIGURE 5.
Topology of AUP1, N and C terminus of AUP1 face the cytoplasm. A, in fixed cells, COS7 cells were cultured in normal growth medium, transfected with N- or C-terminally HA-tagged AUP1, fixed with 3.7% paraformaldehyde in PBS, and permeabilized with either 0.001% digitonin (in order not to permeabilize internal membranes) or 0.1% saponin (to permeabilize all membranes). Cells were stained with anti-protein-disulfide isomerase (PDI) (upper row) and anti-HA (lower row) antibodies. Positive staining for both AUP1 constructs upon digitonin permeabilization indicates cytoplasmic localization of both protein termini, whereas negative staining for the ER luminal protein-disulfide isomerase upon digitonin permeabilization shows that internal ER membranes were intact. B, in living cells, COS7 cells were cultured in 3-cm glass bottom dishes and co-transfected with pDsRed2-ER and either N- or C-terminal EGFP-tagged AUP1. After transfection, medium was supplemented with 50 μm oleate, and cells were grown for another 24 h. Cells were washed and incubated in 70 μm digitonin in KHM buffer for 1 min, and an image was recorded (before). Buffer was exchanged for KHM buffer containing 50 μg/ml proteinase K. Immediately after, images were recorded every 10 s for 2 min. Images taken after 40 s are shown (40s). Note that ER luminal DsRed2 is inaccessible to the protease, and both termini of AUP1 fusion proteins are degraded.
Fluorescence Protease Protection Assay
A431 cells were grown in 3-cm glass bottom dishes and co-transfected with pDsRed2-ER (Clontech) and either pAUP1-EGFP or pEGFP-AUP1. After transfection, medium was supplemented with 50 μm oleate, and cells were further grown for 24 h. Cells were washed three times in KHM buffer (110 mm potassium acetate, 20 mm HEPES, 2 mm MgCl2). The dish was placed onto the microscope stage and incubated in 70 μm digitonin in KHM buffer for 1 min, and an image was recorded (Fig. 5, before). Buffer was exchanged for KHM buffer containing 50 μg/ml proteinase K. Immediately thereafter, images were recorded every 10 s for 2 min. In Fig. 5, images taken after 40 s are shown (40 s proteinase K).
Electron Microscopy
For immuno-EM, A431 cells were incubated overnight with 50 μm oleate and fixed in 2% paraformaldehyde and 0.2% glutaraldehyde in 0.1 m PHEM buffer (60 mm PIPES, 25 mm HEPES, 10 mm EGTA, and 2 mm MgCl2), pH 6.9. Cells were embedded in 10% gelatin, cryoprotected using polyvinylpyrrolidone-sucrose, and snap-frozen onto specimen holders in liquid N2. Thin sections (80 nm) were picked up with a 1:1 mixture of 2.3 m sucrose and 2% methylcellulose. Immunolabeling was performed using the anti-AUP1 antibody and 10-nm gold-conjugated protein A. Grids were viewed using a Leo 922 Omega transmission electron microscope.
RESULTS
AUP1 Is a Highly Conserved Protein
AUP1 is a protein of 410 amino acids, with a high degree of conservation in multicellular organisms (Fig. 1A). AUP1 contains a very long hydrophobic stretch (Fig. 1A, green), an acyltransferase domain as indicated by the presence of three out of four conserved motifs (Fig. 1A, violet) (34), and two additional conserved domains as follows: CUE (coupling of ubiquitin to ER-associated degradation, Fig. 1A, red) and G2BR (G2 binding region; Fig. 1A, orange). CUE domains bind to ubiquitin and promote intramolecular mono-ubiquitination (35–37), whereas the G2BR domain binds to an E2 conjugase (38, 39). The presence of those two domains indicates a direct connection of AUP1 to ubiquitination processes (40).
FIGURE 1.
Domain structure and phylogenetic analysis of the AUP1 protein family. A, AUP1 has 410 amino acids and is highly conserved among multicellular organisms. It contains four conserved domains. In the very N terminus, AUP1 has a long hydrophobic region (green). Close to that, AUP1 contains an acyltransferase domain with three conserved motifs (violet boxes) of bacterial acyltransferases. In addition, AUP1 harbors a CUE domain (red) and a G2BR domain (orange). B, phylogenetic tree and domain organization of the three related proteins hAUP1, hAMFR/gp78, yCue1p as follows: light green, transmembrane (TMD); violet, putative acyltransferase; light blue, E3 ligase; red, ubiquitin binding (CUE); orange, E2 ligase binding (G2BR).
Phylogenetic analyses revealed a close relation of AUP1 to AMFR/gp78 (autocrine motility factor receptor, glycoprotein of 78 kDa) in higher eukaryotes and to Cue1p in the unicellular yeast Saccharomyces cerevisiae (Fig. 1B). All three proteins share the conserved CUE and G2BR domains in their C terminus (Fig. 1B and supplemental Fig. S1A) but differ in their N-terminal regions. Cue1p is a well characterized protein in the yeast ER-associated protein degradation (ERAD) and involved in sterol-dependent degradation of Hmg2p (hydroxymethylglutaryl-CoA reductase), a key enzyme in sterol metabolism (41). Because AMFR/gp78 plays an analogous role in mammalian cells (42), AUP1 might also perform a Cue1p-related function in regulated ubiquitination and degradation of proteins.
AUP1 Is a Ubiquitous Protein and Localizes to LDs
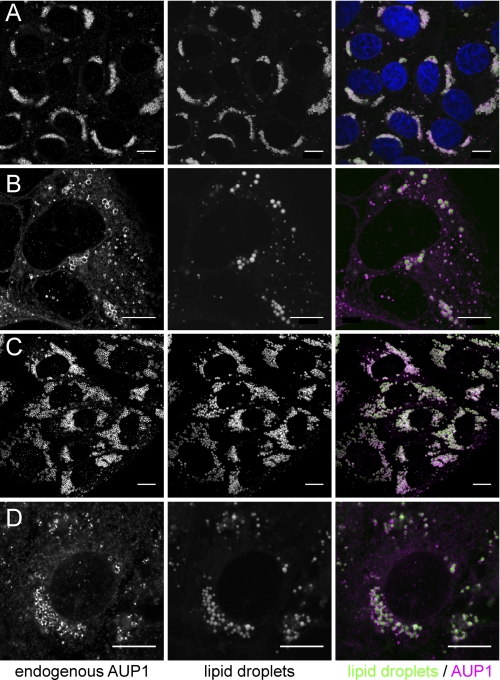
We raised a polyclonal antibody against human AUP1 and studied the subcellular localization of endogenous AUP1 in human Huh7 hepatoma cells, A431 fibroblasts, dog MDCK kidney epithelial cells, and monkey COS7 cells by confocal immunofluorescence microscopy (Fig. 2). In each cell line, a major fraction of the protein (Fig. 2, magenta) was found around LDs (Fig. 2, green), with the remainder on reticular structures likely to represent the ER. The localization and enrichment of AUP1 to LDs were further confirmed by immunoelectron microscopy (Fig. 3).
FIGURE 2.
Localization of endogenous AUP1 in A431, Huh7, MDCK, and COS7 cells. A431 (A), Huh7 (B), MDCK (C), and COS7 cells (D) were grown either with normal FCS (B) or supplemented with oleate (A, 20 μm; C, 100 μm; D, 20 μm), fixed, and stained with anti-AUP1 antibody. Lipid droplets were counterstained with Bodipy 493/503 (bar, 10 μm). A431 cells were additionally stained with DAPI to show the nuclei (top right image, blue).
FIGURE 3.
Localization of endogenous AUP1 in A431 cells by immunoelectron microscopy. Cells were cultured in regular growth medium supplemented with 50 μm oleic acid for 16 h. Cryosections were labeled using anti-AUP1 antibody followed by 10-nm protein A-gold detection. Lipid droplets were defined as large electron-lucent structures that lacked a limiting membrane bilayer with the lipid core often condensed or lost from the section. AUP1 was highly localized to the rim of lipid droplets (arrow heads), frequently found where lipid cores had been lost (black asterisks) and occasionally detected in close proximity to lipid droplets (white asterisks). Mitochondria (m), nuclei, and plasma membrane showed minor labeling only rarely. Bars, 2 μm (A), 200 nm (A′ and A″).
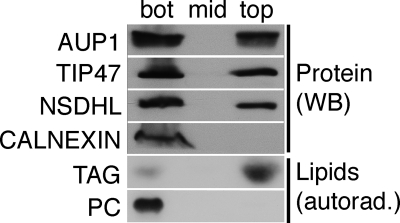
Morphological localization to the vicinity of LDs could represent direct localization to the LD surface but also to adjacent organelles, in particular to ER that frequently surrounds the LDs (4). To address this question, we subjected a homogenate of A431 cells to floatation in a sucrose gradient and analyzed fractions by Western blotting. About 40% of AUP1 was floating with the LD marker proteins NAD(P)H steroid dehydrogenase-like (NSDHL) and TIP47 to the top fraction, which in turn was completely devoid of immunoreactivity for the ER membrane protein calnexin (Fig. 4, Protein). This indicates localization of AUP1 directly to the LD surface, although one cannot exclude localization to a specialized ER domain that floats with LDs but is devoid of the ER marker calnexin. The purity of the LD preparation was further assessed by analysis of lipids that were labeled with radioactive oleic acid (Fig. 4, Lipids). The floating fraction contained about 80% of the radioactive triacylglyceride, but only 1.1% of the radioactive phosphatidylcholine, indicating little, if any, contamination of the LDs with other membranes. The distribution of AUP1 is consistent with the double localization of AUP1 to the surface of the LDs and to the ER, as determined by immunofluorescence and immunoelectron microscopy.
FIGURE 4.
AUP1 localizes to the LD fraction. A431 cells were labeled with radioactive oleic acid and fractionated in a sucrose gradient. Isolated LDs (top), middle (mid) and bottom (bot) fraction, were analyzed by Western blotting (WB). AUP1 localizes, similar to the LD marker proteins NSDHL and TIP47, to the LD and the bottom fraction. In contrast, the ER membrane protein calnexin was not detected in the LD fraction. The purity of the LD preparation was validated by the presence of radioactive triacylglycerol (TAG) and the absence of radioactive phosphatidylcholine (PC).
AUP1 Is a Monotopic Membrane Protein
Like several other integral proteins of LDs (19), AUP1 contains a single, very long hydrophobic stretch (Fig. 1, green) but no N-terminal signal sequence for ER translocation. This suggests a hairpin type of insertion into membrane mono- and bilayers with both termini facing the cytoplasm, as exemplified by caveolin or DGAT2 (43, 44).
To study the topology of the membrane insertion of AUP1, we applied microscopy-based approaches. The use of the very mild permeabilization reagent digitonin during staining of fixed cells allows conservation of internal membranes (45). Although only the plasma membrane was permeabilized, the HA tag of both N- or C-terminally HA-tagged AUP1 was recognized in fixed cells by staining with anti-HA antibody (Fig. 5A, AUP1). In contrast, the ER luminal protein-disulfide isomerase was only detected when cells were fully permeabilized with saponin (Fig. 5A, PDI).
In addition, we performed a fluorescence protease protection assay in living cells (46). COS7 cells were co-transfected with pDsRed2-ER, coding for DsRed2 with an N-terminal signal sequence and a C-terminal KDEL ER-retention motif, resulting in expression of luminal ER resident DsRed2, and N- or C-terminally EGFP-tagged constructs of AUP1. Again, the plasma membrane was permeabilized with digitonin, which did not affect the ER luminal DsRed2 signal or the AUP1-EGFP signal (Fig. 5B, before). Afterward, cells were incubated with proteinase K. Although both N- and C-terminal EGFP tags were degraded rapidly, the ER luminal signal stayed intact (Fig. 5B, 40 s). Results from fixed and living cells showed that both termini of AUP1 face the cytoplasm, providing evidence that AUP1 is another example of a monotopic hairpin membrane protein.
AUP1 Binds to the E2 Ubiquitin Conjugase Ube2g2 and Recruits It to LDs
Close to the C-terminal end, at amino acids 381–408, AUP1 has a short G2BR domain with strong homology to corresponding regions in the mammalian AMFR/gp78 protein and the yeast Cue1p protein (supplemental Fig. S1A and Fig. 1, yellow). For the two latter proteins, this stretch has been shown to mediate binding to the E2 ubiquitin conjugase Ube2g2 and or its yeast orthologue Ubc7p, respectively (38, 47), suggesting a similar interaction for AUP1. We therefore studied direct binding of a recombinant soluble C-terminal fragment AUP1(221–410) to recombinant human GST-Ube2g2 and GST-Ube2g1 by pulldown assays. AUP1(221–410) showed binding to GST-Ube2g2 (Fig. 6A, 1st lane) but no binding to the closely related GST-Ube2g1 or GST alone (Fig. 6A, 2nd and 3rd lanes, respectively).
FIGURE 6.
AUP1 binds Ube2g2 via its G2BR domain in vitro. A, GST pulldown, AUP1 specifically binds to Ube2g2. GST alone or fusion proteins of GST with full-length Ube2g2 or its closest homologue, Ube2g1, were incubated with recombinant His6-AUP1(221–410), comprising the soluble part of AUP1 (including G2BR domain). Complexes were precipitated with glutathione-Sepharose beads and analyzed by SDS-PAGE/Coomassie Brilliant Blue staining. The upper row shows the eluted GST fusion construct, and the lower row shows the co-eluted His6-AUP1(221–410). Note: upper row is a composite of two pieces from the same gel, because the control GST differs in molecular weight from the two fusion proteins. B, GST pulldown, G2BR domain of AUP1 is sufficient to bind Ube2g2. Fusion proteins of GST with wild-type G2BR of AUP1 (G2BR-wt) or with mutated G2BR of AUP1 (G2BR-M1) were incubated with MBP-Ube2g2. Complexes were precipitated with glutathione-Sepharose beads, eluted with free GSH, and analyzed by SDS-PAGE/Coomassie Brilliant Blue staining. Note that the free MBP, which contaminates the MBP-Ube2g2 fusion protein, did not bind to the GST-G2BR fusions.
To show that the G2BR domain of AUP1 alone is sufficient to bind Ube2g2, we also performed GST pulldown assays with the G2BR domain only. GST-G2BR efficiently pulled down MBP-Ube2g2 (Fig. 6B, 3rd lane). Based on the known contact residues of AMFR/gp78 and Ube2g2 (39), we generated mutations in the G2BR region of AUP1 (supplemental Fig. S1B) to interfere with the binding of AUP1 to Ube2g2. GST-G2BR-M1 containing the mutation ARRRFT to EGREDA was unable to pull down MBP-Ube2g2 (Fig. 6B, 4th lane), demonstrating the specificity of the direct binding between the G2BR domain of AUP1 and Ube2g2.
To show that the predicted interaction from in vitro experiments also occurs in a cellular context, we performed an immunoaffinity chromatography of a Triton X-100 detergent lysate from A431 cells on an anti-AUP1 antibody column (supplemental Fig. S2). Eluted bands that bound to the antibody column but not to empty beads were identified by mass spectrometry. Besides immunoglobulin chains, the only identified specifically bound proteins were AUP1 itself and Ube2g2 in an apparently stoichiometric ratio. Notably, no other E2 ubiquitin-conjugase was found, suggesting that Ube2g2 is the only E2 that is bound by AUP1.
To further study the interaction of AUP1 and Ube2g2 in living cells, we co-expressed either full-length AUP1(1–410) or a truncation mutant AUP1(1–362), lacking the G2BR domain, together with HA-tagged Ube2g2. To minimize interference from endogenous AUP1, we performed this experiment in COS7 cells, which have a relatively low expression of endogenous AUP1. In cells expressing full-length AUP1, both proteins co-localized to LDs (Fig. 7A). In contrast, in cells expressing truncated AUP1(1–362) (ΔG2BR), the 3HA-Ube2g2 was found all over the cytoplasm with no enrichment at LDs, although AUP1 was still localized to LDs (Fig. 7A). Similar to the results of in vitro pulldowns, mutations (pAUP1-M1-EGFP) or deletions (pAUP1-M2-EGFP) within the G2BR domain of AUP1 strongly reduced the recruitment of Ube2g2 to LDs in living cells (Fig. 7B). Taken together, AUP1 specifically binds Ube2g2 by its C-terminal G2BR domain and recruits Ube2g2 to LDs in living cells.
FIGURE 7.
AUP1 recruits Ube2g2 to LDs in vivo. A, binding to the G2BR domain of AUP1 recruits Ube2g2 to LDs. COS7 cells were co-transfected with HA-tagged Ube2g2 and either truncated AUP1(1–362), lacking the C-terminal G2BR domain (upper row), or full-length AUP1(1–410) (lower row). Cells were stained with anti-AUP1 (green) and anti-HA to detect Ube2g2 (magenta). Note: truncated AUP1(1–362) localizes to LDs (top left panel) but is unable to recruit Ube2g2, which shows a cytoplasmic staining (top middle panel). In contrast, full-length AUP1(1–410) recruits Ube2g2 to the LDs (lower middle panel). B, mutations in the AUP1 G2BR domain abolish recruitment of Ube2g2 to LDs. COS7 cells were co-transfected with HA-tagged Ube2g2 and either wild-type AUP1-EGFP (AUP1-WT) or AUP1-EGFP bearing mutations (AUP1-M1) or a truncation (AUP1-M2) in the G2BR domain. AUP1 was detected by the GFP fluorescence (lower row), and Ube2g2 was detected by staining with anti-HA (upper row).
DISCUSSION
We identified and characterized AUP1 as an LD protein that provides a molecular connection of LDs to the ubiquitination machinery. Although AUP1 is highly conserved and ubiquitously expressed, little is currently known about the protein. In recent years, AUP1 was found in several LD proteomes (22, 23, 26). Our subcellular fractionation and microscopic localization data demonstrate that AUP1 is a specific component of LDs in several different cell types. The failure to detect AUP1 localization to LDs in former studies (30, 32, 48) is likely due to the cell lines used in those studies and their respective metabolic state. Although some cell lines, e.g. HeLa cells, contain only few and small LDs in the absence of oleate supplementation, our standard cell lines, A431 and Huh7, contain easily detectable LDs under normal growth conditions. Because AUP1 can dually localize to LDs and ER membranes, it will be found on ER membranes in the absence of LDs. Similarly, overexpressed AUP1 tends to localize to the ER because the available LD surface cannot cope with large amounts of AUP1 protein, especially in cells with few or small LDs (see AUP1 in COS7 cells under FCS conditions in Fig. 5A). Another reason for the previous failure to detect AUP1 at LDs may be differences in the fixation and permeabilization procedure before microscopy. It was shown (49, 50) that some LD proteins are not detected on LDs when cells are treated with Triton X-100 for permeabilization.
It has been reported previously that LDs are connected to ubiquitination and protein degradation. Some peripheral LD proteins such as adipophilin and perilipin are known to be ubiquitinated and degraded via the proteasome (51–53). Whether integral LD proteins such as caveolin, DGAT2, or NSDHL use similar mechanisms is unclear. Recently, the importance of E3 ligases for LD turnover was reported, but further molecular components or mechanistic details are lacking (54). Apolipoprotein B accumulates in crescent-shaped structures around LDs in hepatocytes after inhibition of the proteasome or autophagy (7, 55), suggesting a role of LDs in degradation of excess apolipoproteins. In support of this function of LDs, a recent study (56) demonstrated accumulation of hydroxymethylglutaryl-CoA reductase around LDs under conditions of impaired proteasomal degradation.
AUP1 provides a direct molecular connection of LDs to the ubiquitination machinery by binding and recruiting the E2 conjugase Ube2g2 (Fig. 8). Remarkably, Ube2g2 is the E2 conjugase that plays a central role in the ERAD process. Whether the AUP1-Ube2g2 complexes on LDs participate in an ERAD-like process now has to be addressed by further functional studies. Being specifically concentrated on LDs, AUP1 may participate in the regulation of LDs by linking LD turnover to the proteasomal degradation machinery in a cell. Recent data support this concept of AUP1 activity, because AUP1 was found in a complex with Sel1l, a component of the Hrd1-ligase complex, participating in the dislocation of misfolded glycoproteins on the ER (32).
FIGURE 8.
AUP1-Ube2g2 complex at lipid droplets. AUP1 localizes in a monotopic fashion to LDs and binds Ube2g2 via its G2BR domain. Thereby, AUP1 recruits Ube2g2 to the surface of LDs. Blue, phospholipid monolayer; yellow, neutral lipid core. Note that the modules are not drawn to scale.
The assembly of a functional ubiquitination complex at the LD surface should require the presence of an E3 ligase, in addition to the AUP1-Ube2g2 complex. So far, LD proteome analyses have not revealed a candidate E3 ligase. Ube2g2 and its yeast homologue Ubc7p can assemble pre-formed polyubiquitin chains that are transferred to the substrate protein by its cognate E3 ligases AMFR/gp78 or Hrd1p, respectively (47, 57, 58). The formation of polyubiquitin chains on Ube2g2 depends on the G2BR of AMFR/gp78 (59), which turns out to be an allosteric activator of Ube2g2 transfer activity (39). Although the G2BR of AUP1 may functionally replace the G2BR of AMFR/gp78, there is no RING finger domain on AUP1 that might catalyze the actual ubiquitin chain transfer. The missing E3 may either be an (overlooked) LD protein or a cytoplasmic protein or a component of the ER that closely associates with LDs. Further studies are necessary to elucidate its identity.
In summary, we demonstrate that AUP1 is a monotopic membrane protein that localizes to lipid droplets. AUP1 binds the E2-conjugase Ube2g2 via its G2BR domain and recruits it to LDs. Further understanding of AUP1, its control by upstream signaling events, and its role in Ube2g2-dependent ubiquitination will improve our understanding of LD turnover and the role of LDs in protein degradation.
Supplementary Material
Acknowledgments
We gratefully acknowledge support by Dr. Jan Peychl, light microscopy facility; Dr. Anna Shevchenko, mass spectrometry facility; and Dr. Bianca Habermann, bioinformatic facility, at the Max-Planck-Institute of Molecular Cell Biology and Genetics, Dresden, Germany. MDCK cells were kindly provided by Dr. Sabine Buschhorn. We also thank Drs. Kaupp and Irsen for providing the possibility to perform EM analyses at the Center of Advanced European Studies and Research (Caesar), Bonn, Germany.
This work was supported by the German National Academic Foundation (to J. S.), Deutsche Forschungsgemeinschaft Grants TR83 (to C. M.) and SFB645 (to D. L.), and European Union Grant FP7 LipdomicNet (to C. T.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- LD
- lipid droplet
- CUE
- coupling of ubiquitin to endoplasmic-associated degradation
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic-associated protein degradation
- G2BR
- G2-binding region
- GST
- glutathione S-transferase
- MBP
- maltose-binding protein
- NSDHL
- NAD(P)H steroid dehydrogenase-like
- MDCK
- Madin-Darby canine kidney cell
- EGFP
- enhanced GFP.
REFERENCES
- 1. Murphy D. J. (2001) Prog. Lipid Res. 40, 325–438 [DOI] [PubMed] [Google Scholar]
- 2. Londos C., Brasaemle D. L., Schultz C. J., Segrest J. P., Kimmel A. R. (1999) Semin. Cell Dev. Biol. 10, 51–58 [DOI] [PubMed] [Google Scholar]
- 3. Murphy D. J., Vance J. (1999) Trends Biochem. Sci. 24, 109–115 [DOI] [PubMed] [Google Scholar]
- 4. Blanchette-Mackie E. J., Dwyer N. K., Barber T., Coxey R. A., Takeda T., Rondinone C. M., Theodorakis J. L., Greenberg A. S., Londos C. (1995) J. Lipid Res. 36, 1211–1226 [PubMed] [Google Scholar]
- 5. Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. (2002) J. Biol. Chem. 277, 44507–44512 [DOI] [PubMed] [Google Scholar]
- 6. Welte M. A. (2007) Trends Cell Biol. 17, 363–369 [DOI] [PubMed] [Google Scholar]
- 7. Ohsaki Y., Cheng J., Fujita A., Tokumoto T., Fujimoto T. (2006) Mol. Biol. Cell 17, 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martin S., Parton R. G. (2006) Nat. Rev. Mol. Cell Biol. 7, 373–378 [DOI] [PubMed] [Google Scholar]
- 9. Murphy S., Martin S., Parton R. G. (2009) Biochim. Biophys. Acta 1791, 441–447 [DOI] [PubMed] [Google Scholar]
- 10. Turró S., Ingelmo-Torres M., Estanyol J. M., Tebar F., Fernndez M. A., Albor C. V., Gaus K., Grewal T., Enrich C., Pol A. (2006) Traffic 7, 1254–1269 [DOI] [PubMed] [Google Scholar]
- 11. Ozeki S., Cheng J., Tauchi-Sato K., Hatano N., Taniguchi H., Fujimoto T. (2005) J. Cell Sci. 118, 2601–2611 [DOI] [PubMed] [Google Scholar]
- 12. Binns D., Januszewski T., Chen Y., Hill J., Markin V. S., Zhao Y., Gilpin C., Chapman K. D., Anderson R. G., Goodman J. M. (2006) J. Cell Biol. 173, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schrader M. (2001) J. Histochem. Cytochem. 49, 1421–1429 [DOI] [PubMed] [Google Scholar]
- 14. Liu P., Bartz R., Zehmer J. K., Ying Y. S., Zhu M., Serrero G., Anderson R. G. (2007) Biochim. Biophys. Acta 1773, 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le Lay S., Dugail I. (2009) Prog. Lipid Res. 48, 191–195 [DOI] [PubMed] [Google Scholar]
- 16. Guo Y., Cordes K. R., Farese R. V., Jr., Walther T. C. (2009) J. Cell Sci. 122, 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walther T. C., Farese R. V., Jr. (2009) Biochim. Biophys. Acta 1791, 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goodman J. M. (2008) J. Biol. Chem. 283, 28005–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thiele C., Spandl J. (2008) Curr. Opin. Cell Biol. 20, 378–385 [DOI] [PubMed] [Google Scholar]
- 20. Ohsaki Y., Cheng J., Suzuki M., Shinohara Y., Fujita A., Fujimoto T. (2009) Biochim. Biophys. Acta 1791, 399–407 [DOI] [PubMed] [Google Scholar]
- 21. Athenstaedt K., Zweytick D., Jandrositz A., Kohlwein S. D., Daum G. (1999) J. Bacteriol. 181, 6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wan H. C., Melo R. C., Jin Z., Dvorak A. M., Weller P. F. (2007) FASEB J. 21, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brasaemle D. L., Dolios G., Shapiro L., Wang R. (2004) J. Biol. Chem. 279, 46835–46842 [DOI] [PubMed] [Google Scholar]
- 24. Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., Takano T. (2004) Biochim. Biophys. Acta 1644, 47–59 [DOI] [PubMed] [Google Scholar]
- 25. Bartz R., Zehmer J. K., Zhu M., Chen Y., Serrero G., Zhao Y., Liu P. (2007) J. Proteome Res. 6, 3256–3265 [DOI] [PubMed] [Google Scholar]
- 26. Sato S., Fukasawa M., Yamakawa Y., Natsume T., Suzuki T., Shoji I., Aizaki H., Miyamura T., Nishijima M. (2006) J. Biochem. 139, 921–930 [DOI] [PubMed] [Google Scholar]
- 27. Jang W., Weber J. S., Bashir R., Bushby K., Meisler M. H. (1996) Genomics 36, 366–368 [DOI] [PubMed] [Google Scholar]
- 28. Karpisheva K. V., Touber B., Kisliakova T. V., Dobner T. (2002) Tsitologiia 44, 839–845 [PubMed] [Google Scholar]
- 29. Karpisheva K. V., Tauber B., Kisliakova T. V., Dobner T. (2002) Tsitologiia 44, 830–838 [PubMed] [Google Scholar]
- 30. Kato A., Kawamata N., Tamayose K., Egashira M., Miura R., Fujimura T., Murayama K., Oshimi K. (2002) J. Biol. Chem. 277, 28934–28941 [DOI] [PubMed] [Google Scholar]
- 31. Kato A., Oshimi K. (2009) Platelets 20, 105–110 [DOI] [PubMed] [Google Scholar]
- 32. Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Claessen J. H., Mueller B., Spooner E., Pivorunas V. L., Ploegh H. L. (2010) J. Biol. Chem. 285, 20732–20739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lewin T. M., Wang P., Coleman R. A. (1999) Biochemistry 38, 5764–5771 [DOI] [PubMed] [Google Scholar]
- 35. Shih S. C., Prag G., Francis S. A., Sutanto M. A., Hurley J. H., Hicke L. (2003) EMBO J. 22, 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prag G., Misra S., Jones E. A., Ghirlando R., Davies B. A., Horazdovsky B. F., Hurley J. H. (2003) Cell 113, 609–620 [DOI] [PubMed] [Google Scholar]
- 37. Kang R. S., Daniels C. M., Francis S. A., Shih S. C., Salerno W. J., Hicke L., Radhakrishnan I. (2003) Cell 113, 621–630 [DOI] [PubMed] [Google Scholar]
- 38. Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Das R., Mariano J., Tsai Y. C., Kalathur R. C., Kostova Z., Li J., Tarasov S. G., McFeeters R. L., Altieri A. S., Ji X., Byrd R. A., Weissman A. M. (2009) Mol. Cell 34, 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ponting C. P. (2000) Biochem. J. 351, 527–535 [PMC free article] [PubMed] [Google Scholar]
- 41. Hampton R. Y., Bhakta H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12944–12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song B. L., Sever N., DeBose-Boyd R. A. (2005) Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 43. Glenney J. R., Jr., Soppet D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10517–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stone S. J., Levin M. C., Farese R. V., Jr. (2006) J. Biol. Chem. 281, 40273–40282 [DOI] [PubMed] [Google Scholar]
- 45. Plutner H., Davidson H. W., Saraste J., Balch W. E. (1992) J. Cell Biol. 119, 1097–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lorenz H., Hailey D. W., Lippincott-Schwartz J. (2006) Nat. Methods 3, 205–210 [DOI] [PubMed] [Google Scholar]
- 47. Bazirgan O. A., Hampton R. Y. (2008) J. Biol. Chem. 283, 12797–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karpisheva K. V., Tauber B., Kisliakova T. V., Dobner T. (2002) Tsitologiia 44, 846–851 [PubMed] [Google Scholar]
- 49. DiDonato D., Brasaemle D. L. (2003) J. Histochem. Cytochem. 51, 773–780 [DOI] [PubMed] [Google Scholar]
- 50. Ohsaki Y., Maeda T., Fujimoto T. (2005) Histochem. Cell Biol. 124, 445–452 [DOI] [PubMed] [Google Scholar]
- 51. Xu G., Sztalryd C., Lu X., Tansey J. T., Gan J., Dorward H., Kimmel A. R., Londos C. (2005) J. Biol. Chem. 280, 42841–42847 [DOI] [PubMed] [Google Scholar]
- 52. Xu G., Sztalryd C., Londos C. (2006) Biochim. Biophys. Acta 1761, 83–90 [DOI] [PubMed] [Google Scholar]
- 53. Masuda Y., Itabe H., Odaki M., Hama K., Fujimoto Y., Mori M., Sasabe N., Aoki J., Arai H., Takano T. (2006) J. Lipid Res. 47, 87–98 [DOI] [PubMed] [Google Scholar]
- 54. Eastman S. W., Yassaee M., Bieniasz P. D. (2009) J. Cell Biol. 184, 881–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fujimoto T., Ohsaki Y. (2006) Autophagy 2, 299–301 [DOI] [PubMed] [Google Scholar]
- 56. Hartman I. Z., Liu P., Zehmer J. K., Luby-Phelps K., Jo Y., Anderson R. G., DeBose-Boyd R. A. (2010) J. Biol. Chem. 285, 19288–19298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li W., Tu D., Brunger A. T., Ye Y. (2007) Nature 446, 333–337 [DOI] [PubMed] [Google Scholar]
- 58. Ravid T., Hochstrasser M. (2007) Nat. Cell Biol. 9, 422–427 [DOI] [PubMed] [Google Scholar]
- 59. Li W., Tu D., Li L., Wollert T., Ghirlando R., Brunger A. T., Ye Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3722–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.