Abstract
Recent studies show that type II transmembrane serine proteases play important roles in diverse cellular activities and pathological processes. Their expression and functions in the central nervous system, however, are largely unexplored. In this study, we show that the expression of one such member, matriptase (MTP), was cell type-restricted and primarily expressed in neural progenitor (NP) cells and neurons. Blocking MTP expression or MTP activity prevented NP cell traverse of reconstituted basement membrane, whereas overexpression of MTP promoted it. The NP cell mobilization induced by either vascular endothelial growth factor or hepatocyte growth factor was also impaired by knocking down MTP expression. MTP acts upstream of matrix metalloproteinase 2 in promoting NP cell mobility. In embryonic stem cell differentiation to neural cells, MTP knockdown had no effect on entry of embryonic stem cells into the neural lineage. High MTP expression or activity, however, shifts the population dynamics from NP cells toward neurons to favor neuronal differentiation. This is the first report to demonstrate the direct involvement of type II transmembrane serine protease in NP cell function.
Keywords: Cell Surface Enzymes, Differentiation, Embryonic Stem Cell, Neural Stem Cell, Protease, 46C, Sox1, Migration
Introduction
Neurogenesis in the brain involves proliferation of neural stem cells or their progeny, migration of neuroblasts to their final locations, and their ultimate maturation into functional neurons. This multistep process is mediated by a series of sequential cellular interactions. In the normal brain, constitutive neurogenesis is found in two locations: the subventricular zone (SVZ)2 of the lateral ventricle (LV), which gives rise to interneurons in the olfactory bulb, and the dentate gyrus subgranular zone of the hippocampus (HP), which gives rise to neurons in the dentate granule cell circuitry for memory and learning (1, 2). It has been shown that proliferation of neural progenitor (NP) cells in both locations is enhanced after brain injuries, and streams of neuroblasts are attracted by and move toward the injured sites (3–7). However, there is not significant endogeneous neurogenesis to regenerate the damaged areas (8, 9). This problem is partly due to the insufficient numbers of cells surviving or reaching the damaged sites. Nevertheless, this has raised the possibility of inducing neurogenesis because these proliferating cells may be able to replace damaged brain cells. Secreted factors such as cytokines, neurotrophic factors, growth factors, proteoglycan (10), and extracellular proteases (11) have significant influence on neurogenesis under both physiological and pathological conditions (7, 12).
Extracellular and pericellular proteolytic enzymes play important roles in development, tissue homeostasis, and remodeling in all organ systems. Protease cascade of the serine urokinase plasminogen activator/plasminogen and the zinc-dependent metalloproteinases may be responsible for the majority of pericellular proteolysis (13). In the brain, serine proteases including the plasminogen activator system have well established functions in synaptic plasticity (14, 15). Matrix metalloproteinases (MMPs) have been implicated in the pathology and regeneration in the central nervous system (16). Their roles in NP cells have been investigated only recently. Expressed at low levels in the adult brain, MMP expression is stimulated by trauma and can be linked to repair after injury (17). It has been shown that MMP2 and MMP9 expressed by endothelial cells promote NP cell migration (18). More recently, MMP3 and MMP9 were shown to be expressed by NP cells and are involved in migration and differentiation of NP cells (19).
Neurotrophic factors, growth factors, plasminogen activators, and MMPs were synthesized and secreted mostly in their latent forms and require a proteolytic activation process to generate functionally active proteins. The type II transmembrane serine proteases (TTSP) represent candidate activator proteins. Members of this family are usually composed of short cytoplasmic N-terminal domains, a single transmembrane domain, an extracellular domain containing a serine protease domain, and multiple repeats of various domains involved in protein-protein interactions (20). They are ideally positioned to interact with surface proteins, soluble proteins, matrix components, and proteins on adjacent cells. Studies in recent years have shown that TTSPs have diverse roles in cellular activities and pathological processes, including epidermal differentiation (21), hearing (22), immune responses (23), as well as tumor invasion and metastasis (24). Currently, only a handful of TTSPs are reported in the central nervous system (20, 25). The details of their expression and function in the central nervous system or in NP cells, however, are largely unknown. In this work, we examined the expression and function of one TTSP member, matriptase (MTP) in the various cell types in mouse brain NP cells. We further used an in vitro neuronal differentiation culture derived from embryonic stem cells to investigate the function of MTP in NP cells and in neurogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture Reagents
Glasgow modification of Eagle's medium, Dulbecco's modified Eagle's medium (DMEM)/F-12 (50/50), neural basal medium, fetal bovine serum (FBS), knock-out serum replacement, glutamine, sodium pyruvate, N2 supplement, B27 supplement, 2-mercaptoethanol, bovine serum albumin fraction V (BSA-V), and Hank's buffered saline solution were purchased from Invitrogen. Recombinant proteins of leukemia inhibitory factor and MMP inhibitor GM6001 were from Chemicon (Millipore Corp., Billerica, MA). Recombinant proteins of SDF-1α, VEGF, and HGF were purchased from PeproTech Inc. (Rocky Hill, NJ). Poly-d-lysine and growth factor-reduced Matrigel were from BD Biosciences (Bedford, MA). Laminin-1 and recombinant FGF2 were purchased from R&D System, Inc. (Minneapolis, MN). Gelatin and heparin were from Sigma, and FGF2 was from BIOSOURCE (Invitrogen BIOSOURCE Division, Carlsbad, CA).
Reagents and Enzymes for RNA Works
Lipofectamine 2000 transfection reagent, TRIzol reagent, phenol/chloroform (1:1) solution, RNase H, Superscript III reverse transcriptase, oligo(dT)12–18 primers, and dispase were purchased from Invitrogen. Collagenase was purchased from Worthington (Lakewood, NJ). The small interfering RNA (siRNA) against mouse St14 and a control siRNA with no target were purchased from (Qiagen). The MTP overexpression plasmid was a pSPORT6-CMV vector carrying the full-length MTP cDNA. Pro-HGF protein was from Sigma and pro-MMP2 was from R&D Systems.
Antibodies and Chemicals
Mouse monoclonal antibodies to α-actin, βIII-tubulin, neuronal nuclei (NeuN), SSEA1, GalC, nestin, and PSA-NCAM were purchased from Chemicon. Monoclonal antibody to Oct3/4, rabbit polyclonal anti-cMet, and rabbit polyclonal anti-matriptase antibody were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Sheep polyclonal anti-matriptase antibody and anti-chemokine receptor 4 antibody were from R&D Systems. Anti-GFP and anti-CD133 antibodies were from Abcam (Cambridge, UK). Anti-GFAP antibody was purchased from Molecular Probes (Eugene, OR), anti-double cortin antibody was purchased from BD Biosciences, and anti-c-Kit antibody was purchased from eBioscience Inc. (San Diego, CA). FITC-conjugated mouse anti-BrdU antibody was purchased from Roche Applied Science. 7-Amino-actinomycin D (7-AAD) was from BD Biosciences. Paraformaldehyde solution was purchased from Electron Microscopy Sciences (Haffield, PA). All other chemicals were purchased from Sigma.
Embryonic Stem (ES) Cell Culture and Neural Differentiation
Sox1-GFP knock-in mouse ES cells (46C ES cells) (26) obtained from Dr. Austin Smith (University of Edinburgh, United Kingdom) were routinely propagated in 0.1% gelatin-coated Petri dishes without feeder cells in Glasgow modification of Eagle's medium supplemented with 1% FBS, 10% knock-out serum replacement, 0.1 mm 2-mercaptoethanol, 2 mm glutamine, 1 mm sodium pyruvate, and 20 ng/ml of leukemia inhibitory factor. Embryonic stem cell properties were monitored via morphology and Oct3/4 expression as determined by immunofluorescent staining. The cells were never kept in culture over 16 passages. Neuronal commitment was induced by placing 46C ES cells on a gelatin-coated surface at a density of 1–1.5 × 104 cells/cm2 in neuronal differentiation medium. Neuronal differentiation medium is composed of DMEM/F-12 (50/50) (1:1) with neural basal medium supplemented with modified N2, B27, and 50 μg/ml of BSA-V (referred to as N2B27 medium) (26).
For some experiments as indicated, the 46C ES cell-differentiated NP cells were plated on a poly-d-lysine/laminin-coated surface at a density of 1 to 1.5 × 104/cm2 in N2B27 medium for differentiation of mature neurons. To obtain neurospheres, 46C ES cells were dissociated and cultured in uncoated 6-well tissue culture plates at 104 cells/cm2 in N2B27 medium containing 2 mm l-glutamine and 1 mm 2-mercaptoethanol. Neurospheres that expressed Sox1-GFP generally developed by day 4.
Astrocyte Differentiation
NP cells were plated on the poly-d-lysine/laminin-coated surface and cultured in N2B27 medium supplemented with 2% FBS and 50 ng/ml of ciliary neurotrophic factor for 10 days.
Isolation of NP Cells from the SVZ of the Adult Mouse Brain
Adult (8 to 10 weeks of age) male FVB mice were euthanized by CO2 inhalation followed by cervical dislocation. The brains were immediately removed, placed in a mouse brain blocker, and sliced in the coronal direction at 500 μm thickness. LV areas were taken from the brain slices around 1.18 to 0 mm relative to the bregma, as determined from the adult mouse brain atlas (60). The lateral walls (SVZ) of the LV were pinched away under a dissecting scope (Olympus SZ) and placed in DMEM/F-12 (50/50) containing 1 mm N-acetyl-l-cysteine. Tissues were passed through a 25-gauge, then a 30-gauge needle attached to a 1-ml syringe. Collagenase and dispase were added to the broken tissues at final concentrations of 0.62 and 66.7 units/ml, respectively, and tissues were incubated for 30 min at 37 °C. Four volumes of Hank's-buffered saline solution were added, and tissues were further triturated by passing them eight times through a P1000 pipette tip. The dissociated tissue samples were incubated at room temperature for 5 min to allow the connective tissue to settle, and the top half of the supernatant was collected and passed through 35-μm cell strainers. An equal volume of Hank's-buffered saline solution was added to the remaining tissue slurry, and these steps were repeated twice. All collected cell suspensions were pooled together and centrifuged for 5 min at 900 × g, cells were then resuspended and grown in neurospheres in DMEM/F-12 (50/50) supplemented with B27, 2 μg/ml of heparin, and 20 ng/ml of FGF2.
Primary Culture of Mouse Astrocytes and Microglial Cells
The brains of postnatal day 5 FVB mice were dissected, and the meninges were removed. The brains were cut into two hemispheres in the sagittal direction. The neocortex was peeled away from the other parts of the brain tissue, minced into small pieces, and briefly digested with 0.1% trypsin/EDTA in DMEM/F-12 (50/50). After centrifugation at 1,500 × g for 5 min, the tissue pellets were washed twice and further triturated in DMEM/F-12 (50/50) using a pipette tip. The dissociated single cell suspension was then plated in a poly-d-lysine pre-coated T25 flask in DMEM/F-12 (50/50) supplemented with 10% FBS. The culture medium was changed on days 3 and 6 of culture. At 8 days culture, microglial cells were detached from the astrocyte monolayer by shaking at 350 rpm for 20 min. The detached primary microglial cells were collected for culture. Fresh medium was added to the attached astrocyte culture. After shaking overnight at 250 rpm in a 37 °C incubator, medium was removed, and the attached primary astrocytes were kept in culture with medium containing 100 μm cytosine β-d-arabinofuranoside (Sigma) for 3 days. The purity of astrocytes and microglial cells was confirmed by their morphology and expression of GFAP and Iba1, respectively. Typically, more than 90% purity was achieved for both cell types.
Brain Endothelial Cell and Microglial Cell Culture
The mouse brain endothelial cell line bEnd.3 was purchased from the Bioscience Collection and Research Center (Hsinchu, Taiwan, China). Cells were cultured in DMEM supplemented with 4 mm glutamine, 1.5 g/liter of sodium bicarbonate, 4.5 g/liter of glucose, and 10% FBS. The microglial cell line BV2 was routinely cultured in DMEM supplemented with 10% FBS.
Immunofluorescence Staining, BrdU Labeling of Cell Culture, and 7-AAD Flow Cytometric Assay
Cells cultured on coverslips or 8-chamber slides were fixed in 2% paraformaldehyde for 30 min at room temperature. Cells were washed with PBS and then permeabilized with 0.5% Triton X-100. After washing, cells were blocked with 4% horse serum and then incubated with primary antibody. Unbound antibody was removed by three washes of phosphate-buffered saline (PBS). The cells were then incubated with green or red fluorescence-conjugated secondary antibodies. Cell nuclei were counterstained with DAPI, and cells were subsequently mounted and analyzed using epifluorescence (Olympus BX51) or confocal (Leica TCS NT) microscopy. All negative control staining was performed using non-immune IgG of the same subtype.
For BrdU pulse labeling, 10 mm BrdU was added to the day 4 NP cell culture for 2 h. After fixation in 4% paraformaldehyde, cells were permeabilized with Triton X-100. DNA was denatured in ice-cold 0.1 m HCl for 20 min and incubated in 2 m HCl for 1 h at 37 °C. After neutralization in 0.1 m borate buffer, cells were blocked and stained with FITC-conjugated anti-BrdU antibody as described above.
For 7-AAD flow cytometric assay, day 4 NP cells were resuspended in PBS and then incubated with 7-AAD at 50 ng/104 cells for 5 min. Cells were then assayed using a Calibur cytometer (BD Biosciences).
Immunohistochemical Staining of Paraffin-embedded Mouse Brain Sections
Brains were taken from 8–12-week-old FVB males, fixed in 4% paraformaldehyde/PBS fixative solution, and embedded in paraffin as described previously (27). Immunohistochemical staining was performed using the DAKO EnVision+ kit (Dako Cytomation, Inc., Carpinteria, CA) as described (27). For MTP staining, tissue sections were heated 20 min at 95 °C in 10 mm citrate (pH 6.0) for antigen retrieval, incubated with sheep primary antibody, then with rabbit anti-sheep IgG before incubation with the peroxidase-labeled polymer-conjugated anti-rabbit antibody. Bound antibodies were detected with DAB (3′,3′-diaminobenzidine) substrate/chromogen solution. Cell nuclei were counterstained with hematoxylin.
NP Cell Migration Assay
For the migration assay, neurospheres were prepared from 46C ES cells as described under “ES Cell Culture and Neural Differentiation.” On day 2, cells that developed into round spheroids were collected using a cell strainer with a pore size of 40 μm. The collected cell spheroids were either cultured directly with freshly prepared neuronal differentiation medium or transfected with siRNA or DNA plasmid using Lipofectamine 2000 (see next section). After 48 h of incubation, GFP-positive neurospheres 120–150 μm in diameter were hand-picked under the microscope and used in the migration assay. The migration assay was performed in a transwell system with a pore size of 8 μm (Corning Inc., Corning, NY). The upper chamber of the transwell was coated with a thin layer of growth factor-reduced Matrigel/neuronal differentiation medium (2:1). Neurospheres were then added to the coated upper chamber (10 spheres per chamber) and cultured in neuronal differentiation medium. Chemoattracting growth factors were added to the bottom chamber in neuronal differentiation medium. In cultures where protease inhibitors were included, the same concentration of inhibitors was added to both the top and bottom chambers. After 3 days in culture, the number of cells that had traversed to the bottom chamber were counted.
siRNA and Plasmid Transfection
Transfection of siRNA or mammalian expression plasmid was performed using Lipofectamine 2000 following the protocol provided by the manufacturer. Briefly, sufficient siRNA or plasmid DNA was mixed and incubated with Lipofectamine 2000 reagent at room temperature. The DNA or siRNA/Lipofectamine mixtures were then added to the cells followed by incubation. Most GFP-positive NP cells were successfully transfected with siRNA after 48 h of incubation as monitored using fluorescently labeled siRNA. MTP knockdown or overexpression efficiency was checked by RT-PCR.
RNA Isolation, cDNA Synthesis, and PCR
RNA was extracted from brain tissues using TRIzol reagent (Invitrogen) following the manufacturer's protocol. Briefly, brain tissues were ground up in TRIzol using a motorized pellet pestle. After 10 min of centrifugation at 12,000 × g, tissue homogenates were mixed with chloroform by vortexing and then centrifuged. Total RNA was obtained from the aqueous phase by isopropyl alcohol precipitation. Genomic DNA was removed by a 15-min treatment with DNase I (Roche Applied Science) at 37 °C. After heat inactivation, DNase I was removed by phenol/chloroform (1:1) extraction, and RNA was recovered by alcohol precipitation. To extract RNA from cells, cells were directly lysed with TRIzol, and the same procedures as described above were performed. RNA integrity was assessed by electrophoresis, and RNA concentration was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA). The average 260/280 nm ratio (RNA purity) was greater than 1.9. First-strand cDNA was synthesized from 1 μg of RNA using Superscript III reverse transcriptase and oligo(dT) primers at 50 °C for 1 h. cDNA synthesis was stopped by heat inactivation at 70 °C for 15 min, and the RNA in the RNA/DNA hydrid was digested with RNase H. The resulting cDNA was then used in PCR. The gene specific primers were as follows: S15 (mouse ribosomal protein S15), forward, 5′-TTCCGCAAGTTCACCTACC-3′, reverse, 5′-CGGGCCGGCCATGCTTTACG; MTP, forward, 5′-CACTTCCATTATCGGAATGTGCG, reverse, 5′-GGATGTCGCCGGTCAGTATTGGTATCA; MTP3, forward, 5′-CTCATGTTGGTGACACTGAAGTCTCC, reverse, 5′-GGAAGATGCTGCTGTTGCAGGCAGG; MMP2, forward, 5′-ACCATCGAGACCATGCGG, reverse, 5′-CTCCCCCAACACCAGTGC; MMP3, forward, 5′-TTGACGATGATGAACGATGGA, reverse, 5′-CGATCTTCTTCACGGTTGCA; MMP9, forward, 5′-TTCTGCCCTACCCGAGTGGA, reverse, 5′-CATAGTGGGAGGTGCTGTCGG.
Western Blot
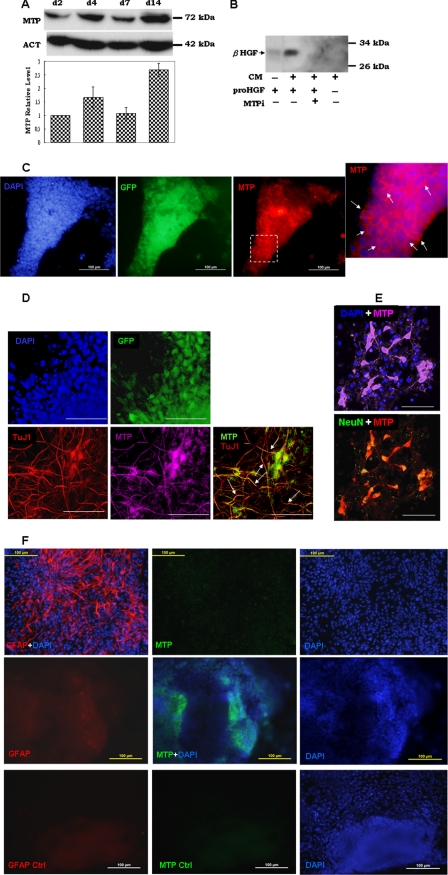
Protein extracts were prepared from brain tissues and cultured cells as described previously (21, 22). Protein concentrations of the cell and tissue extracts were determined by SuperSignal® West Pico protein assay kit (Pierce) using bovine serum albumin as a standard. An equal amount of protein was subjected to SDS-PAGE, followed by Western blotting as described previously (28). For quantitation (graph in Fig. 2A), x-ray films from Western immunoblots were scanned with a Bio-Rad computing densitometer, and the integrated absorbance corresponding to the area of MTP was recorded. The corresponding absorbance of MTP was adjusted to that of loading marker actin. Only the protein bands within the linear range of the x-ray film exposure were taken into quantitative measurement. The graph in Fig. 2A shows the MTP value by densitometry; all values are normalized to that of day 2 point.
FIGURE 2.
Expression of MTP during neural differentiation of mouse ES cells. A, 46C ES cells were seeded on a collagen-coated surface and cultured in neural differentiation culture conditions as described under “Experimental Procedures.” After 2 (d2), 4 (d4), and 7 days (d7) post-culture, cell extracts were prepared and subjected to Western immunoblot with anti-MTP (MTP) or anti-actin (ACT) antibody. The graph shows quantitation of chemiluminescence by densitometry. The value for each point was normalized to that for the day 2 point. Data were collected from two to three independent experiments. B, pro-HGF protein was incubated at room temperature with (+) or without (−) the various components as indicated. CM, conditional medium of Sox1-GFP+ NP cell culture; proHGF, single-chain latent form HGF preparation; MTPi, MTP-specific inhibitor. The lane marked “minus” in the CM row indicates unconditioned culture medium. Samples were boiled in SDS sample buffer and subjected to Western immunoblot with antibody to the β-chain HGF (β-HGF). C, day 4 cultures of NP cells were stained with antibodies to GFP (green) and MTP (red), and then detected with FITC- and TRITC-conjugated secondary antibodies, respectively. Arrows in the enlarged image of MTP staining shows the cell membrane location of this protein. Nuclei were counterstained with DAPI (blue). D, day 7 NP cells were triple stained with antibodies to GFP (green), MTP (purple), and βIII-tubulin (TuJ1) (red). Secondary antibodies used were FITC, Cy5, and TRITC-conjugated immunoglobin for GFP, MTP, and TuJ1, respectively. In MTP-TuJ-1 combined images, MTP is pseudo-colored in green for better viewing. White arrows indicate co-localization of TuJ1 and MTP on neurites. E, day 14 cells were stained with MTP and NeuN and visualized by Cy5 and TRITC-conjugated secondary antibodies, respectively. In the MTP-NeuN combined image, MTP staining was pseudo-colored in red for better viewing. Nuclei in all images were counterstained with DAPI (blue). Images in C are epifluorescence images. F, Sox1-GFP+ NP cells were induced to glial differentiation with ciliary neurotrophic factor for 10 days. Cells were stained with GFAP (red) and MTP (green). Images in the bottom column show background staining using the same subtype of immunoglobin as anti-GFAP (GFAP Ctrl) or anti-MTP (MTP Ctrl). Nuclei were counterstained with DAPI (blue). Images in D and E show three-dimensional Z-stack reconstructions scanned by confocal microscopy at 2-μm increments. The rest of the images were taken using epifluorescent microscopy. Bars: 100 μm in C and F, 50 μm in D and E.
RESULTS
MTP Is Expressed in NP Cells, Neurons, and during Neural Lineage Differentiation by ES Cells
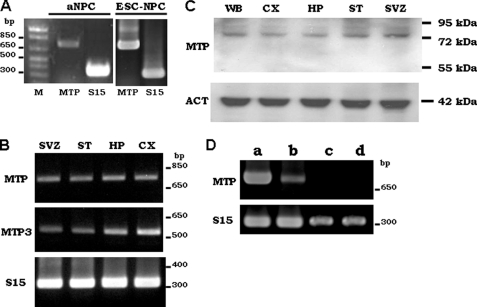
NP cells, both isolated from the SVZ of the adult mouse brain LV (Fig. 1A, aNPC) and differentiated from embryonic stem cells (Fig. 1A, ESC-NPC), expressed MTP mRNA. RT-PCR also detected MTP mRNA in adult brain extracts taken from striatum, neocortex, and the two known neurogenic areas: the SVZ of the LV and the HP (Fig. 1B) along with the expression of MTP3 (Fig. 1B, MTP3), another member of the MTP family that was shown to be expressed in the brain (29). Exponential amplification for both MTP and MTP3 in all the tested tissues occurred between PCR cycles 28–30 (supplemental Fig. S1A), suggesting that the MTP mRNA level in brain is comparable with that of MTP3. Western immunoblot (Fig. 1C) showed that a single protein of 70–72 kDa was detected in tissue extracts taken from whole brain, neocortex, HP, striatum, and SVZ by the MTP-specific antibody. MTP was shown to be present either as the full-length protein of about 95 kDa or as the N-terminal processed protein of 85–72 kDa (30–32). The antibody used recognizes a region in the N terminus of the processed protein. The detection of a single protein of 70–72 kDa in brain extracts indicated that MTP is present mostly in the N-terminal processed form. To further verify the specificity of MTP expression in the central nervous system, its expression in the various types of cells present in the brain were examined. The PCR product of MTP was detected in NP cells between amplification cycles 28 and 30, whereas it was barely detected in bEND cells after the same number of amplification cycles (supplemental Fig. 1B). After five more cycles of amplification, there is some MTP PCR product in bEND cells (Fig. 1D, lane b), but none was detected in the mRNA of either astrocytes (Fig. 1D, lane c) or microglial cells (Fig. 1D, lane d) isolated from the brain.
FIGURE 1.
Expression of MTP in neural progenitor cells. A, NP cells isolated from the SVZ of the LV (aNPC) or differentiated from mouse ES cells (ESC-NPC) were examined by RT-PCR for MTP expression. Both mRNA (B) and protein (C) of MTP were detected in extracts of adult mouse whole brain (WB), neural cortex (CX), hippocampus (HP), striatum (ST), and SVZ of adult mouse brain. D, RT-PCR of MTP in the NP cells differentiated from the 46C-ES cell (first lane), in mouse brain endothelial cells (second lane), in primary astrocytes (third lane), and in primary microglia (fourth lane) isolated from mouse brain. Mouse small ribosome protein 15 (S15) is used as an internal loading control. Actin (ACT) is used as loading control for Western blot.
Immunohistochemical staining of the adult mouse brain showed positive staining of MTP in areas that are enriched for β-tubulin III (TuJ1) or NeuN positive neurons (supplemental Fig. S2). MTP staining, however, was lighter compared with TuJ1 or NeuN staining. The neurons of the neural cortex, the granular neurons in the CA regions and dentate gyrus of HP, and the Purkinje neurons in the cerebellum were all positive for MTP (supplemental Fig. S2). In addition, there is strong MTP immunoreactivity in the lining around the ventricles (supplemental Fig. S2).
To further investigate the expression and possible function of MTP in NP cells, we used an in vitro neural precursor differentiated from ES cells in a feeder-free adherent culture. The Sox1-GFP knock-in ES cell 46C (26) was used in these studies. Sox1 is a mammalian neural progenitor marker; it is first expressed in the neural plate and neuroepithelial cells, but is down-regulated during neuronal differentiation. In 46C ES cells to neural differentiation culture, neural commitment and the subsequent differentiation can be monitored, respectively, by the appearance and disappearance of GFP expression (26). The Sox1-GFP expression also allows isolation of NP cells using fluorescence-activated cell sorting (33). Sox1-GFP expression were detected in a few cells as early as 2 days after 46C ES cells were cultured under neural commitment culture conditions (supplemental Fig. S3, d2, Sox1-GFP). Cells at this time had lost their ES cell identity as shown by loss of Oct4 expression (supplemental Fig. S3, panel d2, Oct4). By days 4 or 5 of neural culture, bright GFP expression was seen in over 75% of the cells (supplemental Fig. S3). These GFP-positive cells expressed neural progenitor markers such as nestin and CD133 (Table 1). PSA-NCAM and double cortin, two proteins that are associated with migrating neuroblasts, were also detected in Sox1-GFP+ NP cells (Table 1). Most of the Sox1-GFP+ cells at this stage were negative for neuron protein β-tubulin III (supplemental Fig. S3, panel d4, TuJ1). By day 7 post-culture, many Sox1-GFP+ cells showed neuronal morphology and expressed early neuron protein β-tubulin III (supplemental Fig. S3, panel d7, TuJ1), but none of the cells expressed the mature neuron marker neuronal nuclei (NeuN) (Table 1). Further differentiation of mature neurons occurred after Sox1-GFP+ NP cells were cultured on poly-d-lysine/laminin. Upon a 2-week culture on poly-d-lysine/laminin, 70–90% of the cells in this culture lost Sox1-GFP expression, and more than 50% of them expressed the mature neuron marker NeuN (supplemental Fig. S3, d14). Thus, ES cells switched identity to NP cells around day 2 in feeder-free adherent culture. By days 4–5, the switch from ES cell identity to NP cell identity was complete, and gave rise to neural progeny with features of neuroblasts. From this time point on, the transit amplifying population was established and became neuron committed by day 7 (d7). Similar observations were described by Abranches et al. (34) recently using a different culture medium.
TABLE 1.
Molecular confirmation of the 46C ES cells and its differentiated NP cells
Expression of these molecules was detected by immunofluorescent staining except for VEGFR2, which was examined by RT-PCR.
| Marker | ES cell culture | NP cell culturea |
|---|---|---|
| Oct3/4 | + | − |
| Nestin | − | + |
| CD133 | Low | + |
| SSEA1 | + | + |
| PSA-NCAM | NAb | + |
| DCX | NAb | low |
| TujI | NAb | +/− |
| NeuN | NAb | − |
| GalC | NAb | +/− c |
| GFAP | − | − |
| CXCR4 | − | + |
| c-Kit | − | − |
| VEGFR2 | NAb | + |
| c-Met | + | + |
a The d5 Sox1-GFP+ NP cells were fixed and stained for indicated protein markers.
b Markers were not tested in these cells.
c Both positive and negative cells were observed.
MTP protein was detected by Western immunoblot in all stages of NP cells (Fig. 2A, d2, d4, and d7) and in their differentiated neurons (Fig. 2A, d14). A 70–72-kDa N-terminal-processed form of MTP was found both in the cell lysate (Fig. 2A) and in the conditioned medium (data not shown). The lower panel in Fig. 2A is a graphic presentation of the changes in the MTP protein level during NP cell differentiation. After normalization to the day 2 culture time point the MTP protein level increased slightly in day 4 NP cells, a time point when the shift from ES to NP identity is completed. After a 14-day culture of Sox1-GFP+ NP cells on poly-d-lysine/laminin, when differentiation of neurons from NP cells is completed, there was more than a 2-fold increase of MTP protein (Fig. 2A, d14). To investigate the activation state of MTP in Sox1-GFP+ NP cells, we checked their ability to activate pro-hepatocyte growth factor (pro-HGF). We showed previously that the single chain pro-HGF is cleavage activated by MTP to α- and β-chain HGF (28). Fig. 2B shows that incubation of the pro-HGF protein with the conditioned medium of Sox1-GFP+ NP cells produced a large amount of β-chain of the activated HGF. Addition of a potent peptide inhibitor of MTP (MTPi), the sunflower seed trypsin inhibitor 1 (27, 35), significantly impaired the generation of β-chain HGF. These data showed that MTP expressed in NP cells is the activated protease.
Immunofluorescent cell staining of day 4 Sox1-GFP+ NP cells showed positive MTP staining on the cell surface (Fig. 2C), confirming the widespread expression of MTP in NP cells. On day 7, when many of the Sox1-GFP+ cells showed neuronal features (Fig. 2D), MTP was found in the TuJ1-positive cells and was co-localized with TuJ1 in neurites (Fig. 2D, arrows). Expression of MTP in neurons was further demonstrated by immunostaining of day 14 cells, which showed positive staining of MTP on the surface of NeuN positive cells (Fig. 2E).
To further verify MTP expression in astrocytes, Sox1-GFP+ NP cells were cultured under glial differentiation conditions and expression of MTP in the differentiated astrocytes was investigated by immunohistochemistry. As shown in Fig. 2F, GFAP positive astrocytes (top panel, GFAP+DAPI) were essentially negative for MTP staining (top panel, MTP). A few cells that expressed MTP (Fig. 2F, middle panel, MTP+DAPI) were completely negative for GFAP expression (Fig. 2F, compare middle panel GFAP with bottom panel GFAP Ctrl).
Taken together, these data demonstrate that MTP is present in the brain. Its expression is generally lower than that of NeuN or TuJ1 and is restricted to NP cells and neurons. The cell type-specific and low abundant expression of MTP could account for the earlier conclusion that MTP is absent in the brain.
MTP Is Required for NP Cells to Migrate Across Reconstituted Basement Membranes
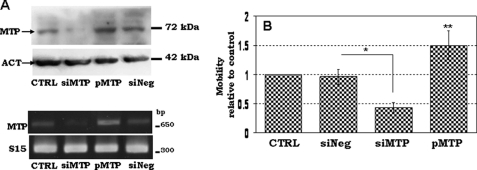
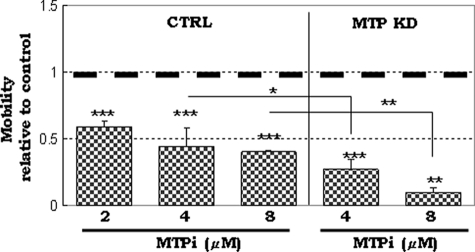
It has been shown that NS/P cells possess migratory ability across the reconstituted basement membrane in vitro (19, 36). MTP has been implicated in cell migration and cancer metastasis (28, 37). We tested if MTP is involved in NP cell mobility across the basement membrane components (EC Matrix) in transwell. We altered the MTP expression level in Sox1-GFP+ NP cells either by knocking down the endogeneous MTP with specific siRNA or by overexpressing the protein ectopically with an expression plasmid carrying the full-length coding region of mouse MTP. Western blotting (Fig. 3A, upper panel) showed that 48 h after transfection with MTP-specific siRNA (siMTP), cells had largely lost their expression of MTP protein, whereas cells transfected with MTP expression plasmid (pMTP) showed increased levels of MTP protein. Transfection with a non-silencing control siRNA (siNeg) or an empty plasmid3 did not change the MTP expression level, demonstrating that changes of MTP expression were not due to the nonspecific effect of the transfection reagent. RT-PCR showed the same results (Fig. 3A, lower panel). Sox1-GFP+ NP cells plated in the top chamber of EC matrix-coated transwells migrated downward toward the bottom chamber (Fig. 3B, CTRL); transfection with a non-silencing control siRNA did not affect the cell ability to traverse the EC matrix (Fig. 3B, siNeg). The number of cells that moved to the bottom chamber decreased by about 60% when transfected with MTP-specific siRNA (Fig. 3B, siMTP). Overexpression of MTP, on the other hand, increased the number of cells in the bottom chamber by about 50% (Fig. 3B, pMTP). The involvement of MTP in NP cell mobility was also demonstrated using the MTP-specific inhibitor (Fig. 4) (23, 31). The inhibitor reduced Sox1-GFP+ NP cell migration in a concentration-dependent manner (Fig. 4, MTPi). Over 40% reduction in cell migration was achieved with 2 μm of the inhibitor, and about 60% reduction in cell migration was seen with 8 μm of the inhibitor, similar to the level of reduction seen in cells transfected with MTP siRNA (Fig. 4). A combination of MTP inhibitor and MTP knockdown (Fig. 4, MTP-KD) resulted in further reduction of migrating cells to less than 10% (Fig. 4, MTP-KD). Apparently, the remaining MTP protein activity due to incomplete knockdown with MTP siRNA was impaired by the inhibitor to achieve the largest reduction of cell migration. These studies demonstrated that MTP, likely its protease activity, is critical for NP cell migration.
FIGURE 3.
The effect of MTP on Sox1-GFP+ NP cell migration across the Matrigel. A, protein assays (upper panel) and RT-PCR assays (lower panel) for MTP in the control day 4 NP cells (CTRL), day 4 NP cells transfected with MTP siRNA (siMTP), no-target control siRNA (siNeg), or MTP expression plasmid (pMTP). Mouse ribosomal protein S15 was used as an internal loading control. B, the mobility of cells prepared in A was assayed in transwells as described under “Experimental Procedures.” Migration of all treatments was compared with that of control that is set at 1. Data were collected from five to seven independent experiments. *, p ≤ 0.05; **, p ≤ 0.01.
FIGURE 4.
The effect of MTP peptide inhibitor on NP cell migration. Control transfected (CTRL) or MTP siRNA-transfected NP cells (MTP KD) were cultured in the presence of 2, 4, or 8 μm of the MTP inhibitor. Data were normalized to cultures without inhibitor (set at 1 shown with dark dotted line). Data were from 3 independent experiments. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
MTP Is Involved in Chemoattractant-induced NP Cell Migration
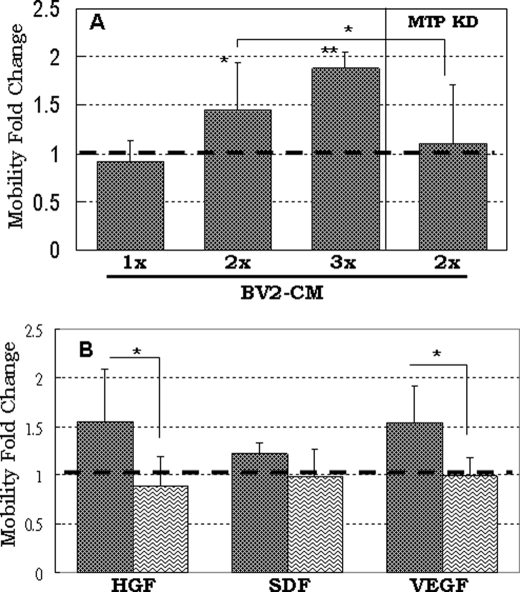
The migration of neuroblasts or neural precursor cells can be induced by a number of factors (7). Expression of many of these factors are induced in response to focal ischemia in the brain and act as chemoattractants to lure NP cells to migrate toward the damaged region (7, 38). Activated microglia and astrocytes are thought to be the source of various chemoattractants. It has been demonstrated that the conditioned medium of activated primary microglia and of the established microglial cell line BV2 can induce neural precursor cell migration (36). Stromal-derived factor-1α (SDF-1α) (39), vascular endothelial factor (VEGF) (40), and stem cell factor (41) are the most studied chemoattractants in NP cell migration. HGF was previously shown to be involved in interneuron (42) and striatal progenitor cell (43) migration. It was recently shown that HGF promotes migration of rostral migratory stream neuroblasts (44) and that HGF is the factor secreted by glioma to attract neural progenitor cells (45). We asked whether MTP is also involved in NP cell migration stimulated by these factors. Fig. 5A shows that the increase in NP cell migration in medium conditioned by BV2 cells is correlated with the amount of conditioned medium that was added (Fig. 5A, control cells). This migration promoting effect was almost completely abolished in cells transfected with MTP-specific siRNA (Fig. 5A, MTP-KD cells), implying that NP cell migration induced by the BV2-secreted factors is mediated by MTP. Sox1-GFP+ NP cells expressed chemokine receptor 4 (receptor for SDF-1α), VEGF receptor 2 and c-Met (receptor for HGF) but not c-Kit (receptor for stem cell factor) (Table 1). Both HGF and VEGF increased cell migration about 1.5-fold; SDF-1α on the other hand had little effect on Sox1-GFP+ NP cell migration (Fig. 5B). Cells transfected with MTP-specific siRNA showed no response to either of these factors (Fig. 5B, wavy line-filled bars). Evidently, MTP is responsible for cell migration induced by HGF, VEGF, and maybe other chemoattractants present in the BV2-conditioned medium.
FIGURE 5.
The involvement of MTP in chemoattractant-induced mobility of Sox-GFP+ NP cells. A, control or MTP siRNA-transfected NP cells (MTP KD) were plated on the top chamber of the transwell. To the bottom chamber, the conditioned medium of BV2 cells equivalent to 0.75 × 106 (1X), 1.5 × 106 (2X), or 2.25 × 106 (3X) cells were added. Data were normalized to control or MTP siRNA-transfected NP cells cultured in the regular neural growth medium (set at 1 shown with dark dotted line). B, non-silencing control siRNA-transfected NP cells (dark dot-filled bars) or MTP siRNA-transfected NP cells (waving line-filled bars) were plated in the top chamber of the transwell. HGF (20 ng/ml), SDF-1α (50 ng/ml), and VEGF (100 ng/ml) were added to the bottom chamber of the culture. Data were normalized to the no-chemokine control culture (set at 1 shown in dark dotted line). Data were from three independent experiments. *, p ≤ 0.05; **, p ≤ 0.01.
MTP Is Involved in Neuronal Differentiation
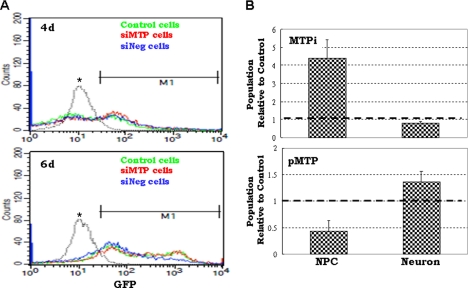
In the ES to neural cell differentiation culture system MTP expression was elevated at two time points: the completion of the switch from ES cell to NP cell identities (day 4 culture) and the complete differentiation of NP cells (day 14 culture) (Fig. 2B). This prompted us to evaluate the effect of MTP on neural specification. For neural commitment, we monitored the appearance of Sox1-GFP+ cells. After transfer to the monolayer neural differentiation medium, the proportion of Sox1-GFP+ cells from the control 46C ES cells (Fig. 6A, green line), MTP siRNA-transfected cells (Fig. 6A, red line), and the negative control siRNA-transfected cells (Fig. 6A, blue line) was monitored by FACS. Four days after the monolayer neural differentiation culture, each cell type generated about 50% GFP+ cells and their proportion was almost identical in all three types of cells (Fig. 6A, upper panel). The GFP+ population increased to about 80% after 6 days in monolayer neural differentiation culture; knock-down of MTP showed no effect on the proportion of GFP+ cells (Fig. 6A, lower panel). Clearly, MTP is not involved in neural lineage entry from ES cells. We next investigated the effect of MTP on changes of cell population between Sox1-GFP+ NP and NeuN+ neurons during the 14-day culture on poly-d-lysine/laminin. In early experiments, we found that siRNA oligonucleotides delivered by transfection reagent did not guarantee prolonged stability or inhibition of MTP expression during 14 days of neural differentiation culture. We therefore used the MTP inhibitor in these studies. In the presence of the inhibitor (Fig. 6B, MTPi), there was a more than 4-fold increase in the Sox1-GFP+ NP population compared with the control treatment (set at 1 as indicated by the dotted line in Fig. 6B), and the NeuN+ population was about 20% lower than the control. Overexpression of MTP, on the other hand, reduced the population of Sox1-GFP+ NP cells to half and increased the population of NeuN+ neurons by almost 50% (Fig. 6B, pMTP). These data demonstrated that high MTP expression or activity shifts the population dynamics between NP cells and neurons to favor neuronal differentiation.
FIGURE 6.
The effect of MTP on Sox-GFP+ NP cell differentiation. A, control 46C ES cells (control cells), 46C ES cells transfected with MTP siRNA (siMTP cells), or transfected with no-target control siRNA (siNeg cells) were transferred to neural differentiation culture for 4 (upper panel) or 6 days (lower panel). The GFP+ population was measured by FACS. M1 is the gate used to quantify the population of Sox1-GFP+ cells. The peak of GFP-negative, undifferentiated ES cells is marked by the asterisk. B, day 6–8 NP cells were plated on poly-d-lysine/laminin-coated coverslips and cultured for 14 days. Cells were fixed and immunostained for GFP and NeuN. Nuclei were stained with DAPI. The ratio of GFP+ NP cells to the DAPI-stained total cell number (labeled as NPC) and the ratio of NeuN+ neurons to the DAPI-stained total cell number (labeled as neurons) were compared. The upper panel graph represents cells cultured in the presence of 2 μm MTP inhibitor (MTPi); the lower panel graph shows cells transfected with the MTP expression plasmid (pMTP). Data were normalized to the corresponding control conditions for each treatment (set at 1 shown in dark dotted lines) and were collected from two independent experiments.
The effect of MTP on neural differentiation is not due to its effect on NP cell proliferation or cell death. As shown under “supplemental materials,” neither total cell number (supplemental Fig. S4A) nor neurosphere formation (supplemental Fig. S4B) was affected by blocking MTP activity. BrdU pulse labeling showed that the number of BrdU-positive cells was not affected by MTP siRNA or MTP overexpression (supplemental Fig. S4C) either. Evidently, MTP did not affect NP cell survival either, because neither the total number of dead cells (supplemental Fig. S4D, with MTPi) nor the amount of 7-AAD incorporation (supplemental Fig. S4D, with siMPT) was affected by treatment with the MTP inhibitor or MTP-specific siRNA.
MTP and MMPs Collaborate in Promoting NP Cell Migration
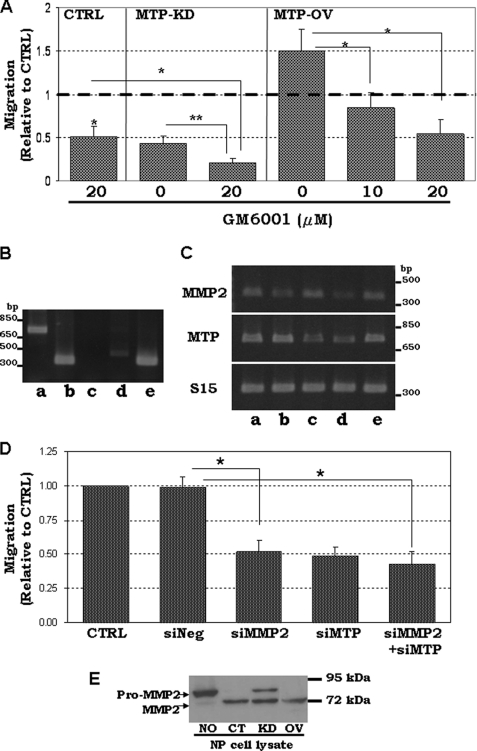
MMP3 and MMP9 have recently been shown to promote neural progenitor cell migration (19). To test if MTP-mediated NP cell migration involves MMP activity, we used the broad spectrum MMP inhibitor GM6001. GM6001 efficiently reduced Sox1-GFP+ NP cell migration (Fig. 7A, CTRL) to a level similar to the cells treated with MTP-specific siRNA (Fig. 7A, MTP-KD with 0 GM6001). As expected, there are GM6001-sensitive MMPs in Sox1-GFP+ NP cells that participate in the migration of these cells. When the same amount of GM6001 was used on cells whose MTP expression was knocked down by siRNA (Fig. 7A, MTP-KD), cell migration was further reduced by about 2-fold. These results clearly showed that both MTP and MMPs participate in Sox1-GFP+ NP cell mobility. When used to treat cells that were transfected with the MTP expression plasmid, 10 μm GM6001 completely reversed the MTP overexpression-induced migration (Fig. 7A, MTP-OV). Addition of 20 μm GM6001 further reduced cell migration to about the same level seen in MTP-knockdown cells despite overexpression of MTP (Fig. 7A). Because MMP2 is expressed at a higher level than the other MMPs tested in Sox1-GFP+ NP cells (Fig. 7B), we examined if MMP2 is required for MTP-mediated cell mobility. MMP2-specific siRNA sufficiently knocked down MMP2 expression (Fig. 7C, MMP2 in lane b) and reduced cell mobility to a level close to that with MTP knockdown (Fig. 7D, siMMP2), even though the MTP expression is not altered (Fig. 7C, MTP in lane b). Double knockdown of both MMP2 and MTP did not result in a much further reduction of cell mobility (Fig. 7D, siMMP2 + siMTP) suggesting a linear relationship between these two molecules in NP cell migration and MTP is likely to be upstream of MMP2. We further checked if activation of MMP2 is affected by MTP. To view pro-MMP2 activation easily, a recombinant pro-MMP2 protein was incubated with lysate of the Sox1-GFP+ NP cell. The Sox1-GFP+ NP cell lysate converted pro-MMP2 to the active MMP2 (Fig. 7C, compare lane NO with lane CT). The lysate of MTP knockdown NP cells, however, had significantly lower MMP2 activation ability (Fig. 7E). The expression of MMP2 mRNA was not affected by either knockdown (Fig. 7C) or overexpression of MTP.4 Collectively, these results indicate that the activity of MTP has a linear relationship to the activity of MMPs in promoting cell migration, and it acts upstream of, at least, MMP2.
FIGURE 7.
The collaboration of MTP with MMPs on NP cell mobility. A, non-transfected (NONE), MTP siRNA-transfected (MTP KD), or MTP expression plasmid-transfected (MTP OV) NP cells were treated with 10 or 20 μm MMP inhibitor (GM6001). Cell mobility was assayed in transwells. B, RT-PCR of MTP (lane a), MMP2 (lane b), MMP3 (lane c), and MMP9 (lane d) in Sox1-GFP+ NP cells. Lane e is S15 as control. C, RT-PCR assay of MTP and MMP2 in cells treated with specific siRNA for MMP2 knockdown (lane b), MTP knockdown (lane c), and MMP2-MTP double knockdown (lane d). Lane a shows cells without siRNA and lane e shows cells treated with a no-target control siRNA. S15 is the internal loading control. D, mobility of the control untreated NP cells (CTRL), NP cells transfected with the non-silencing siRNA (siNeg), MMP2-specific siRNA (siMMP2), MTP-specific siRNA (siMTP), or the combined siRNAs of both MMP2 and MTP (siMMP2+siMTP). E, the recombinant pro-MMP2 protein was incubated overnight at room temperature with buffer (NO), with cell lysate of NP cells (CT), lysates of NP cells transfected with MTP siRNA (KD), or lysates of NP cells transfected with MTP-expressing plasmid (OV) followed by Western immunoblotting with anti-MMP2 antibody. All lanes were assembled from samples on the same Western blot membrane. *, p < 0.05.
DISCUSSION
The most intriguing finding in our studies is the direct involvement of the TTSP family in NP cell functions. We showed that a member of this family, MTP, mediates NP cell migration and neuron differentiation. It participates in NP cell migration, in part, by promoting MMP2 activation.
MTP is present on the surface of a broad range of epithelial cells (46, 47). It is well known for its involvement in cancer progression (48–51) and epidermal differentiation (21). Mice carrying deletion of the MTP encoding gene St14 show dysfunction in multiple epithelial tissues suggesting that MTP plays a global role in maintenance of epithelial homeostasis (52). MTP is an autoactivating protease (30). Proteolytic substrates of MTP uncovered in recent years show that MTP can participate in various physiological functions. These features of MTP place it as an ideal upstream activator of various physiological processes. Early data have suggested that MTP is not expressed in the central nervous system (23, 53). Using PCR-based screening, MTP was found to be expressed in the postnatal day 10 mouse spinal cord (19) and lately MTP was shown in the developing mouse neural tube (54, 55). In this report we further show that MTP is expressed in NP cells, neurons, and the lining around ventricles. MTP may participate in central nervous system functions that have not yet been anticipated.
MTP is often overexpressed in various cancers (46). MTP activates hepatocyte growth factor (28), urokinase plasminogen (28), and MMP3 (56). These activities of MTP suggest its role in cell invasion and migration. Our study is the first to provide evidence that endogenous MTP is crucial to NP cell mobility. One mechanism is the activation of endogenous MMPs. Although our study did not determine this as a direct proteolytic activation by MTP, the presence of MTP is clearly required for MMP2 activation.
MTP was reported in monocytes (57) and peritoneal macrophages (58). Interestingly, microglia, the immunoreactive cells of nonneural lineage in brain, expresses no MTP. We have, however, detected some level of MTP expression in the microglial cell line BV2.5 BV2 cells share some features with the activated microglia such as cell morphology3 and migration promoting activity (Ref. 36 and this work). This implies that MTP can be induced in microglia upon activation and thus may serve in cellular defensive mechanisms. No brain developmental defects were reported in MTP knock-out mice (21), which would not be expected if MTP3 shared some activities with MTP. Alternatively, if MTP functions as a defensive molecule, a defect in MTP expression might not exhibit a significant consequence in brain function unless challenged by injury or stress. Our newest data shows that NP cells lacking MTP are more sensitive to injury-induced cell death,6 which appears to support the role of MTP in cellular defense.
Both VEGF and HGF significantly stimulate GFP+ NP cell motility, whereas SDF-1α induces little response. The little effect of SDF-1α may due to cleavage of this factor by MMP2 expressed in NP cells, which was shown in CD34+ hematopoietic progenitor cells (59). HGF and VEGF used in our studies are mostly the activated forms, thus proteolytic activation of VEGF or HGF by MTP is eliminated from the role of MTP in NP cell migration stimulated by these factors. VEGF and SDF-1α induced MMP3 and MMP9 expression in rat neural precursor cells (19). We6 also found in our system that MMP9 is induced by HGF, whereas the expression of MTP is not affected by either factor. Conceivably, treatment of NP cells with these factors would result in a large amount of MMPs being available for activation by MTP. This could be one way that MTP participates in the chemoattractant-stimulated cell mobility. Alternatively, there are other molecules/mechanisms that are mediated by MTP. It should be noted that there is a putative protein kinase C phosphorylation site in the intracellular tail and that phosphoinositide 3-kinase signaling is involved in the migration of human neural stem cells stimulated by HGF and VEGF (45). Whether these or other signaling molecules are involved in MTP-mediated effects is currently under investigation.
Considering that many proteolytic substrates of MTP also participate in various functions in the central nervous system, MTP could have wide effects on NP or NS cell function in the in vivo situation. Expression of MTP in NP cells provides potentially novel regulation in NP cell interactions with the microenvironment especially under pathological challenges. In summary, our studies provide evidence of the expression and function of MTP in NP cells. This is the first report to show a direct involvement of TTSP in NP cell function. These studies extend our knowledge about the physiological roles of TTSP and reveal a potentially novel regulatory mechanism of neurogeneseis.
Supplementary Material
Acknowledgment
We thank Dr. Austin Smith, University of Edinburgh, United Kingdom, for providing the 46C ES cells.
This work was supported by National Health Research Institutes Grant CS-098-PP-09 (to S.-L. L.), Taiwan, R.O.C.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
S.-L. Lee, unpublished data.
J.-D. Fang and S.-L. Lee, unpublished data.
H.-C. Chou and S.-L. Lee, unpublished observation.
J.-D. Fang and S.-L. Lee, manuscript in preparation.
- SVZ
- subventricular zone
- LV
- lateral ventricle
- HP
- hippocampus
- NP
- neural progenitor
- MMP
- matrix metalloproteinase
- MTP
- matriptase
- 7-ADD
- 7-amino-actinomycin D
- ES
- embryonic stem
- pro-HGF
- pro-hepatocyte growth factor
- SDF-1α
- stromal-derived factor-1α
- TTSP
- type II transmembrane serine protease
- MTPi
- matriptase inhibitor
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Gage F. H. (2000) Science 287, 1433–1438 [DOI] [PubMed] [Google Scholar]
- 2. Lledo P. M., Alonso M., Grubb M. S. (2006) Nat. Rev. Neurosci. 7, 179–193 [DOI] [PubMed] [Google Scholar]
- 3. Arvidsson A., Collin T., Kirik D., Kokaia Z., Lindvall O. (2002) Nat. Med. 8, 963–970 [DOI] [PubMed] [Google Scholar]
- 4. Nakatomi H., Kuriu T., Okabe S., Yamamoto S., Hatano O., Kawahara N., Tamura A., Kirino T., Nakafuku M. (2002) Cell 110, 429–441 [DOI] [PubMed] [Google Scholar]
- 5. Parent J. M., Yu T. W., Leibowitz R. T., Geschwind D. H., Sloviter R. S., Lowenstein D. H. (1997) J. Neurosci. 17, 3727–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rola R., Mizumatsu S., Otsuka S., Morhardt D. R., Noble-Haeusslein L. J., Fishman K., Potts M. B., Fike J. R. (2006) Exp. Neurol. 202, 189–199 [DOI] [PubMed] [Google Scholar]
- 7. Zhang R. L., Zhang Z. G., Chopp M. (2005) Neuroscientist 11, 408–416 [DOI] [PubMed] [Google Scholar]
- 8. Greenberg D. A., Jin K. (2005) Nature 438, 954–959 [DOI] [PubMed] [Google Scholar]
- 9. Ohira K. (2010) Cell. Mol. Life Sci. 1–12 [Google Scholar]
- 10. Galtrey C. M., Fawcett J. W. (2007) Brain Res. Rev. 54, 1–18 [DOI] [PubMed] [Google Scholar]
- 11. Yong V. W., Power C., Forsyth P., Edwards D. R. (2001) Nat. Rev. Neurosci. 2, 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abrous D. N., Koehl M., Le Moal M. (2005) Physiol. Rev. 85, 523–569 [DOI] [PubMed] [Google Scholar]
- 13. Mignatti P., Rifkin D. B. (1993) Physiol. Rev. 73, 161–195 [DOI] [PubMed] [Google Scholar]
- 14. Shiosaka S. (2004) Anat. Sci. Int. 79, 137–144 [DOI] [PubMed] [Google Scholar]
- 15. Melchor J. P., Strickland S. (2005) Thromb. Haemost. 93, 655–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yong V. W. (2005) Nat. Rev. Neurosci. 6, 931–944 [DOI] [PubMed] [Google Scholar]
- 17. Kim H. J., Fillmore H. L., Reeves T. M., Phillips L. L. (2005) Exp. Neurol. 192, 60–72 [DOI] [PubMed] [Google Scholar]
- 18. Wang L., Zhang Z. G., Zhang R. L., Gregg S. R., Hozeska-Solgot A., LeTourneau Y., Wang Y., Chopp M. (2006) J. Neurosci. 26, 5996–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barkho B. Z., Munoz A. E., Li X., Li L., Cunningham L. A., Zhao X. (2008) Stem Cells 26, 3139–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Szabo R., Wu Q., Dickson R. B., Netzel-Arnett S., Antalis T. M., Bugge T. H. (2003) Thromb. Haemost. 90, 185–193 [DOI] [PubMed] [Google Scholar]
- 21. List K., Haudenschild C. C., Szabo R., Chen W., Wahl S. M., Swaim W., Engelholm L. H., Behrendt N., Bugge T. H. (2002) Oncogene 21, 3765–3779 [DOI] [PubMed] [Google Scholar]
- 22. Guipponi M., Vuagniaux G., Wattenhofer M., Shibuya K., Vazquez M., Dougherty L., Scamuffa N., Guida E., Okui M., Rossier C., Hancock M., Buchet K., Reymond A., Hummler E., Marzella P. L., Kudoh J., Shimizu N., Scott H. S., Antonarakis S. E., Rossier B. C. (2002) Hum. Mol. Genet. 11, 2829–2836 [DOI] [PubMed] [Google Scholar]
- 23. Kim M. G., Chen C., Lyu M. S., Cho E. G., Park D., Kozak C., Schwartz R. H. (1999) Immunogenetics 49, 420–428 [DOI] [PubMed] [Google Scholar]
- 24. Jung H., Lee K. P., Park S. J., Park J. H., Jang Y. S., Choi S. Y., Jung J. G., Jo K., Park D. Y., Yoon J. H., Park J. H., Lim D. S., Hong G. R., Choi C., Park Y. K., Lee J. W., Hong H. J., Kim S., Park Y. W. (2008) Oncogene 27, 2635–2647 [DOI] [PubMed] [Google Scholar]
- 25. Stallmach R., Gloor S. M. (2008) Biochem. J. 412, 81–91 [DOI] [PubMed] [Google Scholar]
- 26. Ying Q. L., Stavridis M., Griffiths D., Li M., Smith A. (2003) Nat. Biotech. 21, 183–186 [DOI] [PubMed] [Google Scholar]
- 27. Lee S. L., Huang P. Y., Roller P., Cho E. G., Park D., Dickson R. B. (2010) Mech. Dev. 127, 82–95 [DOI] [PubMed] [Google Scholar]
- 28. Lee S. L., Dickson R. B., Lin C. Y. (2000) J. Biol. Chem. 275, 36720–36725 [DOI] [PubMed] [Google Scholar]
- 29. Szabo R., Netzel-Arnett S., Hobson J. P., Antalis T. M., Bugge T. H. (2005) Biochem. J. 390, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee M. S., Tseng I. C., Wang Y., Kiyomiya K., Johnson M. D., Dickson R. B., Lin C. Y. (2007) Am. J. Physiol. Cell Physiol. 293, C95–105 [DOI] [PubMed] [Google Scholar]
- 31. Kiyomiya K., Lee M. S., Tseng I. C., Zuo H., Barndt R. J., Johnson M. D., Dickson R. B., Lin C. Y. (2006) Am. J. Physiol. Cell Physiol. 291, C40–49 [DOI] [PubMed] [Google Scholar]
- 32. Cho E. G., Kim M. G., Kim C., Kim S. R., Seong I. S., Chung C., Schwartz R. H., Park D. (2001) J. Biol. Chem. 276, 44581–44589 [DOI] [PubMed] [Google Scholar]
- 33. Fukuda H., Takahashi J., Watanabe K., Hayashi H., Morizane A., Koyanagi M., Sasai Y., Hashimoto N. (2006) Stem Cells 24, 763–771 [DOI] [PubMed] [Google Scholar]
- 34. Abranches E., Silva M., Pradier L., Schulz H., Hummel O., Henrique D., Bekman E. (2009) PLoS ONE 4, e6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Long Y. Q., Lee S. L., Lin C. Y., Enyedy I. J., Wang S., Li P., Dickson R. B., Roller P. P. (2001) Bioorg. Med. Chem. Lett. 11, 2515–2519 [DOI] [PubMed] [Google Scholar]
- 36. Aarum J., Sandberg K., Haeberlein S. L., Persson M. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15983–15988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng H., Fukushima T., Takahashi N., Tanaka H., Kataoka H. (2009) Cancer Res. 69, 1828–1835 [DOI] [PubMed] [Google Scholar]
- 38. Belmadani A., Tran P. B., Ren D., Miller R. J. (2006) J. Neurosci. 26, 3182–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stumm R., Höllt V. (2007) J. Mol. Endocrinol. 38, 377–382 [DOI] [PubMed] [Google Scholar]
- 40. Xu Q., Wang S., Jiang X., Zhao Y., Gao M., Zhang Y., Wang X., Tano K., Kanehara M., Zhang W., Ishida T. (2007) Clin. Exp. Pharmacol. Physiol. 34, 624–631 [DOI] [PubMed] [Google Scholar]
- 41. Shen Q., Goderie S. K., Jin L., Karanth N., Sun Y., Abramova N., Vincent P., Pumiglia K., Temple S. (2004) Science 304, 1338–1340 [DOI] [PubMed] [Google Scholar]
- 42. Powell E. M., Mars W. M., Levitt P. (2001) Neuron 30, 79–89 [DOI] [PubMed] [Google Scholar]
- 43. Cacci E., Salani M., Anastasi S., Perroteau I., Poiana G., Biagioni S., Augusti-Tocco G. (2003) J. Neurosci. Res. 74, 760–768 [DOI] [PubMed] [Google Scholar]
- 44. Garzotto D., Giacobini P., Crepaldi T., Fasolo A., De Marchis S. (2008) J. Neurosci. 28, 5901–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kendall S. E., Najbauer J., Johnston H. F., Metz M. Z., Li S., Bowers M., Garcia E., Kim S. U., Barish M. E., Aboody K. S., Glackin C. A. (2008) Stem Cells 26, 1575–1586 [DOI] [PubMed] [Google Scholar]
- 46. Oberst M., Anders J., Xie B., Singh B., Ossandon M., Johnson M., Dickson R. B., Lin C. Y. (2001) Am. J. Pathol. 158, 1301–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oberst M. D., Singh B., Ozdemirli M., Dickson R. B., Johnson M. D., Lin C. Y. (2003) J. Histochem. Cytochem. 51, 1017–1025 [DOI] [PubMed] [Google Scholar]
- 48. Benaud C. M., Oberst M., Dickson R. B., Lin C. Y. (2002) Clin. Exp. Metastasis 19, 639–649 [DOI] [PubMed] [Google Scholar]
- 49. Ito Y., Akinaga A., Yamanaka K., Nakagawa T., Kondo A., Dickson R. B., Lin C. Y., Miyauchi A., Taniguchi N., Miyoshi E. (2006) Glycobiology 16, 368–374 [DOI] [PubMed] [Google Scholar]
- 50. Jin X., Hirosaki T., Lin C. Y., Dickson R. B., Higashi S., Kitamura H., Miyazaki K. (2005) J. Cell. Biochem. 95, 632–647 [DOI] [PubMed] [Google Scholar]
- 51. Sanders A. J., Parr C., Davies G., Martin T. A., Lane J., Mason M. D., Jiang W. G. (2006) J. Exp. Ther. Oncol. 6, 39–48 [PubMed] [Google Scholar]
- 52. List K., Kosa P., Szabo R., Bey A. L., Wang C. B., Molinolo A., Bugge T. H. (2009) Am. J. Pathol. 175, 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. List K., Szabo R., Molinolo A., Nielsen B. S., Bugge T. H. (2006) Am. J. Pathol. 168, 1513–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Szabo R., Hobson J. P., Christoph K., Kosa P., List K., Bugge T. H. (2009) Development 136, 2653–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Camerer E., Barker A., Duong D. N., Ganesan R., Kataoka H., Cornelissen I., Darragh M. R., Hussain A., Zheng Y. W., Srinivasan Y., Brown C., Xu S. M., Regard J. B., Lin C. Y., Craik C. S., Kirchhofer D., Coughlin S. R. (2010) Dev. Cell 18, 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jin X., Yagi M., Akiyama N., Hirosaki T., Higashi S., Lin C. Y., Dickson R. B., Kitamura H., Miyazaki K. (2006) Cancer Sci. 97, 1327–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kilpatrick L. M., Harris R. L., Owen K. A., Bass R., Ghorayeb C., Bar-Or A., Ellis V. (2006) Blood 108, 2616–2623 [DOI] [PubMed] [Google Scholar]
- 58. Bhatt A. S., Welm A., Farady C. J., Vásquez M., Wilson K., Craik C. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5771–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McQuibban G. A., Butler G. S., Gong J. H., Bendall L., Power C., Clark-Lewis I., Overall C. M. (2001) J. Biol. Chem. 276, 43503–43508 [DOI] [PubMed] [Google Scholar]
- 60. Paxinos G., Franklin K. (2001) Mouse Brain Atlas, 2nd Ed., Academic Press, San Diego, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.