Abstract
RNA polymerase II coordinates co-transcriptional events by recruiting distinct sets of nuclear factors to specific stages of transcription via changes of phosphorylation patterns along its C-terminal domain (CTD). Although it has become increasingly clear that proline isomerization also helps regulate CTD-associated processes, the molecular basis of its role is unknown. Here, we report the structure of the Ser(P)5 CTD phosphatase Ssu72 in complex with substrate, revealing a remarkable CTD conformation with the Ser(P)5–Pro6 motif in the cis configuration. We show that the cis-Ser(P)5–Pro6 isomer is the minor population in solution and that Ess1-catalyzed cis-trans-proline isomerization facilitates rapid dephosphorylation by Ssu72, providing an explanation for recently discovered in vivo connections between these enzymes and a revised model for CTD-mediated small nuclear RNA termination. This work presents the first structural evidence of a cis-proline-specific enzyme and an unexpected mechanism of isomer-based regulation of phosphorylation, with broad implications for CTD biology.
Keywords: Phosphatase, Prolyl Isomerase, Protein Structure, RNA Polymerase II, Transcription, CTD, cis-Proline, Ess1, Ssu72
Introduction
The C-terminal domain (CTD)2 of the largest subunit of RNA polymerase II (RNAPII) consists of multiple tandem heptad repeats, with the consensus sequence Y1S2P3T4S5P6S7, that serve as a flexible binding platform for nuclear factors (1). CTD-binding partners influence the initiation, elongation, and termination of transcription as well as a myriad of co-transcriptional processes (2). The recruitment of these activities is tied to the progress of the polymerase by cyclic phosphorylation and dephosphorylation of the CTD repeats. For example, phosphorylation at the Ser5 position (Ser(P)5) predominates at the 5′-end of genes, attracting CTD-binding partners that influence initiation complex formation, mRNA capping, and the transition into elongation (3). As the polymerase moves toward the 3′-end of genes, the level of Ser(P)5 declines, whereas phosphorylation at Ser2 (Ser(P)2) increases, recruiting nuclear factors responsible for elongation, termination, and 3′-end formation (3). The variable phosphorylation patterns within each heptad repeat, and distributions of these patterns across the full domain create a “CTD code” with a staggering potential complexity (4).
In addition to phosphorylation, proline isomerization provides a second mechanism for regulating the association of CTD-binding partners (5). Due to its cyclic side chain, proline can adopt both cis and trans conformations about its peptide bond, creating distinct and interconvertible backbone structures with the cis isomer being energetically disfavored and therefore less populated (6). Each CTD heptad contains 2 proline residues, and both are preceded by serine residues that are critical targets of phosphorylation. Phosphorylation of Ser-Pro motifs in non-CTD peptides has been shown to modestly stabilize the cis form and decrease the rate of isomerization (7). A study with a Ser(P)2 CTD peptide reported a cis population of <30% for the Ser(P)2–Pro3 motif with very slow interconversion of cis and trans isomers, on the order of s−1 (8). Relative to the short timescale of transcriptional events, this slow intrinsic exchange presents two structurally distinct and kinetically isolated binding epitopes for each proline residue in the CTD, adding an additional layer of complexity to the CTD code (4).
The biological importance of CTD proline isomerization is unclear, but hints have been provided by studies of the yeast peptidyl prolyl isomerase Ess1 (Pin1 in humans). Peptidyl prolyl isomerases speed the interconversion of proline isomers by several orders of magnitude to restore cis-trans equilibria at a biologically relevant timescale (6). Regulation of the proline “conformational switch” has been proposed to give these enzymes control over the duration and amplitude of a variety of cellular processes (6). Ess1/Pin1 specifically targets phosphorylated Ser-Pro motifs and influences transcription by RNA polymerase II, likely by regulating the phosphorylation state of the CTD (9). Ess1/Pin1 acts on pCTD peptides in vitro, preferentially binding to the Ser(P)5–Pro6 site over the Ser(P)2–Pro3 site (10, 11). Numerous genetic links between Ess1 and CTD kinases and phosphatases have been reported (5), and abnormal levels of Pin1 activity cause the accumulation of aberrantly phosphorylated forms of the CTD (12). This latter phenotype may be due in part to the influence of Pin1 on the activity of Ser(P)5 CTD phosphatase Fcp1 (12–14). Despite these intriguing findings, there is no detailed understanding of how catalyzed proline isomerization modulates the CTD phosphorylation state.
One of the enzymes linked to Ess1 activity is Ssu72, a CTD phosphatase with specificity for the Ser(P)5 position (15). Despite its distinct substrate, Ssu72 shares many characteristics of the low molecular weight subfamily of protein tyrosine phosphatases (LMW PTPs) including the signature catalytic motif (CX5R) and a similar predicted arrangement of secondary structure elements (16, 17). Biologically, Ssu72 influences all three stages of transcription. Ssu72 interacts genetically and physically with initiation factor TFIIB (18, 19) and has been implicated in gene looping, a proposed mechanism for transcription reinitiation that tethers the promoter and terminator regions of a gene (20–22). During elongation, impaired Ssu72 activity leads to increased RNAPII pausing (23). Finally, Ssu72 is a component of the cleavage and polyadenylation factor complex through its association with Pta1 and is essential for the proper termination of small nuclear RNA (snRNA) transcripts (16, 24–27).
Recently, two studies illuminated an in vivo connection between Ess1 and Ssu72 (28, 29). Impairing Ess1 catalytic activity in yeast cells resulted in a temperature-sensitive phenotype and the accumulation of Ser(P)5 CTD. Both defects were ameliorated by overexpression of Ssu72 (29). A genome-wide analysis of mRNA expression in these cells showed readthrough transcription for a set of genes, mainly snRNAs, with marked similarity to those previously identified in cells with impaired Ssu72 activity (28). The prevailing model explains these results in the following way (28, 29). Phosphorylation at the Ser5 position would cause the CTD to adopt predominantly the cis form of the Ser(P)5–Pro6 motif. Near the end of snRNA transcripts, dephosphorylation of Ser(P)5 would coordinate the exchange of Nrd1 and Pcf11, two factors involved in Nrd1-dependent transcription termination. However, Ssu72 would have isomeric specificity for the less populated trans form of the Ser(P)5–Pro6 motif and would require Ess1 activity to efficiently process the entire pool of available substrate. Consequently, reduced catalysis by Ess1 would lead to an increased level of Ser(P)5 CTD, causing an improper localization of Ser(P)5 CTD-bound Nrd1 and a concomitant blocking of Ser(P)2 CTD-mediated Pcf11 recruitment and resulting in readthrough transcription. This model is supported by the fact that all reported enzymes that target Ser(P)-Pro motifs with isomeric specificity recognize the trans configuration. On the other hand, there is no evidence that the cis-Ser(P)5–Pro6 isomer of the CTD becomes the major population upon phosphorylation (7, 8).
To investigate the mechanism of proline isomer-based regulation of CTD phosphorylation states, we determined the structure of Ssu72 in complex with its Ser(P)5 CTD substrate. The structure reveals an enzyme fold that contains the conserved LMW PTP scaffold and unique additions and a surprising substrate conformation with the Ser(P)5–Pro6 motif of the CTD adopting the cis configuration. We show that the cis isomer is a minor population of substrate in solution and that Ess1 significantly enhances Ssu72 activity by catalyzing rate-limiting cis-trans interconversion. Together, this work presents structural and kinetic evidence for a unique cis-proline-specific enzymatic activity and a fascinating explanation for the in vivo relationship of Ess1 and Ssu72, with broad implications for the regulation of the CTD code and its role in coordinating co-transcriptional events.
EXPERIMENTAL PROCEDURES
Molecular Cloning and Protein Purification
The full-length WT Drosophila melanogaster Ssu72 gene was PCR-amplified from cDNA (Open Biosystems, clone ID: RE29729), digested, and ligated into a pET15b vector (EMD Biosciences, Inc.) between the NdeI and BamHI restriction sites. Point mutants of Ssu72 and Ess1 were prepared using the QuikChange site-directed mutagenesis kit (Stratagene). The correct sequences for all constructs were verified by DNA sequencing. Protein expression and purification procedures are described in the supplemental material.
NMR
Isotopically enriched proteins for NMR studies were overexpressed in M9 minimal media with [15N]NH4Cl and [13C]glucose (Cambridge Isotope Laboratories, Inc.) as the sole nitrogen and carbon sources. NMR experiments were conducted at 30 °C using Varian INOVA 600- or 800-MHz spectrometers. Backbone resonances for C13D/D144N D. melanogaster Ssu72 (dSsu72) were assigned using a 2H/13C/15N-labeled sample with standard three-dimensional triple-resonance experiments.
Crystallization and Structure Determination
The Ssu72-pCTD complex was prepared by incubating a 5:1 molar ratio of pCTD peptide (4.17 mm) to C13D/D144N dSsu72 (0.83 mm) for 30 min on ice. The complex was crystallized by hanging drop vapor diffusion; 1 μl of protein sample was mixed with 1 μl of 22% (w/v) PEG monomethyl ether 550, 100 mm imidazole, pH 6.5, and 150 mm dl-malic acid and suspended over a reservoir of the same solution. Equilibration at 4 °C for ∼5 days produced 50 × 50 × 200-μm rhombic-shaped crystals, which were cryoprotected by dipping into a solution of mother liquor with 25% ethylene glycol before flash-cooling in liquid nitrogen. Methods for data collection, phasing, and model refinement are provided in the supplemental material.
Enzyme Kinetics
Reactions with synthetic pCTD substrate (see Fig. 4, A–D) were followed by measuring phosphate release with a standard Malachite Green reagent (30). For experiments with WT Ess1, 350 μl-reactions were set up in 50 mm HEPES, 20% glycerol, 1 mm EDTA, 2 mm DTT buffer at pH 6.5 (kinetics buffer) with 60 μm pCTD peptide and varying amounts of Ess1. After incubating for 10–15 min, each reaction was started by the addition of 4 μm WT dSsu72 (or 20 μm for the faster timescale reactions in supplemental Fig. S3). 50-μl aliquots were taken each minute and mixed with 50 μl of Malachite Green reagent in a 96-well plate, which was placed in the dark for 30 min to allow for color development. Color was quantified by measuring A630 nm with a SpectraMax microplate reader (Molecular Devices). Absorbance values were converted to phosphate content using a standard curve made with NaxPO4 solutions of known concentrations. The same protocol was followed for experiments with Ess1 mutants, with reactions containing 1 μm of each mutant. Preparation and characterization of the substrate peptide are described in the supplemental material, along with a description of kinetic assays with full-length pCTD substrate (see Fig. 4, E and F).
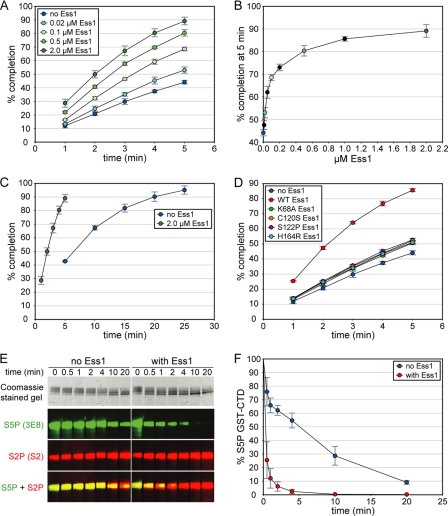
FIGURE 4.
Ess1 stimulates pCTD dephosphorylation by Ssu72. A, the activity of Ssu72 was monitored with various concentrations of the proline isomerase Ess1. The maximum stimulation was ∼2-fold under these conditions. In A–D, each point represents the average from three independent reactions, and error bars denote S.D. B, Ess1 enhancement of Ssu72 activity is saturable. The last points of the reactions in A are plotted versus Ess1 concentration, with black circles representing reactions omitted from A for clarity. C, pCTD dephosphorylation by Ssu72 reaches completion without Ess1. The no Ess1 reaction in A was monitored over 25 min and reached ∼90% completion in 20 min, 4-fold slower than the reaction with 2 μm Ess1. D, four catalytically impaired Ess1 mutants fail to significantly stimulate Ssu72. Reactions contained 1 μm of each Ess1 protein. Residual enhancement corresponds to a 50–100-fold reduction of WT Ess1 activity (compare with A). E and F, Ess1 stimulates Ssu72 dephosphorylation of full-length pCTD. The 26-repeat S. cerevisiae CTD was hyperphosphorylated in vitro and used as a substrate in reactions monitored by Western blotting with Ser(P)5 (S5P)-specific antibodies. As a control, Ser(P)2 (S2P) levels were also measured; the apparent increase in the S2 signal is likely due to a higher affinity of the S2 antibody for singly phosphorylated Ser(P)2 heptads over doubly phosphorylated Ser(P)2/Ser(P)5 heptads, as reported for other Ser(P)2-specific antibodies (44). Ser(P)5 CTD levels were quantified with infrared imaging, and the resulting substrate depletion curves are shown in F. Each point represents the average of three independent reactions, and error bars denote S.D.
RESULTS
Ssu72 Adopts the Conserved LMW PTP Fold with Unique Additions
Previous kinetic experiments suggest that Ssu72 shares the catalytic mechanism of protein tyrosine phosphatases (17, 31), a two-step reaction that requires: 1) the CX5R catalytic motif with the cysteine side chain as a negatively charged thiolate group and 2) an aspartate residue on a flexible loop positioned near the active site (“the aspartate loop”). In the first step, the thiolate of the catalytic cysteine attacks the substrate phosphorous atom to generate a phosphoenzyme intermediate, and in the second step, the aspartate protonates the leaving phosphate to regenerate the enzyme. By screening orthologs, we identified dSsu72 as a highly stable and soluble enzyme with significant sequence similarity to Saccharomyces cerevisiae Ssu72 (Fig. 1A). Mutation of both the catalytic cysteine (C13D) and the catalytic aspartate (D144N) fully abolished the activity of dSsu72, and an NMR titration of 15N-labeled C13D/D144N dSsu72 with Ser(P)5 CTD peptide revealed tight binding (slow exchange on the NMR timescale) and significant perturbations of residues near the active site loop and the aspartate loop (supplemental Fig. S1).
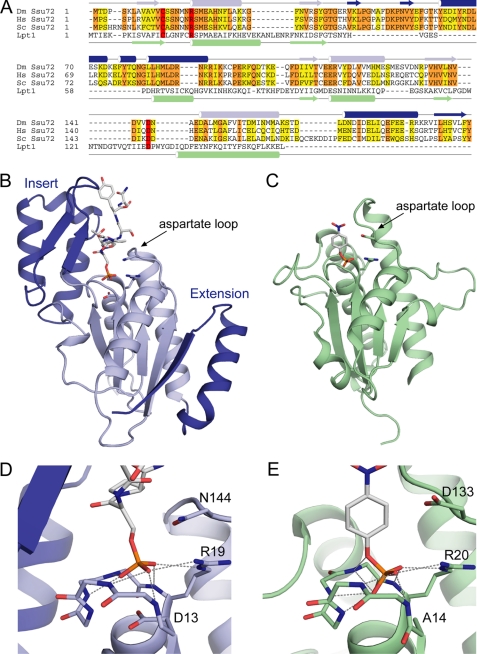
FIGURE 1.
Structure of Ser(P)5 CTD phosphatase Ssu72. A, sequence alignment of Ssu72 orthologues and the LMW PTP Lpt1. Ssu72 proteins from D. melanogaster (Dm Ssu72), human (Hs Ssu72), and S. cerevisiae (Sc Ssu72) are shown with conserved and similar residues highlighted in orange and yellow, respectively. α-Helices and β-strands from the structures of dSsu72 and Lpt1 (PDB code 1D1Q) are denoted with cylinders and arrows, respectively. The secondary structure elements of Ssu72 that comprise the two additions to the LMW PTP scaffold are colored dark blue to match the structure in B. Catalytic residues are highlighted in red. B, ribbon diagram of the dSsu72-pCTD complex. Additions to the LMW PTP fold are colored dark blue. The substrate peptide and catalytic residues Cys13 (mutated to Asp13), Arg19, and Asp144 (mutated to Asn144) are shown as sticks. C, ribbon diagram of the LMW PTP Lpt1. The Lpt1 structure shows a catalytic mutant bound to the substrate p-nitrophenyl phosphate (PDB code 1D1Q). The substrate and catalytic residues Cys14 (mutated to Ala14), Arg20, and Asp133 are shown as sticks. D and E, active site close-ups of substrate-bound dSsu72 (D) and Lpt1 (E). The backbone of the phosphate-binding loops is shown as sticks along with important catalytic residues. Hydrogen bonds are represented by gray dashed lines.
To obtain a high resolution model of the Ssu72-pCTD interaction, we used x-ray crystallography to determine the structure of C13D/D144N dSsu72 in complex with a synthetic, 7-residue Ser(P)5 CTD peptide containing the minimal binding epitope (32) with an additional residue at each end (Ac-PTpSPSYS-NH2, where the pS indicates Ser(P)). Phasing was carried out by molecular replacement using the apoWT dSsu72 structure deposited by the Northeast Structural Genomics Consortium (Protein Data Bank (PDB) code 3FDF),3 and the model was refined at 2.1 Å resolution (Table 1).
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Resolution range (Å) | 50–2.10 (2.18–2.10) |
| Space group | R3 |
| Unit cell dimensions (Å) | 157.5, 157.5, 118.8 |
| Completeness (%) | 99.9 (99.8) |
| Unique reflections | 63898 (6361) |
| Redundancy | 3.9 |
| Rmerge (%) | 6.1 (32.6) |
| I/σI | 23.7 (4.1) |
| Refinement | |
| Rwork/Rfreea (%) | 20.90/24.11 |
| Number of atomsb | |
| Protein | 6360 |
| Peptide | 236 |
| Water | 658 |
| Other moleculesc | 31 |
| B-factors (Å2)d | |
| Protein | 33.38 |
| Peptide | 36.53 |
| Water | 34.55 |
| Other moleculesc | 32.83 |
| r.m.s.d.e from ideal geometry | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.748 |
| Ramachandran plot | |
| Favored (%) | 97.2 |
| Allowed (%) | 99.9 |
a Rfree was calculated with the 5% of the data randomly omitted from refinement.
b Nonhydrogen atoms. Riding hydrogens were used in refinement and are included in the deposited structure.
c Imidazole and a PEG fragment.
d Only nonhydrogen atoms included.
e r.m.s.d., root mean square deviation.
The Ssu72 structure contains the LMW PTP fold and two sizable additions (Fig. 1, A–C, PDB code 3P9Y). The conserved scaffold consists of a central, four-stranded, parallel β-sheet flanked by two α-helices on one side and a third α-helix on the other and is represented in Fig. 1 by the structure of the LMW PTP Lpt1 in complex with its substrate p-nitrophenyl phosphate (33). The first addition to the Ssu72 fold is an ∼60-residue insertion after the second β-strand that forms a subdomain of two β-strands packed against three α-helices. This insert structure sits immediately adjacent to the active site loop and contributes to substrate binding (see below). The corresponding region in the LMW PTP fold contains ∼25 residues and forms a single α-helix that packs against the back of the central β-sheet and is tethered by long loops that interact with substrate. The second addition is an ∼30-residue C-terminal extension that forms a helix-turn-strand motif with the β-strand bonding in an anti-parallel fashion to the central β-sheet. Another distinguishing characteristic of Ssu72 is the length of its aspartate loop. The α-helix following the aspartate loop is one turn shorter in Ssu72, which allows a commensurate shortening of the aspartate loop and increases active site accessibility from the side opposite the insert subdomain.
The active site architecture of Ssu72 is very similar to LMW PTPs, providing structural support for a common mechanism of dephosphorylation. Fig. 1, D and E, show a close-up view of phosphate binding for the Ssu72-pCTD complex and the LMW PTP Lpt1-p-nitrophenyl phosphate complex (33). The conformations of active site loops in these structures are nearly identical, with the backbone amide groups forming a series of hydrogen bonds to the substrate phosphate group and the guanidinium group of the catalytic arginine extending this hydrogen bonding to form a complete circle. Despite the different length of aspartate loops, the positions of the catalytic aspartate side chains (D144N in the Ssu72 structure) are very similar. Although the carboxylate of the Lpt1 aspartate is turned away from the active site, presumably due to electrostatic repulsion with the substrate phosphate group, in the Ssu72 structure, the catalytic D144N side chain is pointed toward the phosphate. Another difference between the active sites is the position of the substrate phosphate. In the Lpt1 structure, the catalytic cysteine is mutated to alanine, and this loss of negative charge allows the phosphate to bind deeper in the active site, close to the position predicted for the phosphoenzyme intermediate (33), whereas the phosphate group in the Ssu72 structure is held ∼0.7 Å higher by the negative charge of the C13D side chain.
Ssu72 Recognizes a cis-Proline Substrate
The electron density for the pCTD peptide is excellent and extends almost completely through both termini (Fig. 2A). While the substrate phosphate group is anchored in the active site with strong hydrogen bonds, the insert subdomain creates a deep and narrow recognition cleft that forces the C-terminal end of the CTD to extend away immediately from the phosphate in a nearly opposite direction. This constrained conformation is made possible by a cis isomer of the Ser(P)5–Pro6 peptide bond (ω = −2.42°) that provides an abrupt turn in the substrate backbone. The uniqueness of the cis-Ser(P)5–Pro6 isomer and its pronounced effect on the substrate conformation are readily apparent by comparison with the structure of Ser(P)5 CTD bound to the CTD phosphatase Scp1 (PDB code 2GHT) in Fig. 2, C and D (34). In the Scp1 complex, the trans isomer of the Ser(P)5–Pro6 bond provides an extended backbone configuration, in contrast to the sharp kink produced by the cis-Ser(P)5–Pro6 isomer.
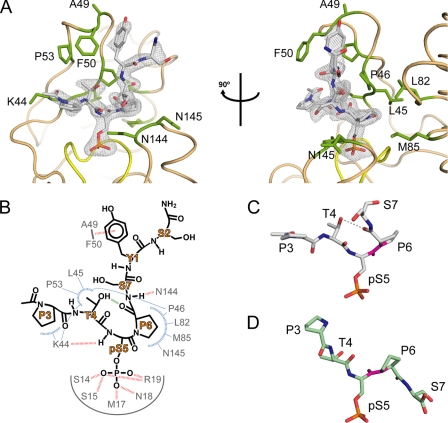
FIGURE 2.
Binding of cis-proline pCTD substrate by Ssu72. A, orthogonal views of pCTD conformation and binding. The enzyme structure is shown in tan with yellow highlighting for the active site loop, and residues that form the substrate recognition surface are labeled and shown as green sticks. The gray mesh represents the Fo − Fc omit map density (contoured at 3.0σ) surrounding the substrate. B, schematic of the Ssu72-pCTD interaction. Hydrogen bonds and van der Waal contacts are shown as red and blue dashed lines, respectively. The green line represents an intramolecular hydrogen bond in the pCTD substrate. C and D, examples of cis and trans substrate configurations. The conformations of Ser(P)5 CTD peptides in complex with dSsu72 (C) and phosphatase Scp1 (D) are shown with the Ser(P)5–Pro6 peptide bond highlighted in magenta. For clarity, only residues Pro3 to Ser7 are shown. The cis isomer is stabilized by an intramolecular hydrogen bond (gray dashed line) and causes a severe kink in the CTD backbone.
To achieve its unusual isomeric specificity, Ssu72 must stabilize the high energy cis-proline substrate conformation (Fig. 2B). This is accomplished in part by facilitating the formation of an intramolecular hydrogen bond between the substrate Thr4 side-chain hydroxyl and the Pro6 backbone carbonyl. The Thr4 side chain is positioned by its methyl group binding in a hydrophobic pocket formed by the side chains of Lys44, Pro46, and Pro53 and the backbone of Leu45. Hydrogen bonding with the Thr4 hydroxyl forces the Pro6 residue to adopt the cis isomer, which is further stabilized by strong van der Waal interactions between the Pro6 ring and a hydrophobic patch of Ssu72 formed by the side chains of Pro46, Leu82, and Met85 and the backbone of Asn145. Between the Thr4 and Pro6 residues, the Ser(P)5 backbone amide forms a hydrogen bond with the Lys44 backbone carbonyl of Ssu72, while the Ser(P)5 side chain sits in the active site as described above. At the N-terminal end of the substrate peptide, the Pro3 residue makes minor van der Waal contacts with the Lys44 side chain of Ssu72. Toward the C-terminal end, additional isomer-specific binding energy is provided by an intermolecular hydrogen bond between the substrate Ser7 main-chain amide group and the Asn144 backbone carbonyl. While the side chains of the Ser7 and Ser2 residues are largely solvent-exposed, the phenol ring of the intervening Tyr1 side chain forms an aromatic-amide stacking interaction with the Ala49–Phe50 peptide bond of Ssu72 (35), causing a reorientation of the loop between the two β-strands of the insert subdomain in the complex when compared with the apo structure (supplemental Fig. S2).
Ess1-catalyzed Proline Isomerization Stimulates Ssu72 Activity
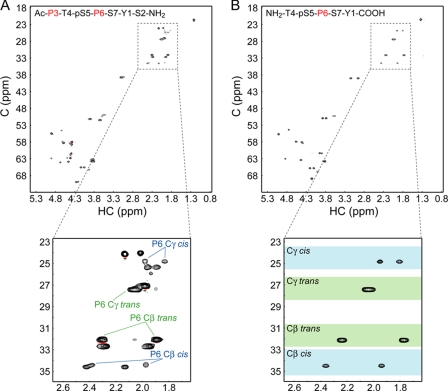
A previous study of peptides with Ser(P)-Pro motifs showed that the cis isomer is less favored than the trans form, with populations of 12–20% depending on the adjacent residues (7). Similar work with a Ser(P)2 CTD peptide reported a cis population under 30% for the Ser(P)2–Pro3 isomer (8). Although these results suggest that phosphorylation of the Ser5–Pro6 CTD motif is unlikely to change the minority status of the cis isomer, no direct measurements have been reported. To address this point, we measured the fractions of cis- and trans-Ser(P)5–Pro6 in our substrate, a synthetic peptide with the sequence Ac-PTpSPSYS-NH2 (where the pS indicates Ser(P)), which is the same peptide used for crystallization. A natural abundance 13C-heteronuclear single quantum correlation spectrum shows two sets of signals for each proline Cβ and Cγ atom, corresponding to the cis and trans forms (Fig. 3A). However, the substrate peptide contains 2 prolines, making it difficult to unambiguously assign peaks to the Pro3 and Pro6 residues. By collecting the same experiment with a shorter peptide containing only the Pro6 residue (Fig. 3B), we were able to assign the Pro6 resonances and readily map them to the longer peptide. Populations were calculated using the average ratio of cis and trans peak volumes for the Pro6 Cβ and Cγ atoms. This gave a fractional population of 12.4% for the cis isomer, verifying that the Ser(P)5–Pro6 motif is predominantly in the trans configuration.
FIGURE 3.
Measurement of proline isomer populations for Ser(P)5 CTD substrate. A and B, natural abundance 13C-heteronuclear single quantum correlation spectra were collected for the substrate peptide (A) and a shorter pCTD peptide with only one proline residue (B). Close-up views depict the proline β- and γ-region in each spectrum. Colored boxes indicate the range of chemical shifts (mean ± S.D.) for each proline atom in either the cis (blue) or the trans (green) form (43). pS5, Ser(P)5. C, carbon; HC, proton.
Given that Ssu72 recognizes the minor population of cis-Ser(P)5–Pro6 isomers, the extremely slow interconversion of cis-trans-proline (s−1) may present a rate-limiting step for dephosphorylation. To test this possibility, we examined the effect of catalyzed proline isomerization on Ssu72 activity. Reactions were monitored by measuring phosphate release with a Malachite Green-based assay. In a reaction with 60 μm pCTD and 4 μm WT dSsu72, less than half of the input substrate was dephosphorylated in 5 min (Fig. 4A). To speed the interconversion of proline isomers, we added increasing amounts of Ess1, a CTD proline isomerase that acts on Ser(P)5–Pro6 motifs. Ess1 significantly stimulated the activity of Ssu72, with a 2 μm Ess1 concentration causing an ∼2-fold increase in reaction completion over 5 min. The increase in Ssu72 activity was saturable (Fig. 4B), and a concentration of 10 μm Ess1 was sufficient to maximize the completion percentage at all time points (data not shown). To ensure that Ssu72 was capable of completely dephosphorylating the pCTD substrate in the absence of Ess1, we followed the reaction without Ess1 over the course of 25 min (Fig. 4C), which showed that ∼90% of substrate is dephosphorylated in 20 min, 4-fold slower than the reaction with 2 μm Ess1. This behavior implies a kinetic effect on dephosphorylation, with Ess1-catalyzed isomerization improving the availability of cis-proline substrate for Ssu72 until the rate of isomer interconversion exceeds the rate of pCTD dephosphorylation.
To confirm that catalyzed isomerization is the cause of Ssu72 stimulation, we performed identical experiments with four catalytically impaired Ess1 mutants. Three of these mutations are in the catalytic domain of Ess1 (C120S, S122P, and H164R), whereas the fourth disrupts phosphate binding in the WW domain (K68A). All four mutations significantly decreased the enhancement of Ssu72 activity (Fig. 4D). To ensure that this result is not caused by reduced thermostability, we measured melting temperatures for each Ess1 mutant by circular dichroism (data not shown). Only the H164R mutation caused a large decrease in stability (melting temperature of 32 °C) that may contribute to its impaired stimulation. Together, these studies demonstrate that Ess1 enhances Ssu72 activity by catalyzing proline isomerization of the pCTD substrate.
Finally, to evaluate the effect of Ess1 on Ssu72 activity toward its natural substrate, we performed in vitro reactions using the full-length, 26-repeat CTD from S. cerevisiae. A GST-S. cerevisiae CTD-His fusion construct was hyperphosphorylated with the yeast CTD kinase CTDK-1 (36), and dephosphorylation by Ssu72 was monitored by measuring substrate depletion with a Ser(P)5-specific antibody in reactions with and without 100 μm Ess1 (Fig. 4, E and F). As in the reactions with synthetic pCTD substrate, Ess1 greatly facilitates dephosphorylation by Ssu72, decreasing the time required to reach ∼90% completion by 20-fold (from 20 to 1 min).
DISCUSSION
Ssu72 as a New Family of LMW Phosphatases
The CTD phosphatase Ssu72 is an intriguing enzyme. Despite being built on the scaffold of a protein tyrosine phosphatase, its activity is directed at phosphorylated serine residues in the 5th position of the CTD heptad, with isomeric specificity for the cis configuration of the Ser(P)5–Pro6 peptide bond. Our complex structure also suggests a conserved catalytic mechanism, which agrees well with prior kinetic studies (17). Of the two additions to the LMW PTP scaffold, the insert subdomain plays a major role in the unique substrate specificity of Ssu72, contributing nearly all of the residues that form the non-phosphate substrate recognition surface and severely restricting access to the active site. Very recently, Xiang et al. (37) reported a crystal structure of human Ssu72 in complex with pCTD substrate and symplekin (homologous to Pta1 in yeast), a scaffold protein in the cleavage and polyadenylation complex, which shows that the C-terminal extension of Ssu72 forms the symplekin-binding site. The Ssu72 fold, pCTD conformation, and effect of catalyzed proline isomerization on dephosphorylation are consistent with the data presented in this study.
Substrate Specificity of Ssu72
Prior kinetic work on Ssu72 has demonstrated a strict specificity for the Ser(P)5 position of the CTD heptad (15, 32), in contrast to CTD phosphatases Scp1 and Fcp1, which have only preferential activity for Ser(P)5 and Ser(P)2, respectively (38, 39). Interestingly, both potential sites are Ser(P)-Pro motifs, meaning that 2 of the 4 residues recognized by Ssu72 are identical in the Ser(P)2 and Ser(P)5 substrates. Therefore, discrimination must be based on the Thr4 and distal Tyr1 positions. For a Ser(P)2 substrate, the Tyr1 position would be substituted by serine, causing a decrease in binding energy from the loss of the aromatic amide stacking interaction. However, the Thr4 position is likely the more important determinant of substrate specificity. The Thr4 residue would be replaced by tyrosine in a Ser(P)2 substrate, which would disrupt substrate recognition in two major ways. First, the favorable hydrophobic interactions with the insert subdomain would be lost, and the accommodation of the large phenol group would require a significant reconfiguration of the substrate backbone to prevent steric clashes. Second, this substrate would be unable to replicate the intramolecular hydrogen bond we observe between the Thr4 hydroxyl and the Pro6 backbone carbonyl, making the already unfavorable cis conformation even less energetically stable.
Although the exact degree of isomeric specificity of Ssu72 is unclear, our structural and kinetic studies demonstrate a strong preference for the cis-Ser(P)5–Pro6 conformation. First, modeling the trans-Ser(P)5–Pro6 isomer in the Ssu72-pCTD complex structure leads to large steric clashes between the pCTD backbone and the side chains of the C-terminal α-helix of the insert subdomain. Also, if the activity toward cis and trans isomers were comparable, there would be no stimulation of dephosphorylation by Ess1. Finally, additional support for strong cis-proline specificity comes from a kinetic study that measured activity toward a set of Ser(P)5 CTD substrates with an alanine substitution at each heptad position (32). These experiments showed that CTD positions Thr4, Pro6, and Tyr1 of the following heptad were critical for substrate recognition, a result that is consistent with the intermolecular contacts in our complex structure. Importantly, the largest decrease in Ssu72 activity was observed with the P6A mutant substrate, an ∼30-fold reduction when compared with wild type. This likely reflects not only the loss of binding energy for the proline ring but also the reduced propensity of the Ser(P)5–Ala6 peptide bond to adopt a cis configuration.
Regulation of Isomer-specific Enzymes
To the best of our knowledge, Ssu72 is the first example of an enzyme with specificity for the cis isomer of proline. Isomeric specificity for trans-Ser(P)-Pro motifs has been previously established for two enzymes. The serine/threonine phosphatase PP2A dephosphorylates trans-Ser(P)-Pro motifs in its substrate Cdc25C and Tau proteins and shows increased activity in reactions that include Pin1 (40). Likewise, the serine/threonine kinase ERK2 phosphorylates trans-Ser-Pro motifs in the RNase T1 substrate and is also stimulated by proline isomerase activity (41). Importantly, catalyzed proline isomerization only increases the activity of these trans-specific enzymes toward Ser(P)-Pro motifs that adopt the cis conformation in the structure of their protein. For example, ERK2 phosphorylates two sites in RNase T1, cis-Ser54–Pro55 and trans-Ser72–Pro73; however, only phosphorylation at Ser54–Pro55 is enhanced by the addition of peptidyl prolyl isomerases. This illustrates a crucial point: catalyzed proline isomerization is an effective regulatory mechanism of phosphorylation/dephosphorylation only when the target motif exists predominantly in the opposite isomeric configuration required for catalysis. In this case, the isomer-specific enzyme can only process a small fraction of the total substrate, and the extremely slow, uncatalyzed cis-trans isomerization prevents re-equilibration of the non-processed fraction. In the opposite scenario, the majority of substrate is already in the correct isomeric form and can be readily processed, with catalyzed proline isomerization having little influence.
The effect of proline isomerization on CTD-associated activities depends greatly on the CTD conformation during the transcription cycle. All structural studies of CTD-binding proteins have revealed binding that is, in fact, isomer-specific but preferential for trans-Ser(P)-Pro motifs. In the case of Pcf11, the CTD-interacting domain discriminates in favor of 3 consecutive trans-prolines, including one Ser(P)2–Pro3 motif (8). In the structure of CTD phosphatase Scp1 in complex with Ser(P)2/Ser(P)5 CTD, both the Ser(P)2–Pro3 and the Ser(P)5–Pro6 motifs adopt the trans configuration, with binding to either cis form seemingly prevented by steric clashes (34). For both of these proteins, the regulation of binding or catalysis by proline isomerization would demand a CTD conformation with a minority trans population. This is especially true for non-enzymatic proteins such as Pcf11, which may only require one correctly configured binding site out of dozens of CTD repeats. However, our measurements with a Ser(P)5 CTD peptide and prior work with a Ser(P)2 CTD peptide (8) show that cis-proline is the less populated isomer. Given the lack of evidence for any stable full-length CTD structure, it seems likely that cis-prolines constitute the isomeric minority in vivo as well and that proteins with specific recognition of cis-proline are the major targets of regulation by isomerization.
The timescale of transcriptional events is another critical determinant of the effect of Ess1 on CTD-associated activities. We observed only an ∼2-fold enhancement of Ssu72-catalyzed product accumulation in reactions with synthetic pCTD substrate as a significant amount of non-catalyzed proline isomerization occurs over the course of the 5-min reactions. A similar level of stimulation was reported for the PP2A phosphatase described above in reactions on the same timescale (40). However, at a faster timescale, the effect of Ess1 on Ssu72 activity is greater, as shown in supplemental Fig. S3 in 1.25-min reactions monitored at 15-s increments. Although the exact in vivo timescale of Ssu72 activity is unknown, the effect of impaired Ess1 activity on Nrd1 localization and Pcf11 recruitment is largely confined to the 3′-end of genes (28), suggesting that Ser(P)5 CTD dephosphorylation by Ssu72 occurs mainly during termination, which may take only tens of seconds (42). This scenario is well illustrated by the kinetic assays with full-length pCTD in Fig. 4, E and F, where the reaction without Ess1 takes 20 min to dephosphorylate ∼90% of the initial pCTD substrate. In contrast, the reaction with Ess1 reaches ∼90% completion in 1 min, on par with the estimated timescale of transcription termination.
A Revised Model for Ess1-regulated Ssu72 Activity
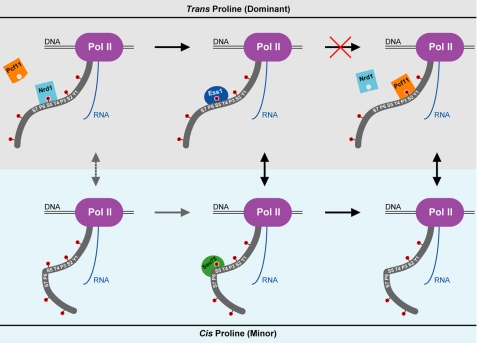
Our kinetic experiments provide an excellent interpretation of recent in vivo studies of Ess1 (28, 29). We show that the stimulation of Ssu72 activity by Ess1-catalyzed cis-trans interconversion of Ser(P)5–Pro6 motifs in the CTD heptad is severely reduced with catalytically impaired Ess1 mutants, which explains the in vivo accumulation of Ser(P)5 CTD in ess1 cells. Interestingly, overexpression of Ssu72 suppressed this phenotype in an A144T ess1 strain, but not an H164R ess1 strain (29). It is possible that different levels of residual activity or thermostability of the Ess1 mutants are responsible for this discrepancy. In contrast, overexpression of Ssu72 rescues the temperature sensitivity of both ess1 strains (29), suggesting that improper regulation of CTD phosphorylation is the main cause of this phenotype. As described in the Introduction, the proposed proline isomer-based regulatory scheme that explains these results requires trans specific activity by Ssu72 (28, 29). Our results compel a simple but unexpected adjustment of this model: reversing the isomeric specificity of Ssu72 (Fig. 5). Interestingly, genetic evidence suggests that this model may be partially bypassed by overexpression of Ser(P)5 CTD phosphatase Fcp1 in yeast (16).
FIGURE 5.
A new model for Ess1-regulated Ssu72 activity. RNAPII (Pol II) is shown near the termination of an snRNA transcript. For clarity, only one CTD heptad is labeled. The upper (gray) and lower (light blue) panels represent the trans and cis forms, respectively, of the Ser(P)5–Pro6 motif. Dephosphorylation cannot proceed through the trans form (red cross) due to the cis-proline substrate specificity of Ssu72. Instead, Ess1 catalyzes the interconversion of isomers, which is the rate-limiting step for Ssu72 activity. The minor pathway is indicated with gray arrows, with the dashed line representing non-catalyzed isomerization that occurs very slowly comparison with transcriptional events. The change in CTD phosphorylation facilitates proper termination by promoting the release of Nrd1 and recruiting Pcf11.
Implications of Proline Isomerization in CTD Biology
Ssu72 provides an exciting validation of proline isomers as a critical component of the CTD code but also raises many new questions about how this regulatory mechanism operates. For example, it is unclear whether the regulation of Ssu72 is a general property of RNAPII transcription or a gene-specific one. Although impaired Ess1 and Ssu72 activity causes readthrough transcription of a small subset of genes consisting mainly of snRNA that undergo Nrd1-dependent termination (16, 28), it is possible this phenotype is only apparent for short transcripts that require a sudden drop in Ser(P)5 levels for proper termination. It will also be interesting to see what other CTD-binding partners specifically recognize cis-proline and how their functions are regulated by catalyzed isomerization. Although these proteins may only regulate the CTD phosphorylation state, there may also be non-enzymatic CTD-binding factors with cis-proline recognition whose association with the transcribing polymerase is regulated by Ess1/Pin1 to fine-tune the recruitment of their co-transcriptional activities.
Supplementary Material
Acknowledgments
Crystal screening was performed at the Duke University Medical Center X-ray Crystallography Shared Resource. Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found on the SER-CAT web page. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract Number W-31-109-Eng-38. We thank Louis Metzger and Jeffrey Boyles for enlightening discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM079376 (to P. Z.) and GM040505 (to A. L. G.). This work was also supported by a Kamin Fellowship (to J. W. W.-A.).
The atomic coordinates and structure factors (code 3P9Y) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S3.
A. P. Kuzin, Y. Chen, J. Seetharaman, F. Forouhar, Y. Chinag, Y. Fang, K. Cunningham, L.-C. Ma, R. Xiao, J. Liu, M. C. Baran, T. B. Acton, B. Rost, G. T. Montelione, J. F. Hunt, and L. Tong, unpublished results.
- CTD
- C-terminal domain
- pCTD
- phosphorylated CTD
- RNAPII
- RNA polymerase II
- LMW PTP
- low molecular weight protein tyrosine phosphatase
- snRNA
- small nuclear RNA.
REFERENCES
- 1. Meinhart A., Kamenski T., Hoeppner S., Baumli S., Cramer P. (2005) Genes Dev. 19, 1401–1415 [DOI] [PubMed] [Google Scholar]
- 2. Phatnani H. P., Greenleaf A. L. (2006) Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 3. Buratowski S. (2009) Mol. Cell 36, 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buratowski S. (2003) Nat. Struct. Biol. 10, 679–680 [DOI] [PubMed] [Google Scholar]
- 5. Shaw P. E. (2007) EMBO Rep. 8, 40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu K. P., Finn G., Lee T. H., Nicholson L. K. (2007) Nat. Chem. Biol. 3, 619–629 [DOI] [PubMed] [Google Scholar]
- 7. Schutkowski M., Bernhardt A., Zhou X. Z., Shen M., Reimer U., Rahfeld J. U., Lu K. P., Fischer G. (1998) Biochemistry 37, 5566–5575 [DOI] [PubMed] [Google Scholar]
- 8. Noble C. G., Hollingworth D., Martin S. R., Ennis-Adeniran V., Smerdon S. J., Kelly G., Taylor I. A., Ramos A. (2005) Nat. Struct. Mol. Biol. 12, 144–151 [DOI] [PubMed] [Google Scholar]
- 9. Xu Y. X., Manley J. L. (2004) Cell Cycle 3, 432–435 [PubMed] [Google Scholar]
- 10. Gemmill T. R., Wu X., Hanes S. D. (2005) J. Biol. Chem. 280, 15510–15517 [DOI] [PubMed] [Google Scholar]
- 11. Verdecia M. A., Bowman M. E., Lu K. P., Hunter T., Noel J. P. (2000) Nat. Struct. Biol. 7, 639–643 [DOI] [PubMed] [Google Scholar]
- 12. Xu Y. X., Hirose Y., Zhou X. Z., Lu K. P., Manley J. L. (2003) Genes Dev. 17, 2765–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palancade B., Marshall N. F., Tremeau-Bravard A., Bensaude O., Dahmus M. E., Dubois M. F. (2004) J. Mol. Biol. 335, 415–424 [DOI] [PubMed] [Google Scholar]
- 14. Kops O., Zhou X. Z., Lu K. P. (2002) FEBS Lett. 513, 305–311 [DOI] [PubMed] [Google Scholar]
- 15. Krishnamurthy S., He X., Reyes-Reyes M., Moore C., Hampsey M. (2004) Mol. Cell 14, 387–394 [DOI] [PubMed] [Google Scholar]
- 16. Ganem C., Devaux F., Torchet C., Jacq C., Quevillon-Cheruel S., Labesse G., Facca C., Faye G. (2003) EMBO J. 22, 1588–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meinhart A., Silberzahn T., Cramer P. (2003) J. Biol. Chem. 278, 15917–15921 [DOI] [PubMed] [Google Scholar]
- 18. Sun Z. W., Hampsey M. (1996) Mol. Cell. Biol. 16, 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu W. H., Pinto I., Chen B. S., Hampsey M. (1999) Genetics 153, 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh B. N., Hampsey M. (2007) Mol. Cell 27, 806–816 [DOI] [PubMed] [Google Scholar]
- 21. Ansari A., Hampsey M. (2005) Genes Dev. 19, 2969–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Sullivan J. M., Tan-Wong S. M., Morillon A., Lee B., Coles J., Mellor J., Proudfoot N. J. (2004) Nat. Genet. 36, 1014–1018 [DOI] [PubMed] [Google Scholar]
- 23. Reyes-Reyes M., Hampsey M. (2007) Mol. Cell. Biol. 27, 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Steinmetz E. J., Brow D. A. (2003) Mol. Cell. Biol. 23, 6339–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim M., Vasiljeva L., Rando O. J., Zhelkovsky A., Moore C., Buratowski S. (2006) Mol. Cell 24, 723–734 [DOI] [PubMed] [Google Scholar]
- 26. Ghazy M. A., He X., Singh B. N., Hampsey M., Moore C. (2009) Mol. Cell. Biol. 29, 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nedea E., He X., Kim M., Pootoolal J., Zhong G., Canadien V., Hughes T., Buratowski S., Moore C. L., Greenblatt J. (2003) J. Biol. Chem. 278, 33000–33010 [DOI] [PubMed] [Google Scholar]
- 28. Singh N., Ma Z., Gemmill T., Wu X., Defiglio H., Rossettini A., Rabeler C., Beane O., Morse R. H., Palumbo M. J., Hanes S. D. (2009) Mol. Cell 36, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krishnamurthy S., Ghazy M. A., Moore C., Hampsey M. (2009) Mol. Cell. Biol. 29, 2925–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baykov A. A., Evtushenko O. A., Avaeva S. M. (1988) Anal. Biochem. 171, 266–270 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Z. Y. (2003) Prog. Nucleic Acid Res. Mol. Biol. 73, 171–220 [DOI] [PubMed] [Google Scholar]
- 32. Hausmann S., Koiwa H., Krishnamurthy S., Hampsey M., Shuman S. (2005) J. Biol. Chem. 280, 37681–37688 [DOI] [PubMed] [Google Scholar]
- 33. Wang S., Tabernero L., Zhang M., Harms E., Van Etten R. L., Stauffacher C. V. (2000) Biochemistry 39, 1903–1914 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y., Kim Y., Genoud N., Gao J., Kelly J. W., Pfaff S. L., Gill G. N., Dixon J. E., Noel J. P. (2006) Mol. Cell 24, 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tóth G., Watts C. R., Murphy R. F., Lovas S. (2001) Proteins 43, 373–381 [DOI] [PubMed] [Google Scholar]
- 36. Phatnani H. P., Jones J. C., Greenleaf A. L. (2004) Biochemistry 43, 15702–15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiang K., Nagaike T., Xiang S., Kilic T., Beh M. M., Manley J. L., Tong L. (2010) Nature 467, 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yeo M., Lin P. S., Dahmus M. E., Gill G. N. (2003) J. Biol. Chem. 278, 26078–26085 [DOI] [PubMed] [Google Scholar]
- 39. Hausmann S., Shuman S. (2002) J. Biol. Chem. 277, 21213–21220 [DOI] [PubMed] [Google Scholar]
- 40. Zhou X. Z., Kops O., Werner A., Lu P. J., Shen M., Stoller G., Küllertz G., Stark M., Fischer G., Lu K. P. (2000) Mol. Cell 6, 873–883 [DOI] [PubMed] [Google Scholar]
- 41. Weiwad M., Werner A., Rücknagel P., Schierhorn A., Küllertz G., Fischer G. (2004) J. Mol. Biol. 339, 635–646 [DOI] [PubMed] [Google Scholar]
- 42. Femino A. M., Fay F. S., Fogarty K., Singer R. H. (1998) Science 280, 585–590 [DOI] [PubMed] [Google Scholar]
- 43. Schubert M., Labudde D., Oschkinat H., Schmieder P. (2002) J. Biomol. NMR 24, 149–154 [DOI] [PubMed] [Google Scholar]
- 44. Chapman R. D., Heidemann M., Albert T. K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007) Science 318, 1780–1782 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.