Abstract
Calnexin (CANX) and calreticulin (CALR) are homologous lectin chaperones located in the endoplasmic reticulum and cooperate to mediate nascent glycoprotein folding. In the testis, calmegin (CLGN) and calsperin (CALR3) are expressed as germ cell-specific counterparts of CANX and CALR, respectively. Here, we show that Calr3−/− males produced apparently normal sperm but were infertile because of defective sperm migration from the uterus into the oviduct and defective binding to the zona pellucida. Whereas CLGN was required for ADAM1A/ADAM2 dimerization and subsequent maturation of ADAM3, a sperm membrane protein required for fertilization, we show that CALR3 is a lectin-deficient chaperone directly required for ADAM3 maturation. Our results establish the client specificity of CALR3 and demonstrate that the germ cell-specific CALR-like endoplasmic reticulum chaperones have contrasting functions in the development of male fertility. The identification and understanding of the maturation mechanisms of key sperm proteins will pave the way toward novel approaches for both contraception and treatment of unexplained male infertility.
Keywords: ADAM, ADAMTS, Chaperone, Chaperonin, Endoplasmic Reticulum (ER), Extracellular Matrix Proteins, Gene Knockout, Glycoprotein Biosynthesis, Lectin, Reproduction, Sperm, Fertilization
Introduction
Membrane and secretory proteins are cotranslationally translocated into the endoplasmic reticulum (ER)3 lumen, where numerous molecular chaperones and folding enzymes assist their maturation (1, 2). Failures in protein folding and quality control can result in compromised cellular function and diseases such as diabetes, cystic fibrosis, and amyloidosis (3–5). ER membrane-bound calnexin (CANX) and luminal calreticulin (CALR) are well characterized homologous lectin chaperones that transiently interact with newly synthesized glycoproteins and play critical roles in calcium homeostasis and the process of protein maturation and degradation (6–10). Despite their extensive homology, CANX and CALR show some differences in their activity and client specificity, as demonstrated by CANX- and CALR-deficient cell lines (11). The distinct phenotypes of knock-out mice (Calr−/− is embryonic lethal (12), whereas Canx−/− is semilethal after birth (13)) reinforce their different functions. Because CANX is membrane-tethered and CALR is soluble in the lumen, the contrasting function might be explained mainly by their different topological constraints (11, 14). Recent studies have suggested, however, that client specificity is due in part to glycan-independent polypeptide recognition (15–18).
Calmegin (CLGN) and calsperin (CALR3) have been reported to be testis-specific homologs of CANX and CALR, respectively (19, 20). We have shown previously that CLGN is a CANX-like lectin chaperone during spermatogenesis and is required for the assembly of ADAM (a disintegrin and metalloproteinase) sperm fertilizing proteins ADAM1A/ADAM2 and ADAM1B/ADAM2 (21, 22). The ADAM1B/ADAM2 complex, also known as fertilin, was first identified as a protein required for sperm-egg fusion but was later proved to be dispensable for fertilization using gene-disrupted mice (23, 24). On the contrary, ER-resident ADAM1A/ADAM2 is critical for subsequent ADAM3 maturation and sperm fertilizing ability (24, 25). This explains why Clgn−/− mature sperm lose ADAM3 expression and, subsequently, fertilizing ability (26, 27).
CALR3 was first identified as a CALR homolog through BLASTP analysis, and its testis-specific expression was demonstrated by Northern blot analysis (20). However, the biochemical and physiological functions of CALR3 and its relationship with CALR and CLGN remain unknown. In this study, we generated Calr3−/− mice and describe the indispensable role of CALR3 in the development of sperm fertilizing ability. In contrast to CLGN, CALR3 does not have broad lectin-like activity but directly associates with ADAM3 and is required for its maturation. We therefore conclude that the testis-specific calreticulin homologs CLGN and CALR3 have distinct but cooperative roles in the maturation of ADAM3 during spermatogenesis and subsequent sperm fertilizing ability.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit antisera against the CALR C-terminal peptide (EEDEKEEDEEESPGQAKDEL) and the CALR3 C-terminal peptide (MGKFHRHWHLSRFHRQGEL) were raised for this study (supplemental Fig. S1). Antisera against the CANX C-terminal and CLGN C-terminal peptides were described previously (22). Antisera against MFGE8 (SED1) (28) and ZAN (zonadhesin) (29) were kindly provided by Drs. Barry Shur and Daniel Hardy, respectively. Monoclonal antibodies were described previously (TRA369 for CLGN (30) and 1D5 for ACE (26)), established in this study (KS107–158 for ADAM1B, KS107–190 for ADAM2, and KS64–125 for IZUMO1), or purchased (7C1 for ADAM3 (Chemicon), 7C5 for ZP3R (SP56; QED Bioscience), and SC53029 for α-tubulin (Santa Cruz Biotechnology)).
Immunoblotting
Immunoblot analysis was performed as described previously (26). Briefly, sperm from the epididymis and vas deferens were collected and incubated in lysis buffer containing 1% Triton X-100 for 20 min on ice. The various tissues were excised and homogenized in lysis buffer and then placed on ice for 1 h. These extracts were centrifuged, and the supernatants were collected. Proteins were separated by SDS-PAGE under reducing or nonreducing conditions and transferred to PVDF membranes (Millipore). After blocking, blots were incubated with primary antibodies overnight at 4 °C and then incubated with horseradish peroxidase-conjugated secondary antibodies. Detection was performed using an ECL Western blotting detection kit (GE Healthcare).
Immunohistochemistry
Testes were collected from adult mice and fixed in 4% paraformaldehyde/PBS overnight at 4 °C, cryopreserved in graded 10–30% sucrose, and embedded in a Tissue-Tek O.C.T. compound (Sakura Finetechnical Co.). Frozen sections (8 μm) were mounted on aminopropyltriethoxysilane-coated glass slides. After washing with PBS, slides were blocked with 10% newborn calf serum/PBS for 1 h and incubated with primary antibodies in 10% newborn calf serum/PBS at 4 °C overnight. After washing with PBS, the slides were incubated with secondary antibodies with Alexa Fluor 488 or 546 (Invitrogen) in 10% newborn calf serum/PBS for 2 h. After washing with PBS, the slides were observed under an Olympus IX-70 fluorescence microscope with a Nipkow disk confocal scanner unit (CSU-X1, Yokogawa Electric) and a cooled charge-coupled device camera (CoolSNAP HQ, Photometrics). Device control and image capture were performed with MetaMorph software (Molecular Devices).
Immunoprecipitation
Immunoprecipitation was performed by magnetic bead separation (MACS separation, Miltenyi Biotec) according to the manufacturer's protocol with minor modifications. Briefly, wild-type or Calr3−/− testis was solubilized with 1 ml of ice-cold lysis buffer containing 10 mm Tris-HCl (pH 8.0), 50 mm NaCl, 1% Triton X-100, and 1% protease inhibitor mixture (Nacalai Tesque). Lysates were centrifuged at 15,300 × g for 10 min at 4 °C. One mg of testis lysate was incubated with 2 μl of each antiserum and 50 μl of Protein A-conjugated MicroBeads (MACS Protein A MicroBeads, Miltenyi Biotec) for 2 h at 4 °C. The samples were applied to columns in the magnetic field of the μMACS separator, and the columns were rinsed four times with 200 μl of wash buffer (1 m NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mm Tris-HCl (pH 8.0)) and once with 100 μl of low salt wash buffer (50 mm Tris-HCl (pH 8.0) and 1% Nonidet P-40). Finally, 70 μl of preheated (96 °C) 1× SDS sample buffer was applied to the columns, and eluate fractions were collected for analysis by SDS-PAGE.
Endoglycosidase H Treatment
For deglycosylation, the testis lysates prepared as described above were treated with endoglycosidase H (Roche Applied Science) at 37 °C overnight according to the manufacturer's instructions. The treated lysates were used for immunoprecipitation assays.
Trypsin Treatment
Testes from mutant mice were homogenized in lysis buffer containing 50 mm NaCl, 10 mm Tris-HCl (pH 7.5), 1% Triton X-100, and 20 mm iodoacetamide (Nacalai Tesque) to prepare 1 mg/ml lysate. The lysate was incubated with 0, 0.25, 0.75, 1.25, and 2.5 μg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (Sigma) for 30 min at 4 °C. Trypsin digestion was terminated by the addition of 200 μg/ml soybean trypsin inhibitor (Sigma). Samples were analyzed by reducing or nonreducing SDS-PAGE and immunoblotted with anti-ADAM3 antibody. For cell-surface proteins, live germ cells were collected from testes. The seminiferous tubules were minced with a razor blade to release germ cells. The cell suspension in PBS was filtered through a nylon mesh, the cells were collected by centrifugation at 2000 rpm for 5 min, and the cell pellet was resuspended in PBS. The germ cells (5 × 107 cells/ml) were incubated with 0, 20, and 40 μg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin for 5 min at 20 °C and then homogenized in lysis buffer.
Fertility Test and Sperm Migration Assay
Three B6D2F1 females were caged with each male for 1 month and kept another 3 weeks to observe pregnancy. Four males were examined for each group. Sperm migration from the uterus into the oviduct was examined as described previously (22). Briefly, oviducts with the proximal part of the uterus were excised 2 h after copulation and fixed with 4% paraformaldehyde/PBS. Frozen sections containing the uterotubal junction were stained with hematoxylin and eosin and observed under the microscope.
In Vitro Fertilization
Eggs collected from superovulated females, 14 h after human chorionic gonadotropin injection, were treated with hyaluronidase (1 mg/ml) for 5 min to remove cumulus cells and then placed in a 200-μl TYH medium (44) drop covered with paraffin oil. Epididymal sperm were collected from 3-month-old male mice and incubated in TYH medium for 2 h for capacitation. Capacitated sperm were added to the drop containing eggs at a final concentration of 2 × 105 sperm/ml. After 8 h of co-incubation, the formation of pronuclei was observed under a Hoffman modulation contrast microscope. To assess the zona pellucida binding ability, cumulus-free eggs were incubated with capacitated sperm for 20 min prior to observation. To assess the fusion ability, the zona pellucida was dissolved in acidic Tyrode's solution for 30 s to 1 min, and the eggs were preloaded with Hoechst 33342 (1 μg/ml) by incubation for 10 min and washing them before insemination. After 30 min of co-incubation with capacitated sperm, the eggs were fixed in 0.25% glutaraldehyde and observed under a fluorescence microscope with ultraviolet excitation (31). Partial zona dissection was performed with a piezo-micromanipulator with a glass capillary needle (diameter of 5–10 μm) (32). Fertilized eggs were counted at the two-cell stage and then transferred into the oviduct of pseudopregnant females.
Statistical Analysis
All values are the means ± S.D. of at least three independent experiments. Statistical analyses were performed using Student's t test. Differences were considered significant at p < 0.05.
RESULTS
Haploid-specific Expression of CALR3
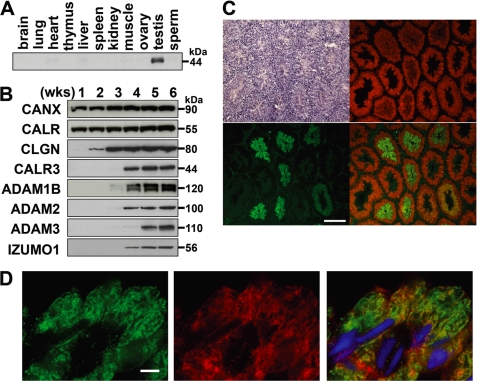
We raised anti-CALR3 antibodies and showed that expression of CALR3 was testis-specific (Fig. 1A). We next determined the differentiation stage-specific expression of CALR3 in mouse testis (Fig. 1, B–D). Whereas constitutive expression of CALR and CANX started at birth, CLGN and CALR3 appeared at 2 and 3 weeks of age, respectively (Fig. 1B). In mouse, spermatogenesis begins at birth, and germ cells enter and exit meiosis at approximately 11 and 21 days of age (33). Therefore, the onset of CLGN and CALR3 expression was considered to be meiotic and post-meiotic, respectively. It is notable that CLGN expression preceded that of CALR3, mirroring the offset expression of ADAM1B/ADAM2 and ADAM3 (Fig. 1B). When we stained testicular sections with antibodies (Fig. 1, C and D), post-meiotic expression of CALR3 was reconfirmed, whereas CLGN expression faded after meiosis. The stage-specific expression and co-localization of CLGN and CALR3 suggested that they may have independent yet cooperative functions.
FIGURE 1.
Haploid-specific expression of CALR3 and chaperone activity. A, CALR3 was detected only in the testis when various tissues were examined by Western blot analysis. B, testis lysates collected from mice of different ages were examined by Western blotting. CALR3 appeared as a faint signal at 3 weeks (wks) of age, whereas CLGN was detected at 2 weeks of age. It should be noted that ADAM1 is composed of two independent proteins, ADAM1A and ADAM1B (43). C, immunofluorescence staining of a testicular section with anti-CLGN (shown in red) and anti-CALR3 (shown in green) antibodies. Sequential sections were stained with hematoxylin and eosin (upper left panel). CALR3 was detected in elongating spermatids, whereas CLGN was detected in pachytene spermatocytes to elongating spermatids. D, co-localization of CLGN and CALR3 was observed in elongating spermatids. Green, CALR3; red, CLGN; blue, Hoechst 33258. Scale bars = 200 μm (C) and 4 μm (D).
Generation of Calr3 Knock-out Mice
To analyze the physiological function of CALR3 in vivo, we generated Calr3−/− mice. The targeting vector was constructed by substituting exons 3 and 4 with a neomycin resistance gene cassette in reverse orientation relative to the CALR3 transcriptional unit (supplemental Fig. S2A). The targeting event in embryonic stem cells and the germ line transmission were confirmed by PCR analysis (supplemental Fig. S2B). Intercrosses between heterozygous F1 mice yielded offspring that segregated in a Mendelian distribution: 28 wild-type mice (+/+) and 51 heterozygous (+/−) and 26 homozygous (−/−) mutant mice. No overt developmental abnormalities were observed in Calr3−/− mice. In Calr3−/− testis, Calr3 mRNA was not detected (supplemental Fig. S2C). Apart from the lack of CALR3, we found no significant differences in the protein amount of CANX, CALR, CLGN, and the Hsp70 ER chaperone HSPA5 (Bip/GRP78) (supplemental Fig. S2D). When we observed testicular sections, disruption of CALR3 in the testis caused no deleterious effect on spermatogenesis (supplemental Fig. S2E).
Male Infertility in Calr3 Knock-out Mice
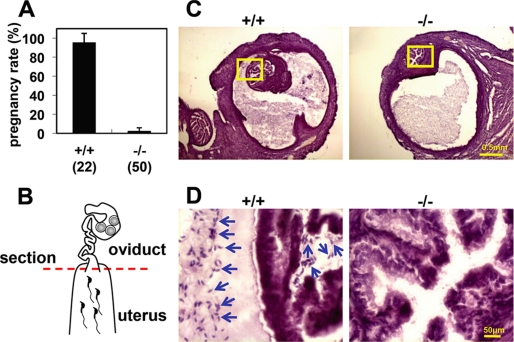
Whereas Calr3−/− females were as fertile as wild-type females, Calr3−/− males were almost sterile despite normal mating behavior, with successful sperm ejaculation and vaginal plug formation. Only one litter with two pups was obtained from 50 copulations with four Calr3−/− males, whereas 21 litters from 22 copulations were obtained from four wild-type males (Fig. 2A and supplemental Fig. S3). Even though ejaculated Calr3−/− sperm in the uterus showed a normal appearance and motility, their migration into the oviduct was severely impaired (Fig. 2, B–D). Almost no Calr3−/− sperm were found in the uterotubal junction even 2 h after copulation. Therefore, impaired sperm migration into the oviduct is the primary cause of Calr3−/− male sterility.
FIGURE 2.
Impaired sperm migration from the uterus into the oviduct in Calr3−/− mice. A, pregnancy rate (pregnancy/vaginal plug formation) presented as a percentage. Calr3−/− males copulated normally but failed to induce pregnancy. Four males were examined in each group. The total number of plugs observed is indicated in parentheses. B–D, sperm migration assay. The oviduct with the uterus was removed 2 h after coitus, and frozen sections containing the uterotubal junction (red dashed line in B) were prepared. Although wild-type sperm were observed in the uterine part and the uterotubal junction, Calr3−/− sperm were observed only in the uterine part and not in the uterotubal junction (C and D). The boxes in C are magnified in D. Arrows indicate sperm. The sections were stained with hematoxylin and eosin.
Defective Sperm Binding to the Zona Pellucida
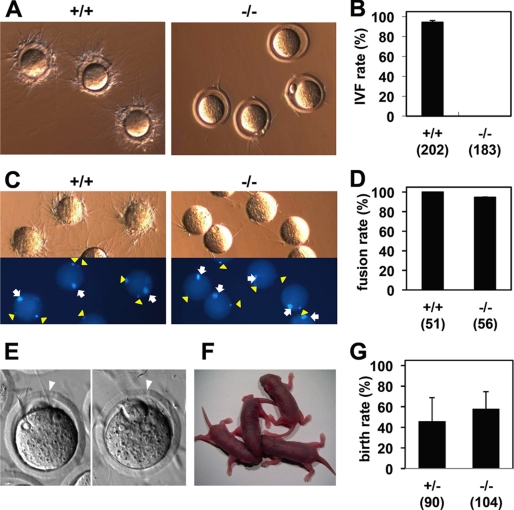
We next examined sperm fertilizing ability in vitro. Epididymal sperm collected from Calr3−/− mice showed a normal morphology and viability in the in vitro fertilization medium. When sperm parameters were analyzed with the computer-aided sperm analysis system (SMAS, Kaga Solution Network Co., Ltd.), there were no significant differences in motility, velocity, and linearity over 2 h (supplemental Table S1). However, when the sperm were co-incubated with unfertilized eggs, Calr3−/− sperm could not bind to the zona pellucida despite frequent collisions with the eggs (Fig. 3A and supplemental Movies S1 and S2). As a result, in vitro fertilization efficiency was drastically decreased in Calr3−/− sperm (94.2 ± 1.8% with +/+ and 0% with −/− sperm (n = 3), p < 0.001) (Fig. 3B).
FIGURE 3.
Impaired sperm binding to the zona pellucida in Calr3−/− mice. A, wild-type sperm successfully bound to the egg zona pellucida, but Calr3−/− sperm failed to adhere despite frequent collisions (supplemental Movies S1 and S2). B, Calr3−/− sperm were not able to fertilize eggs in vitro. The average fertilization rate from three independent experiments is presented. The total number of eggs examined is indicated in parentheses. IVF, in vitro fertilization. C and D, zona–free eggs preloaded with Hoechst 33342 were incubated with +/+ or Calr3−/− sperm, and fused sperm were observed under a fluorescence microscope. Arrows indicate MII chromosomes, and arrowheads indicate fused sperm (C). The average fusion rate (fused eggs/examined eggs) from three independent experiments is presented in D. The number of eggs examined is indicated in parentheses. E–G, successful fertilization was observed with Calr3−/− sperm after partial zona dissection and in vitro fertilization. Arrowheads indicate the slit made by partial zona dissection (E). The offspring of Calr3−/− males obtained after partial zona dissection, in vitro fertilization, and embryo transfer are shown in F. The average birth rate (pups/transplanted eggs) after transferring embryos subjected to partial zona dissection and in vitro fertilization (n = 3) is shown in G.
Genomic Integrity of Calr3 Knock-out Sperm
When zona pellucida binding and penetration were bypassed by removal of the zona pellucida, Calr3−/− sperm were able to fuse with 94.6 ± 0.2% of eggs (100% with +/+ sperm, n = 3) (Fig. 3, C and D). Because solubilized zona remnants coat the egg plasma membrane and attract +/+ sperm, less binding was observed with Calr3−/− sperm, similar to that seen with Clgn−/− sperm (32). Zona removal tends to lead to polyspermy (Fig. 3C), so we performed partial zona dissection prior to in vitro fertilization to decrease the risk of polyspermy and examined the developmental ability of eggs fertilized with Calr3−/− sperm (Fig. 3, E–G). Healthy and fertile offspring developed from eggs fertilized in this way, indicating the genomic integrity of Calr3−/− sperm. As we expected, all the offspring were +/− mutants and showed normal fecundity.
Transgenic Rescue of Male Infertility in Calr3 Knock-out Mice
To exclude the possibility that the phenotype was caused by a side effect of genetic and/or epigenetic interference from the mutant Calr3 locus, at which there are three other genes starting within 3 kbp upstream and downstream of Calr3 (supplemental Fig. S2A), we generated transgenic mouse lines expressing Calr3 on the Calr3−/− background (supplemental Fig. S3). The recovery of male fertility was observed in two independent transgenic lines (litter size of 8.6 ± 2.7 and 9.0 ± 2.9), demonstrating that the CALR3 disruption by itself diminished the sperm fertilizing ability.
Calreticulin Family Chaperones and Client Specificity
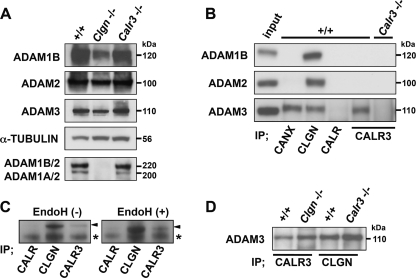
Because CALR3 is absent in mature sperm (Fig. 1A), the sperm defects are likely to be secondary effects of CALR3 disruption during spermatogenesis. Previously, we have demonstrated that ADAM2 is specifically recognized by CLGN, but not by CANX, and that the heterodimerization of ADAM1A/ADAM2 and ADAM1B/ADAM2 is diminished with CLGN disruption (22). In contrast, heterodimerization was not disrupted in Calr3−/− testis (Fig. 4A). Unlike CLGN, CALR3 did not associate with ADAM1B and ADAM2 in wild-type testis (Fig. 4B). Using a co-immunoprecipitation assay, we found that CALR3 and CANX, but not CALR, specifically associated with ADAM3 (Fig. 4B), demonstrating chaperone and client selectivity.
FIGURE 4.
Interactions between ADAM proteins and ER chaperones. A, Western blot analysis of testis lysates separated by SDS-PAGE under reducing conditions (upper four panels) and nonreducing conditions (lower panel). Comparable amounts of ADAM1B, ADAM2, and ADAM3 were detected in +/+, Clgn−/−, and Calr3−/− testes under reducing conditions. When ADAM1A/ADAM2 and ADAM1B/ADAM2 heterodimers were probed by anti-ADAM2 antibody under nonreducing conditions (22–24), ADAM1/ADAM2 heterodimerization was absent in Clgn−/− testis but was not impaired in Calr3−/− testis. B, immunoprecipitates (IP) with anti-CANX, anti-CALR, anti-CLGN, and anti-CALR3 antibodies from testis lysates (100 μg) were probed with the indicated antibodies. Testis lysates (10 μg) were loaded as the input control. Whereas CLGN associated with ADAM1B, ADAM2, and ADAM3, CALR3 associated with only ADAM3. C, testis lysates digested with endoglycosidase H (EndoH) were immunoprecipitated with the indicated antibodies and then probed with anti-ADAM3 antibody. Endoglycosidase H treatment changed the molecular mass of ADAM3 but did not affect the association with CLGN and CALR3. Arrowheads indicate ADAM3. Asterisks indicate the nonspecific signal. D, testis lysates from Clgn−/− and Calr3−/− mice were immunoprecipitated with anti-CALR3 and anti-CLGN antibodies and then probed with anti-ADAM3 antibody.
Lectin-independent CALR3 Function upon ADAM3 Maturation
To investigate whether CALR3 can interact with other client proteins, we performed biochemical studies on HepG2 transfectants in the presence of the glucosidase inhibitor castanospermine. Although CALR clearly interacted with putative clients in a lectin-dependent manner, CALR3 was not a lectin chaperone for general nascent proteins (supplemental Fig. S4). We next examined whether endoglycosidase H treatment of ADAM3 influences its interaction with CLGN and CALR3 and found that the treatment did not interfere with CLGN/ADAM3 and CALR3/ADAM3 interactions (Fig. 4C). This suggested that ADAM3 is a lectin-independent client of CLGN and CALR3. We further examined whether the CLGN-mediated maturation of ADAM3 is a prerequisite for CALR3 interaction and found that both CLGN/ADAM3 and CALR3/ADAM3 interactions were not disturbed in Calr3−/− and Clgn−/− testis, respectively (Fig. 4D).
ADAM3 Transport from the ER to the Sperm Surface
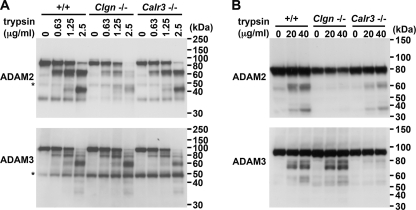
To investigate the maturation status/conformation of ADAM3 in Clgn−/− and Calr3−/− mice, we tested the trypsin sensitivity of ADAM3 during spermatogenesis. When cellular extracts were trypsinized, a similar digestion pattern of ADAM3 was observed in all of the wild-type, Clgn−/−, and Calr3−/− testicular cells (Fig. 5A). However, when live spermatogenic cells were trypsinized, ADAM3 was similar in the wild-type and Clgn−/− cells but was absent in the Calr3−/− cells (Fig. 5B). In contrast, ADAM2 was unaffected in Calr3−/− spermatogenic cells but was absent in Clgn−/− cells. The data in Fig. 5B indicate that ADAM3 remains in the secretory pathway in the absence of CALR3, whereas it is transported onto the sperm surface in the absence of CLGN. The absence of ADAM3 in mature sperm of the Calr3−/− mouse was unique among the various sperm fertilizing proteins analyzed (Fig. 6), reinforcing the view that CALR3 does not function as a general chaperone but is client-specific.
FIGURE 5.
Trypsin sensitivity of ADAM2 and ADAM3 during spermatogenesis in +/+, Clgn−/−, and Calr3−/− mice. A, testis lysates (5 μg) were incubated with tosylphenylalanyl chloromethyl ketone-treated trypsin and then subjected to Western blot analysis with the indicated antibodies. Asterisks indicate the IgG that reacted with the secondary antibody. B, live germ cells collected from testes were trypsinized and then analyzed by Western blotting. The lysate prepared from 5 × 105 cells was loaded in each lane. ADAM2 was not digested in the Clgn−/− germ cells, whereas ADAM3 was not digested in the Calr3−/− germ cells.
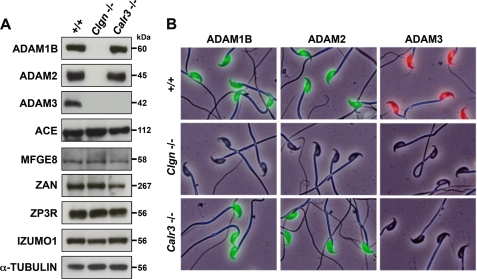
FIGURE 6.
Disappearance of ADAM3 from mature Calr3−/− sperm. A, Western blot analysis of sperm lysates. ADAM3 was not detected in Calr3−/− sperm, whereas other sperm fertilizing proteins remained. ACE, angiotensin-converting enzyme. B, immunofluorescence staining of sperm with antibodies against ADAM1B, ADAM2, and ADAM3. Fresh sperm collected from the epididymis of mutant mice were fixed in 4% paraformaldehyde and then immunostained with the following antibodies: KS107–57 (rat anti-mouse ADAM1B), KS107–190 (rat anti-mouse ADAM2), and rabbit anti-mouse ADAM3 (25). All ADAM proteins were detected in the head region of control +/+ sperm. Whereas Clgn−/− sperm lack all ADAM proteins, Calr3−/− sperm lack only ADAM3.
DISCUSSION
In this study, we have shown that CALR3 is a male haploid-specific homolog of a ubiquitously expressed ER lectin-like chaperone, CALR. Whereas CLGN is a lectin chaperone during meiosis (21), our data suggest that CALR3 lacks broad lectin activity and no longer functions as a general chaperone for nascent N-glycoproteins. Because the essential amino acids required for oligosaccharide binding in CALR are well conserved in CALR3 (34), the lectin deficiency is likely due to differences in the divergent P domain (20, 35). However, CALR3 directly associates with ADAM3 and controls its maturation. Lectin-deficient chaperone activity was also reported in CALR (18). Thus, we propose that CALR3 is a bona fide molecular chaperone capable of selectively interacting with ADAM3.
Our data indicate that CLGN and CALR3 have distinct and specific roles in the maturation of ADAM proteins, similar to that observed for CANX and CALR in the maturation of influenza HA protein (11). For the assembly of ADAM1A/ADAM2 and ADAM1B/ADAM2, CLGN was essential, but CALR3 was not. For ADAM3 quality control, CALR3 was essential for ADAM3 transport onto the sperm surface, but CLGN was not (Fig. 5). This result adds significantly to our understanding of the distinct roles of CLGN and CALR3 in the quality control of ADAM3. The different chaperone requirements could be explained by the stage-dependent expression, the client specificity, and/or the topological difference that CLGN is membrane-anchored whereas CALR3 is soluble.
The defects found in Calr3−/− sperm are very similar to those in sperm from mice lacking CLGN (21), ADAM1A (25), ADAM2 (23), ADAM3 (27), or angiotensin-converting enzyme (36). Because ADAM3 is the only protein commonly disrupted or displaced in these sperm (27, 37), we believe that ADAM3 plays a central role in both sperm migration into the oviduct and sperm binding to the zona. This is supported by the fact that the ADAM3 directly binds to ZP3 zona pellucida protein (38).
Recently, a testis-specific homolog of protein-disulfide isomerase (PDILT) was reported (39, 40). Because ubiquitously expressed protein-disulfide isomerases such as ERp57 cooperate together with ER lectin chaperones and help in protein disulfide bond formation (41), it is possible that CALR3 cooperates with PDILT or with other protein-disulfide isomerases during quality control in testes. The elucidation of PDILT function will help us to understand why the testis ER requires specific quality control and how the sperm fertilizing proteins mature during spermatogenesis. It is interesting to note that a plant-specific CALR homolog, also called CRT3, is largely tissue-specific, being expressed mainly in the leaves. Plant CRT3 has been implicated in the specialized quality control of the leucine-rich repeat receptor protein EFR, a protein involved in innate immunity (42). Thus, both plant leaves and mammalian testis have converged upon the same strategy, namely to release a calreticulin homolog from an essential role in calcium homeostasis to mediate distinct quality control processes in the ER. This also suggests that CALR engineering to improve the production of hard to fold glycoproteins may be achievable without adverse effects on the quality control of bystander proteins.
We conclude that the testis-specific chaperones CLGN and CALR3 regulate sperm fertilizing ability by playing essential but contrasting functions in ADAM3 maturation. Taken together with the observation that the defective binding of sperm to the zona is reminiscent of unexplained male infertility, manipulation of the testis-specific CLGN/CALR3 ER quality control system could be exploited to develop novel therapeutic approaches for both contraception and treatment of male infertility.
Supplementary Material
Acknowledgments
We thank Y. Maruyama and A. Kawai for technical assistance in generating Calr3−/− mice.
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Table S1, Movies S1 and S2, and additional references.
- ER
- endoplasmic reticulum
- CANX
- calnexin
- CALR
- calreticulin
- CLGN
- calmegin
- CALR3
- calsperin.
REFERENCES
- 1. Ellgaard L., Helenius A. (2003) Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 2. Trombetta E. S., Parodi A. J. (2003) Annu. Rev. Cell Dev. Biol. 19, 649–676 [DOI] [PubMed] [Google Scholar]
- 3. Sitia R., Braakman I. (2003) Nature 426, 891–894 [DOI] [PubMed] [Google Scholar]
- 4. Cohen F. E., Kelly J. W. (2003) Nature 426, 905–909 [DOI] [PubMed] [Google Scholar]
- 5. Hammarström P. (2009) FEBS Lett. 583, 2579–2580 [DOI] [PubMed] [Google Scholar]
- 6. Helenius A., Aebi M. (2004) Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 7. Römisch K. (2005) Annu. Rev. Cell Dev. Biol. 21, 435–456 [DOI] [PubMed] [Google Scholar]
- 8. Molinari M. (2007) Nat. Chem. Biol. 3, 313–320 [DOI] [PubMed] [Google Scholar]
- 9. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 10. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 11. Molinari M., Eriksson K. K., Calanca V., Galli C., Cresswell P., Michalak M., Helenius A. (2004) Mol. Cell 13, 125–135 [DOI] [PubMed] [Google Scholar]
- 12. Coppolino M. G., Woodside M. J., Demaurex N., Grinstein S., St-Arnaud R., Dedhar S. (1997) Nature 386, 843–847 [DOI] [PubMed] [Google Scholar]
- 13. Denzel A., Molinari M., Trigueros C., Martin J. E., Velmurgan S., Brown S., Stamp G., Owen M. J. (2002) Mol. Cell. Biol. 22, 7398–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wada I., Imai S., Kai M., Sakane F., Kanoh H. (1995) J. Biol. Chem. 270, 20298–20304 [DOI] [PubMed] [Google Scholar]
- 15. Ihara Y., Cohen-Doyle M. F., Saito Y., Williams D. B. (1999) Mol. Cell 4, 331–341 [DOI] [PubMed] [Google Scholar]
- 16. Swanton E., High S., Woodman P. (2003) EMBO J. 22, 2948–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brockmeier A., Williams D. B. (2006) Biochemistry 45, 12906–12916 [DOI] [PubMed] [Google Scholar]
- 18. Ireland B. S., Brockmeier U., Howe C. M., Elliott T., Williams D. B. (2008) Mol. Biol. Cell 19, 2413–2423 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19. Watanabe D., Okabe M., Hamajima N., Morita T., Nishina Y., Nishimune Y. (1995) FEBS Lett. 368, 509–512 [DOI] [PubMed] [Google Scholar]
- 20. Persson S., Rosenquist M., Sommarin M. (2002) Gene 297, 151–158 [DOI] [PubMed] [Google Scholar]
- 21. Ikawa M., Wada I., Kominami K., Watanabe D., Toshimori K., Nishimune Y., Okabe M. (1997) Nature 387, 607–611 [DOI] [PubMed] [Google Scholar]
- 22. Ikawa M., Nakanishi T., Yamada S., Wada I., Kominami K., Tanaka H., Nozaki M., Nishimune Y., Okabe M. (2001) Dev. Biol. 240, 254–261 [DOI] [PubMed] [Google Scholar]
- 23. Cho C., Bunch D. O., Faure J. E., Goulding E. H., Eddy E. M., Primakoff P., Myles D. G. (1998) Science 281, 1857–1859 [DOI] [PubMed] [Google Scholar]
- 24. Kim E., Yamashita M., Nakanishi T., Park K. E., Kimura M., Kashiwabara S., Baba T. (2006) J. Biol. Chem. 281, 5634–5639 [DOI] [PubMed] [Google Scholar]
- 25. Nishimura H., Kim E., Nakanishi T., Baba T. (2004) J. Biol. Chem. 279, 34957–34962 [DOI] [PubMed] [Google Scholar]
- 26. Yamaguchi R., Yamagata K., Ikawa M., Moss S. B., Okabe M. (2006) Biol. Reprod. 75, 760–766 [DOI] [PubMed] [Google Scholar]
- 27. Yamaguchi R., Muro Y., Isotani A., Tokuhiro K., Takumi K., Adham I., Ikawa M., Okabe M. (2009) Biol. Reprod. 81, 142–146 [DOI] [PubMed] [Google Scholar]
- 28. Ensslin M. A., Shur B. D. (2003) Cell 114, 405–417 [DOI] [PubMed] [Google Scholar]
- 29. Hickox J. R., Bi M., Hardy D. M. (2001) J. Biol. Chem. 276, 41502–41509 [DOI] [PubMed] [Google Scholar]
- 30. Watanabe D., Sawada K., Koshimizu U., Kagawa T., Nishimune Y. (1992) Mol. Reprod. Dev. 33, 307–312 [DOI] [PubMed] [Google Scholar]
- 31. Inoue N., Okabe M. (2008) Methods Mol. Biol. 475, 335–345 [DOI] [PubMed] [Google Scholar]
- 32. Yamagata K., Nakanishi T., Ikawa M., Yamaguchi R., Moss S. B., Okabe M. (2002) Dev. Biol. 250, 348–357 [PubMed] [Google Scholar]
- 33. Clermont Y., Trott M. (1969) Fertil. Steril. 20, 805–817 [DOI] [PubMed] [Google Scholar]
- 34. Martin V., Groenendyk J., Steiner S. S., Guo L., Dabrowska M., Parker J. M., Müller-Esterl W., Opas M., Michalak M. (2006) J. Biol. Chem. 281, 2338–2346 [DOI] [PubMed] [Google Scholar]
- 35. Michalak M., Groenendyk J., Szabo E., Gold L. I., Opas M. (2009) Biochem. J. 417, 651–666 [DOI] [PubMed] [Google Scholar]
- 36. Hagaman J. R., Moyer J. S., Bachman E. S., Sibony M., Magyar P. L., Welch J. E., Smithies O., Krege J. H., O'Brien D. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2552–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ikawa M., Inoue N., Benham A. M., Okabe M. (2010) J. Clin. Invest. 120, 984–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim E., Baba D., Kimura M., Yamashita M., Kashiwabara S., Baba T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18028–18033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Lith M., Hartigan N., Hatch J., Benham A. M. (2005) J. Biol. Chem. 280, 1376–1383 [DOI] [PubMed] [Google Scholar]
- 40. van Lith M., Karala A. R., Bown D., Gatehouse J. A., Ruddock L. W., Saunders P. T., Benham A. M. (2007) Mol. Biol. Cell 18, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oliver J. D., van der Wal F. J., Bulleid N. J., High S. (1997) Science 275, 86–88 [DOI] [PubMed] [Google Scholar]
- 42. Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., Zipfel C., Jones J. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nishimura H., Kim E., Fujimori T., Kashiwabara S., Kuroiwa A., Matsuda Y., Baba T. (2002) Gene 291, 67–76 [DOI] [PubMed] [Google Scholar]
- 44. Toyoda Y., Yokoyama M., Hoshi T. (1971) Jpn. J. Anim. Reprod. 16, 147–151 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.