Abstract
The matriptase-prostasin proteolytic cascade is essential for epidermal tight junction formation and terminal epidermal differentiation. This proteolytic pathway may also be operative in a variety of other epithelia, as both matriptase and prostasin are involved in tight junction formation in epithelial monolayers. However, in polarized epithelial cells matriptase is mainly located on the basolateral plasma membrane whereas prostasin is mainly located on the apical plasma membrane. To determine how matriptase and prostasin interact, we mapped the subcellular itinerary of matriptase and prostasin in polarized colonic epithelial cells. We show that zymogen matriptase is activated on the basolateral plasma membrane where it is able to cleave relevant substrates. After activation, matriptase forms a complex with the cognate matriptase inhibitor, hepatocyte growth factor activator inhibitor (HAI)-1 and is efficiently endocytosed. The majority of prostasin is located on the apical plasma membrane albeit a minor fraction of prostasin is present on the basolateral plasma membrane. Basolateral prostasin is endocytosed and transcytosed to the apical plasma membrane where a long retention time causes an accumulation of prostasin. Furthermore, we show that prostasin on the basolateral membrane is activated before it is transcytosed. This study shows that matriptase and prostasin co-localize for a brief period of time at the basolateral plasma membrane after which prostasin is transported to the apical membrane as an active protease. This study suggests a possible explanation for how matriptase or other basolateral serine proteases activate prostasin on its way to its apical destination.
Keywords: Endocytosis, Intracellular Trafficking, Plasma Membrane, Protein Sorting, Serine Protease, HAI-1, Activation, Matriptase, Prostasin, Transcytosis
Introduction
The trypsin-like membrane serine protease matriptase is essential for maintenance of multiple types of epithelia. Conditional ablation of the St14 gene coding for matriptase in intestine, kidney, and lung of adult mice results in weight loss, severe decline in health and death within 2 weeks, caused by organ dysfunction associated with increased permeability and loss of tight junctions (1). Knock down of matriptase by siRNA in a cell model of the intestinal epithelium caused a leaky barrier, impaired ability to develop transepithelial electrical resistance (TEER)2 and enhanced paracellular permeability through regulation of tight junction proteins (2). Together these data suggest a key role for matriptase in epithelial barrier function and tight junction assembly.
Prostasin (also known as CAP1 and PRSS8) is a GPI-anchored trypsin-like serine protease. Prostasin is co-expressed with matriptase in most epithelial tissues including the epidermis, kidney, and colon (3). Prostasin proteolytic activity has also been suggested to promote the development of functional tight junctions, TEER and paracellular permeability (4–6). Unlike matriptase, which undergoes efficient auto-activation, the prostasin zymogen is not able to auto-activate, and formation of active prostasin requires activation site cleavage by other trypsin-like serine proteases. Strong data suggest that matriptase acts upstream of prostasin in a zymogen cascade in the epidermis. The severe epidermal defects of matriptase deficiency appear to be a consequence of lack of active prostasin. Only the inactive form of prostasin is found in matriptase-deficient mice and matriptase-deficient and prostasin-deficient mice have nearly identical phenotypes with compromised epidermal tight junction formation and no terminal epidermal differentiation (6–11). Furthermore, it has been shown in vitro that the serine protease domain of matriptase is directly able to cleave the zymogen-form of prostasin, to generate proteolytically active prostasin (11). Matriptase-dependent activation of prostasin was recently demonstrated in a human organotypic skin model and in matriptase-deficient human epidermis (6, 12).
The plasma membrane of a polarized epithelial cell is divided into an apical and a basolateral plasma membrane domain separated by tight junctions. The tight junctions prevent diffusion of membrane proteins between the two membrane domains. In polarized epithelial cells there are two pathways for newly synthesized proteins to reach the apical plasma membrane: The direct pathway from the trans Golgi network directly to the apical plasma membrane and the indirect pathway, where newly synthesized proteins via the basolateral plasma membrane are endocytosed and transcytosed to the apical plasma membrane (13). It has been reported that matriptase is mainly located at the basolateral plasma membrane in rat enterocytes and other polarized epithelial cells (14, 15). However, proteolytically shed matriptase in complex with HAI-1 was purified from human milk suggesting an apical secretion (16). Conversely, the matriptase substrate, prostasin, is mainly located at the apical plasma membrane of polarized epithelial cells (17, 18).
The present study aims to determine where in the polarized epithelial cell active matriptase interacts with its substrate prostasin, in order to explain how a basolateral protease can cleave and activate an apicallylocated substrate. Matriptase is synthesized as an inactive, single-chain zymogen. Its activation requires two sequential endoproteolytic cleavages. The first proteolytic processing cleavage occurs after Gly-149, yet the processed form remains tightly associated with the membrane.
Matriptase is, subsequently (and dependent on the first cleavage) cleaved after Arg-614 in the serine protease domain to gain proteolytic activity. Shortly after activation, matriptase forms a complex with HAI-1, whereby matriptase is enzymatically inhibited. Hence, substrates should be present at the same location as matriptase activation takes place.
We present biochemical data showing that a matriptase-prostasin zymogen cascade is indeed possible in polarized epithelial cells. We show that matriptase is cleaved to its active form on the basolateral plasma membrane, subsequently inhibited by HAI-1 followed by endocytosis. Importantly, we find prostasin present on the basolateral plasma membrane during matriptase activation. Furthermore, the basolateral prostasin is active and transcytosed to the apical plasma membrane where it accumulates. These results demonstrates that matriptase and prostasin may functionally interact in polarized epithelial cells, despite their, respective, basolateral and apical locations at steady state.
EXPERIMENTAL PROCEDURES
Cell Culture
Caco-2 cells were grown in minimal essential medium supplemented with 2 mm l-glutamine, 10% fetal bovine serum, 1× nonessential amino acids, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37 °C in an atmosphere of 5% CO2. For all experiments, 2 × 106 cells were seeded into 0.4 μm-pore-size 24 mm Transwell® filter chamber (Corning) allowing separate access to the apical and basolateral plasma membrane. Cells were grown until day 11 postconfluence before they were used for experiments. The tightness of filter-grown cells was assayed by filling the inner chamber to the brim and allowing it to equilibrate overnight. The cell culture medium was changed every day.
Biotinylation, Internalization, and Biotin-removal
Caco-2 cells grown on transwell filters were washed three times with ice-cold PBS+ (PBS supplemented with 0.7 mm CaCl2 and 0.25 mm MgCl2) on both apical and basolateral side. The cells were biotin-labeled for 30 min at 4 °C, either from the apical or the basolateral side, with 1 mg/ml EZ-linkTM Sulfo-NHS-SS-Biotin (Pierce) dissolved in PBS+. After biotin labeling the cells were washed twice with ice-cold PBS+. Residual biotin was quenched for 5 min at 4 °C with 50 mm glycine in PBS+ and the cells were washed again with PBS+. For internalization experiments, preheated media (serum-free MEM medium containing 20 mm NaHCO3, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen)) was added and the cells were incubated at 37 °C to regain normal trafficking. For endocytosis and transcytosis experiments, surface biotin was removed after incubation as indicated using the non-membrane permeable reducing agent glutathione (Sigma Aldrich). Surface exposed biotin was removed from either apical or basolateral side with 16 mg/ml glutathione in 75 mm NaCl, 75 mm NaOH, 1 mm EDTA, and 0.5% BSA in H2O under gentle agitation for 2 × 20 min. Cells were washed with PBS+ and residual glutathione was inactivated with 5 mg/ml iodoacetamide (Sigma Aldrich) in PBS+ for 5 min. A set of parallel samples was left without the glutathione reduction to monitor the total amount of biotinylated protein present through the course of the experiment. Cells were washed twice in PBS+, and lysed in PBS containing 1% Triton X-100, 0.5% deoxycholate and protease inhibitors (10 mg/liter benzamidine, 2 mg/liter pepstatin A, 2 mg/liter leupeptin, 2 mg/l antipain, and 2 mg/liter chymostatin). For inhibitor-Sepharose pull-downs, protease inhibitors were omitted from the lysis buffer.
Monomeric Avidin/Streptavidin Precipitation of Biotinylated Proteins
Lysates from biotin-labeled cells were centrifuged at 20,000 × g for 20 min to pellet the insoluble material. The supernatant was transferred to clean eppendorf tubes with either Pierce® monomeric avidin agarose (Pierce) (120 μl/24 mm filter) or Pierce® streptavidin agarose (50 μl/24 mm filter), prepared as described by manufacturer. After overnight incubation at 4 °C with end-over-end rotation, the agarose was washed three times with 50 mm Tris-HCl, pH 6.25. Biotinylated proteins were eluted from the monomeric avidin agarose with 4 mm biotin (Pierce) in PBS for 30 min followed by addition of SDS sample buffer. For elution from streptavidin-agarose, the samples were boiled in SDS sample buffer.
Western Blot
The 2× SDS sample buffer (125 mm Tris-HCl, 25 mm EDTA, pH 6.8, 4% SDS, 5% glycerol, 0.01% bromphenol blue) did not contain any reducing agent and the samples were not boiled prior to SDS-PAGE to prevent protein complexes from dissociating, unless otherwise specified (0.1 m dithiothreitol, DTT). All lysates were incubated with the SDS sample buffer for 10 min at room temperature before gel loading. The proteins were separated on 7% acrylamide gels made in the laboratory and transferred to Immobilon-P PVDF membranes (Millipore). The membranes were blocked with 10% nonfat dry milk in PBS containing 0.1% Tween-20 (PBST) for 1 h at room temperature. The individual PVDF membranes were probed with primary antibodies diluted in 1% nonfat dry milk in PBST at 4 °C overnight. The next day the membranes were washed 3× with PBST and the binding of primary antibodies was followed by recognition with secondary horseradish peroxidase (HRP)-conjugated secondary antibodies (Pierce). After 3× wash with PBST, the signal was developed using the ECL reagent Super Signal West Femto Maximum Sensitivity Substrate (Pierce), according to the protocol supplied by the manufacturer and visualized with a Fuji LAS1000-camera (Fujifilm, Sweden AB). For graphs, the free online software ImageJ (created by Wayne Rasband, NIH, Bethesda) was used to quantify the bands on the blot. Values in the graph represent the sum of all the bands visualized on the gel. Furthermore, the graph represents the mean of three independent experiments and is presented with the standard deviation.
Antibodies
The antibodies used were monoclonal mouse anti-human antibodies M32 and M69 (19). The antibody M32 detects all forms of matriptase, including zymogen, activated form, and complexes. Under the conditions used in this study, the antibody M69 only detected the matriptase-HAI-1 complex and M69 reactive material is hereafter referred to as the matriptase-HAI-1 complex. The other antibodies used were polyclonal rabbit anti-human matriptase raised against the serine protease domain of matriptase (Cat. no. IM1014, Calbiochem), mouse anti-human HAI-1 antibody M19 (19), and mouse anti-human prostasin antibody (Cat. no. 612173, BD Transduction Laboratories). M32, M69, and polyclonal goat anti-human HAI-1 (cat. no. AF1048, R&D) antibodies were used for immunocytochemistry.
Protease Pull-down with Protease Inhibitor-coupled Sepharose 4B
The protease inhibitors aprotinin (5 mg/ml Sepharose), leupeptin (5 mg/ml Sepharose), and soybean trypsin inhibitor (5 mg/ml Sepharose) were immobilized to CNBr-activated SepharoseTM 4B (GE Healthcare), as specified by the manufacturer's instructions. Caco-2 cells were biotin-labeled and biotinylated proteins were pulled down with monomeric avidin agarose. The biotinylated proteins were gently eluted with 4 mm biotin in 50 mm Tris-HCl, pH 8.5. The biotin-eluate was separated from the avidin-agarose and incubated with 60 μl protease inhibitor-coupled Sepharose in 50 mm Tris-HCl, pH 8.75 at 37 °C for 30 min. The inhibitor Sepharose was washed 3× with 50 mm Tris-HCl, pH 6.5. Proteases were eluted from the inhibitor-coupled Sepharose using 0.1 m glycine, pH 2.4. Samples were neutralized with 1 m Tris immediately after elution. The eluates were added to SDS sample buffer and analyzed by Western blotting.
Gelatin Zymography
Biotinylated monomeric avidin agarose-purified proteins were separated on a 7% SDS-polyacrylamide gel containing 0.1% gelatin. The proteins were re-natured by washing the gelatin gel 2× 30 min in 2.5% Triton X-100 in H2O. To wash out excess Triton X-100, the gel was washed 2 × 10 min in H2O and thereafter incubated in 50 mm Tris-HCl, pH 8.75 overnight at 37 °C. Gelatinolytic bands on the gel were visualized by Coomassie Brilliant Blue staining.
Immunofluorescence
Caco-2 cells grown on filters were fixed for 20 min in 4% paraformaldehyde in PBS (Bie & Berntsen) at room temperature. The following was performed at 4 °C. Cells were permeabilized with 0.05% Triton X-100 in PBS for 20 min. Unspecific staining was blocked with PBS containing 3% BSA (PBS/BSA) for 30 min. Cells were incubated with primary antibody diluted in PBS/BSA for 1.5 h, washed 3× in PBS followed by incubation with relevant Alexa Fluor-conjugated secondary antibodies (Invitrogen) for 1 h. Where indicated the nuclei of the cells were visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. The cells were finally mounted with Prolong Gold mounting medium (Invitrogen) and subjected to laser scanning confocal microscopy using the Leica TCS SP2 system and Zeiss LS700 system.
RESULTS
Steady-state Distribution of Matriptase, HAI-1, and Prostasin
Where in the cell matriptase cleaves its substrates is not well understood, however it is fundamental to understanding how matriptase maintains epithelial integrity. Trafficking and activation of endogenous matriptase, prostasin, and HAI-1 was therefore studied in Caco-2 cells-a human colon epithelial cell line. Upon reaching confluence, Caco-2 cells spontaneously differentiate into a tight monolayer of polarized cells with an apical and a basolateral plasma membrane, separated by tight junctions. Matriptase expression increases during Caco-2 differentiation (2) consistent with the higher levels of matriptase at the intestinal villous tip (20). The amount of matriptase-HAI-1 complex also increases during differentiation and reaches a plateau around day 7–10 in differentiating Caco-2 cells (data not shown). For that reason, Caco-2 cells at day 11 post-confluence were used for the following experiments.
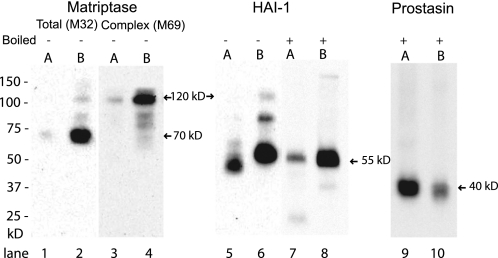
We first investigated the steady state distribution of matriptase, HAI-1, and prostasin between the apical and the basolateral plasma membrane domains. Surface biotinylation experiments showed that the majority of matriptase was present among basolateral membrane proteins and most prevalent in a 70 kDa form corresponding to the extracellular domain of matriptase cleaved at Gly-149 (Fig. 1, lanes 1 and 2), representing either zymogen matriptase or enzymatically active matriptase. A small fraction of the basolateral matriptase was detected in a 120 kDa form (Fig. 1, lane 2). The 120 kDa form was identified as matriptase-HAI-1 complex by detection with both the M69 and anti-HAI-1 antibodies (Fig. 1, lanes 4 and 6). Two matriptase forms below the 120 kDa complex were also detected on the basolateral plasma membrane (Fig. 1, lanes 2 and 4). This could possibly be degradation products of the matriptase-HAI-1 complex or activated matriptase in complex with other inhibitors (21).
FIGURE 1.
Matriptase is located on the basolateral membrane whereas prostasin is apically located. Caco-2 cells grown on Transwell filters were biotin-labeled from either the apical (A) (lanes 1, 3, 5, 7, and 9) or basolateral (B) side (lanes 2, 4, 6, 8, and 10). Biotinylated proteins were precipitated with monomeric avidin, separated by SDS-PAGE and analyzed by Western blot with antibodies against total matriptase (M32) (lanes 1 and 2), matriptase-HAI-1 complex (M69) (lanes 3 and 4), the inhibitor HAI-1 (lanes 5–8) and prostasin (lanes 9 and 10) −/+ boiling of samples. Samples analyzed with the anti-prostasin antibody were boiled and reduced with DTT. The positions of molecular weight markers are indicated on the left. Protein and complex are marked with arrows and size on the right. The majority of the plasma membrane-bound matriptase was located on the basolateral membrane in a 70 kDa form, as detected with the antibody M32 (lane 2). A small fraction of the basolateral plasma membrane-bound matriptase was found in a 120 kDa form (lane 2). This form was also detected by the M69 (lane 4) and anti-HAI-1 (lane 6) antibodies. HAI-1 was found both on apical and basolateral plasma membrane (lanes 5–8). Apical HAI-1 (lane 5) migrated faster on the SDS-PAGE than the basolateral HAI-1 (lane 6) when samples were not boiled but displayed the same size after boiling of the samples (lanes 7 and 8). Plasma membrane-bound prostasin was located mainly on the apical side (lane 9), although minor amounts were detected on the basolateral membrane (lane 10). Results shown are representative of five independent experiments.
Full-length HAI-1 has an estimated molecular weight of 55 kDa and was detected on both the apical and the basolateral plasma membrane (Fig. 1, lanes 5 and 6). Apically located HAI-1 displayed a higher mobility than basolaterally located HAI-1 in the presence of SDS (Fig. 1, lanes 5 and 6) but the same mobility when the samples were boiled (Fig. 1, lanes 7 and 8). This could indicate a conformational difference making apical HAI-1 more resistant to denaturation with SDS. Further studies are needed to elucidate the nature of the two forms. A HAI-1 complex around 85 kDa was also detected on the basolateral plasma membrane (Fig. 1, lane 6). An 85 kDa HAI-1 complex with matriptase as well as prostasin has previously been reported (22, 23).
Prostasin was located mainly on the apical plasma membrane although minor amounts could be detected on the basolateral plasma membrane (Fig. 1, lanes 9 and 10). These data are consistent with the existing literature showing the basolateral localization of matriptase and apical localization of prostasin in a polarized cell (15, 18).
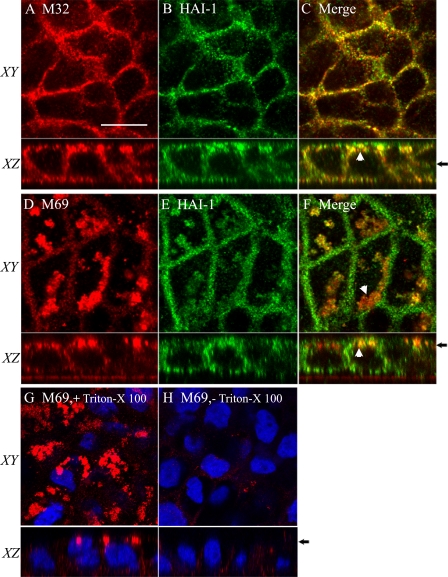
Next, we investigated the subcellular steady state distribution of matriptase and HAI-1 in Caco-2 cells by immunocytochemistry (Fig. 2). Caco-2 cells were grown on Transwell filters before fixation, permeabilization, and immunolabeling. Matriptase and HAI-1 were both detected on the basolateral plasma membrane (Fig. 2, A–F). Interestingly, matriptase and HAI-1 were also detected in structures near the apical plasma membrane (Fig. 2F). Only weak detection of matriptase-HAI-1 complex was observed on the basolateral plasma membrane (Fig. 2D). Surprisingly the majority of the matriptase-HAI-1 complex was detected in structures near the apical plasma membrane (Fig. 2, D–F). To further investigate the location of the matriptase-HAI-1 complex in the apical region of the cells, Caco-2 cells were immunolabeled with and without permeabilization of the plasma membrane (Fig. 2, G and H). Without permeabilization, almost no matriptase-HAI-1 complex could be detected (Fig. 2H), however, with permeabilization a distinct labeling of the vesicular structures was clearly visible below the apical plasma membrane (Fig. 2G), confirming an intracellular localization of the structures containing the matriptase-HAI-1 complex. These data are consistent with our biotinylation experiments, as we were only able to label matriptase on the basolateral plasma membrane (Fig. 1, lanes 1 and 3).
FIGURE 2.
Matriptase-HAI-1 complex accumulates in intracellular structures. Caco-2 cells grown on Transwell filters, were fixed, permeabilized, and immunolabeled with antibodies against total matriptase (M32) and HAI-1 (A–C) or matriptase-HAI-1 complex (M69) and HAI-1 (D–F). Using a confocal scanning microscope, images were taken in the XY plane showing a single section through the monolayer and the XZ plane showing a cross section of the monolayer. The position of the plane of the XY section is indicated on the XZ plane (black arrows) on the right. The scale bar represents 10 μm. A–C, Matriptase was detected on the basolateral plasma membrane of the Caco-2 cells co-localizing with HAI-1. Matriptase and HAI-1 was also co-localizing in structures near the apical membrane (white arrowheads). D–F, matriptase-HAI-1 complex was observed in structures near the apical plasma membrane (white arrowheads). Low amounts of matriptase-HAI-1 complex were detected on the basolateral plasma membrane. HAI-1 was detected both on apical and basolateral plasma membranes as well as in the apical structures co-localizing with matriptase. G and H, Caco-2 cells were treated with or without Triton X-100 prior to immunolabeling with the antibody M69. G, in the cells permeabilized with Triton X-100, a distinct detection of matriptase-HAI-1 complex was observed in apical vesicular structures. H, in cells without permeabilization, the vesicular structures with matriptase-HAI-1 complex were not detected. Only a weak basolateral signal was observed. The nuclei were visualized by DAPI staining shown in blue. Results shown are representative of three independent experiments.
The Matriptase-HAI-1 Complex Is Generated during Endocytosis
Our steady state experiments show that a large portion of matriptase is present in a 70 kDa form on the basolateral plasma membrane. Furthermore, the experiments show that large amounts of matriptase-HAI-1 complex are detected in intracellular structures. This suggests that matriptase is endocytosed from the plasma membrane and inhibited by HAI-1 during this process.
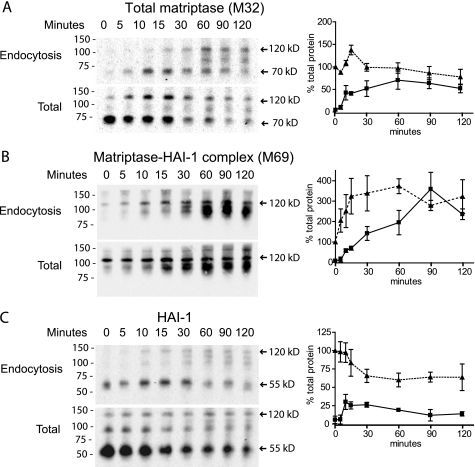
We examined the endocytosis of basolateral plasma membrane bound matriptase and HAI-1 using biotinylation and internalization techniques (see “Experimental Procedures”). The endocytosis experiments showed that matriptase is very efficiently, and almost completely, endocytosed within 60 min from the basolateral membrane (Fig. 3A). Furthermore, a 3-fold increase in the signal of matriptase-HAI-1 complexes was detected within the first 90 min of incubation, showing that the 70 kDa matriptase forms a complex with HAI-1 during the endocytosis (Fig. 3B). Several matriptase-HAI-1 complexes between 95 and 120 kDa were generated from the 70 kDa form during the endocytosis, as detected by both the matriptase and HAI-1 antibodies (Fig. 3, A–C). After 90 min, all of the surface-labeled matriptase was found in complex with HAI-1 and was located exclusively intracellularly (Fig. 3B).
FIGURE 3.
Matriptase in complex with HAI-1 is generated during the efficient endocytosis of matriptase from the basolateral plasma membrane. Caco-2 cells were grown on Transwell filters, and proteins on the basolateral plasma membrane were biotinylated at 4 °C using a cleavable biotinylation reagent, s-NHS-SS-biotin. The labeled cells were incubated at 37 °C for the time indicated (0–120 min). After incubation, the proteins remaining on the plasma membrane were biotin-stripped, using the membrane non-permeant reducing agent glutathione, leaving only endocytosed proteins biotinylated (endocytosis panels). Biotin reduction was omitted in a parallel set of samples to monitor the degradation of the biotinylated proteins over time (total panels). Biotinylated proteins were precipitated with monomeric avidin agarose, and the avidin pull-downs were analyzed with SDS-PAGE and Western blotting using the antibodies (A) M32 against total matriptase, (B) M69 against matriptase-HAI-1 complex, and (C) M19 against HAI-1. The positions of molecular markers (kDa) are indicated on the left. Proteins and complexes are indicated with arrows and size. Quantification of bands from Western blots was done using the software ImageJ. A graphic presentation of three independent endocytosis experiments is shown on the right hand side. The dotted line equals total protein and solid line equals endocytosed protein. The standard deviation is shown with error bars. A, matriptase was endocytosed from the basolateral plasma membrane within 60 min. Approximately 80% of matriptase was endocytosed at 60 min as indicated on the graph. B, during the 120-min incubation there was a 3-fold increase in the matriptase-HAI-1 complexs. C, free 55 kDa HAI-1 was partially endocytosed within 15 min. (C, endocytosis panel). Results shown are representative of three independent experiments.
The 55 kDa HAI-1 had an endocytosis pattern different from HAI-1 in complex with matriptase. The 55 kDa HAI-1 was only partially endocytosed, with the largest intracellular pool seen after 15 min. This suggests that HAI-1, when not in complex with matriptase, is recycling to the plasma membrane from early endosomes (Fig. 3C). This type of recycling of HAI-1 has been shown previously in MDCK cells (24).
A HAI-1 complex around 85 kDa was observed in the Total panel from time point 0 to 15 min, which was not detected by the matriptase antibodies (Fig. 3, compare C to A and B, respectively). This could possibly be a HAI-1-prostasin complex. A similar complex has previously been reported in keratinocytes (22). Together these data show that matriptase is efficiently endocytosed from the basolateral side and forms a complex with HAI-1 during this process.
Matriptase Zymogen Is Cleaved to the Active Protease on the Surface of the Basolateral Plasma Membrane
We have up to now showed that matriptase is present predominantly in a 70 kDa form on the basolateral plasma membrane and is efficiently endocytosed forming a complex with HAI-1 that accumulates in intracellular structures, making it a prerequisite that matriptase activation occurs prior to endocytosis. Whether matriptase activation occurs before arrival or at the plasma membrane is not well defined.
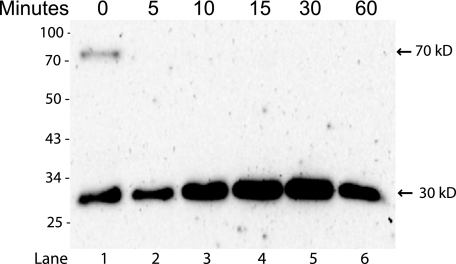
We examined the activation cleavage of basolateral plasma membrane bound matriptase using surface biotinylation and an incubation assay. Under reducing conditions, matriptase zymogen is a 70 kDa protein while enzymatically-active matriptase separates into two fragments; the stem domain and the 30 kDa protease domain. The antibody IM1014 reacts with the serine protease domain of both zymogen and activated matriptase under reducing conditions. This experiment showed that minor amounts of the 70 kDa matriptase zymogen were present on the basolateral plasma membrane together with 30 kDa cleaved matriptase serine protease domain (Fig. 4, lane 1). The basolateral matriptase zymogen was rapidly cleaved as only the 30 kDa band could be detected after just 5 min of incubation (Fig. 4, lane 2). A slight increase in the 30 kDa band intensity was observed up to 15 min after labeling, most likely caused by a higher affinity of IM1014 antibody for the cleaved 30 kDa protease domain than the 70 kDa form. These data suggest that matriptase is fast and efficiently cleaved to its active form on the basolateral cell surface.
FIGURE 4.
Matriptase is cleaved to the two chain form on the basolateral cell surface. Caco-2 cells were grown on Transwell filters, biotinylated from the basolateral side at 4 °C and incubated up to 60 min at 37 °C after labeling. Biotinylated proteins were pulled down with streptavidin-agarose, boiled, reduced and analyzed by SDS-PAGE and Western blotting using the antibody IM1014. The positions of molecular markers (kDa) are indicated on the left. Bands are indicated with arrows and size. At time 0 the matriptase is detected as a 70 kDa band, representing the non-cleaved zymogen and a 30 kDa band representing the cleaved protease domain (lane 1). After 5 min of incubation the zymogen could no longer be detected and an increase in the 30 kDa serine protease domain was detected (lanes 2–6). Results shown are representative of two independent experiments.
Active Matriptase Is Present on the Basolateral Plasma Membrane
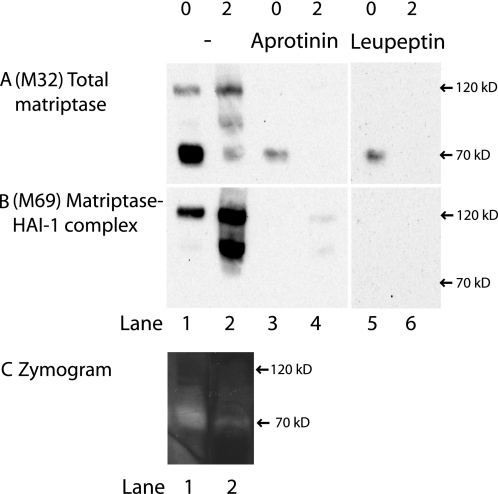
It has previously been reported that the period of time for active matriptase to act on its substrates is very limited as matriptase activation is tightly coupled to inhibition by HAI-1. This window of action is assumed to be in between the proteolytic cleavage creating the fully active protease and the rapid complex formation with its inhibitor HAI-1. Our previous experiments suggest that both matriptase activation and inhibition takes place on the basolateral membrane and we would therefore expect to find free active matriptase on the basolateral plasma membrane. To address this, we investigated whether matriptase at the basolateral plasma membrane was able to bind to the general serine protease inhibitors, aprotinin and leupeptin, to which we expect only the active protease to bind. We were able to purify matriptase from the basolateral plasma membrane, with both aprotinin- and leupeptin-coupled Sepharose (Fig. 5A, lanes 3 and 5). No matriptase was purified with the negative control soybean trypsin inhibitor-Sepharose or uncoupled Sepharose (data not shown). The inhibitor-bound fraction of matriptase was not detected by the antibody detecting matriptase-HAI-1 complexes (Fig. 5B, lanes 3 and 5).
FIGURE 5.
Matriptase is active on the basolateral plasma membrane but not intracellularly. Caco-2 cells on Transwell filters were surface-biotinylated from the basolateral side at 4 °C. Some were lysed immediately after biotinylation (0) and some were incubated for 2 h at 37 °C to transform all biotin-labeled matriptase into activated matriptase in complex with HAI-1 (2). Biotinylated proteins were precipitated with monomeric avidin and gently eluted with biotin. The avidin pull-downs were divided into three groups: No further treatment (lanes 1 and 2), pull-down with aprotinin-Sepharose (lanes 3 and 4) and pull-down with leupeptin-Sepharose (lanes 5 and 6). The samples were analyzed by SDS-PAGE and Western blot with antibodies against total matriptase (M32) and matriptase-HAI-1 complex (M69). A, only at time 0 was it possible to pull-down M32-detectable matriptase with both aprotinin and leupeptin. B, M69-detectable matriptase was pulled down with the monomeric avidin (B, lanes 1 and 2), but was lost with additional inhibitor pull-down (B, lanes 3–6). C, two avidin-purified fractions showed gelatinolytic properties with a band around 70 kDa (C, lanes 1 and 2), matching the size and pattern of A, lanes 1 and 2. Results shown are representative of two independent experiments.
We wanted to verify that the purified matriptase detected using the inhibitor-coupled Sepharose indeed was free active matriptase and not activated matriptase dissociated from the matriptase-HAI-1 complexes. To test this, a sample containing mostly matriptase-HAI-1 complex was exposed to the inhibitor-coupled Sepharose. No detectable matriptase from the matriptase-HAI-1 complex was purified with the inhibitor Sepharose (Fig. 5, lanes 4 and 6). This verifies that it is free active matriptase that is being pulled down and that the matriptase-HAI-1 complex is not dissociated during our cell extraction procedure.
Finally, to test the proteolytic activity of matriptase, the two biotinylated fractions, containing 70 kDa matriptase and matriptase-HAI-1 complexes, respectively, were analyzed for their ability to display gelatinolytic activity at pH 8.75, where matriptase has optimal enzymatic activity (25) (Fig. 5C). The fraction with the 70 kDa matriptase displayed one gelatinolytic band around 70 kDa at pH 8.75, corresponding to the size of free active matriptase (Fig. 5C, lane 1). The fraction containing mostly matriptase-HAI-1 complex, showed only weak gelatinolytic activity around 70 kDa (Fig. 5C, lane 2). Thus, the gelatinolytic activity pattern matches the one observed for matriptase immunoreactivity with the antibody M32 (Fig. 5A) implying that the gelatinolytic activity observed is caused by matriptase.
Active Prostasin Is Present on Both the Apical and the Basolateral Plasma Membrane
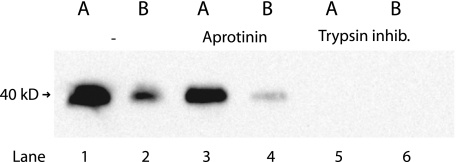
All of our data suggest that matriptase is active and able to cleave relevant substrates in a short period of time on the basolateral plasma membrane before it forms a complex with HAI-1 and is endocytosed. If prostasin is indeed a substrate for matriptase in polarized epithelial cells, we would expect to find the cleaved form of prostasin on the basolateral membrane. To test this, we utilized the fact that prostasin is a serine protease and only the cleaved form is proteolytically active and thereby able to bind serine protease inhibitors such as aprotinin. Our experiment showed that prostasin on both the apical and the basolateral plasma membrane was able to bind to aprotinin (Fig. 6, lanes 3 and 4). This suggests that prostasin is active on both the apical and the basolateral side of the cell.
FIGURE 6.
Active prostasin is present on the basolateral as well as on the apical plasma membrane. Caco-2 cells on Transwell filters were surface biotinylated from either the apical or basolateral side at 4 °C. Biotinylated proteins were precipitated with monomeric avidin and gently eluted with biotin. The avidin pull-downs were divided into three: No further treatment (lanes 1 and 2), pull-down with aprotinin-Sepharose (lanes 3 and 4) and pull-down with trypsin inhibitor as negative control (lanes 5 and 6). The pull-down fractions were analyzed by SDS-PAGE and Western blotting. Prostasin was purified from both apical plasma membrane (lane 1) and basolateral plasma membrane (lane 2). The majority of the apical prostasin could be pulled down with the serine protease inhibitor aprotinin (lane 3). A small but significant fraction of basolateral prostasin was also able to bind aprotinin and hence was in its active form (lane 4). No prostasin binding was observed when using the negative control trypsin-coupled Sepharose (lanes 5 and 6). Results shown are representative of three independent experiments.
Matriptase Co-localizes with Prostasin on the Basolateral Plasma Membrane, Subsequently Prostasin Is Transcytosed to the Apical Plasma Membrane
We know from the surface labeling experiment that the majority of membrane-bound prostasin at steady state is located on the apical side (Fig. 1, lane 9) whereas only a minor fraction is located on the basolateral membrane in Caco-2 cells (Fig. 1, lane 10). From our experiments, we also know that prostasin is not only found in its active form on the apical plasma membrane but also on the basolateral plasma membrane. This could indicate that prostasin is transported to the basolateral plasma membrane to get activated by matriptase.
We wanted to investigate the endocytic transport of the apical and basolateral prostasin. Initially, we tested if the two membrane fractions were endocytosed using the same experimental setup as for matriptase and HAI-1 endocytosis. We found that prostasin was not endocytosed from the apical plasma membrane but was completely endocytosed from the basolateral plasma membrane within 60 min (data not shown). This made us question if prostasin is initially transported to the basolateral membrane and activated by matriptase before it is re-routed by transcytosis to the apical membrane. A long residence time at the apical plasma membrane combined with a short residence time at the basolateral plasma membrane would give a steady state accumulation at the apical plasma membrane. We, therefore, tested if the basolaterally endocytosed fraction was transcytosed to the apical membrane.
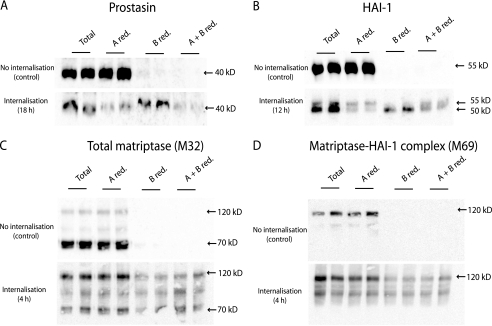
Transcytosis for several incubation times was investigated (1, 2, 4, 8, 12, and 18 h) The time points most clearly demonstrating transcytosis are shown in Fig. 7. Transcytosis of prostasin was detectable after 8 h (data not shown) and most of prostasin was transcytosed from the basolateral to the apical plasma membrane after 18 h (Fig. 7A). HAI-1 was also transcytosed, however, less efficiently than prostasin (Fig. 7B). Our results strongly suggest that the slow migrating form of HAI-1 at the basolateral plasma membrane is the precursor for the faster migrating HAI-1 present on the apical plasma membrane (Fig. 7B).
FIGURE 7.
Prostasin and HAI-1 are transcytosed from the basolateral to the apical plasma membrane. Caco-2 cells were surface biotinylated from the basolateral side at 4 °C. The labeled cells were incubated at 37 °C for the time indicated in the internalization panel (4, 12, or 18 h as indicated). After incubation the proteins remaining on the plasma membrane were biotin-stripped, using the membrane non-permeant reducing agent glutathione, from either apical (A red.), basolateral (B red.), or both sides (A + B red.). Biotin reduction was omitted in a set of samples to monitor the total amount of biotinylated protein remaining after the incubation (Total). The experiment was performed in duplicates. Biotinylated proteins were precipitated with monomeric avidin agarose, separated with SDS-PAGE and analyzed by Western blotting. A, prostasin could be biotin-stripped from the apical side (A red.) after 18 h of internalization, as a decrease in signal was observed compared with the total samples. This shows that prostasin has moved from the basolateral to the apical side. When surface proteins were biotin-stripped from the basolateral side no decrease in signal was observed (B red.), once again showing that no biotinylated prostasin was left on the basolateral side after 18 h. To clarify if some of the prostasin was located intracellularly, the cells were biotin stripped from both apical and basolateral side (A + B red.). Biotin-stripping from both apical and basolateral side removed the majority of the biotinylated prostasin suggesting that all of prostasin had within the 18 h transferred from the basolateral to the apical plasma membrane. B, HAI-1 was transcytosed from the basolateral to the apical plasma membrane within 12 h. Most of HAI-1 could be biotin stripped from the apical side after incubation (A red.) where a major fraction was left after basolateral reduction (B. red) showing that a large fraction of biotinylated HAI-1 had moved from the basolateral to the apical plasma membrane. It was demonstrated that the faster migrating form of HAI-1 resides at the apical plasma membrane 12 h after biotinylation and the slower migrating form remained at the basolateral plasma membrane, as these could be biotin-stripped from their respective sites (A red. compared with B red., respectively). C, after just 4 h of incubation most biotinylated matriptase was no longer detectable in the cell. The remaining matriptase was mainly found in the 120 kDa complex with HAI-1. A fraction of the matriptase-HAI-1 complex could be biotin stripped from the basolateral side (B red.) but not the apical side (A red.). D, M69 detectable matriptase showed the same amount of matriptase-HAI-1 complex at time 0 as for 4 h of incubation. A fraction of the complex was reducible from the basolateral side after 4 h of incubation (B red.). Results shown are representative of three independent experiments.
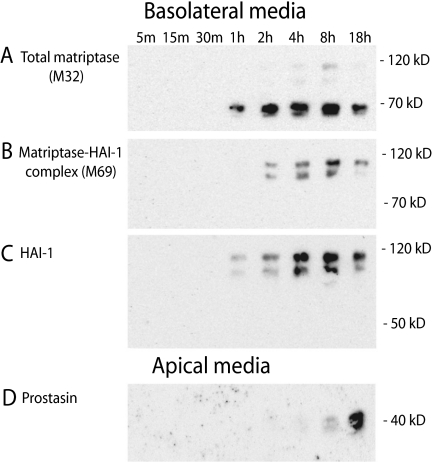
Matriptase-HAI-1 complex has been purified from milk, suggesting that matriptase is secreted from the apical plasma membrane (16). However, we were unable to detect transcytosis of matriptase from the basolateral to the apical plasma membrane, as matriptase has a shorter half-life in the cell than HAI-1 and prostasin (Fig. 7, C and D). The remaining biotin-labeled matriptase was found in the matriptase-HAI-1 complexes between 95 and 120 kDa after 4 h incubation (Fig. 7, C and D). These complexes were also detected with the anti-HAI-1 antibody after 4 h incubation (data not shown). Instead, basolaterally biotin-labeled matriptase could be detected in the basolateral media after only 1 h and matriptase accumulated in the media over the 18 h (Fig. 8, A and B). Both the 70 kDa form as well as two forms of the matriptase-HAI-1 complex around 110 and 85 kDa were detected in the media. Only HAI-1 in complex with matriptase was detected in the basolateral media (Fig. 8C), whereas no free HAI-1 could be detected. None of the biotin-labeled matriptase or HAI-1 could be detected in the apical media (data not shown). Basolateral prostasin could not be detected in the basolateral media (data not shown) but after 18 h of incubation after the biotinylation, prostasin appeared in the apical media (Fig. 8D). This is consistent with the finding that prostasin is transcytosed from the basolateral to the apical plasma membrane within this time frame. Thus, we were able to show that prostasin on the basolateral plasma membrane is transcytosed to the apical plasma membrane from where it is shed to the media. This suggests that prostasin is routed via the basolateral plasma membrane where it is activated before it is transcytosed to the apical membrane, thereby providing a mechanism for activation of the prostasin zymogen in polarized epithelial cells by matriptase or other basolaterally located serine proteases, despite the separate steady-state localization of the proteases.
FIGURE 8.
Matriptase is shed into the basolateral media while basolateral prostasin is transcytosed and shed into the apical media. Caco-2 cells were surface-biotinylated from the basolateral side at 4 °C. Cells were incubated at 37 °C after labeling and apical and basolateral media was collected at 5, 15, 30 min, 1, 2, 4, 8, and 18 h to reveal any shedding of the biotin-labeled matriptase, HAI-1 and prostasin. Biotinylated proteins from the media were pulled down with monomeric avidin agarose, separated with SDS-PAGE and analyzed by Western blotting. A, matriptase was released to the basolateral media within 1 h as three forms: a 70 kDa form and two proteolytically shed complexes at 85 and 110 kDa. B, two complexes of size 85 and 110 kDa could be detected with the M69 antibody within 1–2 h, suggesting these to be matriptase-HAI-1 complexes. C, no free 55 kDa HAI-1 was found in the media, only HAI-1 in the two complexes at 85 and 110 kDa was detected in the basolateral media after 1 h and accumulated up to 18 h. D, basolateral prostasin was detected in the apical media after 18 h incubation, confirming transcytosis before secretion into the apical media. Results shown are representative of two independent experiments.
DISCUSSION
It has been shown that matriptase and prostasin are constitutively expressed and co-localize in most epithelia including the tissues affected by matriptase ablation, suggesting a possible global role for a matriptase-prostasin cascade in epithelial homeostasis (26). In a polarized cell, matriptase is at steady-state concentrated and localized mainly at the basolateral plasma membrane together with HAI-1 (15, 24) while prostasin is mainly concentrated and localized to the apical plasma membrane (17, 18). Our data provide a possible solution to how matriptase can activate prostasin despite their two different subcellular localizations, by showing that the two molecules meet en route.
Matriptase is mainly located on the basolateral plasma membrane where it is activated, inhibited, endocytosed in complex with its inhibitor HAI-1 and is accumulated in intracellular structures. Observation of the transmembrane N-terminal fragment of matriptase in intracellular compartments has previously been described in rat enterocytes (14). We show here that prostasin is mainly located on the apical plasma membrane in its active form. Interestingly, we find a small fraction of prostasin on the basolateral plasma membrane co-localizing with matriptase, making it possible for protease and substrate to interact and activation to occur. In agreement with this, we show that part of the basolateral prostasin is active by its ability to bind to general serine protease inhibitors. The basolateral prostasin is endocytosed and transcytosed to the apical plasma membrane, where it accumulates.
Both matriptase and prostasin has been shown to be able to activate the epithelial sodium channel (ENaC) in Xenopus oocytes (5, 27, 28). ENac is an epithelial membrane-bound sodium channel located in the apical membrane of polarized cells and is required for normal epidermal differentiation (29, 30). Because matriptase is targeted to the basolateral membrane where it is activated and rapidly inhibited it is more likely that prostasin which co-localizes with ENaC on the apical membrane is a candidate activator in polarized cells. Both matriptase and prostasin have been coupled to maintenance of functional tight junctions in a variety of epithelial cell types. In a Caco-2 model, loss of matriptase was associated with enhanced expression and incorporation of the pore-forming protein claudin-2 at tight junctions (2). Prostasin has been reported to regulate tight junctions, paracellular permeability and TEER by a protease activity-dependent mechanism in renal cells (4, 5). This means that the two proteases are both involved in cascades important for tight junction formation in polarized epithelia.
Increasing evidence suggests that serine proteases contribute in a complex way to the regulation of intestinal integrity and barrier function. Protease inhibitors have been shown to suppress the formation of tight junctions in gastrointestinal cell lines suggesting that proteolytic activity is necessary for a functional epithelial barrier (31). The epithelial barrier is crucial in the gastrointestinal tract and is often compromised in inflammatory bowel diseases like Crohn disease and ulcerative colitis. The sodium channel ENaC is down-regulated in inflammatory bowel diseases and has been proposed as a therapeutic target (32–34). It will be important for future studies to identify the role of the matriptase-prostasin cascade and the substrates involved in this pathway to investigate the role of matriptase in inflammatory diseases of the gastrointestinal tract.
Together, our data show that active matriptase and its substrate prostasin co-localize at the basolateral plasma membrane and hereby provide a venue for a matriptase-prostasin zymogen cascade to be initiated in polarized epithelial cells. Furthermore, we show how prostasin is transported after activation to the apical membrane to co-localize with its primary substrate ENaC.
Acknowledgment
We thank Dr. Mary Jo Danton for critically reading the manuscript.
This work was supported, in whole or in part, by the NIDCR Intramural Research Program and by National Institutes of Health Grant R01-CA-123223. This work was also supported by The Harboe Foundation, The Augustinus Foundation, The Brothers Hartmanns Foundation, The A.P. Møllers Foundation for the Advancement of Medical Science, The Cluster of Cell Biology at the University of Copenhagen, and the Lundbeck Foundation.
- TEER
- transepithelial electrical resistance
- HAI-1
- hepatocyte growth factor activator inhibitor 1
- ENaC
- epithelial sodium channel.
REFERENCES
- 1. List K., Kosa P., Szabo R., Bey A. L., Wang C. B., Molinolo A., Bugge T. H. (2009) Am. J. Pathol. 175, 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buzza M. S., Netzel-Arnett S., Shea-Donohue T., Zhao A., Lin C. Y., List K., Szabo R., Fasano A., Bugge T. H., Antalis T. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4200–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu J. X., Chao L., Chao J. (1995) J. Biol. Chem. 270, 13483–13489 [DOI] [PubMed] [Google Scholar]
- 4. Steensgaard M., Svenningsen P., Tinning A. R., Nielsen T. D., Jorgensen F., Kjaersgaard G., Madsen K., Jensen B. L. (2010) Acta Physiol. 200, 347–349 [DOI] [PubMed] [Google Scholar]
- 5. Verghese G. M., Gutknecht M. F., Caughey G. H. (2006) Am. J. Physiol. Cell Physiol. 291, C1258–C1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leyvraz C., Charles R. P., Rubera I., Guitard M., Rotman S., Breiden B., Sandhoff K., Hummler E. (2005) J. Cell Biol. 170, 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. List K., Szabo R., Wertz P. W., Segre J., Haudenschild C. C., Kim S. Y., Bugge T. H. (2003) J. Cell Biol. 163, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ovaere P., Lippens S., Vandenabeele P., Declercq W. (2009) Trends Biochem. Sci. 34, 453–463 [DOI] [PubMed] [Google Scholar]
- 9. Szabo R., Kosa P., List K., Bugge T. H. (2009) Am. J. Pathol. 179, 2015–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. List K., Haudenschild C. C., Szabo R., Chen W., Wahl S. M., Swaim W., Engelholm L. H., Behrendt N., Bugge T. H. (2002) Oncogene 21, 3765–3779 [DOI] [PubMed] [Google Scholar]
- 11. Netzel-Arnett S., Currie B. M., Szabo R., Lin C. Y., Chen L. M., Chai K. X., Antalis T. M., Bugge T. H., List K. (2006) J. Biol. Chem. 281, 32941–32945 [DOI] [PubMed] [Google Scholar]
- 12. Alef T., Torres S., Hausser I., Metze D., Türsen U., Lestringant G. G., Hennies H. C. (2009) J. Invest. Dermatol. 129, 862–869 [DOI] [PubMed] [Google Scholar]
- 13. Le Bivic A., Quaroni A., Nichols B., Rodriguez-Boulan E. (1990) J. Cell Biol. 111, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsuzuki S., Murai N., Miyake Y., Inouye K., Hirayasu H., Iwanaga T., Fushiki T. (2005) Biochem. J. 388, 679–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J. K., Lee M. S., Tseng I. C., Chou F. P., Chen Y. W., Fulton A., Lee H. S., Chen C. J., Johnson M. D., Lin C. Y. (2009) Am. J. Physiol. Cell Physiol. 279, C459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin C. Y., Anders J., Johnson M., Dickson R. B. (1999) J. Biol. Chem. 274, 18237–18242 [DOI] [PubMed] [Google Scholar]
- 17. Chen M., Chen L. M., Lin C. Y., Chai K. X. (2008) Biochim. Biophys. Acta 1785, 896–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Selzer-Plon J., Bornholdt J., Friis S., Bisgaard H. C., Lothe I. M., Tveit K. M., Kure E. H., Vogel U., Vogel L. K. (2009) BMC. Cancer 9, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin C. Y., Wang J. K., Torri J., Dou L., Sang Q. A., Dickson R. B. (1997) J. Biol. Chem. 272, 9147–9152 [PubMed] [Google Scholar]
- 20. Satomi S., Yamasaki Y., Tsuzuki S., Hitomi Y., Iwanaga T., Fushiki T. (2001) Biochem. Biophys. Res. Commun. 287, 995–1002 [DOI] [PubMed] [Google Scholar]
- 21. Tseng I. C., Chou F. P., Su S. F., Oberst M., Madayiputhiya N., Lee M. S., Wang J. K., Sloane D. E., Johnson M., Lin C. Y. (2008) Am. J. Physiol. Cell Physiol. 295, C423–C431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y. W., Wang J. K., Chou F. P., Chen C. Y., Rorke E. A., Chen L. M., Chai K. X., Eckert R. L., Johnson M. D., Lin C. Y. (2010) J. Biol. Chem. 285, 31755–31762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin C. Y., Tseng I. C., Chou F. P., Su S. F., Chen Y. W., Johnson M. D., Dickson R. B. (2008) Front Biosci. 13, 621–635 [DOI] [PubMed] [Google Scholar]
- 24. Godiksen S., Selzer-Plon J., Pedersen E. D., Abell K., Rasmussen H. B., Szabo R., Bugge T. H., Vogel L. K. (2008) Biochem. J. 413, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beliveau F., Desilets A., Leduc R. (2009) FEBS J. 276, 2213–2226 [DOI] [PubMed] [Google Scholar]
- 26. List K., Hobson J. P., Molinolo A., Bugge T. H. (2007) J. Cell. Physiol. 213, 237–245 [DOI] [PubMed] [Google Scholar]
- 27. Vallet V., Chraibi A., Gaeggeler H. P., Horisberger J. D., Rossier B. C. (1997) Nature 389, 607–610 [DOI] [PubMed] [Google Scholar]
- 28. Andreasen D., Vuagniaux G., Fowler-Jaeger N., Hummler E., Rossier B. C. (2006) J. Am. Soc. Nephrol. 17, 968–976 [DOI] [PubMed] [Google Scholar]
- 29. Myerburg M. M., Harvey P. R., Heidrich E. M., Pilewski J. M., Butterworth M. (2010) Am. J. Respir. Cell Mol. Biol. 43, 712–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mauro T., Guitard M., Behne M., Oda Y., Crumrine D., Komuves L., Rassner U., Elias P. M., Hummler E. (2002) J. Invest. Dermatol. 118, 589–594 [DOI] [PubMed] [Google Scholar]
- 31. Bacher A., Griebl K., Mackamul S., Mitreiter R., Mückter H., Ben-Shaul Y. (1992) Exp. Cell Res. 200, 97–104 [DOI] [PubMed] [Google Scholar]
- 32. Farkas K., Yeruva S., Rakonczay Z., Jr., Ludolph L., Molnar T., Nagy F., Szepes Z., Schnur A., Wittmann T., Hubricht J., Riederer B., Venglovecz V., Lazar G., Kiraly M., Zsembery A., Varga G., Seidler U., Hegyi P. (2010) Inflamm. Bowel. Dis. in press [DOI] [PubMed] [Google Scholar]
- 33. Sullivan S., Alex P., Dassopoulos T., Zachos N. C., Iacobuzio-Donahue C., Donowitz M., Brant S. R., Cuffari C., Harris M. L., Datta L. W., Conklin L., Chen Y., Li X. (2009) Inflamm. Bowel. Dis. 15, 261–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zeissig S., Bergann T., Fromm A., Bojarski C., Heller F., Guenther U., Zeitz M., Fromm M., Schulzke J. D. (2008) Gastroenterology 134, 1436–1447 [DOI] [PubMed] [Google Scholar]