Abstract
Toll-like receptors (TLRs) and nucleotide-binding domain, leucine-rich repeat (NLR) proteins are two major forms of innate immune receptors that trigger inflammatory responses by various biological mechanisms such as cytokine production, recruitment of inflammatory cells, or activation of adaptive immunity. Although the innate immune system is designed to fight against infectious pathogens, excessive activation of TLR or NLR signaling pathways may lead to unwarranted inflammation with hazardous outcomes, including septic shock or inflammatory diseases. As part of the search for effective therapeutics to regulate these responses, here we show that a novel aminosaccharide compound, named DFK1012, inhibits immune responses caused by TLR and NLR activation. Treatment with DFK1012, but not its derivatives DFK845 or DFK846, strongly inhibited pro-inflammatory cytokine production upon stimulation via either TLR or NLR proteins in macrophages. Importantly, we have not observed cytotoxicity in any range of its working concentration. Treatment with DFK1012 did not interfere with TLR- or NLR-induced activation of p38 and JNK, phosphorylation/degradation of IκB, and subsequent nuclear translocation of NF-κB subunit p65, suggesting that the inhibitory activity of DFK1012 is not due to the suppression of downstream signaling. Indeed, DFK1012 did not impair transcription of pro-inflammatory cytokine genes but rather promoted post-translational degradation of pro-inflammatory cytokines. Therefore, DFK1012 is a novel anti-inflammatory compound that drives proteolysis of proinflammatory cytokines induced by TLR and NLR stimulation. DFK1012 may represent a novel class of potential therapeutic agents aimed at the treatment of inflammatory disorders.
Keywords: Inflammation, Innate Immunity, Macrophage, NF-kappa B, Signal Transduction
Introduction
The innate immune system serves as the first line of host defense against infectious agents by detecting the presence of microbial infection through germ line-encoded pattern recognition receptors (1). Membrane-bound Toll-like receptors (TLRs)2 as well as cytoplasmic nucleotide-binding domain and leucine-rich repeat (NBD-LRRs or NLRs) protein families are important groups of pattern recognition receptors that provide immediate immune responses by recognizing different but overlapping microbial components, frequently referred to as pathogen-associated molecular patterns (PAMPs) (2). Mammalian TLRs detect components from various microorganisms, including bacteria, viruses, and protozoa (3–18). Signaling of TLRs utilizes an adaptor MyD88, except for TLR3, which requires an adaptor TIR-domain-containing adapter-inducing interferon-β (TRIF), leading to the activation of signaling cascades such as MAP kinases and NF-κB. These events result in the transcriptional activation of immune response genes and lead to the up-regulation of surface co-stimulatory molecules on antigen-presenting cells and the secretion of pro-inflammatory cytokines such as IL-6, IL-8, IL-12, and TNF-α (19–22).
Similar to TLRs, NLR proteins have been shown to play an important role in the recognition and control of bacterial infection (23, 24). For example, Nod2 recognizes muramyl dipeptide (MDP), an active moiety of the bacterial cell wall peptidoglycan, and transcriptionally activates immune response genes via a serine/threonine kinase, Rip2 (25–28), which activates the NF-κB and MAP kinase signaling pathways (27–29).
The production of inflammatory mediators is tightly regulated by a variety of mechanisms at the transcriptional, post-transcriptional, and post-translational levels. For example, the transcription of TNF-α, a key pro-inflammatory cytokine, is regulated by MAP kinase and NF-κB signaling, and inflammatory stimuli also stabilize TNF-α mRNA through AU-rich elements at the 3′-untranslated region (30). Furthermore, TNF-α protein is translated as a precursor, which is transported to the plasma membrane via recycling vesicles (31, 32). Maturation and secretion of TNF-α are dependent on cleavage by TNF-α-converting enzyme (TACE) (33). A substantial amount of TNF-α precursor remains uncleaved and is degraded through the lysosomal pathway (34). Although synthesis and/or release of pro-inflammatory mediators are tightly controlled, dysregulation or overproduction of these mediators may cause septic shock or other inflammatory disorders (35). Studies using animal models have revealed that TLRs play an important role in the pathogenesis of chronic inflammatory disease such as lupus, experimental autoimmune encephalomyelitis, and chronic colitis (36–42). Recent genome-wide linkage studies have also identified many factors that link the polymorphisms of TLRs and NLRs to inflammatory diseases. Polymorphisms of TLR2 are associated with the pathogenesis of acute rheumatic fever and lepromatous leprosy, and a polymorphism of TLR4 is linked to inflammatory bowel diseases (43–45). Gain-of-function mutations in the NOD2 gene are associated with the pathogenesis of Blau syndrome and early onset sarcoidosis (46–48). Given the vast number of diseases linked to excessive inflammation, the regulation of TLR and NLR responses is an important target for the control or prevention of various inflammatory disorders.
In this study, we show that a novel aminosaccharide compound, DFK1012, blocks responses upon stimulation through either TLRs or NLRs. We generated various derivatives of MDP on which biotin was appended. We found that one of such compounds, DFK1012, strongly inhibited cytokine production by macrophages upon TLR or NLR stimulation at the post-translational level. This serendipitous discovery may lead to the development of a novel class of anti-inflammatory compounds that have a unique mechanism of action.
EXPERIMENTAL PROCEDURES
Cell Culture
RAW 264.7 cells were maintained in DMEM (Invitrogen) containing 10% (v/v) heat-inactivated FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin at 37 °C under 5% CO2. Bone marrow-derived macrophages were prepared as described (27). Briefly, mouse bone marrow was obtained by flushing the tibia and femur of C57BL/6 mouse (Taconic) with DMEM supplemented with 10% heat-inactivated FBS (Invitrogen). Bone marrow cells were cultured in 10 ml of DMEM, supplemented with 10% FBS, glutamine (both from Invitrogen), and 30% L929 cell supernatant containing M-CSF, at an initial density of 1 × 106 cells/ml in 100-mm Petri dishes (BD Biosciences) at 37 °C in humidified 5% CO2 for 6 days. Cells were harvested with cold PBS (Invitrogen), washed, resuspended in DMEM supplemented with 10% FBS, and used at a density of 2 × 105 cells/ml.
Reagents
MDP (N-acetylmuramyl-l-alanyl-d-isoglutamine) was purchased from Sigma-Aldrich and resuspended in endotoxin-free water. DFK845, DFK846, and DFK1012 were synthesized by Eisai Research Institute (Andover, MA). Lipopolysaccharide (LPS) was from Enzo Life Sciences (Plymouth Meeting, PA). Phosphorothioate-modified CpG oligonucleotide DNA (TCCATGACGTTCCTGACGTT) was synthesized by Integrated DNA Technologies (Coralville, IA). The proteasome inhibitor, MG-132, was obtained from Calbiochem (Darmstadt, Germany). Rabbit polyclonal anti-poly(ADP-ribose) polymerase (PARP), anti-IκBα, anti-p65, anti-FITC-p65, and goat polyclonal anti-actin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-active caspase-3, anti-p-ERK, anti-p-SAPK/JNK, anti-p-p38, anti-p-Rip2, and mouse monoclonal anti-p-IκBα antibodies were from Cell Signaling (Beverly, MA). Rat polyclonal anti-Nod2 and rabbit polyclonal anti-Rip2 were from eBioscience (San Diego, CA). Rabbit polyclonal anti-iNOS was from Abcam (Cambridge, MA), and goat anti-mouse IL-1β was from R&D Systems. Goat anti-rabbit/mouse/goat secondary antibodies conjugated with horseradish peroxidase were from GE Healthcare. Hoechst 33342 (trihydrochloride, trihydrate) was from Invitrogen. 3-(4,5-Dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) and chloroquine were from Sigma-Aldrich.
Determination of Cytokine Secretion
Cytokine levels in culture supernatants were determined using ELISA kits for IL-6 (Pharmingen), TNF-α (R&D Systems), and IL-12 p40 (Pharmingen), according to the manufacturer's instructions.
MTT and LDH Release Assay
Cell viability was measured by MTT and LDH assay. MTT solution was added to cells in 96-well plates to a final concentration of 0.5 mg/ml, and the cells were incubated at 37 °C for 1 h. After removing culture medium, 50 μl of DMSO were added, and the optical density of each well was read at 570 nm. LDH release assay was performed by using a non-radioactive cytotoxicity assay kit (Promega, Madison, WI) according to the manufacturer's instructions.
Preparation of Cytoplasmic and Nuclear Extracts
Cells were washed twice with 1× PBS and allowed to equilibrate for 5 min in ice-cold cytoplasmic extraction buffer consisting of 10 mm Tris-HCl (pH 7.8), 10 mm KCl, l.5 mm EDTA, 0.5 mm DTT. Cells were lysed on ice for 5 min in 0.4% Nonidet P-40/cytoplasmic extraction buffer/protease inhibitor mixture. Cells were gently scraped with a rubber policeman. Following centrifugation at 3500 rpm for 5 min, the supernatants (cytoplasmic extracts) were collected and snap-frozen on dry ice. The nuclear pellets were washed in detergent-free cytoplasmic extraction buffer containing a protease inhibitor mixture (Roche Diagnostics) and then suspended in nuclear extraction buffer (New England Biolabs) consisting of 20 mm Tris-HCl (pH 7.8), 150 mm NaCl, 50 mm KCl, 1.5 mm EDTA, 5 mm DTT, 1 mm Na2VO4, and a protease inhibitor mixture. After vigorous vortexing at maximum speed and incubation for 10 min on ice, the solution was clarified by centrifugation at 15,000 rpm for 10 min, and the supernatant (nuclear extract) was collected and snap-frozen on dry ice before storage at −70 °C. The protein concentration was determined by Bradford assay.
Western Blot Analysis
Twenty micrograms of protein were resolved by 4–12% gradient SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. The membranes were blocked with 5% skim milk-PBS-0.1% Tween 20 for 1 h before being incubated overnight at 4 °C with primary antibodies in 5% skim milk-PBS, 0.1% Tween 20. The membranes were then washed three times in 1× PBS-0.1% Tween 20 and incubated with horseradish peroxidase-conjugated secondary antibodies diluted in 5% skim milk-PBS, 0.1% Tween 20 for 1 h. After successive washes, the membranes were developed using the SuperSignal West Pico chemiluminescent kit (Thermo Scientific).
Immunofluorescent Staining for NF-κB
Cells grown in 35-mm dishes were fixed in methanol and incubated with rabbit polyclonal anti-p65 antibody diluted 1:100 in 3% BSA for 24 h. The cells were subsequently incubated with rhodamine isothiocyanate-conjugated goat anti-rabbit immunoglobulin G antibody diluted 1:100 in 3% BSA for 30 min. After being mounted with 50% glycerol, the slides were analyzed with a fluorescence microscope (Nikon, ECLIPSE TE300).
Flow Cytometry Analysis
For the NF-κB translocation assay, nuclei were prepared by incubating the cells in 1% Triton X-100 (pH 7.4) (10 mm HEPES, 320 mm sucrose, 5 mm MgCl2) for 10 min at 4 °C. Nuclei were washed twice with PBS-BSA (1% BSA) and stained with either FITC-conjugated anti-NF-κB p65 antibody or FITC-conjugated monoclonal immunoglobulin isotype control (Santa Cruz Biotechnology) for 30 min at 4 °C. After incubation, the nuclei were washed with PBS-BSA and counterstained with 1 mg/ml propidium iodide. Nuclei were gated on the basis of propidium iodide staining, after doublet elimination by area versus width measurement on a FACSCalibur using CELLQuest software. Voltage settings for FITC parameters were performed with isotype control. For intracellular cytokine staining, allophycocyanin (APC) anti-mouse TNF-α antibody was obtained from BioLegend (San Diego, CA). Cells were fixed and permeabilized using a permeabilization/fixation buffer (eBioscience) for 20 min at room temperature and then incubated with anti-CD16/CD32 antibody for 15 min on ice. Following Fc block, the cells were stained with TNF-α antibody diluted in permeabilization buffer for intracellular staining. Stained cells were washed, resuspended in 1% paraformaldehyde PBS solution, and analyzed by FACSCalibur (BD Biosciences) followed by analysis using FlowJo software.
Real-time Quantitative PCR
RNA was isolated using TRIzol reagent (Invitrogen) and ethanol-precipitated. cDNA synthesis was performed using the qScript Flex cDNA synthesis kit (Quanta Biosciences) according to the manufacturer's instructions. RNA expression was quantified on the 7300 real-time PCR system (Applied Biosystems) using the PerfeCTa SYBR Green supermix with ROX (Quanta Biosciences). Primer pairs used in the quantitative PCR analysis were as follows: β-actin, forward, 5′-GCTGTGCTGTCCCTGTATGCCTCT-3′, β-actin, reverse, 5′-CTTCTCAGCTGTGGTGGTGAAGC-3′; Interleukin-6 (IL-6), forward, 5′-CCAGAAACCGCTATGAAGTTCC-3′, IL-6, reverse, 5′-TTGTCACCAGCATCAGTCCC-3′; Tumor necrosis factor-α (TNF-α), forward, 5′-ACAGAAAGCATGATCCGCG-3′, TNF-α, reverse, 5′-GCCCCCCATCTTTTGGG-3′; Interleukin-1β (IL-1β), forward, 5′-GCCTCGTGCTGTCGGACC-3′, IL-1β, reverse, 5′-TGTCGTTGCTTGGTTCTCCTTG-3′.
RESULTS
DFK1012 Inhibits the Production of Inflammatory Cytokines Induced by TLR and NLR Activation
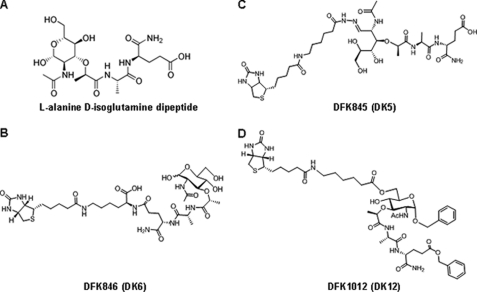
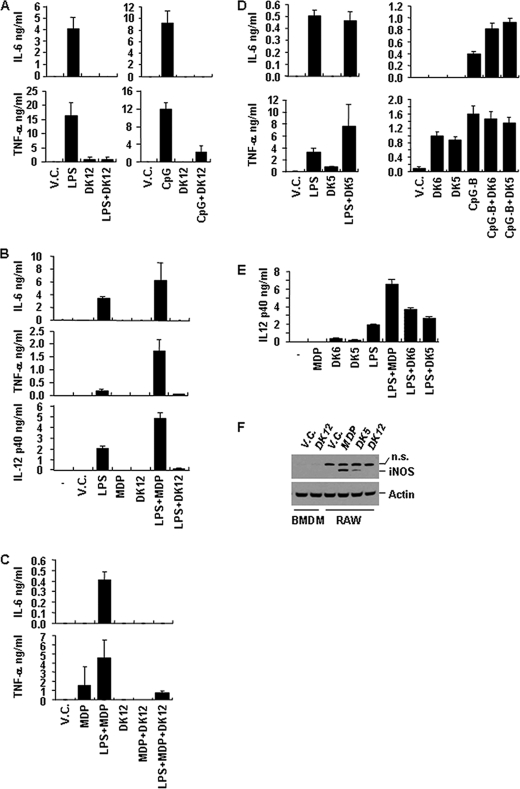
Using MDP as a lead compound, we generated various biotinylated derivatives, including DFK1012 (Fig. 1). To determine whether these compounds influence the expression of pro-inflammatory cytokines, we stimulated a murine macrophage cell line (RAW264.7) with LPS (TLR4 ligand) or CpG-B oligodeoxynucleotide (TLR9 ligand) in the presence or absence of DFK1012 and determined levels of IL-6 and TNF-α in culture medium by ELISA. DFK1012 strongly inhibited the production of IL-6 and TNF-α induced by both LPS and CpG-B oligodeoxynucleotide, indicating that DFK1012 inhibits TLR4 and TLR9 responses (Fig. 2A). Furthermore, DFK1012 has an inhibitory effect on primary macrophages because DFK1012 treatment also abolished the production of IL-6, TNF-α, and IL-12 p40 in bone marrow-derived macrophages (BMDM) stimulated with LPS (Fig. 2B). In contrast, the parent molecule MDP enhanced the production of IL-6, TNF-α, and IL-12 p40 following LPS stimulation (Fig. 2, B and C), which is consistent with an earlier report on the synergism between MDP and TLR signaling (27). Indeed, MDP enhanced the production of IL-6 and TNF-α induced by LPS (Fig. 2, B and C). These results indicate that, despite their shared structure, DFK1012 may have very different biochemical properties (Fig. 1, A and D). To examine the potency of DFK1012, we titrated the compound for its blocking effect. DFK1012 was able to block LPS-induced IL-6 production from RAW cells even at a concentration of 15 nm (data not shown). Interestingly, DFK1012 also inhibited TNF-α production upon Nod2 activation in MDP-stimulated RAW 264.7 cells (Fig. 2C). Therefore, DFK1012 can inhibit responses elicited by TLRs and Nod2 activation. To determine the specificity of the inhibitory effect of DFK1012, we generated two derivatives of DFK1012 by changing the biotin and spacer position on MDP. DFK845 was generated using an analogue of biotin hydrazide that contains a hexanoic acid extension to label an aldehyde of the open form of muramic acid, forming a hydrazone bond. DFK846 synthesis results from amide bond formation between the amine group of a biotinylated lysine and the carboxylic acid of d-isoglutamine (Fig. 1, B and C). In contrast to the strong inhibitory activity of DFK1012, the two derivatives DFK845 and DFK846 did not suppress the production of IL-6 and TNF-α upon LPS or CpG-B stimulations (Fig. 2, D and E). Instead, DFK845 and DFK846 enhanced cytokine production in response to CpG-B stimulation in RAW264.7 cells (Fig. 2D) and increased the secretion of IL-12 p40 in LPS-treated BMDM (Fig. 2E). Moreover, treatment with MDP or DFK845, but not DFK1012, increased the level of iNOS, an NF-κB-dependent gene product (Fig. 2F). These data suggest that DFK845 and DFK846, like MDP, induce pro-inflammatory effects, whereas DFK1012 has a strong inhibitory effect on responses induced by TLR or NLR simulation.
FIGURE 1.
Chemical structure of MDP, DFK846, DFK845, and DFK1012.
FIGURE 2.
DFK1012 inhibits IL-6 and TNF-α production induced by the TLR4 ligand (LPS), TLR9 ligand (CpG-B), or Nod2 ligand (MDP). A–E, cells were stimulated in the following conditions, and levels of IL-6, TNF-α, and IL-12 p40 were determined by ELISA. A, RAW264.7 cells were stimulated with LPS (1 ng/ml) or CpG-B (1 μm) in the presence or absence of DFK1012 (DK12, 30 nm) for 24 h. V.C., vehicle control (DMSO). B, BMDM were stimulated with LPS (0.1 ng/ml) in the presence or absence of DFK1012 or MDP (100 μg/ml) for 24 h. C, RAW264.7 cells were stimulated with LPS (0.5 ng/ml) plus MDP (100 μg/ml) in the presence or absence of DFK1012 (30 nm) for 24 h. D, RAW264.7 cells were treated with LPS (0.5 ng/ml) or CpG-B (1 μm) in the presence or absence of DFK845 (DK5, 100 μg/ml) or DFK846 (DK6, 100 μg/ml) for 24 h. E, BMDM were stimulated with LPS (1 ng/ml) in the presence or absence of DFK845 (100 μg/ml) or DFK846 (100 μg/ml) for 24 h. Data represent the mean ± S.D. of triplicates. Data are representative of three independent experiments with similar results. F, BMDM and RAW264.7 cells were treated with MDP, DFK1012, or DFK845 for 18 h. Cell extracts were subjected to Western blot analysis for iNOS and actin. n.s., nonspecific.
The Anti-inflammatory Effect of DFK1012 Is Not Due to Cytotoxicity
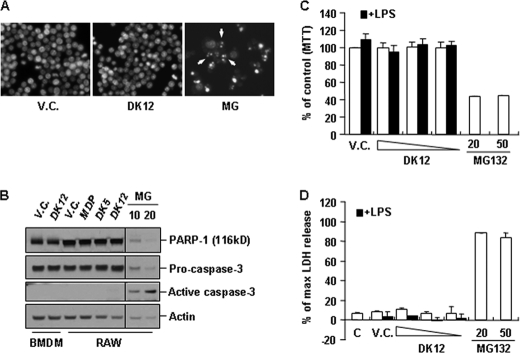
One possible explanation for the decrease in production of inflammatory cytokines upon DFK1012 treatment could be changes in cell viability. To determine whether DFK1012 adversely affects cell survival, RAW264.7 cells were incubated with DFK1012 or 20 μm MG-132 as a positive control to induce apoptosis. Upon MG-132 treatment, cells underwent characteristic apoptotic changes in their morphology, including cell shrinkage and chromatin compaction (Fig. 3A). However, DFK1012 treatment did not induce any morphological changes, indicating that DFK1012 does not cause apoptotic cell death (Fig. 3A). Caspase activation and caspase-mediated proteolysis such as PARP-1 (poly(ADP-ribose) polymerase-1) cleavage, along with the fragmentation of cellular DNA, are hallmarks of apoptosis and contribute to the irreversibility of the cell death process (49). MG-132-treated RAW264.7 cells exhibited proteolysis of PARP-1 and active caspase-3 as expected (Fig. 3B), but DFK1012 treatment did not induce PARP-1 cleavage or the generation of active caspase-3 in BMDM and RAW264.7 cells (Fig. 3B), again indicating that DFK1012 does not induce apoptosis. Furthermore, MTT and LDH release assays were used to verify that DFK1012 did not exhibit any effect on cell viability, even in the presence of LPS (Fig. 3, C and D). These results indicate that the inhibitory effect of DFK1012 on proinflammatory cytokine production is not due to cytotoxicity.
FIGURE 3.
The anti-inflammatory effect of DFK1012 is not due to cytotoxicity. A, RAW 264.7 cells were stimulated with DFK1012 (DK12, 30 nm) or MG-132 (MG, 20 μm) for 24 h. Cells were fixed in methanol and then stained with 2 μg/ml Hoechst 33342 to label nuclear DNA. Cells were examined by fluorescence microscopy (×400 in magnification). V.C., vehicle control (DMSO). B, RAW 264.7 cells were incubated with DFK1012 (30 nm), MDP (200 μg/ml), DFK845 (DK5, 160 μg/ml), or MG-132 (10 or 20 μm) for 18 h. Total cellular extracts were subjected to Western blot analysis for PARP-1, caspase-3, and actin. C and D, RAW 264.7 cells were treated with DFK1012 (30, 15, and 7.5 nm) or MG-132 (20 and 50 μm) in the presence (black bar) or absence (white bar) of LPS for 24 h. Cell viability was determined by MTT assay (C) and LDH release assay (D). Data represent the mean ± S.D. of triplicates. Data are representative of three independent experiments with similar results. % of max, percentage of maximum; C, medium.
DFK1012 Does Not Inhibit IκBα Degradation and MAP Kinase Activation upon LPS or MDP Stimulation
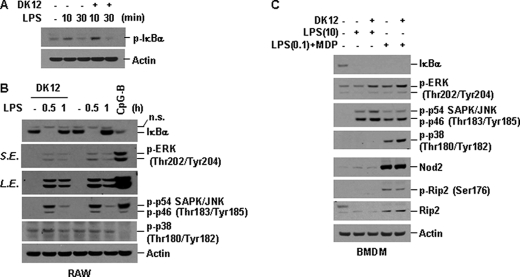
Because TLR and NLR signaling pathways activate transcription factors such as NF-κB and AP-1 that are required for cytokine production, one plausible mechanism by which DFK1012 inhibits pro-inflammatory cytokine production is in interfering with the signaling pathways involving NF-κB or MAP kinases. To examine this possibility, RAW264.7 cells were stimulated with LPS in the presence or absence of DFK1012, and the activation of NF-κB and MAPKs was assessed by Western blotting for phosphorylated and total IκBα, an inhibitor of NF-κB, and phosphorylation of MAPKs, including JNK, p38, and ERK. In LPS-stimulated RAW264.7 cells, IκBα was phosphorylated and degraded 10 and 30 min after stimulation, respectively, and returned to baseline levels at 1 h, representing the degradation of IκBα and subsequent activation of NF-κB, which in turn transcriptionally activates and up-regulates IκBα (Fig. 4, A and B) (50). DFK1012 did not inhibit phosphorylation and degradation of IκBα in LPS-stimulated RAW264.7 cells (Fig. 4, A and B). DFK1012 also did not affect the phosphorylation, and thus activation, of the MAP kinase p38 and JNK (Fig. 4B). Phosphorylation of ERK was only slightly affected by DFK1012, suggesting that DFK1012 may perturb ERK activation upon TLR4 stimulation (Fig. 4B).
FIGURE 4.
DFK1012 does not block phosphorylation and degradation of IκBα and activation of MAP kinase by LPS or MDP stimulation. A and B, RAW264.7 cells were pretreated with DFK1012 (DK12) for 1 h and then stimulated with LPS (10 ng/ml) for the indicated periods in the presence or absence of DFK1012. Cells were treated with CpG-B (1 μm) for 30 min as a positive control. Total cellular extracts were subjected to Western blot analysis for phosphorylated IκBα (p-IκBα), IκBα, phosphorylated ERK (p-ERK), phosphorylated p54 and p46 SAPK/JNK (p-SAPK/JNK), phosphorylated p38 (p-p38), and actin. n.s., nonspecific; L.E., long exposure; S.E., short exposure. C, BMDM were pretreated with or without DFK1012 for 1 h and then stimulated with LPS (10 ng/ml) for 30 min (lanes 1–3). Cells were pretreated with LPS (0.1 ng/ml) for 5 h and then stimulated with MDP (100 μg/ml) for 30 min in the presence or absence of DFK1012 (lanes 4 and 5) Total cellular extracts were subjected to Western blot analysis for IκBα, p-ERK, p-SAPK/JNK, p-p38, Nod2, phosphorylated Rip2 (p-Rip2), total Rip2, and actin. Data are representative of three independent experiments with similar results.
Next we examined whether DKF1012 could interfere with Nod2 signaling. Without priming with either TLR ligands or IFNγ, BMDM express a low level of Nod2 protein and only weakly respond to MDP stimulation (data not shown). Therefore, BMDM were primed with a low dose of LPS to induce Nod2 expression and then subsequently challenged with MDP. In BMDM pre-primed with a low dose of LPS, Nod2 as well as Rip2 were up-regulated (Fig. 4C). DFK1012 failed to inhibit IκBα degradation and the activation of p38, JNK, and ERK upon MDP stimulation (Fig. 4C). These results indicate that DFK1012 does not interfere with the activation of NF-κB and MAP kinases, including ERK, upon Nod2 activation. In contrast, treatment with DFK1012 derivatives DFK845 and DFK856 alone induced IκBα degradation, again implying that DFK845 and DFK856 are stimulatory compounds, in sharp contrast to the inhibitory function of DFK1012 (data not shown). In summary, the TLR and NLR cytoplasmic signaling pathways are predominantly unaffected by DFK1012.
DFK1012 Does Not Block Nuclear Translocation of the NF-κB Subunit p65
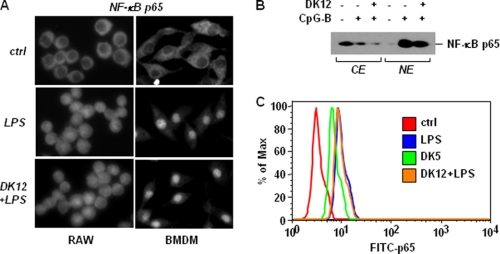
The activation process of NF-κB involves the phosphorylation of IκBα by IκB kinases (IKKs) and the subsequent degradation and nuclear translocation of NF-κB (51). To test whether DFK1012 has any effect on the translocation of NF-κB into the nucleus in response to TLR stimulation, RAW264.7 cells and BMDM were treated with LPS in the presence or absence of DFK1012 and then examined for the subcellular localization of p65 by immunofluorescent staining. Incubation with LPS caused nuclear translocation of p65, as demonstrated by a clear nuclear staining, in contrast to the cytoplasmic distribution in unstimulated cells (Fig. 5A). DFK1012-treated RAW264.7 cells and BMDM also exhibited nuclear staining of p65 after LPS stimulation, suggesting that DFK1012 does not inhibit the nuclear translocation of p65 (Fig. 5A). To confirm that nuclear p65 levels were not affected by DFK1012, we subjected cytoplasmic and nuclear extracts to Western blot analysis. Although the majority of p65 was located in the cytoplasm of control cells without TLR stimulation, stimulation with CpG-B led to an increase of p65 protein in the nucleus, which was not blocked by DFK1012 treatment (Fig. 5B). To assess the translocation of p65 in a more quantitative manner, levels of translocated p65 in RAW264.7 cells stimulated with LPS were analyzed using flow cytometry after staining of the isolated nuclei with FITC-labeled anti-p65 antibody. Translocation of p65 after LPS stimulation is indicated by increased FITC-p65 staining, which was not impaired by treatment with DFK1012 (Fig. 5C). However, in accordance with the effect of DFK845 on cytokine production and IκBα degradation, DFK845 treatment induced the translocation of p65 into the nucleus, again indicating that DFK845 is an active but not an inhibitory compound (Fig. 5C). Overall, these findings indicate that the anti-inflammatory effect of DFK1012 is not due to a blocking effect on NF-κB nuclear translocation.
FIGURE 5.
DFK1012 does not block NF-κB subunit p65 nuclear translocation upon LPS or CpG-B stimulation. A, RAW 264.7 cells or BMDM were pretreated with DFK1012 (DK12, 30 nm) for 1 h and then stimulated with 1 ng/ml of LPS for 30 min. Cells were fixed and permeabilized for 5 min. Immunofluorescent staining for p65 was performed using anti-p65 antibody followed by Alexa Fluor 594 detection antibody. Cells were analyzed using fluorescence microscope. B, BMDM were pretreated with DFK1012 (30 nm) for 1 h and then stimulated with CpG-B (1 μm) for 30 min. Equal amounts of nuclear and cytoplasmic protein extracts were subjected to Western blot analysis for p65. CE, cytoplasmic extracts; NE, nuclear extracts. C, RAW 264.7 cells were pretreated with DFK1012 (30 nm) for 1 h and then stimulated with LPS (10 ng/ml) for 30 min or treated with DFK845 (DK5) for 90 min. Cells were incubated in Hepes-Triton buffer (pH 7.4) to isolate pure nuclei and analyzed for p65/NF-κB localization by flow cytometry after staining with FITC-labeled anti-p65 antibody. Data are representative of three independent experiments with similar results. ctrl, control.
DFK1012 Induces Lysosomal Proteolysis of Inflammatory Cytokine at the Post-translational Level
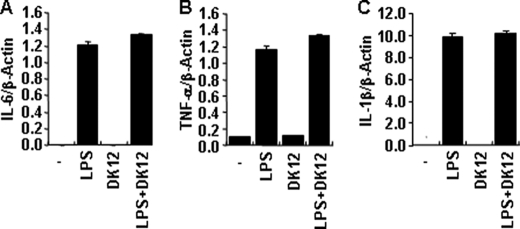
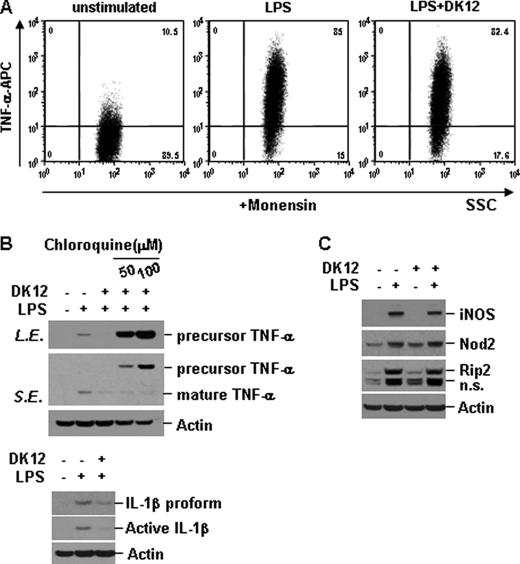
Because signaling downstream of TLR and NLR proteins was largely intact even in the presence of DFK1012, we examined whether the inhibition of proinflammatory cytokine production is due to regulation at the transcriptional or translational level. First, we examined whether DFK1012 could block the induction of mRNA levels of IL-6, TNF-α, and IL-1β by quantitative real-time PCR. Upon LPS stimulation in RAW macrophage cells, transcripts of IL-6, TNF-α, and IL-1β were induced, and this induction was not significantly altered in the presence of DFK1012 (Fig. 6, A–C). Next we examined whether DFK1012 inhibits the synthesis of pro-inflammatory cytokines. We stimulated RAW cells with LPS for 8 h in the presence of monensin, which prevents protein transport to the Golgi complex and leads to accumulation of proteins in the endoplasmic reticulum, and intracellular levels of TNF-α were examined by flow cytometry. As shown in Fig. 7A, the level of TNF-α retained in cells in the presence of monensin was not altered by DFK1012, indicating that DFK1012 does not affect translation or post-translational steps prior to the trafficking of the protein through the ER-Golgi. However, in the absence of monensin, DFK1012 reduced intracellular protein levels of both precursor and mature forms of TNF-α after 12 h of co-treatment with LPS and DFK1012 (Fig. 7B), suggesting altered post-translational regulation, probably post-Golgi. The reduction of precursor TNF-α by DFK1012 was completely blocked by treatment with chloroquine, an inhibitor of lysosomal acidification (Fig. 7B). These results demonstrate that DFK1012 inhibits cytokine secretion by promoting post-translational degradation through the lysosomal pathway. In addition to TNF-α, IL-1β expression is regulated at the protein level because Western blotting showed that DFK1012 treatment suppressed IL-1β expression (Fig. 7B), whereas quantitative PCR showed unchanged IL-1β transcript levels (Fig. 6C). To examine whether the synthesis of other proteins induced by TLR signaling is impaired by DFK1012, RAW cells were stimulated with LPS in the presence or absence of DFK1012, and the expression of LPS-inducible proteins, iNOS, Nod2, and Rip2, was examined by Western blot analysis. There was no significant alteration in the expression of these proteins by DFK1012 treatment (Fig. 7C). Taken together with the severe reduction in secreted proinflammatory cytokine levels as assessed by ELISA (Fig. 2, A–C), these results indicate that the major target of DFK1012 involves post-translational degradation.
FIGURE 6.
DFK1012 does not block induction of IL-6, TNF-α, and IL-1β gene transcripts. A–C, RAW264.7 cells were treated with LPS (10 ng/ml) in the presence or absence of DFK1012 (DK12) for 4 h. The expression of IL-6, TNF-α, and IL-1β was examined by quantitative real-time PCR. Data were normalized to the expression of the β-actin gene. Data represent the mean ± S.D. of triplicates.
FIGURE 7.
DFK1012 triggers post-translational degradation of TNF-α through the lysosomal pathway. A, RAW264.7 cells were treated with LPS (10 ng/ml) in the presence or absence of DFK1012 (DK12) for 8 h in the presence of monensin (2 μm). The expression of intracellular TNF-α was analyzed by flow cytometry. APC, allophycocyanin. B, RAW cells were stimulated with LPS in the presence or absence of DFK1012 or chloroquine (50 and 100 μm) for 12 h. The expression of intracellular TNF-α or IL-1β was analyzed by Western blot analysis. n.s., nonspecific; L.E., long exposure; S.E., short exposure. C, RAW264.7 cells were stimulated with LPS (10 ng/ml) in the presence or absence of DFK1012 for 18 h. Total cellular extracts were subjected to Western blot analysis for iNOS, Nod2, Rip2, and actin. Data are representative of three independent experiments with similar results.
DISCUSSION
In this study, we demonstrate that DFK1012, a novel aminosaccharide compound, can induce a strong anti-inflammatory effect by blocking TLR and NLR responses. DFK1012, but not its derivatives, DFK845 and DFK846, showed an inhibitory effect on LPS and CpG-B-induced production of pro-inflammatory cytokines via TLR4 and TLR9, respectively (Fig. 2, A and B). DFK1012 also suppressed the release of cytokines upon Nod2 activation after stimulation with MDP (Fig. 2C). Therefore, DFK1012 targets a common mediator in MyD88-dependent TLR and Nod2 signaling. DFK1012 treatment did not suppress LPS- or MDP-induced degradation of IκBα or activation of p38 and SAPK/JNK, although ERK activation downstream of TLR4 was slightly affected by the compound in RAW264.7 cells (Fig. 4, A and B). The strong anti-inflammatory effect of DFK1012 cannot be fully explained by reduced ERK activation in RAW264.7 cells because ERK activation was not affected in DFK1012-treated BMDM (Fig. 4C), where cytokine production was also inhibited. Furthermore, we demonstrated that DFK1012 also did not block p65 nuclear translocation induced by LPS stimulation (Fig. 5). Interestingly, induction of pro-inflammatory cytokines at transcriptional as well as translational levels was largely unimpaired by DFK1012. (Figs. 6, A and B, and 7A). Indeed, DFK1012 did not significantly change intracellular TNF-α levels in the presence of a secretion inhibitor, monensin, up to 8 h after co-stimulation (Fig. 7A). However, both precursor and mature forms of TNF-α completely disappeared after 12 h of stimulation. Moreover, the inhibition of lysosome acidification totally restored DFK1012-induced loss of intracellular TNF-α level, suggesting that the observed loss of TNF-α is due to post-translational degradation through the endo/lysosome (Fig. 7B).
DFK1012 has chemical similarities to MDP, a ligand that activates Nod2. Two derivatives of DFK1012, DFK845 and DFK846, mimicked the enhancing effect of MDP on TLR-induced cytokine production (Fig. 2, D and E, and data not shown). In contrast, DFK1012 reduced both TLR-dependent and Nod2-dependent cytokine production (Fig. 2, A–C). Interestingly, this inhibitory effect of DFK1012 is not due to competition with MDP as an antagonist. DFK1012 did not block NF-κB and MAP kinase signaling downstream of MDP stimulation, suggesting that the inhibitory effect of DFK1012 is not due to impaired Nod2 activation. Furthermore, DFK1012 blocks cytokine production by TLR stimulation, not only by Nod2 stimulation (Fig. 2, A and B). Therefore, the strong anti-inflammatory activity of DFK1012 is not caused by an antagonistic inhibitory function. Although analogues of MDP have been reported to limit inflammation in murine models of septic shock (52), this is the first report to demonstrate that DFK1012, a novel aminosaccharide compound similar to MDP, suppresses TLR- and NLR-induced cytokine production from innate immune cells.
Surprisingly, DFK1012 does not induce any cytotoxicity at any working concentration we tested. Despite a strong inhibition of cytokine production, DFK1012 treatment does not impair cell viability. We have used four different approaches to assess the effect of DFK1012 on cell viability, including nuclei staining with Hoechst 33342, Western blot analysis for the cleavage of PARP-1 and caspase-3, and MTT and LDH assays. All of our experiments have indicated that DFK1012 does not induce any sign of cytotoxicity (Fig. 3, A–D), which suggests that DFK1012 does not affect the expression of proteins required for cell survival. Furthermore, DFK1012 targets the post-translational regulation of pro-inflammatory cytokines, and not other immune response genes, including LPS-induced iNOS, Nod2, and Rip2 (Fig. 7C). This specificity of DFK1012 for the suppression of inflammatory cytokines would be a tremendous practical advantage for its use as a therapeutic agent.
Excessive immune responses are detrimental to the host, and thus proper regulation of immune responses is critical for maintaining immune homeostasis and preventing inflammatory disorders and septic shock (35). Following recent rapid advances in the field of innate immunity, innate immune receptors have become attractive targets in the development of anti-inflammatory drugs (53). For example, inhibitors for TLR7 and TLR9 have been studied for the treatment of autoimmune diseases (54), and the TLR4 antagonist, eritoran tetrasodium (E5564), is under clinical trial for the treatment of septic shock patients (55). Potential immunomodulators also include inhibitors of TLR signaling molecules, such as p38α MAPK (56, 57), MK2 (58), or IRAK4 (59–61). As a strong inhibitor of cytokine production, usage of DFK1012 or related molecules may have some advantages over treatment with Infliximab, an anti-TNF-α antibody, or other related anti-inflammatory drugs targeting inflammatory cytokines. First, because DFK1012 is a small compound with a relatively simple structure, modifications to improve its efficacy and safety are relatively easy. Second, DFK1012 inhibits the production of multiple pro-inflammatory cytokines from various TLR and NLR stimuli, rather than a particular cytokine or signaling pathway, which may prove more useful in disease settings characterized by the excessive production of many pro-inflammatory cytokines. Third, unlike the antibody-based drug, Infliximab, which requires intravenous administration, various routes could be used for future derivatives of DFK1012. DFK1012 thus has a great deal of therapeutic potential.
In summary, this study demonstrates that DFK1012 strongly inhibits pro-inflammatory cytokine production in response to TLR and NLR stimulation. In light of its low cytotoxicity, DFK1012 is an attractive candidate for future drug development targeting inflammatory diseases.
Acknowledgments
We thank Tanja Petnicki-Ocwieja and Torsten Meissner for helpful discussions; Amelia Chen and Andrea Dearth for general assistance; and Amy Li for critical reading.
This work was supported by the Eisai Research Institute under the Sponsored Research Agreement (Dana-Farber Cancer Institute Agreement Number Reference 2696/Eisai Research Institute CRF 734).
- TLR
- Toll-like receptor
- NLR
- nucleotide-binding domain, leucine-rich repeat protein
- MDP
- muramyl dipeptide
- LDH
- lactose dehydrogenase
- PARP
- poly(ADP-ribose) polymerase-2
- iNOS
- inducible NOS
- MTT
- 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- DMSO
- dimethyl sulfoxide
- BMDM
- bone marrow-derived macrophages
- p
- phosphorylated.
REFERENCES
- 1. Medzhitov R., Janeway C. A., Jr. (1997) Cell 91, 295–298 [DOI] [PubMed] [Google Scholar]
- 2. Kawai T., Akira S. (2009) Int. Immunol. 21, 317–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alexopoulou L., Thomas V., Schnare M., Lobet Y., Anguita J., Schoen R. T., Medzhitov R., Fikrig E., Flavell R. A. (2002) Nat. Med. 8, 878–884 [DOI] [PubMed] [Google Scholar]
- 4. Takeuchi O., Sato S., Horiuchi T., Hoshino K., Takeda K., Dong Z., Modlin R. L., Akira S. (2002) J. Immunol. 169, 10–14 [DOI] [PubMed] [Google Scholar]
- 5. Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 6. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 7. Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 8. Qureshi S. T., Larivière L., Leveque G., Clermont S., Moore K. J., Gros P., Malo D. (1999) J. Exp. Med. 189, 615–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. (1999) J. Immunol. 162, 3749–3752 [PubMed] [Google Scholar]
- 10. Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A. (2001) Nature 410, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 11. Takeuchi O., Kawai T., Mühlradt P. F., Morr M., Radolf J. D., Zychlinsky A., Takeda K., Akira S. (2001) Int. Immunol. 13, 933–940 [DOI] [PubMed] [Google Scholar]
- 12. Diebold S. S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. (2004) Science 303, 1529–1531 [DOI] [PubMed] [Google Scholar]
- 13. Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. (2004) Science 303, 1526–1529 [DOI] [PubMed] [Google Scholar]
- 14. Lund J. M., Alexopoulou L., Sato A., Karow M., Adams N. C., Gale N. W., Iwasaki A., Flavell R. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5598–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hemmi H., Kaisho T., Takeuchi O., Sato S., Sanjo H., Hoshino K., Horiuchi T., Tomizawa H., Takeda K., Akira S. (2002) Nat. Immunol. 3, 196–200 [DOI] [PubMed] [Google Scholar]
- 16. Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. (2000) Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 17. Zhang D., Zhang G., Hayden M. S., Greenblatt M. B., Bussey C., Flavell R. A., Ghosh S. (2004) Science 303, 1522–1526 [DOI] [PubMed] [Google Scholar]
- 18. Yarovinsky F., Zhang D., Andersen J. F., Bannenberg G. L., Serhan C. N., Hayden M. S., Hieny S., Sutterwala F. S., Flavell R. A., Ghosh S., Sher A. (2005) Science 308, 1626–1629 [DOI] [PubMed] [Google Scholar]
- 19. Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C. A., Jr. (1998) Mol. Cell 2, 253–258 [DOI] [PubMed] [Google Scholar]
- 20. Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. (1999) Immunity 11, 115–122 [DOI] [PubMed] [Google Scholar]
- 21. Akira S., Takeda K., Kaisho T. (2001) Nat. Immunol. 2, 675–680 [DOI] [PubMed] [Google Scholar]
- 22. Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 23. Wilmanski J. M., Petnicki-Ocwieja T., Kobayashi K. S. (2008) J. Leukoc. Biol. 83, 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ting J. P., Willingham S. B., Bergstralh D. T. (2008) Nat. Rev. Immunol. 8, 372–379 [DOI] [PubMed] [Google Scholar]
- 25. Girardin S. E., Boneca I. G., Carneiro L. A., Antignac A., Jéhanno M., Viala J., Tedin K., Taha M. K., Labigne A., Zähringer U., Coyle A. J., DiStefano P. S., Bertin J., Sansonetti P. J., Philpott D. J. (2003) Science 300, 1584–1587 [DOI] [PubMed] [Google Scholar]
- 26. Chamaillard M., Hashimoto M., Horie Y., Masumoto J., Qiu S., Saab L., Ogura Y., Kawasaki A., Fukase K., Kusumoto S., Valvano M. A., Foster S. J., Mak T. W., Nuñez G., Inohara N. (2003) Nat. Immunol. 4, 702–707 [DOI] [PubMed] [Google Scholar]
- 27. Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 28. Kobayashi K., Inohara N., Hernandez L. D., Galán J. E., Núñez G., Janeway C. A., Medzhitov R., Flavell R. A. (2002) Nature 416, 194–199 [DOI] [PubMed] [Google Scholar]
- 29. Li J., Moran T., Swanson E., Julian C., Harris J., Bonen D. K., Hedl M., Nicolae D. L., Abraham C., Cho J. H. (2004) Hum. Mol. Genet. 13, 1715–1725 [DOI] [PubMed] [Google Scholar]
- 30. Zhang T., Kruys V., Huez G., Gueydan C. (2002) Biochem. Soc. Trans. 30, 952–958 [DOI] [PubMed] [Google Scholar]
- 31. Jue D. M., Sherry B., Luedke C., Manogue K. R., Cerami A. (1990) Biochemistry 29, 8371–8377 [DOI] [PubMed] [Google Scholar]
- 32. Murray R. Z., Kay J. G., Sangermani D. G., Stow J. L. (2005) Science 310, 1492–1495 [DOI] [PubMed] [Google Scholar]
- 33. Black R. A., Rauch C. T., Kozlosky C. J., Peschon J. J., Slack J. L., Wolfson M. F., Castner B. J., Stocking K. L., Reddy P., Srinivasan S., Nelson N., Boiani N., Schooley K. A., Gerhart M., Davis R., Fitzner J. N., Johnson R. S., Paxton R. J., March C. J., Cerretti D. P. (1997) Nature 385, 729–733 [DOI] [PubMed] [Google Scholar]
- 34. Solomon K. A., Covington M. B., DeCicco C. P., Newton R. C. (1997) J. Immunol. 159, 4524–4531 [PubMed] [Google Scholar]
- 35. Fukata M., Vamadevan A. S., Abreu M. T. (2009) Semin. Immunol. 21, 242–253 [DOI] [PubMed] [Google Scholar]
- 36. Leadbetter E. A., Rifkin I. R., Hohlbaum A. M., Beaudette B. C., Shlomchik M. J., Marshak-Rothstein A. (2002) Nature 416, 603–607 [DOI] [PubMed] [Google Scholar]
- 37. Ehlers M., Fukuyama H., McGaha T. L., Aderem A., Ravetch J. V. (2006) J. Exp. Med. 203, 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Christensen S. R., Shupe J., Nickerson K., Kashgarian M., Flavell R. A., Shlomchik M. J. (2006) Immunity 25, 417–428 [DOI] [PubMed] [Google Scholar]
- 39. Pisitkun P., Deane J. A., Difilippantonio M. J., Tarasenko T., Satterthwaite A. B., Bolland S. (2006) Science 312, 1669–1672 [DOI] [PubMed] [Google Scholar]
- 40. Marta M., Andersson A., Isaksson M., Kämpe O., Lobell A. (2008) Eur. J. Immunol. 38, 565–575 [DOI] [PubMed] [Google Scholar]
- 41. Kobayashi M., Kweon M. N., Kuwata H., Schreiber R. D., Kiyono H., Takeda K., Akira S. (2003) J. Clin. Invest. 111, 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rakoff-Nahoum S., Hao L., Medzhitov R. (2006) Immunity 25, 319–329 [DOI] [PubMed] [Google Scholar]
- 43. Berdeli A., Celik H. A., Ozyürek R., Dogrusoz B., Aydin H. H. (2005) J. Mol. Med 83, 535–541 [DOI] [PubMed] [Google Scholar]
- 44. Kang T. J., Chae G. T. (2001) FEMS Immunol. Med. Microbiol. 31, 53–58 [DOI] [PubMed] [Google Scholar]
- 45. Franchimont D., Vermeire S., El Housni H., Pierik M., Van Steen K., Gustot T., Quertinmont E., Abramowicz M., Van Gossum A., Devière J., Rutgeerts P. (2004) Gut. 53, 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miceli-Richard C., Lesage S., Rybojad M., Prieur A. M., Manouvrier-Hanu S., Häfner R., Chamaillard M., Zouali H., Thomas G., Hugot J. P. (2001) Nat. Genet. 29, 19–20 [DOI] [PubMed] [Google Scholar]
- 47. Rosé C. D., Doyle T. M., McIlvain-Simpson G., Coffman J. E., Rosenbaum J. T., Davey M. P., Martin T. M. (2005) J. Rheumatol. 32, 373–375 [PubMed] [Google Scholar]
- 48. Kanazawa N., Okafuji I., Kambe N., Nishikomori R., Nakata-Hizume M., Nagai S., Fuji A., Yuasa T., Manki A., Sakurai Y., Nakajima M., Kobayashi H., Fujiwara I., Tsutsumi H., Utani A., Nishigori C., Heike T., Nakahata T., Miyachi Y. (2005) Blood 105, 1195–1197 [DOI] [PubMed] [Google Scholar]
- 49. Soldani C., Scovassi A. I. (2002) Apoptosis 7, 321–328 [DOI] [PubMed] [Google Scholar]
- 50. Ghosh S., Hayden M. S. (2008) Nat. Rev. Immunol. 8, 837–848 [DOI] [PubMed] [Google Scholar]
- 51. Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 52. Wardowska A., Dzierzbicka K., Szaryńska M., Dabrowska-Szponar M., Wiśniewska K., Myśliwski A., Trzonkowski P. (2009) Vaccine 27, 369–374 [DOI] [PubMed] [Google Scholar]
- 53. Schmidt C. (2006) Nat. Biotechnol. 24, 230–231 [DOI] [PubMed] [Google Scholar]
- 54. Barrat F. J., Coffman R. L. (2008) Immunol. Rev. 223, 271–283 [DOI] [PubMed] [Google Scholar]
- 55. Tidswell M., Tillis W., Larosa S. P., Lynn M., Wittek A. E., Kao R., Wheeler J., Gogate J., Opal S. M. (2010) Crit. Care Med. 38, 72–83 [DOI] [PubMed] [Google Scholar]
- 56. Hill R. J., Dabbagh K., Phippard D., Li C., Suttmann R. T., Welch M., Papp E., Song K. W., Chang K. C., Leaffer D., Kim Y. N., Roberts R. T., Zabka T. S., Aud D., Dal Porto J., Manning A. M., Peng S. L., Goldstein D. M., Wong B. R. (2008) J. Pharmacol. Exp. Ther. 327, 610–619 [DOI] [PubMed] [Google Scholar]
- 57. Munoz L., Ralay Ranaivo H., Roy S. M., Hu W., Craft J. M., McNamara L. K., Chico L. W., Van Eldik L. J., Watterson D. M. (2007) J. Neuroinflammation 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Anderson D. R., Meyers M. J., Vernier W. F., Mahoney M. W., Kurumbail R. G., Caspers N., Poda G. I., Schindler J. F., Reitz D. B., Mourey R. J. (2007) J. Med. Chem. 50, 2647–2654 [DOI] [PubMed] [Google Scholar]
- 59. Powers J. P., Li S., Jaen J. C., Liu J., Walker N. P., Wang Z., Wesche H. (2006) Bioorg. Med. Chem. Lett. 16, 2842–2845 [DOI] [PubMed] [Google Scholar]
- 60. Wang Z., Liu J., Sudom A., Ayres M., Li S., Wesche H., Powers J. P., Walker N. P. (2006) Structure 14, 1835–1844 [DOI] [PubMed] [Google Scholar]
- 61. Buckley G. M., Fosbeary R., Fraser J. L., Gowers L., Higueruelo A. P., James L. A., Jenkins K., Mack S. R., Morgan T., Parry D. M., Pitt W. R., Rausch O., Richard M. D., Sabin V. (2008) Bioorg. Med. Chem. Lett. 18, 3656–3660 [DOI] [PubMed] [Google Scholar]