Abstract
Intracellular pathogen sensor, NOD2, has been implicated in regulation of wide range of anti-inflammatory responses critical during development of a diverse array of inflammatory diseases; however, underlying molecular details are still imprecisely understood. In this study, we demonstrate that NOD2 programs macrophages to trigger Notch1 signaling. Signaling perturbations or genetic approaches suggest signaling integration through cross-talk between Notch1-PI3K during the NOD2-triggered expression of a multitude of immunological parameters including COX-2/PGE2 and IL-10. NOD2 stimulation enhanced active recruitment of CSL/RBP-Jk on the COX-2 promoter in vivo. Intriguingly, nitric oxide assumes critical importance in NOD2-mediated activation of Notch1 signaling as iNOS−/− macrophages exhibited compromised ability to execute NOD2-triggered Notch1 signaling responses. Correlative evidence demonstrates that this mechanism operates in vivo in brain and splenocytes derived from wild type, but not from iNOS−/− mice. Importantly, NOD2-driven activation of the Notch1-PI3K signaling axis contributes to its capacity to impart survival of macrophages against TNF-α or IFN-γ-mediated apoptosis and resolution of inflammation. Current investigation identifies Notch1-PI3K as signaling cohorts involved in the NOD2-triggered expression of a battery of genes associated with anti-inflammatory functions. These findings serve as a paradigm to understand the pathogenesis of NOD2-associated inflammatory diseases and clearly pave a way toward development of novel therapeutics.
Keywords: Cyclooxygenase (COX) Pathway, Inflammation, Innate Immunity, Interleukin, Notch Pathway, Pattern Recognition Receptor
Introduction
Macrophages as an important component of the innate immune system often reside beneath the epithelium or mucosal surface of various organs that are entry points of diverse pathogens (1). In this perspective, signaling cascades triggered through pattern recognition receptors signify as critical events regulating the activity of macrophages in terms of cytokine or chemokine production, receptor expression, and directed migration to secondary lymphoid organs for subsequent antigen presentation (2). These attributes often have a major influence in determining the overall strength of the immune response. Among pattern recognition receptors, NOD-like receptors (NLR)3 represent a novel class of intracellular pathogen recognition receptors and among NLRs, nucleotide oligomerization domain (NOD) 2 is expressed specifically in monocytes/macrophages, T cells, and intestinal or mucosal epithelial cells, in addition to other cells. The rate-limiting step in NOD2 activation requires oligomerization of the NOD domain effectuating in formation of a robust signaling platform, a critical necessity in binding of effector kinases and adaptor proteins including receptor-interacting protein 2 (RIP2) and transforming growth factor-β-activated kinase 1 (TAK1), which subsequently facilitates activation of NF-κB thus activating a wide range of immunomodulatory genes (2–4).
Despite relevance in regard to regulation of inflammatory responses or local immune homeostasis, the NOD2 contribution to adaptive immunity remains to be elucidated. Albeit a integral component of bacterial peptidoglycan, MDP, a well characterized ligand for NOD2, is known for its adjuvant properties; interestingly, unlike many TLR agonists, its engagement with NOD2 receptor preferentially drives Th2 skewed immune responses, which are generally associated with the anti-inflammatory M2 phenotype of macrophages (5–7). Importantly, natural mutations in the NOD2 gene have been reported to be associated with genetic predisposition to severe inflammatory diseases like Crohn disease, asthma, chronic inflammatory bowel disease etc. (8–10). Thus because of its wide implications, our aim is to understand the nature of the molecular signaling events that control the NOD2-triggered immune responses.
Notch signaling effectuates important roles not only in the development of the immune system, but also critically in cell fate judgments among fully differentiated immune cells including macrophages or dendritic cells (11–14). The Notch pathway is characterized by juxtacrine ligand-receptor interactions between adjacent cells, and macrophages are known to express Notch ligands as well as receptors, thus possessing the capacity to both induce and respond to Notch signals. Upon productive engagement of Notch receptor with its cognate ligand-like Jagged1 on cell membrane of adjacent macrophages, the γ-secretase complex generates release of the Notch intracellular domain (NICD/Cleaved Notch) by proteolytic processing from the cell membrane. NICD upon translocation to the nucleus binds and complexes with CSL/RBP-Jk, a DNA-binding protein that normally associates with Notch target gene promoters along with several co-repressors. This results in coordinated recruitment of transcriptional co-activators like mastermind, p300, and/or CBP (CREB-binding protein) with simultaneous displacement of co-repressors, leading to CSL/RBP-Jk-dependent transcription of target genes (15, 16). Interestingly, because of significant cross-talk among diverse signaling platforms, CSL/RBP-Jk independent activation of its target genes by Notch has also been documented (17–20). However, the importance of the functional significance of Notch signaling in cytosolic immune surveillance mediated by NLR-like NOD2 remains unexplored.
Herein, we demonstrate a potential role for NOD2 in activating Notch1 signaling by virtue of directed regulation of Jagged1 expression in macrophages. Importantly, signaling perturbations or genetic approaches implicated a critical role of cross-talk among members of Notch1-phosphoinositide 3-kinase (PI3K) in NOD2-mediated anti-inflammatory immune responses, including the expression of cyclooxygenase-2 (COX-2) and IL-10, in macrophages. Correlative evidence demonstrates that this mechanism operates in vivo as MDP, a known NOD2 agonist, triggered the expression of activated Notch1, Jagged1, or its target gene COX-2 in brain or splenocytes of mice. Importantly, we demonstrate that the NOD2-driven activation of the Notch1-PI3K signaling cascade is instrumental in protection of macrophages against TNF-α or IFN-γ-triggered apoptosis. Briefly, the current investigation identifies a novel role for Notch1 signaling in orchestrating functional attributes of NOD2 activation in an immunological milieu as well as in modulation of a distinct set of innate immunological parameters including COX-2 in innate immune cells.
EXPERIMENTAL PROCEDURES
Cells and Mice
Peritoneal macrophages were isolated from peritoneal exudates of C57BL/6 or iNOS−/− C57BL/6 mice maintained at the central animal facility, Indian Institute of Science. The RAW 264.7 mouse macrophage cell line was cultivated in DMEM (Sigma) supplemented with 10% heat inactivated FBS (Sigma). All studies involving macrophages were performed utilizing mice peritoneal macrophages with the exception of transfection experiments, which were performed on RAW 264.7 macrophages. Mouse bone marrow-derived dendritic cells were generated as described previously (21). All studies were carried out with approval from the Institutional Ethics Committee for animal experimentation as well as from the Institutional Biosafety Committee.
Reagents and Antibodies
General laboratory chemicals were purchased from Sigma or Merck. Lipopolysaccharide-free MDP was procured from Sigma. Anti-β-actin, anti-iNOS, and anti-PGE2 antibodies were obtained from Sigma. Anti-COX-2 and anti-proliferating cell nuclear antigen were purchased from Calbiochem. HRP-conjugated anti-rabbit IgG and anti-mouse IgG antibodies were obtained from Jackson ImmunoResearch Laboratories. The anti-Thr70 phospho-eukaryotic initiation factor 4E-binding protein 1 (4EBP1), anti-4EBP1, anti-Thr180/Tyr182 phospho-p38 MAPK, anti-p38 MAPK, anti-Thr202/Tyr204 phospho-ERK1/2, anti-ERK1/2, anti-NF-κB p65, anti-cleaved Notch1 or anti-NICD (Val1744), anti-Tyr458 p85/Tyr199 p55 phospho-PI3K, anti-p85 PI3K, anti-Jagged1, anti-βII Ser660 phospho-PKC, anti-Ser176 phospho-RIP2, anti-Tyr416 Src, anti-Src, and anti-Ser412 phospho-TAK1 antibodies were purchased from Cell Signaling Technology.
Treatment with Pharmacological Reagents
All pharmacological reagents were obtained from Calbiochem and were reconstituted in sterile DMSO (Sigma) and used at concentrations detailed under supplemental Table S1. 1% DMSO was used as the vehicle control. In all experiments with inhibitors, a tested concentration was used after careful titration experiments assessing the viability of the macrophages using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. In experiments with inhibitors, the cells were treated with a given inhibitor for 60 min before experimental treatment. Nitric oxide donor, SIN-1 (Sigma), was used at concentration of 20 μm.
RNA Isolation and Quantitative Real Time PCR
Total RNA from treated macrophages was isolated by the TRIzol method (Sigma) as per the manufacturer's protocol, and treated with RNase-free DNase (Promega). The cDNA synthesis kit (Fermentas) was used for reverse transcription according to the manufacturer's protocol. A real time PCR amplification (Applied Biosystems) using SYBR Green PCR mix (Finnzymes, Finland) was performed for quantification of target gene expression. All reactions were repeated at least three times independently to ensure the reproducibility of the results. Amplification of the GAPDH housekeeping gene was used as internal control. Primer sequences used in the current study are detailed in supplemental Table S2.
Immunoblotting Analysis
Cells were washed with PBS and scrapped off the culture dish and collected by centrifugation. Cell pellets were lysed in RIPA buffer consisting of 50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 1 mm PMSF, 1 μg/ml of each aprotinin, leupeptin, pepstatin, 1 mm Na3VO4, 1 mm NaF, and incubated on ice for 30 min. Whole cell lysate was collected by centrifuging lysed cells at 13,000 × g for 10 min at 4 °C. Equal protein amounts from each cell lysate was subjected to SDS-PAGE and transferred onto PVDF membranes (Millipore) by semidry Western blotting (Bio-Rad). Nonspecific binding was blocked with 5% nonfat dry milk powder in TBST (20 mm Tris-HCl (pH 7.4), 137 mm NaCl, and 0.1% Tween 20) for 60 min. The blots were incubated overnight at 4 °C with primary antibodies diluted in TBST with 5% BSA. After washing with TBST, blots were incubated with anti-rabbit or anti-mouse IgG secondary antibodies conjugated to HRP for 2 h. After further washing in TBST, the immunoblots were developed with an enhanced chemiluminescence detection system (PerkinElmer Life Sciences) as per the manufacturer's instructions.
Transfection Studies
RAW 264.7 cells were transfected with 100 nm siRNA using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Transfection efficiency was more than 50% throughout the experiments as determined by counting the number of siGLO Lamin A/C positive cells in a microscopic field using a fluorescent microscope. 72 h post-transfection the cells were treated as indicated and processed for expression analysis. All siRNAs were obtained from Dharmacon as siGENOMETM SMARTpool reagents, which contains a pool of four different double-stranded RNA oligonucleotides (siRNA). RAW 264.7 macrophages were transiently transfected with AKT or p85 dominant-negative (D/N) cDNA constructs using low Mr polyethylenimine (Sigma).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assays were carried out as described previously (22). Peritoneal macrophages were left untreated or treated as indicated for 6 h. The cells were fixed with 1.42% formaldehyde for 15 min at room temperature followed by inactivation of formaldehyde with addition of 125 mm glycine. Chromatin extracts containing DNA fragments with an average size of 500 bp were immunoprecipitated using anti-NF-κB or anti-NICD antibodies for NF-κB and RBP-Jk ChIP, respectively. Purified DNA was analyzed by quantitative PCR using the SYBR Green method (Finnzymes). Regions with NF-κB and RBP-Jk binding sites in the mouse COX-2 promoter were amplified using primer pairs, NF-κB forward, 5′-ttaaccggtagctgtgtgcgt-3′, reverse, 5′-tctccggtttcctcccagt-3′; and RBP-Jk forward, 5′-tcccgtgaaaagagttgctga-3′, reverse, 5′-ttcatggaactaccctccgg-3′. 28S rRNA was used as control in the PCR and the primers were forward, 5′-ctgggtataggggcgaaagac-3′ and reverse, 5′-ggccccaagacctctaatcat-3′. All results were normalized either by the respective input values or by amplification of 28S rRNA. All ChIP experiments were repeated at least three times.
In Vivo Challenge of Mice with MDP or SIN-1
C57BL/6 or iNOS−/− C57BL/6 mice utilized in the current investigation were 5–6 weeks old and each in vivo experiment involved three animals per group. For intracranial or intravenous challenge, 5 μg of MDP or 20 μg of SIN-1 were resuspended in 50 μl of sterile PBS and then inoculated using 1-ml syringes and a 26-gauge needle. Control mice were injected with 50 μl of sterile PBS using the same protocol. Before intracranial inoculation, mice were anesthetized with intraperitoneal injection of 80 μl of ketamine (6 mg). Experimental mice were sacrificed after 3 or 6 days of inoculations in the case of intravenous and intracranial inoculation, respectively, and splenocytes or brain tissues were harvested.
Immunohistochemistry
Microtome sections (4 μm) were cut from formalin-fixed, decalcified, and paraffin-embedded tissue samples. These paraffin-embedded sections were first deparafinized, followed by antigen retrieval with boiling 10 mm citrate buffer (pH 6.0) in a microwave for 10 min, treated with 1% H2O2 for 10 min, incubated with 0.1 m glycine for 15 min, and blocked with 5% BSA for 1 h at room temperature. Primary antibodies were incubated overnight and HRP-conjugated secondary antibodies for 90 min. The horseradish peroxidase reaction was detected with 0.05% diaminobenzidine and 0.03% H2O2. Sections were counterstained with hematoxylin, dehydrated, and mounted. Stained tissue sections were analyzed with a Leica DMLB microscope (Leica Microsystems, Wetzlar, Germany). All experiments were performed with appropriate isotype-matched control antibodies.
Nuclear and Cytosolic Subcellular Fractionation
Macrophages were harvested by centrifugation and gently resuspended in ice-cold Buffer A (10 mm HEPES (pH 7.9), 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, and 0.5 mm PMSF). After incubation on ice for 15 min, cell membranes were disrupted with 10% Nonidet P-40, and the nuclear pellets were recovered by centrifugation 13,000 × g for 15 min at 4 °C. The supernatants from this step were used as cytosolic extracts. Nuclear pellets were lysed with ice-cold Buffer C (20 mm HEPES (pH 7.9), 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, and 1 mm PMSF) and nuclear extracts were collected after centrifugation at 13,000 × g for 20 min at 4 °C.
Enzyme Immunoassay for PGE2
Enzyme immunoassays for quantitation of PGE2 were carried out in 96-well microtiter plates (Nunc) utilizing the culture supernatant of treated samples. Culture supernatants were incubated in assay plates overnight at 4 °C followed by three washes with PBS. After blocking with 1% BSA for 1 h at 37 °C, wells were incubated with anti-PGE2 antibodies for 6 h at 37 °C followed by washing with PBS. The plates were further incubated with HRP-labeled anti-rabbit secondary antibody for 1 h at 37 °C followed by development with 3,3′,5,5′-tetramethylbenzidine. The absorbance was measured at 492 nm using an ELISA reader (Molecular Devices).
Measurement of IL-10 Levels
Enzyme immunoassay for IL-10 in cell-free culture supernatants was carried out using the IL-10 ELISA kit (Peprotech Asia, Israel) following the manufacturer's instructions. Briefly, assay plates were coated with anti-IL-10 capture antibodies overnight at 4 °C followed by incubation with 1% BSA for 1 h at 37 °C. After three washes with PBST (0.05% Tween 20), plates were incubated with cell-free culture supernatants for 2 h followed by three washes with PBST and incubation with anti-IL-10 detection antibodies for 2 h at 37 °C. The plates were further incubated with streptavidin-HRP antibodies for 1 h at 37 °C followed by development with 3,3′,5,5′-tetramethylbenzidine. The absorbance was measured in an ELISA reader (Molecular Devices) at 492 nm.
MTT Assay
1.5 × 103 macrophages/well were seeded in a 96-well plate. After 12 h of plating, the cells were treated as indicated followed by addition of MTT (20 μl of 5 mg/ml) 3 h prior to completion of the experiment. MTT is a tetrazolium salt that is converted by living cells into blue formazan crystals. The cell-free supernatant was removed from the wells 3 h after MTT addition and 200 μl of DMSO was added to dissolve the formazan crystals and the absorbance was measured at 550 nm in an ELISA reader (Molecular Devices).
Statistical Analysis
Levels of significance for comparison between samples were determined by Student's t test. The data in the graphs are expressed as the mean ± S.E. Graphpad Prism 3.0 software (Graphpad Software) was used for all statistical analysis.
RESULTS
NOD2 Receptor Engagement Results in Activation of Notch1 Signaling through Induced Jagged1 Expression
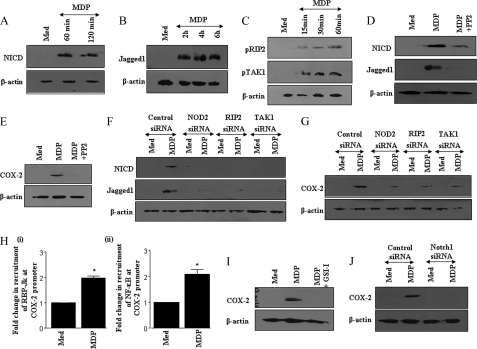
NOD2 receptor-mediated signaling could elicit a spectrum of cellular immune responses that constitute cytosolic innate immune surveillance, including the production of multifaceted effector PGE2, a product of inducible COX-2 enzymatic activity (23). Herein, we show that engagement of the NOD2 receptor by its cognate ligand MDP culminates in the robust activation of Notch1 signaling as evidenced by induced formation of the cleavage product of Notch1 (NICD) (Fig. 1A). Importantly, MDP induced Jagged1 expression, a Notch1 receptor ligand, is critical for the NOD2-driven Notch1 signaling (Fig. 1B). As a “proof of concept,” NOD2 engagement resulted in the activation of effector kinases downstream of NOD2, including RIP2 and TAK1, in a time-dependent manner (Fig. 1C). To assess the importance of NOD2 and RIP2 in activation of Notch1 signaling, expression levels of NICD and Jagged1 expression were quantified in the presence of the pharmacological inhibitor of RIP2, PP2. Although PP2 is known to inhibit Src family protein kinases, it has been recently reported that it inhibits RIP2 more potently (24). Furthermore, utilized concentrations of PP2 failed to inhibit phorbol myristate acetate-triggered activation of Src in primary macrophages (supplemental Fig. S1A). In addition, we could not detect activation of Src upon stimulation of NOD2, ruling out any possibility of a nonspecific effect of PP2 on Src family kinases (supplemental Fig. S1B). In this regard, inhibition of the RIP2 kinase by PP2 abrogated the MDP driven activation of Notch1 signaling as quantified by NICD formation, Jagged1 expression, and expression analysis of Notch1 signaling target genes, COX-2 and Hes1 (Fig. 1, D and E, data not shown). Furthermore, siRNA-mediated knockdown of NOD2, RIP2, or TAK1 severely compromised the NOD2-driven expression of NICD, Jagged1, or COX-2 (Fig. 1, F and G, and supplemental Fig. S2, A–C). From bioinformatic analysis, we identified six CSL/RBP-Jk and five NF-κB-binding consensus in mouse COX-2 promoter (25). As shown in Fig. 1H, NOD2 engagement results in enhanced recruitment of CSL/RBP-Jk or NICD and NF-κB to COX-2 promoter signifying the involvement of the Notch1-driven signaling events upon NOD2 stimulation. Concomitantly, signaling perturbations with the Notch1 activation inhibitor, γ-secretase inhibitor I (GSI-I), as well as siRNA-mediated knockdown of Notch1 or CSL/RBP-Jk clearly established the involvement of Notch1 in NOD2-triggered expression of Notch1 target genes COX-2 or Hes1 (Fig. 1, I and J, and supplemental Fig. S2D, data not shown). Furthermore, stable expression of NICD in RAW 264.7 macrophages resulted in augmented expression of Notch1 target genes Hes1 or COX-2 (data not shown) (22). Intriguingly, recent studies have suggested that MDP as a NOD2 agonist has the potential to drive Th2 responses in dendritic cells (5, 6). Furthermore, when analyzed, activation of the NOD2 receptor by its cognate ligand MDP resulted in induced formation of NICD, expression of Jagged1, and a concomitant increase in expression of the Notch1 target gene COX-2 in bone marrow-derived dendritic cells (supplemental Fig. S3A). Importantly, pretreatment of bone marrow-derived dendritic cells with the RIP2 kinase inhibitor abolished the NOD2-triggered activation of Notch1 signaling (supplemental Fig. S3A). These findings strongly implicate a critical positive role for Notch1-mediated CSL/RBP-Jk recruitment in the NOD2-driven anti-inflammatory immune responses in macrophages as well as dendritic cells.
FIGURE 1.
Stimulation of NOD2 by MDP activates Jagged1 expression and Notch1 signaling. A and B, mouse macrophages were treated with 200 ng/ml of MDP for the indicated time periods and expression of NICD (A) or Jagged1 (B) was analyzed by immunoblotting. C, activation of RIP2 and TAK1 by MDP. D, macrophages were treated with the RIP2 kinase inhibitor, PP2, prior to treatment with MDP followed by analysis of the protein levels of NICD and Jagged1 by immunoblotting. E, RIP2 kinase inhibitor, PP2, curtailed MDP induced COX-2 expression. F, siRNA-mediated knockdown of NOD2, RIP2, and TAK1 compromised the MDP-triggered activation of Notch1 and Jagged1 expression. G, reduction in MDP-induced COX-2 expression upon siRNA-mediated knockdown of NOD2, RIP2, and TAK1. H, recruitment of (i) RBP-Jk/CSL or (ii) NF-κB at COX-2 promoter was analyzed by the ChIP with antibodies specific to NF-κB or NICD in MDP-treated cell lysates. The active recruitment of NF-κB or CSL/RBP-Jk was assessed by quantitative real time PCR (mean ± S.E., n = 3). I, pretreatment with GSI-I abolished MDP-induced expression of the Notch1 target gene, COX-2. J, transient transfection of Notch1 siRNA significantly abrogated MDP-triggered COX-2 expression in RAW 264.7 cells. The data represents results obtained from three independent experiments. Med, medium; *, p < 0.05 versus medium.
NOD2 Receptor Triggering Effectuated the Signaling Cross-talk among Notch1 and PI3K Pathways
Notch-specific genetic signatures are critical not only in the development of innate immunity, but also in imparting survival benefits to a wide range of cell types (26, 27). In this perspective, Notch1 signaling-mediated survival effects frequently necessitate participation of members of the PI3K pathway (27, 28). Importantly, delineation of the mechanisms involved in NOD2-driven activation of the PI3K pathway, if any, remains obscure.
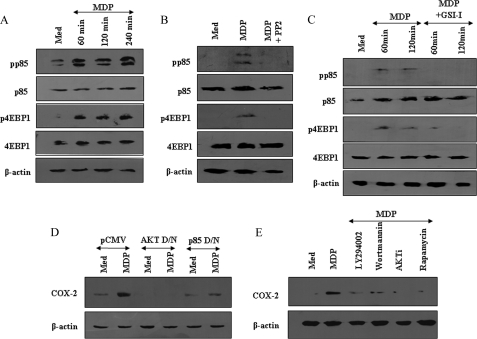
To elucidate the involvement of NOD2 in PI3K signaling activation, macrophages were pulsed with MDP and as shown in Fig. 2A, NOD2 engagement triggered phosphorylation of the p85 subunit of PI3K, 4EBP1, an important member of the PI3K pathway. As described, stimulation of NOD2 resulted in activation of RIP2 and TAK1 in a time-dependent manner (Fig. 1C). In this perspective, to investigate the role for RIP2 kinase, NOD2-mediated activation of the PI3K pathway was assessed upon pre-treatment of macrophages with the pharmacological inhibitor PP2. As demonstrated, inhibition of the RIP2 kinase by PP2 abrogated the NOD2-driven activation of PI3K as quantified by phosphorylation of the p85 subunit of PI3K or 4EBP1 (Fig. 2B). Extensive cross-talk among members of the Notch1 and PI3K signaling cascades in terms of genetic interactions, physical association between specific members, or association to complex comprising unique cofactors have been documented (22, 25, 27, 28). In this regard, blockade of Notch1 signaling activation by GSI-I resulted in marked inhibition of NOD2-driven activation of PI3K (Fig. 2C). Furthermore, in concurrence with these results, inhibition of PI3K by the p85 dominant-negative construct or pharmacological inhibitor LY294002 and wortmannin; AKT by the dominant-negative construct or AKT inhibitor; and mTOR by rapamycin significantly attenuated NOD2-triggered Notch1-PI3K-mediated COX-2 expression (Fig. 2, D and E). These observations clearly implicate NOD2-driven activation of Notch1 signaling in regulating the PI3K pathway thus exerting direct effects in modulating specific molecular signatures in macrophages.
FIGURE 2.
Cross-talk of Notch1 signaling and PI3K pathway during MDP-triggered activation of NOD2 signaling. A, MDP triggers phosphorylation of p85 and 4EBP1 as analyzed by immunoblotting. B, inhibition of the RIP2 kinase by PP2 abolishes MDP-triggered phosphorylation of PI3K pathway members p85 and 4EBP1. C, pharmacological inhibition of Notch1 activation by GSI-I-abrogated MDP-triggered phosphorylation of p85 and 4EBP1. D, transient transfection of AKT dominant-negative (AKT D/N) or p85 dominant-negative (p85 D/N) constructs significantly reduced MDP-induced COX-2 expression in RAW 264.7 macrophages as assessed by immunoblotting. E, pharmacological intervention of PI3K signaling modulates MDP-triggered COX-2 expression. The blots are representative of three independent experiments. Med, medium.
Requirement of iNOS/NO in NOD2-driven Notch1-PI3K Signaling
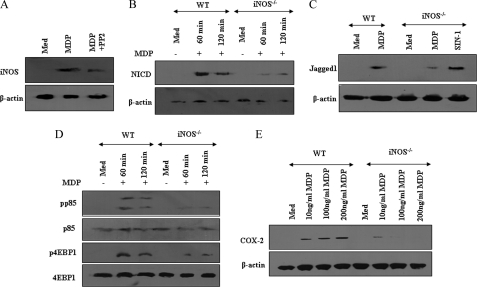
Recent studies have suggested that NOD/RIP2 signaling is crucial to iNOS/NO production in response to bacterial infection, thus contributing to bacterial clearance and host innate immune response. Interestingly, recent reports have rendered the role of iNOS/NO in activation of Notch1 signaling (22, 29). In this perspective, we addressed whether NOD2 triggered expression of iNOS/NO to participate in activation of Notch1-PI3K signaling. As shown in Fig. 3A, NOD2 stimulation by MDP resulted in the robust iNOS expression and inhibition of RIP2 kinase by the PP2 compromised ability of NOD2 to induce iNOS expression. To validate the importance of NOD2-triggered iNOS expression in induction of NICD and Jagged1 expression, macrophages derived from wild type (WT) and iNOS−/− mice, treated with or without MDP, were analyzed. As represented in Fig. 3, B and C, iNOS null macrophages demonstrated a marked inhibition in NOD2-driven Notch1 activation and Jagged1 expression compared with WT macrophages. Importantly, iNOS deficiency caused a defect in the ability of NOD2 to trigger Jagged1 expression and Notch1 activation was not due to the general inability of cells to function, because analogues of iNOS downstream mediators, SNAP or SIN-1 (NO donor), could augment Jagged1 and NICD expression in iNOS-deficient macrophages comparable with that in WT macrophages (Fig. 3, B and C, data not shown). Significantly, NOD2-triggered activation of the PI3K pathway was severely diminished in iNOS null macrophages compared with WT macrophages as evidenced by phosphorylation of the p85 subunit of PI3K or 4EBP1 (Fig. 3D). Accordingly, the NOD2-Notch1-PI3K signaling driven COX-2 expression was strikingly attenuated in iNOS null macrophages (Fig. 3E). Noticeably, varying the concentrations of MDP could not induce the expression of COX-2 in iNOS-deficient macrophages. These findings conjecture the critical participation of the iNOS/NO pathway in the NOD2-driven Jagged1 expression and activation of Notch1 signaling in macrophages (Fig. 3E).
FIGURE 3.
Involvement of iNOS in NOD2-mediated activation of Notch1-PI3K signaling. A, PP2 abolishes MDP-triggered expression of iNOS in mouse macrophages. B, mouse macrophages derived from WT or iNOS−/− mice were treated with MDP for the indicated time periods and protein levels of NICD were analyzed by immunoblotting. C, macrophages derived from WT mice were treated with MDP, whereas that from iNOS−/− mice were either treated with MDP or with SIN-1 and expression levels of Jagged1 were analyzed by immunoblotting. D and E, reduced (D) phosphorylation of p85 and 4EBP1 and (E) expression of COX-2 upon NOD2 stimulation by MDP in macrophages derived from iNOS−/− mice in comparison to WT mice. The data are representative of three independent experiments. Med, medium.
iNOS/NO Axis Is an Essential Link in NOD2-driven Notch1 Signaling in Vivo
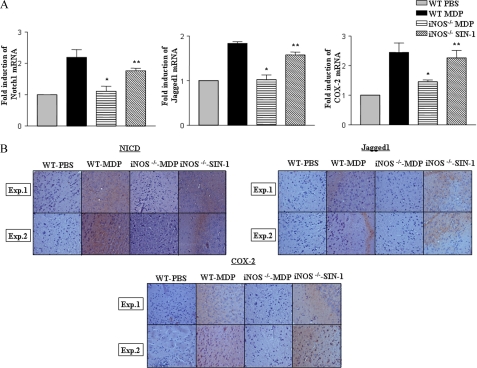
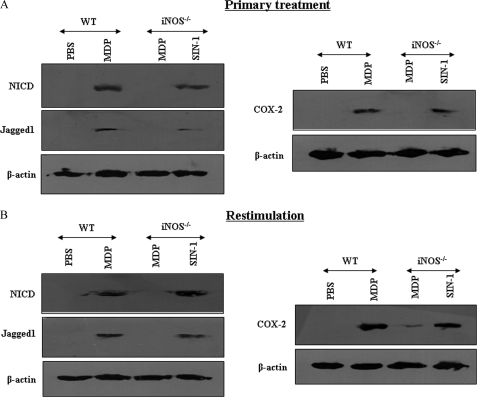
To bring relevance to the biology of NOD2 in vivo, a suggested murine model to study the central nervous system and systemic inflammation was utilized (30, 31). In this perspective, mice were challenged with MDP by an intracranial or intravenous route and attempts were made to validate the importance of iNOS in induction of Notch1 signaling in vivo. In accordance with results obtained with macrophages, iNOS deficiency in iNOS−/− mice severely compromised potential of the NOD2 agonist to trigger augmented expression of NICD, Jagged1, and COX-2 compared with WT mice as evaluated by quantitative real time PCR and immunohistochemistry based quantifications in the brain sections (Fig. 4, A and B). Importantly, NO donor (SIN-1) treatment of iNOS−/− mice in vivo rescued activation of Notch1, expression of Jagged1, as well as Notch1 target genes, COX-2 and Hes1 (Fig. 4, A and B, data not shown). Furthermore, results presented in Fig. 5A demonstrate that NOD2 engagement triggers the activation of Notch1 signaling in splenocytes derived from WT but not from iNOS−/− mice as evidenced by expression levels of NICD, Jagged1, and the Notch1 target gene, COX-2. However, intravenous inoculation of iNOS−/− mice in vivo with the NO donor (SIN-1) rescued activation of Notch1 signaling (Fig. 5A). To carry out recall experiments, mice were challenged with MDP followed by restimulation of the splenocytes ex vivo. As presented in Fig. 5B, ex vivo restimulation of NOD2 triggered the augmented expression of NICD, Jagged1, as well as COX-2. In agreement with previous results (Fig. 5A), the NO donor (SIN-1) augmented activation of Notch1 signaling in ex vivo restimulated splenocytes derived from iNOS−/− mice (Fig. 5B). These results strongly suggest that NO acts as an essential mediator of NOD2-dependent Notch1 signaling activation and is accountable for expression of specific anti-inflammatory molecular signatures like COX-2 in immune cells.
FIGURE 4.
Essential role for iNOS/NO in NOD2-triggered activation of Notch1 signaling in vivo. A and B, expression analysis of (A) Notch1, Jagged1, and COX-2 by quantitative real time PCR (mean ± S.E., n = 3) or (B) NICD, Jagged1, and COX-2 by IHC in brain sections of WT mice injected with MDP and iNOS−/− mice injected with MDP or SIN-1 intracranially. Hematoxylin (blue) was used for nuclear staining. IHC, immunohistochemistry; Exp., experiment; *, p < 0.05 versus WT MDP; **, p < 0.05 versus iNOS−/− MDP.
FIGURE 5.
Requirement of the iNOS/NO signaling axis during NOD2-mediated activation of Notch1 signaling in vivo. A, splenocytes from WT mice, injected with MDP, and iNOS−/− mice intravenously injected with MDP or SIN-1 were assessed for expression levels of NICD, Jagged1, and COX-2. B, in a recall experiment, MDP and SIN-1 augmented the expression of NICD, Jagged1, and COX-2 upon restimulation of splenocytes ex vivo from WT and iNOS−/− mice, respectively. Data are representative of two independent experiments.
iNOS/NO Is Dispensable for the NOD2-driven PKC-MAPK Signaling Axis
The onset and robust expansion of innate immune responses initiated by innate immune receptors often involves regulatory effector proteins that act either upstream or downstream of activation of MAPK (32). Interestingly, PKC is suggested to act as a critical regulatory kinase in the initiation of diverse innate immune responses (33). Furthermore, identification of signaling partners of the NOD2-iNOS-Notch1-PI3K axis that are critical in driving the expression of anti-inflammatory molecular signatures, assumes novel significance. In this perspective, we assessed the involvement of PKC and MAPK during NOD2-induced COX-2 expression. As shown in supplemental Fig. S4A, NOD2 stimulation triggered activation of PKC, ERK1/2, and p38 MAPK in a time-dependent manner. The inhibition of RIP2 kinase by PP2 abrogated the NOD2 driven activation of PKC, ERK1/2, and p38 MAPK as quantified by phosphorylation of the respective kinases (Fig. 6A). Interestingly, the pan-PKC inhibitor RO31-8220 or chelerythrine significantly blocked the ability of NOD2 to trigger COX-2 expression, signifying the important role for PKC in NOD2-driven signaling events (Fig. 6B). Additionally, NOD2 engagement by MDP markedly triggered the phosphorylation of various PKC isoforms (data not shown). To identify a role, if any, for the specific PKC isoform, we utilized well defined inhibitors for PKCα, PKCζ, PKCβ, PKCδ, and PKCϵ. As shown, utilized inhibitors markedly diminished NOD2-induced COX-2 expression, albeit to varying degrees (Fig. 6B). Expectedly, blockade of ERK1/2 or p38 MAPK, but not JNK1/2, during NOD2-driven signaling cascades, significantly reduced the NOD2-stimulated COX-2 expression (Fig. 6C). Intriguingly, NOD2-driven activation of PKC regulated activation of ERK1/2 and p38 MAPK downstream (Fig. 6D).
FIGURE 6.
Nitric oxide-dependent and -independent signaling cascades regulate NOD2-triggered COX-2 expression. A, mouse macrophages were pretreated with PP2 followed by treatment with MDP and analysis of the activation status of PKC, ERK1/2, and p38 MAPK by immunoblotting. B, pan-PKC inhibitors RO31-8220 and chelerythrine as well as isoform-specific inhibitors of PKC abolished the MDP-triggered COX-2 expression. C, analysis of the MDP-triggered COX-2 expression upon pretreatment of macrophages with ERK1/2 inhibitor, U0126; p38 MAPK inhibitor; JNK1/2 inhibitor, SP600125. D, pretreatment with the pan-PKC inhibitor chelerythrine abrogated MDP-triggered activation of ERK1/2 and p38 MAPK. E, analysis of MDP-induced activation of PKC, ERK1/2, and p38 MAPK in macrophages derived from WT and iNOS−/− mice. F, nuclear localization of p65 NF-κB in MDP-treated WT macrophages, with or without pretreatment with PP2, GSI-I, or LY294002, or chelerythrine, U0126, or p38 MAP kinase inhibitor as well as in macrophages derived from iNOS−/− mice. The blots are representative of three independent experiments. Med, medium.
As described in Figs. 3–5, we have shown that iNOS/NO acts as a key regulator of NOD2-mediated Notch1 signaling activation as well as expression of Jagged1 and the Notch1 target gene, COX-2. In this regard, we explored whether NOD2-mediated activation of PKC, ERK1/2, or p38 MAPK requires the involvement of iNOS/NO by utilizing peritoneal macrophages derived from WT and iNOS−/− mice. The data presented in Fig. 6E suggests that the ability of NOD2 to trigger phosphorylation of PKC, ERK1/2, and p38 MAPK remains unaltered in iNOS null macrophages in comparison to WT suggesting iNOS activity-independent activation of PKC or MAPK during NOD2 stimulation.
Signaling Integration through Cross-talk of iNOS-dependent and -Independent Signaling Cascades during NOD2-triggered Activation of NF-κB
Several pro- or anti-inflammatory genes' promoters including COX-2 contain many cis-acting consensus elements for NF-κB and Notch1-mediated activation of its target genes often involves the active recruitment of transcription factor NF-κB (22, 25). In this perspective, we show that NOD2 stimulation drives nuclear translocation of NF-κB in time-dependent manner with a concomitant decrease in the cytosolic levels of NF-κB (supplemental Fig. S4B). Accordingly, nuclear translocation of NF-κB was abrogated upon inhibition of NOD2 signaling by PP2 (Fig. 6F). Expectedly, interference in Notch1 signaling (GSI-I), activation of PI3K (LY294002), PKC (chelerythrine), ERK1/2 (U1026), or p38 MAPK (p38 MAP kinase inhibitor) reversed the NOD2 potential to trigger nuclear translocation of NF-κB from the cytosol (Fig. 6F). Blocking the NF-κB signaling pathway using Bay 11-7082, a specific inhibitor of IκBα phosphorylation, markedly diminished the induced expression of NOD2/Notch1 responsive molecular signature, COX-2 (supplemental Fig. S4C). Interestingly, the ability of NOD2 to trigger nuclear translocation of NF-κB was compromised in iNOS null macrophages implicating iNOS activity dependence in NOD2-mediated activation of NF-κB (Fig. 6F).
Signaling Cohorts of NOD2 Regulate Expression of Anti-inflammatory Cytokine IL-10
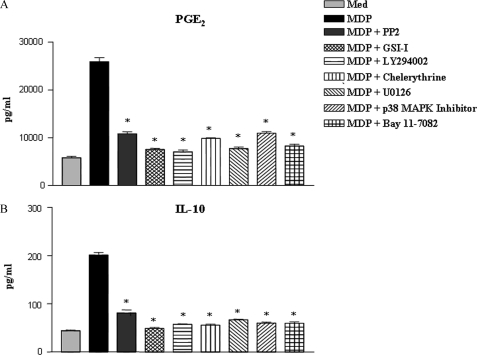
IL-10 is known to be up-regulated following Notch1 activation by Jagged1 (34). Interestingly, COX-2 is known to regulate IL-10 levels in macrophages by virtue of induced secretion of PGE2 (35). Importantly, NOD2 mutation associated with Crohn disease fails to activate IL-10 expression (10). In this regard, our data suggests that NOD2 activated the Notch1-PI3K and PKC-MAPK signaling axis integrate together to activate NF-κB, which drives the activation/expression of the factors involved in IL-10 expression. In this regard, functional activation of NOD2 in macrophages led to enhanced secretion of PGE2 (25910 ± 827 pg/ml). Furthermore, signaling perturbation experiments involving inhibition of RIP2, Notch1, PI3K, PKC, ERK1/2, p38 MAPK, and NF-κB diminished the NOD2-triggered PGE2 levels (Fig. 7A). Conversely, pharmacological inhibition of RIP2, Notch1, PI3K, PKC, ERK1/2, p38 MAPK, or NF-κB led to abrogation of NOD2-triggered IL-10 expression, although up to different levels (Fig. 7B and supplemental Fig. S5A). Accordingly, pharmacological inhibition of RIP2 kinase in bone marrow-derived dendritic cells abrogated NOD2-triggered secretion of IL-10 and PGE2 (supplemental Fig. S5, B and C).
FIGURE 7.
Critical role for NOD2-mediated activation of the Notch1-PI3K and PKC-MAPK signaling axis in anti-inflammatory responses. A and B, pretreatment of macrophages with PP2, GSI-I, or LY294002, or chelerythrine, U0126, or p38 MAP kinase inhibitor or Bay 11-7082 abrogates MDP triggered secretion of PGE2 (A) and IL-10 (B) (mean ± S.E., n = 3). Med, medium; *, p < 0.05 versus MDP.
NOD2-triggered Notch1 Signaling Attenuates TNF-α or IFN-γ-mediated Apoptosis
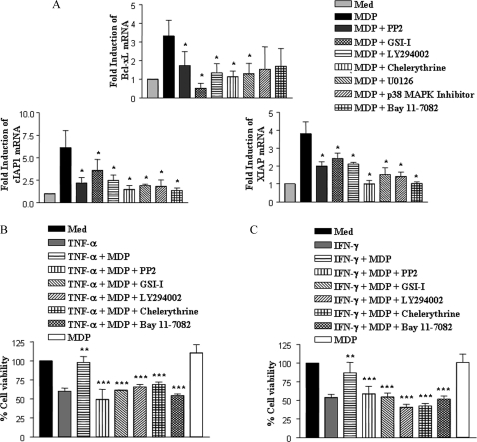
Pro-inflammatory cytokines including TNF-α and IFN-γ are known to induce macrophage cell death and tissue damage associated with various inflammatory diseases (36). In this regard, NOD2 as well as Notch1 are known to promote survival of a wide range of cellular systems (26, 27, 37). In this perspective, we explored the role of NOD2-mediated activation of Notch1 and other signaling cohorts in induced expression of a set of anti-apoptotic genes by quantitative real time PCR. As demonstrated, NOD2 stimulation triggered induced expression of Bcl-xL, cIAP1, and XIAP (Fig. 8A). However, we could not detect any enhancement in expression of another anti-apoptotic gene, Bcl2, conjecturing specificity of the system (data not shown). Signaling perturbation experiments involving inhibition of RIP2, Notch1, PI3K, PKC, and NF-κB limited the potential of NOD2 to trigger Bcl-xL, cIAP1, and XIAP expression (Fig. 8A). Conversely, inhibition of Notch1-PI3K or PKC-MAPK signaling axis-mediated activation of NF-κB compromised the ability of NOD2 to protect macrophages against TNF-α- and IFN-γ-induced cell death (Fig. 8, B and C). Together, these results eloquently suggest that NOD2-driven Notch1-PI3K activation, in conjunction with the PKC-MAPK signaling axis, exert cooperative regulation of a distinct set of effector functions in macrophages.
FIGURE 8.
Signaling integration through cross-talk of the Notch1-PI3K and PKC-MAPK axis is critical for NOD2-mediated resolution of inflammation. A, quantitative real time PCR analysis for Bcl-xL, cIAP1, and XIAP expression upon stimulation of NOD2 with MDP, with or without pretreatment with PP2, GSI-I, or LY294002 or chelerythrine, U0126, or p38 MAP kinase inhibitor or Bay 11-7082. B and C, MTT assay performed on TNF-α (B) (200 ng/ml) or IFN-γ (C) (1000 ng/ml) treated macrophages shows a significant decrease in apoptotic cells upon NOD2 stimulation with MDP. Pharmacological inhibition of NOD2 (PP2), Notch1 (GSI-I), PI3K (LY294002), PKC (chelerythrine), or NF-κB (Bay 11-7082) abrogated the suppressive effect of MDP. The data are represented as mean ± S.E. from three independent experiments. Med, medium; *, p < 0.05 versus MDP; **, p < 0.05 versus TNF-α or IFN-γ; ***, p < 0.05 versus TNF-α + MDP or IFN-γ + MDP.
DISCUSSION
Activated macrophages have long been classified as the most potent inflammatory cells in the body. However, recent reports have suggested pivotal anti-inflammatory functions of macrophages upon activation by different stimuli (38). In this regard, receptors belonging to the pattern recognition receptor family, in particular TLRs and NLRs, are critical innate molecular sensors of surreptitious entry of intruding pathogens and make a platform for initiation and execution for efficient immune responses (2). In this perspective, although TLRs are largely believed to be pro-inflammatory, NLRs like NOD2 have been implicated in regulation of anti-inflammatory responses as well as polarization of T cells toward the skewed Th2 phenotype (3, 6, 8). Intriguingly, certain disease conditions characterized by heightened pro-inflammatory responses, including Crohn disease, Blau syndrome, and chronic inflammatory bowel disease are associated with mutations in the functional NOD2 receptor, suggesting polymorphism in NOD2 predisposes the individual for a plethora of inflammatory diseases (9). However, information about the signaling cascades regulating a battery of genes associated with NOD2-mediated cellular functions is still imprecisely understood.
COX-2 has long been validated as a pro-inflammatory molecule and thus has been the target of many anti-inflammatory drugs; however, recent reports have suggested a dual role for COX-2 in the initiation as well as resolution of inflammation (39–42). It has been advocated that COX-2 is critical for production of electrophilic oxo-derivatives from the omega-3 fatty acids, which can act as peroxisome proliferator-activated receptor-γ agonist and inhibit production of pro-inflammatory cytokines (39, 41). Furthermore, 15-deoxyΔ12–14PGJ2 (15d-PGJ2), a metabolite of PGD2 and crucial regulator of anti-inflammatory responses through peroxisome proliferator-activated receptor-γ, is elevated through the COX-2 pathway in an animal model of acute inflammation (40, 42). In addition, the biosynthetic product of COX-2 activity, PGE2, is known to regulate the expression levels of IL-10 (35). IL-10, originally described as a cytokine synthesis inhibitory factor, is a pleiotropic molecule with a crucial role in preventing inflammatory and autoimmune pathologies (43). It is well established that IL-10 potently inhibits the production of soluble mediators and cytokines, which hold the capacity to activate as well as sustain inflammatory responses. By acting on innate immune cells including macrophages and dendritic cells, IL-10 inhibits the development of Th1 responses. Interestingly, IL-10-deficient mice develop disease pathologies in the murine model of inflammatory bowel disease (44). Importantly, a recent study demonstrated that COX-2−/− mice, but not wild type mice developed Crohn disease-like pathology associated with cellular infiltration, and decreased IL-10 expression in mice model of inflammatory bowel disease (45). These observations eloquently advocate a paradigm shift in understanding about the regulatory role of COX-2 during inflammation and assign novel anti-inflammatory functions to COX-2.
In the current investigation, we demonstrate that the cytoplasmic immune surveillance receptor, NOD2, is involved in initiating innate immune signaling events that are important for a diverse array of effector functions of the macrophages. We observed that NOD2 receptor engagement resulted in activation of Notch1 signaling, which required induced expression of the Notch1 ligand, Jagged1. We further demonstrate that perturbations in expression of RIP2 or TAK1, which are essentials for NOD2-mediated signaling, markedly diminishes Notch1 signaling activation, and modulates the expression of anti-inflammatory mediators like IL-10 and COX-2/PGE2 as well as suppression of Notch1 signaling-mediated attenuation of TNF-α or IFN-γ-triggered apoptosis in macrophages. These results suggest a multidimensional role of NOD2 in regulation of a variety of cellular functions. Interestingly, the ability of the NOD2 receptor to trigger robust immunomodulatory molecular signatures was associated with enhanced iNOS expression and NO production.
Several reports have suggested a critical link between NOD2 and iNOS or NO production and it has been demonstrated that NOD2 could trigger NO production in macrophages (46). Importantly, a recent report suggests that infection of RIP2 null mice with Chlamydophila pneumoniae resulted in increased mortality due to a defect in generation of effective NO production. This clearly ascribes a significant and dominant role for NOD2/RIP2 signaling in induced expression of iNOS/NO, despite the presence of the intact TLR2 signaling module in RIP2-deficient mice or TLR2 agonists in C. pneumoniae (47). NO act as a crucial molecular signal during diverse pathophysiological conditions and NO/iNOS-mediated regulation of immunomodulatory gene expression involves multiple pathways in macrophages (22). Differential levels of NO flux depending on the state of activation as well as initiation of specific signaling pathways often account for elicitation of differential macrophage responses (48). In this perspective, the local concentration and duration of NO exposure serve as significant elements in regulation of a variety of key genes involved in immune responses and macrophage transition from execution of cellular apoptosis or matrix degradation to cellular proliferation or matrix regeneration, processes required for contraction of inflammatory responses and wound healing (49). In the current study, we identified NO as a pathological link between NOD2 and Notch1 that may be pertinent to expression of key anti-inflammatory mediators like COX-2/PGE2, IL-10, or anti-apoptotic molecular signatures like Bcl-xL, XIAP, and cIAP1. Upon NOD2/RIP2 signaling activation, the expression of Jagged1 is directly regulated by NO, thus leading to positive regulation of Notch1 activation. We further demonstrate that the NOD2-driven expression of COX-2 requires Notch1-mediated recruitment of CSL/RBP-Jk and NF-κB to the COX-2 promoter. Importantly, data from in vivo challenge experiments in mice clearly implicate NO as a vital link in NOD2-triggered activation of Notch1 signaling in vivo as specified by significantly enhanced expression levels of activated Notch1, Jagged1, or COX-2 in wild type mice compared with iNOS null mice.
Cellular responses of macrophages upon activation with a wide variety of stimuli are often suggested to involve extensive cross-talk between PI3K-AKT, PKC, MAPK, and NF-κB signaling cascades (20, 22, 50). In this perspective, NOD2-triggered expression of COX-2 involved PKC-dependent activation of ERK1/2 and p38 MAPK. Interestingly, the ability of NOD2 to induce activation of PKC, ERK1/2, and p38 MAPK remained unaltered in iNOS null macrophages, conjecturing a role for iNOS/NO independent mechanisms in regulation of the PKC-MAPK signaling axis during NOD2 activation. In addition, signaling perturbation data suggest that NOD2 triggering brings signaling integration between iNOS/NO-Notch1-PI3K and the PKC-MAPK signaling axis to activate NF-κB, which plays a crucial role in regulation of a multitude of genes associated with modulation of inflammation including COX-2, IL-10, Bcl-xL, cIAP1, and XIAP.
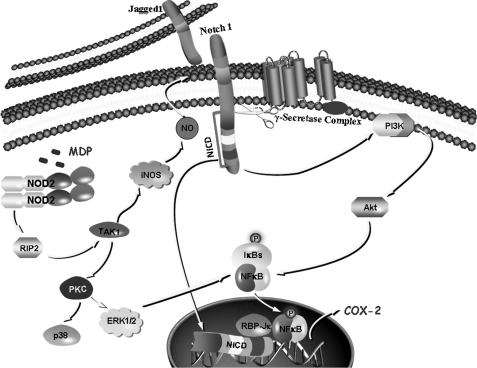
In conclusion, our study reveals that iNOS/NO is an essential link in NOD2-mediated activation of Notch1-PI3K signaling, which can modulate a defined set of macrophage effector functions. However, the PKC-MAPK signaling axis holds the capacity to effectuate NOD2-mediated cellular functions independent of iNOS activity. This integrated cross-talk of iNOS/NO, Notch1, PI3K, PKC, and MAPK downstream to NOD2 can be of importance during diverse pro-inflammatory pathological conditions like Crohn disease and inflammatory bowel disease, as these signaling cohorts regulate the expression of a multitude of genetic signatures crucial for mounting appropriate anti-inflammatory responses including COX-2, IL-10, Bcl-xL, cIAP1, and XIAP (Fig. 9). Because, prevention of exaggerated macrophage activation or stimulation of a population of anti-inflammatory macrophages holds the key to minimize pro-inflammatory pathologies; our study provides new insights into the understanding about the role of NOD2 in these pathologies and clearly paves a way toward the development of novel therapeutics.
FIGURE 9.
Model depicting NOD2-driven activation of the Notch1-PI3K and PKC-MAPK signaling integration axis.
Supplementary Material
Acknowledgments
We thank the Central Animal facility, Indian Institute of Science, for providing mice for experimentation. We thank Mainak Dasgupta and Dr. Utpal Nath for help during the course of the current investigation.
This work was supported by funds from the Indian Council of Medical Research (ICMR), the Department of Biotechnology (DBT); a collaborative grant (to the Indian Institute of Science and Karolinska Institute) from VINNOVA (The Swedish Governmental Agency for Innovation Systems) and DBT, India; the Department of Science and Technology (DST); and the Council for Scientific and Industrial Research (to K. N. B.) and by infrastructure support from ICMR (Center for Advanced Study in Molecular Medicine), DST (Fund for Improvement of Science and Technology Infrastructure in Universities and Higher Educational Institutions), and University Grants Commission (special assistance).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
- NLR
- NOD-like receptors
- COX-2
- cyclooxygenase-2
- 4EBP1
- eukaryotic initiation factor 4E-binding protein 1
- GSI-I
- γ-secretase inhibitor I
- MDP
- muramyl dipeptide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NICD
- Notch intracellular domain
- NOD
- nucleotide oligomerization domain
- RIP2
- receptor-interacting protein 2
- SIN-1
- 3-morpholinosydnonimine
- TAK1
- transforming growth factor-β-activated kinase 1
- TLR
- Toll-like receptor.
REFERENCES
- 1. Rosenberger C. M., Finlay B. B. (2003) Nat. Rev. Mol. Cell Biol. 4, 385–396 [DOI] [PubMed] [Google Scholar]
- 2. Kawai T., Akira S. (2009) Int. Immunol. 21, 317–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen G., Shaw M. H., Kim Y. G., Nuñez G. (2009) Annu. Rev. Pathol. 4, 365–398 [DOI] [PubMed] [Google Scholar]
- 4. Franchi L., Park J. H., Shaw M. H., Marina-Garcia N., Chen G., Kim Y. G., Núñez G. (2008) Cell Microbiol. 10, 1–8 [DOI] [PubMed] [Google Scholar]
- 5. Butler M., Chaudhary R., van Heel D., Playford R., Ghosh S. (2007) J. Crohns Colitis 1, 106–115 [DOI] [PubMed] [Google Scholar]
- 6. Magalhaes J. G., Fritz J. H., Le Bourhis L., Sellge G., Travassos L. H., Selvanantham T., Girardin S. E., Gommerman J. L., Philpott D. J. (2008) J. Immunol. 181, 7925–7935 [DOI] [PubMed] [Google Scholar]
- 7. Mantovani A., Sica A., Locati M. (2005) Immunity 23, 344–346 [DOI] [PubMed] [Google Scholar]
- 8. Eckmann L., Karin M. (2005) Immunity 22, 661–667 [DOI] [PubMed] [Google Scholar]
- 9. Henckaerts L., Vermeire S. (2007) Inflamm. Bowel Dis. 13, 235–241 [DOI] [PubMed] [Google Scholar]
- 10. Philpott D. J., Girardin S. E. (2009) Nat. Immunol. 10, 455–457 [DOI] [PubMed] [Google Scholar]
- 11. Fung E., Tang S. M., Canner J. P., Morishige K., Arboleda-Velasquez J. F., Cardoso A. A., Carlesso N., Aster J. C., Aikawa M. (2007) Circulation 115, 2948–2956 [DOI] [PubMed] [Google Scholar]
- 12. Maillard I., Fang T., Pear W. S. (2005) Annu. Rev. Immunol. 23, 945–974 [DOI] [PubMed] [Google Scholar]
- 13. Monsalve E., Pérez M. A., Rubio A., Ruiz-Hidalgo M. J., Baladrón V., García-Ramírez J. J., Gómez J. C., Laborda J., Díaz-Guerra M. J. (2006) J. Immunol. 176, 5362–5373 [DOI] [PubMed] [Google Scholar]
- 14. Schroeder T., Kohlhof H., Rieber N., Just U. (2003) J. Immunol. 170, 5538–5548 [DOI] [PubMed] [Google Scholar]
- 15. Bray S. J. (2006) Nat. Rev. Mol. Cell Biol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 16. Ehebauer M., Hayward P., Martinez-Arias A. (2006) Sci STKE 2006, cm7. [DOI] [PubMed] [Google Scholar]
- 17. Dumont E., Fuchs K. P., Bommer G., Christoph B., Kremmer E., Kempkes B. (2000) Oncogene 19, 556–561 [DOI] [PubMed] [Google Scholar]
- 18. Hori K., Cholewa-Waclaw J., Nakada Y., Glasgow S. M., Masui T., Henke R. M., Wildner H., Martarelli B., Beres T. M., Epstein J. A., Magnuson M. A., Macdonald R. J., Birchmeier C., Johnson J. E. (2008) Genes Dev. 22, 166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakhai H., Siveke J. T., Klein B., Mendoza-Torres L., Mazur P. K., Algül H., Radtke F., Strobl L., Zimber-Strobl U., Schmid R. M. (2008) Development 135, 2757–2765 [DOI] [PubMed] [Google Scholar]
- 20. Narayana Y., Balaji K. N. (2008) J. Biol. Chem. 283, 12501–12511 [DOI] [PubMed] [Google Scholar]
- 21. Lutz M. B., Kukutsch N., Ogilvie A. L., Rössner S., Koch F., Romani N., Schuler G. (1999) J. Immunol. Methods 223, 77–92 [DOI] [PubMed] [Google Scholar]
- 22. Bansal K., Narayana Y., Patil S. A., Balaji K. N. (2009) J. Leukocyte Biol. 85, 804–816 [DOI] [PubMed] [Google Scholar]
- 23. Maeda S., Hsu L. C., Liu H., Bankston L. A., Iimura M., Kagnoff M. F., Eckmann L., Karin M. (2005) Science 307, 734–738 [DOI] [PubMed] [Google Scholar]
- 24. Windheim M., Lang C., Peggie M., Plater L. A., Cohen P. (2007) Biochem. J. 404, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bansal K., Kapoor N., Narayana Y., Puzo G., Gilleron M., Balaji K. N. (2009) PLoS One 4, e4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beverly L. J., Felsher D. W., Capobianco A. J. (2005) Cancer Res. 65, 7159–7168 [DOI] [PubMed] [Google Scholar]
- 27. Sade H., Krishna S., Sarin A. (2004) J. Biol. Chem. 279, 2937–2944 [DOI] [PubMed] [Google Scholar]
- 28. Gutierrez A., Look A. T. (2007) Cancer Cell 12, 411–413 [DOI] [PubMed] [Google Scholar]
- 29. Ishimura N., Bronk S. F., Gores G. J. (2005) Gastroenterology 128, 1354–1368 [DOI] [PubMed] [Google Scholar]
- 30. Goralski K. B., Abdulla D., Sinal C. J., Arsenault A., Renton K. W. (2005) Am. J. Physiol. Gastrointest. Liver Physiol. 289, G434–443 [DOI] [PubMed] [Google Scholar]
- 31. Nieuwenhuijzen G. A., Haskel Y., Lu Q., Berg R. D., van Rooijen N., Goris R. J., Deitch E. A. (1993) Ann. Surg. 218, 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dong C., Davis R. J., Flavell R. A. (2002) Annu. Rev. Immunol. 20, 55–72 [DOI] [PubMed] [Google Scholar]
- 33. Masek K. S., Fiore J., Leitges M., Yan S. F., Freedman B. D., Hunter C. A. (2006) J. Cell Sci. 119, 4565–4573 [DOI] [PubMed] [Google Scholar]
- 34. Sharma V. M., Calvo J. A., Draheim K. M., Cunningham L. A., Hermance N., Beverly L., Krishnamoorthy V., Bhasin M., Capobianco A. J., Kelliher M. A. (2006) Mol. Cell. Biol. 26, 8022–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Betz M., Fox B. S. (1991) J. Immunol. 146, 108–113 [PubMed] [Google Scholar]
- 36. Kimura M., Haisa M., Uetsuka H., Takaoka M., Ohkawa T., Kawashima R., Yamatsuji T., Gunduz M., Kaneda Y., Tanaka N., Naomoto Y. (2003) Cell Death Differ. 10, 718–728 [DOI] [PubMed] [Google Scholar]
- 37. Rahman M. K., Midtling E. H., Svingen P. A., Xiong Y., Bell M. P., Tung J., Smyrk T., Egan L. J., Faubion W. A., Jr. (2010) J. Immunol. 184, 7247–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang X., Mosser D. M. (2008) J. Pathol. 214, 161–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen C. (2010) Nat. Chem. Biol. 6, 401–402 [DOI] [PubMed] [Google Scholar]
- 40. Gilroy D. W., Colville-Nash P. R., Willis D., Chivers J., Paul-Clark M. J., Willoughby D. A. (1999) Nat. Med. 5, 698–701 [DOI] [PubMed] [Google Scholar]
- 41. Groeger A. L., Cipollina C., Cole M. P., Woodcock S. R., Bonacci G., Rudolph T. K., Rudolph V., Freeman B. A., Schopfer F. J. (2010) Nat. Chem. Biol. 6, 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seibert K., Lefkowith J., Tripp C., Isakson P., Needleman P. (1999) Nat. Med. 5, 621–622 [DOI] [PubMed] [Google Scholar]
- 43. Harizi H., Gualde N. (2006) Cell Mol. Immunol. 3, 271–277 [PubMed] [Google Scholar]
- 44. Saraiva M., O'Garra A. (2010) Nat. Rev. Immunol. 10, 170–181 [DOI] [PubMed] [Google Scholar]
- 45. Watanabe J., Lin J. A., Narasimha A. J., Shahbazian A., Ishikawa T. O., Martin M. G., Herschman H. R., Reddy S. T. (2010) Am. J. Physiol. Gastrointest. Liver Physiol. 298, G842–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tötemeyer S., Sheppard M., Lloyd A., Roper D., Dowson C., Underhill D., Murray P., Maskell D., Bryant C. (2006) J. Immunol. 176, 4804–4810 [DOI] [PubMed] [Google Scholar]
- 47. Shimada K., Chen S., Dempsey P. W., Sorrentino R., Alsabeh R., Slepenkin A. V., Peterson E., Doherty T. M., Underhill D., Crother T. R., Arditi M. (2009) PLoS Pathog. 5, e1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Duffield J. S. (2003) Clin. Sci. 104, 27–38 [DOI] [PubMed] [Google Scholar]
- 49. MacMicking J., Xie Q. W., Nathan C. (1997) Annu. Rev. Immunol. 15, 323–350 [DOI] [PubMed] [Google Scholar]
- 50. Narayana Y., Bansal K., Sinha A. Y., Kapoor N., Puzo G., Gilleron M., Balaji K. N. (2009) Mol. Immunol. 46, 2947–2954 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.