Abstract
Mitochondrial cytochrome P450 11A1 (CYP11A1 or P450 11A1) is the only known enzyme that cleaves the side chain of cholesterol, yielding pregnenolone, the precursor of all steroid hormones. Pregnenolone is formed via three sequential monooxygenation reactions that involve the progressive production of 22R-hydroxycholesterol (22HC) and 20α,22R-dihydroxycholesterol, followed by the cleavage of the C20-C22 bond. Herein, we present the 2.5-Å crystal structure of CYP11A1 in complex with the first reaction intermediate, 22HC. The active site cavity in CYP11A1 represents a long curved tube that extends from the protein surface to the heme group, the site of catalysis. 22HC occupies two-thirds of the cavity with the 22R-hydroxyl group nearest the heme, 2.56 Å from the iron. The space at the entrance to the active site is not taken up by 22HC but filled with ordered water molecules. The network formed by these water molecules allows the “soft” recognition of the 22HC 3β-hydroxyl. Such a mode of 22HC binding suggests shuttling of the sterol intermediates between the active site entrance and the heme group during the three-step reaction. Translational freedom of 22HC and torsional motion of its aliphatic tail are supported by solution studies. The CYP11A1–22HC co-complex also provides insight into the structural basis of the strict substrate specificity and high catalytic efficiency of the enzyme and highlights conserved structural motifs involved in redox partner interactions by mitochondrial P450s.
Keywords: Cholesterol, Cholesterol-binding Protein, Cholesterol Metabolism, Crystal Structure, Cytochrome P450, Enzyme Mechanisms, Enzyme Structure, Hormones, Membrane Proteins
Introduction
Steroid hormones are essential for life. In vertebrates, they are all synthesized from pregnenolone, which is in turn formed from cholesterol via a three-step process catalyzed by cytochrome P450 11A1 (CYP11A1) (1). During the first step, cholesterol is converted to 22R-hydroxycholesterol (22HC),3 the second step produces 20α,22R-dihydroxycholesterol (20,22DHC), and the third step involves the cleavage of the C20-C22 bond in 20,22DHC to yield pregnenolone (supplemental Fig. S1A) (2). Sterol intermediates do not accumulate during the conversion of cholesterol to pregnenolone and bind much more tightly to CYP11A1 than cholesterol, suggesting that they remain in the active site until all three oxidative steps are completed (1, 3, 4). All enzymatic steps take place in the inner mitochondrial membrane of steroidogenic tissues and require a total of 3 mol of O2, 3 mol of NADPH, and six electrons transferred separately from NADPH to adrenodoxin reductase, adrenodoxin (Adx), and CYP11A1 (supplemental Fig. S1B). CYP11A1 is an important enzyme whose deficiency leads to lipoid congenital adrenal hyperplasia, a lethal disease if untreated (5). Although this P450 has been the subject of extensive studies (6), the key amino acid residues involved in substrate recognition and catalysis have not yet been identified, and the mechanism by which CYP11A1 carries out three sequential monooxygenation reactions with the intermediates not leaving the enzyme active site remains unclear.
The significance of CYP11A1 extends beyond involvement in biosynthesis of steroid hormones. This hemoprotein is among the only four enzymes in vertebrates (CYP7A1, CYP27A1, CYP46A1, and CYP11A1) that initiate biotransformation of cholesterol in different organs and thus play important roles in the maintenance of cholesterol homeostasis (7). Cholesterol-metabolizing P450s share low amino acid sequence identity (<25%) and produce different cholesterol metabolites (5) yet bind cholesterol tightly and represent a unique system to study enzyme adaption to physiological requirements of different organs for cholesterol turnover rate. Previously, we determined crystal structures of ligand-free and sterol-bound CYP46A1 (8), a microsomal monooxygenase that controls cholesterol turnover in the brain (9). In this study, we structurally characterize sterol-bound CYP11A1. The crystal structure reveals specific interactions between the enzyme and the intermediate substrate 22HC and implies a unique mechanism for substrate retention underlying the three-step catalysis. Comparison with CYP46A1 reveals the diversity of cholesterol-binding modes by different cholesterol hydroxylases. Comparison with CYP24A1, the first structurally characterized mitochondrial P450 (10), elucidates the mechanism of closure of the active site, demonstrates a similar orientation for P450 insertion into the lipid bilayer, and confirms the role of conserved structural elements in interaction with the redox partner, Adx.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
Full-length mature bovine CYP11A1 with a His4 tag at the C terminus was expressed as described previously (11) and purified using five consecutive chromatography steps on DEAE-cellulose, nickel-agarose, hydroxylapatite, Adx-Sepharose, and SP-Sepharose with 50 μm 22HC and 5 mm CHAPS present in all the buffers. During the last chromatography step, CHAPS was replaced with 2.8 mm octyl pentaethylene glycol ether (C8E5). Crystals of CYP11A1 were grown at 18 °C overnight in sitting drops after 1 μl of CYP11A1 solution was mixed with 1 μl of precipitant solution. The P450 solution contained ∼23 mg/ml CYP11A1, 50 mm potassium phosphate buffer (pH 7.2), 20% glycerol, 0.1 m NaCl, 0.1% C8E5, 50 μm 22HC, and 1 mm EDTA. The precipitant solution was 14% PEG 1000, 20% glycerol, 12% JEFFAMINE ED-2001, 0.1 m MES (pH 7.0), and 10% isopropyl alcohol.
Structure Determination
The structure was determined by molecular replacement using rat mitochondrial CYP24A1 (Protein Data Bank code 3K9V) as a search model and 2.8-Å resolution data for CYP11A1 in space group P32212. The structure was refined against a 2.5-Å resolution data set for CYP11A1 in space group P21 (supplemental Table S1 and Figs. S2 and S3). Coordinates and structure factors have been deposited with the Research Collaboratory for Structural Bioinformatics (Protein Data Bank code 3MZS).
Solution Studies
Reduced CO difference spectra were recorded as described (12) under anaerobic conditions in 50 mm potassium phosphate buffer (pH 7.2) containing an oxygen-scavenging system (60 mm glucose, 2000 units/ml catalase, and 30 units/ml glucose oxidase). The buffer was degassed and bubbled with nitrogen prior to the addition of the oxygen-scavenging system and CYP11A1. Reduced Adx was obtained by addition of an equimolar amount of sodium dithionite to oxidized protein. When reduced Adx was mixed with reduced CYP11A1, the latter was reduced prior to addition of the former by addition of an equimolar amount of sodium dithionite.
RESULTS AND DISCUSSION
Binding of 22HC to CYP11A1
The CYP11A1–22HC complex crystallized in space group P21 with four copies in the asymmetric unit (a D2 symmetric tetramer with root mean square deviation ∼0.5 Å between component monomers) (supplemental Table S1). The overall fold and details of the active site are similar in all copies, including well defined electron density for 22HC allowing unambiguous fitting of the substrate (supplemental Figs. S2 and S3). Ninety-two H2O molecules were resolved, most within the active site cavity.
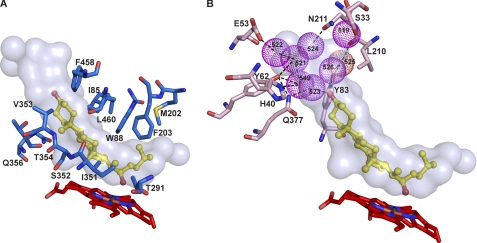
The CYP11A1 structure displays the canonical P450 fold with two additional helices, A′ and K″, which are also present in mitochondrial P450 CYP24A1 (10). The active site cavity in CYP11A1 is closed and resembles a banana-shaped tunnel (Fig. 1A). 22HC occupies only two-thirds of the volume of the tunnel, with the sterol β-surface facing the distal side of the heme. The aliphatic tail of 22HC is centered over the heme iron, and the 3β-hydroxyl is directed toward the aqueous cavity. Thirty-nine residues from 14 different secondary structural elements define the active site cavity in CYP11A1. Yet only 12 amino acid residues are within 4 Å of 22HC. The aliphatic tail of 22HC makes van der Waals contacts with the side chains of Ile-351 (β1–4 strand region), Thr-291 (I helix), Phe-203 (F helix), and Met-202 (F helix). The sterol α-face is surrounded by the side chains of Ile-85 (B-B′ loop), Leu-460 (β4-1/4-2 loop), and Trp-88 (B′ helix), and the β-face is defined by Gln-356, Thr-354, Val-353, and Ser-352 (all residues are from the β1–4 strand region). In addition, Phe-458 (β4–1 strand) restrains the edge of sterol ring A. Collectively, the active site amino acid residues form a cavity whose shape is in good agreement with previous studies on CYP11A1 substrate specificity. The enzyme active site was shown to have strict requirements for the planarity of the sterol backbone, extra space beyond the 3β-hydroxyl and aliphatic tail, and limited space at carbon atoms 4–6 and 17 (13–22).
FIGURE 1.
Views of the CYP11A1 active site illustrating interactions with 22HC. A, amino acid residues (marine) within 4 Å of 22HC. B, array of structural water molecules (violet spheres) hydrogen-bonded to the 22HC 3β-hydroxyl and to the residues (pink) at the entrance to the active site. For clarity, residues interacting with 22HC are omitted in this panel. The heme group is depicted in red, and 22HC is depicted in yellow. The nitrogen, oxygen, sulfur, and iron atoms are in blue, red, yellow, and orange, respectively. The enclosed volume of the active site is shown as a semitransparent surface.
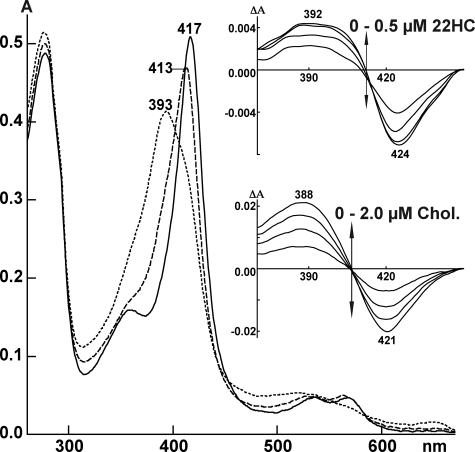
In the crystal structure, the 22R-hydroxyl is located above the heme tetrapyrrole with the Fe-O22 distance (2.56 Å) within the reported errors for a coordinate bond (23), yet, in solution, 22HC elicits a partial type 1 spectral response in the CYP11A1 absolute spectrum and predominantly a type 1 response in the difference spectrum (Fig. 2), indicative of pentacoordinate iron with no distal coordination. This apparent discrepancy could be explained as follows. The CYP11A1–22HC complex is sampling more than one spin state population, some characterized by the distal iron coordination, whereas others are not. The population of these spin states would conform to Boltzmann statistics and is temperature-dependent. Because the x-ray and spectral data were collected at two different temperatures (100 K and ambient, respectively), the coordinated state is captured in the crystal structure, whereas at ambient temperature, the ligand is more mobile, affording the partial type 1 spectra. If this is the case, coordination of the heme iron explains a much tighter binding of 22HC relative to cholesterol and its stereoisomer 22S-hydroxycholesterol (3, 4) but does not support the existence of an active site residue that interacts with the 22R-hydroxyl by hydrogen bonding proposed by previous studies (4). There are no amino acid residues or water molecules closer than 4.0 Å from the 22R-hydroxyl in the crystal structure of CYP11A1.
FIGURE 2.
Absolute and difference (insets) spectra of CYP11A1 in the absence (solid line) and presence of 22HC (dashed line) and cholesterol (dotted line). Absolute spectra were recorded using 5 μm CYP11A1 with the concentrations of 22HC and cholesterol being 50 μm. Both sterols were added from 10 mm stocks in 45% aqueous 2-hydroxypropyl-β-cyclodextrin. The buffer was 50 mm potassium phosphate (pH 7.2) containing 1 mm EDTA. Difference spectra in the insets were recorded using 0.4 μm CYP11A1. 22HC was added from 0.1 mm stock in 4.5% aqueous 2-hydroxypropyl-β-cyclodextrin, and cholesterol (Chol.) was added from 0.5 mm stock 4.5% aqueous 2-hydroxypropyl-β-cyclodextrin.
A remarkable feature of the CYP11A1 co-complex is the nature of the interactions with the sterol 3β-hydroxyl. Although this group has no direct contacts with the protein, it forms two hydrogen bonds with crystallographically observed waters Wat-523 and Wat-526, which are hydrogen-bonded to the side chain of Gln-377 and Wat-525, respectively (Fig. 1B). Gln-377 participates in an extensive hydrogen bond network with the Tyr-62 hydroxyl, Wat-549, Wat-521, Wat-522, Glu-53, and the His-40 imidazole, whereas Wat-525 interacts with the Leu-210 carbonyl and Tyr-83 hydroxyl (waters are found in similar positions in all four monomers) (supplemental Fig. S4). We propose that these water-mediated interactions with substrate are the mechanism by which CYP11A1 recognizes substrate; hence, we term it “soft” recognition. Portions of the enclosed space at the active site entrance not exhibiting ordered solvent are presumed to contain water, as they are also surrounded by polar residues. Altogether, the volume unoccupied by 22HC at the active site entrance accounts for ∼28% of the total volume of the active site cavity (625 Å3), which is sufficient to accommodate up to 15 waters at one time. The presence of the water array at the active site entrance in all four copies suggests that hydration of this part of the active site is important for CYP11A1 function. In particular, the lack of specific recognition of the 3β-hydroxyl and the presence of water-mediated interactions with the protein instead suggest that the substrate can exhibit mobility while remaining bound within the enzyme. The shape of the active site implies that substrate movement is translational such that the intermediates move toward or away from the heme. Translational freedom of the entire substrate, together with torsional changes in the aliphatic tail, could position the C20 or C22 atoms for stereospecific sequential oxidation reactions.
Mechanistic Implications of the CYP11A1 Structure
The CYP11A1–22HC complex likely reflects sterol binding at the end of the first monooxygenation cycle (supplemental Fig. S1B). During the next cycle, CYP11A1 must be reduced by its redox partner, Adx, and the aliphatic tail of 22HC must change position so that molecular oxygen can bind to the heme iron. We propose that interaction with reduced Adx induces conformational changes in the P450 that push 22HC away from the heme iron, displacing the intermediate toward the entrance to the active site. This translational mode of movement of 22HC would create the space above the iron necessary for the binding of molecular oxygen. During subsequent electron transfer from reduced Adx and oxygen binding to the iron, 22HC would remain displaced toward the entrance of the active site. However, after formation of the reactive oxyferryl intermediate and dissociation of Adx, movement of 22HC back toward the heme could position the aliphatic tail for hydroxylation at C20. Similar motions of 20,22DHC could occur during the third monooxygenation cycle, leading to formation of pregnenolone.
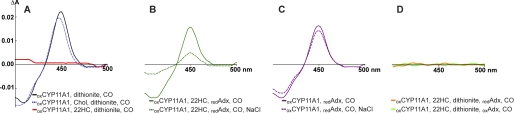
To test our hypothesis about sterol shuttling during the catalytic cycle, we capitalized on the ability of reduced P450s to form a complex with CO that has a characteristic absorption maximum at 450 nm (supplemental Fig. S1B) (12). A solution containing ligand-free CYP11A1 and an oxygen-scavenging system was reduced chemically with sodium dithionite and divided into two cuvettes, and CO was bubbled through one cuvette. The recorded difference spectrum showed a peak at 450 nm (Fig. 3A, black line), indicating that reduced CYP11A1 has space near the heme iron to bind CO, as expected. When repeated with an equimolar amount of 22HC added to CYP11A1 prior to reduction and bubbling with CO, no peak at 450 nm was observed in the CO spectrum (Fig. 3A, red line). This result is consistent with the close proximity of the C22 hydroxyl group to iron (Fig. 1), leaving no space near the iron for CO to bind, and with prior studies by others showing that the Kd value for CO increases >300-fold when CYP11A1 interacts with 22HC (24). A control experiment with cholesterol (10-fold molar excess over CYP11A1) added prior to sodium dithionite and bubbling with CO showed a peak at 450 nm, thus demonstrating that it is the 22R-hydroxyl that blocks CO access to the heme iron in the CYP11A1–22HC complex (Fig. 3A, dashed blue line). In the next series of experiments, CYP11A1 was reduced with Adx. The enzyme was first sequentially mixed with equimolar 22HC and a 5-fold molar excess of reduced Adx, followed by CO bubbling of one of the cuvettes. The CO spectrum was restored (Fig. 3B, green solid line) as compared with the chemically reduced CYP11A1–22HC complex (Fig. 3A, red line), indicating that binding of reduced Adx alters the position of 22HC and creates space above the iron for CO. Then 1 m NaCl was added to both cuvettes to induce dissociation of Adx from CYP11A1 (interactions of Adx with CYP11A1 are mainly electrostatic (25)). The peak at 450 nm decreased significantly (Fig. 3B, green dashed line) yet did not disappear completely. After correcting for dilution, the decrease in absorbance at 450 nm was 60%. Three control experiments were then carried out. The first control experiment was with addition of 1 m NaCl to the solution of CYP11A1 mixed with reduced Adx but no 22HC present. This experiment showed only a decrease in the 450 nm peak corresponding to the dilution (Fig. 3C, solid and dashed magenta lines). In the second and third control experiments, CYP11A1 in complex with 22HC was reduced with sodium dithionite, followed by addition of reduced and oxidized Adx, respectively, and CO bubbling. No peaks at 450 nm were observed (Fig. 3D, orange and green lines), indicating a lack of complex formation. However, according to the calculated Kd values of reduced and oxidized Adx for cholesterol-bound reduced CYP11A1 (250 nm and 120 nm, respectively), the formation of these complexes should occur under the experimental conditions used (26). It is possible that the affinities of reduced and oxidized Adx are different for cholesterol-bound CYP11A1 relative to 22HC-bound P450. Also, these affinities should be determined experimentally to assess the values calculated previously (26).
FIGURE 3.
Binding of CO to reduced CYP11A1 as assessed by difference spectroscopy (Fe2+-CO versus Fe2+-P450 spectrum). By utilizing different assay conditions, we show that blockage of CO binding by 22HC is relieved by Adx binding, and that upon release of Adx, 22HC is able to significantly displace bound CO. The same concentration of P450 (0.25 μm) was used in each experiment, and the order in which reagents were mixed is indicated; the ox and red subscripts specify whether CYP11A1 and Adx are in oxidized and reduced states, respectively. A, reduction of CYP11A1 with sodium dithionite in the absence (black line) and presence (red line) of equimolar 22HC and a 10-fold molar excess of cholesterol (Chol; dashed blue line). B, reduction of CYP11A1 with reduced Adx in the presence of equimolar 22HC (solid green line) and in the presence of equimolar 22HC followed by addition of 1 m NaCl (dashed green line). C, control experiment in which CYP11A1 was treated either with reduced Adx only (solid magenta line) or with reduced Adx followed by addition of 1 m NaCl (dashed magenta line). D, control experiment in which the CYP11A1–22HC complex was reduced with sodium dithionite, followed by addition of reduced (orange line) or oxidized (green line) Adx.
To summarize, by using CO as a probe for the space above the heme iron, we demonstrate that binding of Adx indeed shifts the 22HC molecule, which can then displace bound CO upon dissociation of Adx. These results provide evidence for motion of 22HC both away from and toward the heme, in support of soft recognition. Our hypothesis is in agreement with previous studies showing mutually facilitated binding of cholesterol and Adx to CYP11A1 as assessed by the absorbance changes in the Soret region (27) and studies by electron paramagnetic resonance indicating reorientation of cholesterol in the active site of oxidized CYP11A1 upon interaction with reduced Adx (28). Yet our proposed model is not consistent with a study on oxygen interaction with the reduced CYP11A1, which demonstrates that oxygen can bind to the P450 without binding of Adx (24). We note, however, that the steric clash of bound CO with 22HC would be considerably more severe than for O2 because of the linear coordination of CO (Fe-C-O angle of 180°) versus the bent coordination of O2 (Fe-O-O angle of 109°).
Comparison of Bovine CYP11A1 Bound to 22HC and Human CYP11A1 Bound to 20,22DHC
While this manuscript was in preparation, the coordinates of the 2.5-Å crystal structure of human CYP11A1 in complex with 20,22DHC (Protein Data Bank code 3NA0) were released by the Structural Genomic Consortium (University of Toronto). Superposition of this structure onto our structure of bovine CYP11A1 bound to 22HC (code 3MZS) shows a root mean square deviation of 0.95 Å for all atoms of 462 aligned residues (3872 atoms of 3NA0 and 3904 atoms of 3MZS). The Cα atom traces of the two structures are very similar, as are the positions of the substrates and substrate contact residues (supplemental Fig. S5, A–D) Similarities also extend to the shape (supplemental Fig. S5C) and volume (624 and 625 Å3 in 3NA0 and 3MZS, respectively) of the active site and the presence and position of ordered water molecules at the entrance to the active site, six waters in 3NA0 and seven waters in 3MZS (supplemental Fig. S5D).
Like the 3β-hydroxyl in 22HC, the 3β-hydroxyl in 20,22DHC has no direct contacts with the protein and is hydrogen-bonded to two H2O molecules, Wat-42 and Wat-599, which are water-bridged to the side chains of His-78, Tyr-100, and Gln-416 and the Glu-91 carbonyl (supplemental Fig. S5D), analogs of His-40, Tyr-62, Gln-377, and Glu-53, which participate in the interactions with the 3β-hydroxyl in 22HC. (The two CYP11A1 structures have different numbering of amino acid residues; numbering in 3NA0 is based on the sequence of the precursor protein, whereas that in 3MZS reflects the sequence of the mature protein.) The Val-96 carbonyl also participates in the interactions with the water array in 3NA0. Its analog in 3MZS does not have water contacts; instead, the Leu-210 and Asn-211 carbonyls are involved. The 20α- and 22R-hydroxyls in 20,22DHC are at 3.3 and 3.6 Å, respectively, from the iron and, similar to 22R-hydroxyl in 22HC, are not at a hydrogen bond distance (2.8–3.2 Å) to any of the amino acid residues or waters in the active site. Close similarity of the 22HC- and 20,22DHC-bound CYP11A1 structures supports the proposed soft recognition mechanism and sterol shuttling during the three-step catalysis.
The 3NA0 structure also has a 40-amino acid residue piece of Adx near the proximal side of the P450 molecule, presumably because CYP11A1 is in complex with Adx. However, discussion of these aspects must await the publication that accompanies the 3NA0 structure, as we need to know how this complex was obtained and whether it is functional, i.e. can perform cholesterol hydroxylation.
Comparison of 22HC-bound CYP11A1 and Cholesterol 3-Sulfate-bound CYP46A1
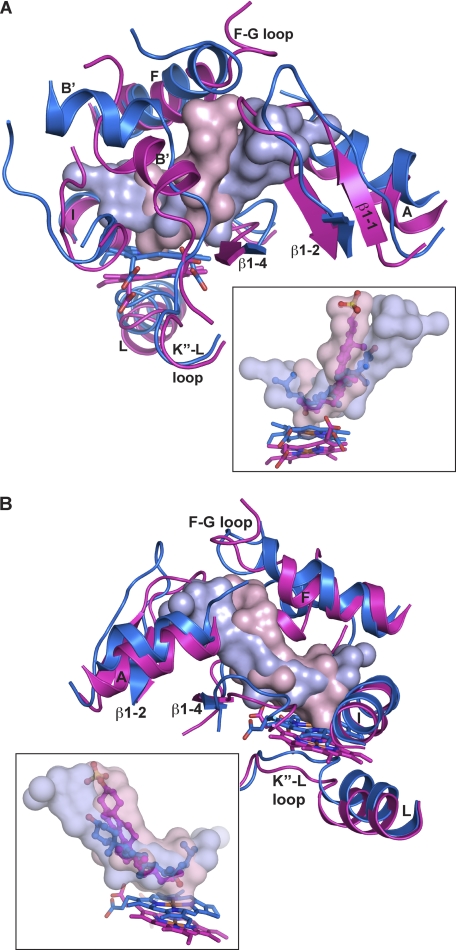
Both CYP11A1 and CYP46A1 oxidize the sterol aliphatic tail. The sites of cholesterol hydroxylation by the two enzymes are different, however, as are their tissue distribution, catalytic efficiencies, degrees of substrate specificity, and cellular location (5). Also, in CYP46A1, the hydroxylation product, 24S-hydroxycholesterol, is released from the enzyme after the monooxygenation reaction and then can re-enter the active site for further hydroxylation (29). Structural superposition reveals that the overall shape of the active site and orientation of the substrate are similar in the two P450s, yet in CYP11A1, the active site is longer, the sterol aliphatic chain is closer to the heme, and the angle between the sterol backbone and the heme plane is more acute (Fig. 4).
FIGURE 4.
Superimposed views of the active sites in CYP11A1 and CYP46A1 illustrating positioning of the secondary structural elements (A and B) and sterol substrates (insets). Secondary structural elements and the heme group are colored in marine in CYP11A1 and magenta in CYP46A1. The solvent-occupied volume of the active site is shown either as a solid or semitransparent surface in light blue in CYP11A1 and light pink in CYP46A1. 22HC is in blue, and cholesterol 3-sulfate is in magenta. Coloring of atoms is the same as in Fig. 1.
Three structural elements contribute to these differences. One is the B′ helix, which is rotated ∼90° in CYP11A1 (Fig. 4A). This rotation shifts the active site tunnel and consequently the 22HC backbone (Fig. 4, A and B, insets). The helix A-β1 sheet loop (residues 53–61) moves outward (up to 6 Å), leading to elongation of the active site cavity in CYP11A1 compared with CYP46A1 (Fig. 4, A and B, insets). As a result, none of the residues lining the entrance to the active site cavity can interact with the 3β-hydroxyl of 22HC, and ordered water molecules fill this extension of the active site to allow for soft recognition. In contrast, in the cholesterol 3-sulfate-CYP46A1 complex, the active site cavity conforms much more to the substrate, and the sterol carbon 3 sulfate group forms hydrogen bonds directly with protein residues (8) consistent with the single oxidation per turnover. The surplus volume in the vicinity of the aliphatic tail of CYP11A1 (Fig. 4, A and B, insets), which accommodates conformational changes during sequential hydroxylation reactions, also arises from repositioning of the B′ helix. Overall, the volumes of the active site cavities in CYP11A1 versus CYP46A1 are 625 Å3 versus 522 Å3. In CYP11A1, the K″-L loop that lies below the heme plane and contains the cysteine residue that coordinates the iron is also shifted inward by ∼2 Å together with the heme (Fig. 4, A and B), bringing the latter closer to the substrate aliphatic chain. The proximity of the aliphatic chain to the site of catalysis likely contributes to the greater efficiency of CYP11A1 (kcat/Km = 4.05 min−1 μm−1) compared with CYP46A1 (0.02 min−1 μm−1) (5).
Structural Comparison of 22HC-bound CYP11A1 and Substrate-free CYP24A1
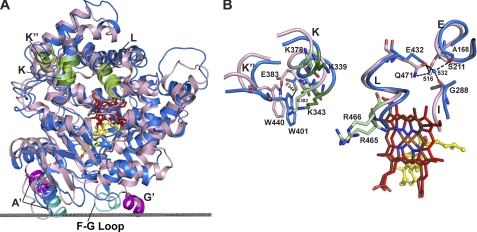
Among P450s, CYP11A1 belongs to the separate evolutionary branch of enzymes that reside in mitochondria and catalyze highly specific reactions in the biosynthesis and degradation of hormones (30). The first structurally characterized mitochondrial P450 was CYP24A1 (10), which performs the multistep oxidation of the side chain of 1α,25-dihydroxyvitamin D3, the hormonal form of vitamin D (31). CYP11A1 is the second mitochondrial P450 whose structure has been determined. Specificity of the enzymatic reactions, similarity of the substrates, multiple sites of sequential hydroxylation, and interactions with the same redox partner and membrane suggest conservation of tertiary structural interactions in the two mitochondrial P450s. Despite <25% of amino acid sequence identity, the root mean square deviation between CYP11A1 and CYP24A1 for 403 residue pairs based on secondary structure alignment is 2.25 Å. Superposition shows significant shifts in the position of the F and G helices, F-G loop, A′ and A helices, and N-terminal β-sheet domain, which reflect closed (substrate-bound) and open (substrate-free) conformations of the active site in CYP11A1 and CYP24A1, respectively (supplemental Fig. S6). Despite this difference, the positions of putative membrane insertion sequences involving the F-G loop and the A′ helix, identified based on computational analysis (10, 32), are similar in CYP11A1 and CYP24A1 (supplemental Fig. S6 and Fig. 5A). Therefore, the orientation of P450s with respect to the membrane suggests a similar mode of substrate diffusion from within the lipid bilayer to the active site, as implied from the locations of detergent molecules in the open form of CYP24A1 (10). Overall, there is excellent correlation between the membrane-interacting residues in CYP11A1 identified by computational analysis (Asn-19, Trp-21, Leu-22, Tyr-25, Leu-216, Leu-219, and Phe-220) and results of site-directed mutagenesis and mass spectrometry studies (33–35).
FIGURE 5.
Superpositioning of CYP11A1 (marine) and CYP24A1 (light pink) illustrating interactions with the membrane and Adx. A, comparison of the overall fold at the proximal side showing the positions of the membrane insertion sequences (cyan in CYP11A1 and magenta in CYP24A1) and of the secondary structural elements that could participate in Adx binding in mitochondrial P450s (K, K″, and L helices, in green). The gray dotted line separates the cytosol (above) and the lipid bilayer (below) with respect to CYP11A1. B, conserved positively charged residues (dark and light green in CYP11A1 and CYP24A1, respectively) and tertiary structure interactions that govern specificity for the redox partner in mitochondrial P450s. One such interaction involves the invariant glutamic acid in the K helix (Glu-383), which hydrogen bonds with the invariant tryptophan in the K″ helix (Trp-440) in CYP24A1 (10); in CYP11A1, this interaction exists as the Glu-344–Trp-401 hydrogen bond. In addition, Glu-432 in CYP11A1 and its counterpart Gln-471 in CYP24A1, at the N terminus of the L helix, are hydrogen-bonded to the spatially overlaid structural Wat-532 and Wat-516, respectively, which in turn interact with residues in the I (Gly-288) and E (Ala-168) helices in CYP11A1 and with Ser-211 in the E helix in CYP24A1. Coloring of atoms is the same as in Fig. 1.
Binding of the common redox partner Adx occurs on the proximal face of the P450 molecule and is mediated by positively charged amino acid residues (36–38). These residues are located in one or more of the three secondary structural elements: in the K helix containing the conserved (K/R)(A/G)xxKE motif, in the K″ helix containing the invariant PxRWL motif, and at the N terminus of the L helix containing the highly conserved GRRxAEx(Q/E) sequence (10). Two residues, Lys-339 and Lys-343, from the K helix motif, have been unambiguously shown to interact with Adx in CYP11A1 (36, 39, 40), and four residues, Lys-378 and Lys-382 from the K helix and Arg-465 and Arg-466 from the L helix, are predicted to bind Adx in CYP24A1 (10). These conserved basic residues and their conserved secondary structural elements are positioned similarly in CYP11A1 and CYP24A1 because of conservation of the tertiary structure interactions (Fig. 5, A and B). It is thus the conserved tertiary structure interactions that likely underlie the specificity of mitochondrial P450s for their shared redox partner, Adx.
In summary, we have determined the structure of mitochondrial P450 CYP11A1 in complex with the first reaction intermediate, 22HC, of its three-step catalytic cycle. The structure reveals an enlarged active site cavity containing a cluster of ordered H2O molecules and a lack of specific contacts to the 3β-hydroxyl of cholesterol, implying mobility of the intermediates during turnover. Displacement of 22HC away from the heme, induced by Adx binding, and movement back toward the heme upon Adx release are demonstrated by difference spectroscopy in the presence of CO and are consistent with the structure. This soft mode of substrate recognition, allowing positional and conformational changes in intermediates as they remain trapped in the active site, provides a mechanistic explanation for the stereospecific sequential hydroxylation reactions catalyzed by CYP11A1. Comparative analysis of CYP11A1 with CYP46A1, which catalyzes a single hydroxylation of cholesterol, reveals how repositioning of two elements of the P450 fold, the B′ helix and helix A-β1 sheet loop region, enlarges the active site tunnel and allows for capture of up to 15 H2O molecules that surround the 22HC 3β-hydroxyl group. Comparison with CYP24A1 illustrates the mechanism of closure of the active site channel upon binding substrate and establishes common features for mitochondrial P450 membrane insertion and redox partner interaction. Together, these results provide important mechanistic insight into the conversion of cholesterol to pregnenolone, a process of fundamental biological and medical significance.
Supplementary Material
Acknowledgment
We thank C. Charvet for calculation of the active site volumes and volumes of the substrates.
This work was supported in part by United States Public Health Service Grants GM62882 and AG024336 (to I. A. P.), EY019718 (to D. T. L.), and EY0090339 (to K. P.). Portions of this work were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, by the Biomedical Technology Program of the National Center for Research Resources, National Institutes of Health, and by NIGMS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S6, Table S1, and references.
The atomic coordinates and structure factors (code 3MZS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- 22HC
- 22R-hydroxycholesterol
- 20,22DHC
- 20α,22R-dihydroxycholesterol
- Adx
- adrenodoxin
- Wat
- water.
REFERENCES
- 1. Simpson E. R., Boyd G. S. (1967) Eur. J. Biochem. 2, 275–285 [DOI] [PubMed] [Google Scholar]
- 2. Burstein S., Middleditch B. S., Gut M. (1975) J. Biol. Chem. 250, 9028–9037 [PubMed] [Google Scholar]
- 3. Orme-Johnson N. R., Light D. R., White-Stevens R. W., Orme-Johnson W. H. (1979) J. Biol. Chem. 254, 2103–2111 [PubMed] [Google Scholar]
- 4. Lambeth J. D., Kitchen S. E., Farooqui A. A., Tuckey R., Kamin H. (1982) J. Biol. Chem. 257, 1876–1884 [PubMed] [Google Scholar]
- 5. Pikuleva I. A. (2006) Drug Metab. Dispos. 34, 513–520 [DOI] [PubMed] [Google Scholar]
- 6. Guengerich F. P. (2005) in Cytochrome P450 (Ortiz de Montellano P. R. ed) 3rd Ed., pp. 377–530, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 7. Pikuleva I. A. (2006) Pharmacol. Ther. 112, 761–773 [DOI] [PubMed] [Google Scholar]
- 8. Mast N., White M. A., Bjorkhem I., Johnson E. F., Stout C. D., Pikuleva I. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9546–9551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Russell D. W., Halford R. W., Ramirez D. M., Shah R., Kotti T. (2009) Annu. Rev. Biochem. 78, 1017–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Annalora A. J., Goodin D. B., Hong W. X., Zhang Q., Johnson E. F., Stout C. D. (2010) J. Mol. Biol. 396, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pikuleva I. A., Mackman R. L., Kagawa N., Waterman M. R., Ortiz de Montellano P. R. (1995) Arch. Biochem. Biophys. 322, 189–197 [DOI] [PubMed] [Google Scholar]
- 12. Omura T., Sato R. (1964) J. Biol. Chem. 239, 2370–2378 [PubMed] [Google Scholar]
- 13. Trout E. C., Jr., Arnett W. (1971) Proc. Soc. Exp. Biol. Med. 136, 469–472 [DOI] [PubMed] [Google Scholar]
- 14. Kobayashi S., Ichii S. (1969) J. Biochem. 66, 51–56 [DOI] [PubMed] [Google Scholar]
- 15. Morisaki M., Duque C., Takane K., Ikekawa N., Shikita M. (1982) J. Steroid Biochem. 16, 101–105 [DOI] [PubMed] [Google Scholar]
- 16. Aringer L., Eneroth P., Nordström L. (1979) J. Steroid Biochem. 11, 1271–1285 [DOI] [PubMed] [Google Scholar]
- 17. Sheets J. J., Vickery L. E. (1983) J. Biol. Chem. 258, 1720–1725 [PubMed] [Google Scholar]
- 18. Degenhart H. J., Alsema G. J., Hoogerbrugge J., Wolthers B. G., Kaptein R. (1984) J. Steroid Biochem. 21, 447–451 [DOI] [PubMed] [Google Scholar]
- 19. Burstein S., Letourneux Y., Kimball H. L., Gut M. (1976) Steroids 27, 361–382 [DOI] [PubMed] [Google Scholar]
- 20. Heyl B. L., Tyrrell D. J., Lambeth J. D. (1986) J. Biol. Chem. 261, 2743–2749 [PubMed] [Google Scholar]
- 21. Morisaki M., Duque C., Ikekawa N., Shikita M. (1980) J. Steroid Biochem. 13, 545–550 [DOI] [PubMed] [Google Scholar]
- 22. Lambeth J. D. (1986) Endocr. Res. 12, 371–392 [DOI] [PubMed] [Google Scholar]
- 23. Shaik S., Kumar D., de Visser S. P., Altun A., Thiel W. (2005) Chem. Rev. 105, 2279–2328 [DOI] [PubMed] [Google Scholar]
- 24. Tuckey R. C., Kamin H. (1983) J. Biol. Chem. 258, 4232–4237 [PubMed] [Google Scholar]
- 25. Lambeth J. D., Kriengsiri S. (1985) J. Biol. Chem. 260, 8810–8816 [PubMed] [Google Scholar]
- 26. Lambeth J. D., Pember S. O. (1983) J. Biol. Chem. 258, 5596–5602 [PubMed] [Google Scholar]
- 27. Lambeth J. D., Seybert D. W., Kamin H. (1980) J. Biol. Chem. 255, 138–143 [PubMed] [Google Scholar]
- 28. Tsubaki M., Hiwatashi A., Ichikawa Y., Fujimoto Y., Ikekawa N., Hori H. (1988) Biochemistry 27, 4856–4862 [DOI] [PubMed] [Google Scholar]
- 29. Mast N., Norcross R., Andersson U., Shou M., Nakayama K., Bjorkhem I., Pikuleva I. A. (2003) Biochemistry 42, 14284–14292 [DOI] [PubMed] [Google Scholar]
- 30. Nelson D. R. (1998) Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 121, 15–22 [DOI] [PubMed] [Google Scholar]
- 31. Omdahl J. L., Morris H. A., May B. K. (2002) Annu. Rev. Nutr. 22, 139–166 [DOI] [PubMed] [Google Scholar]
- 32. Lomize M. A., Lomize A. L., Pogozheva I. D., Mosberg H. I. (2006) Bioinformatics 22, 623–625 [DOI] [PubMed] [Google Scholar]
- 33. Pikuleva I. A. (2004) Mol. Cell. Endocrinol. 215, 161–164 [DOI] [PubMed] [Google Scholar]
- 34. Headlam M. J., Wilce M. C., Tuckey R. C. (2003) Biochim. Biophys. Acta 1617, 96–108 [DOI] [PubMed] [Google Scholar]
- 35. Pikuleva I. A., Mast N., Liao W. L., Turko I. V. (2008) Lipids 43, 1127–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wada A., Waterman M. R. (1992) J. Biol. Chem. 267, 22877–22882 [PubMed] [Google Scholar]
- 37. Pikuleva I. A., Cao C., Waterman M. R. (1999) J. Biol. Chem. 274, 2045–2052 [DOI] [PubMed] [Google Scholar]
- 38. Urushino N., Yamamoto K., Kagawa N., Ikushiro S., Kamakura M., Yamada S., Kato S., Inouye K., Sakaki T. (2006) Biochemistry 45, 4405–4412 [DOI] [PubMed] [Google Scholar]
- 39. Tuls J., Geren L., Millett F. (1989) J. Biol. Chem. 264, 16421–16425 [PubMed] [Google Scholar]
- 40. Tsubaki M., Iwamoto Y., Hiwatashi A., Ichikawa Y. (1989) Biochemistry 28, 6899–6907 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.