Abstract
The molecules involved in vertebrate tendon formation during development remain largely unknown. To date, only two DNA-binding proteins have been identified as being involved in vertebrate tendon formation, the basic helix-loop-helix transcription factor Scleraxis and, recently, the Mohawk homeobox gene. We investigated the involvement of the early growth response transcription factors Egr1 and Egr2 in vertebrate tendon formation. We established that Egr1 and Egr2 expression in tendon cells was correlated with the increase of collagen expression during tendon cell differentiation in embryonic limbs. Vertebrate tendon differentiation relies on a muscle-derived FGF (fibroblast growth factor) signal. FGF4 was able to activate the expression of Egr genes and that of the tendon-associated collagens in chick limbs. Egr gene misexpression experiments using the chick model allowed us to establish that either Egr gene has the ability to induce de novo expression of the reference tendon marker scleraxis, the main tendon collagen Col1a1, and other tendon-associated collagens Col3a1, Col5a1, Col12a1, and Col14a1. Mouse mutants for Egr1 or Egr2 displayed reduced amounts of Col1a1 transcripts and a decrease in the number of collagen fibrils in embryonic tendons. Moreover, EGR1 and EGR2 trans-activated the mouse Col1a1 proximal promoter and were recruited to the tendon regulatory regions of this promoter. These results identify EGRs as novel DNA-binding proteins involved in vertebrate tendon differentiation by regulating type I collagen production.
Keywords: Collagen, Connective Tissue, Development, Differentiation, Extracellular Matrix, Limb, Chick Embryo, Mouse Embryo, Tendon
Introduction
Vertebrate tendons are specialized dense connective tissues mainly composed of collagens that connect muscle to bone. Tendon repair following injuries or during aging is a clinical challenge because tendons are only repaired slowly and partially. The establishment of new strategies for tendon repair is prevented by a limited understanding of tendon development and consequently awaits a better understanding of tendon development. An important goal is to understand the molecular mechanisms involved in regulating collagen expression, production, and assembly during tendon development to be able to design new therapies for tendon repair.
Tendons consist of elongated fibroblasts named tenocytes that produce an abundant extracellular matrix composed of >90% collagen. The major collagen component of mature tendon is type I collagen, a heterotrimeric molecule composed of two α1 chains and one α2 chain that are genetically distinct and encoded by Col1a1 and Col1a2, respectively (1–3). Type I collagen molecules self-assemble into highly organized parallel collagen fibrils and then into fibers, which provide the tensile strength of tendon. Other collagens, such as the fibrillar collagens III and V and the nonfibrillar collagens named FACITs (fibril-associated collagens with interrupted triple helices), and collagens XII and XIV are important for collagen fibril formation, growth, and integrity of tendons. In addition to collagens, tendons contain various matrix components contributing to the properties of tendons such as proteoglycans, elastic matrix components, and glycoproteins. Most of the available mice deficient for these tendon components display abnormal collagen structure in tendons but also in other tissues (reviewed in Refs. 1–4). Tendon limb progenitors start to differentiate into tenocytes by synthesizing specific components of the extracellular matrix, at embryonic day 7 (E7)5 in chick and at E14.5 in mouse limbs (5–7). The molecular mechanisms responsible for the differentiation of mesenchymal cells into tenocytes are largely unknown. Similarly, the molecules involved in the regulation of collagen genes during embryonic tendon development remain to be identified. It is notable that very few DNA-binding proteins have been identified as being involved in tendon formation. The bHLH transcription factor scleraxis (SCX) is expressed in the tendon progenitor and differentiated cells and has been shown to be involved in embryonic tendon differentiation (7, 8). Scx is required for the embryonic expression of Col14a1 and tenomodulin of the force-transmitting and intermuscular tendons (7, 9, 10). In addition, SCX is also involved in the regulation of the tendon-specific activity of the mouse Col1a1 promoter (11). Recently, the Mohawk (Mkx) homeobox gene has been shown to be involved in tendon formation, by regulating type I collagen production (12, 13).
Tendons have the same embryological origin as cartilage and bone in vertebrates. Limb tendons originate from the lateral plate as cartilage and bone. Classical studies in avian embryos and analysis of muscleless limbs from Pax3 mutant mice have highlighted that limb tendons initiate their development independently of muscles, but muscles are required for further tendon development (8, 14–17). Interestingly, similar interactions between tendons and muscles are observed in Drosophila (18, 19). Although Drosophila tendon cells differ in their ectodermal embryonic origin when compared with the mesodermal origin of vertebrate tendon cells, Drosophila tendon cells are specified independently of muscle cells, but muscle cells are required for further tendon cell differentiation (18, 20). Drosophila tendon precursor cells are characterized by the expression of the transcription factor Stripe, an Egr (early growth response)-like transcription factor. Analysis of loss- and gain-of-function Stripe mutant phenotypes shows that Stripe is a key regulator of tendon cell specification and differentiation (20–23). Stripe shows sequence homologies with members of the vertebrate Egr family of transcription factors, including Egr1, Egr2, Egr3, and Egr4. No tendon phenotype has been reported to date in any of the Egr mutant mice (see Refs. 24–26 for recent references). Based on the Stripe requirement for Drosophila tendon assembly, we investigated the involvement of Egr1 and Egr2 genes in vertebrate tendon formation.
EXPERIMENTAL PROCEDURES
Chick Embryos and Mouse Lines
Fertilized chick eggs from commercial sources were incubated at 38 °C. Chick embryos were staged according to Hamburger and Hamilton (HH) stages (27), before E4 and then to days in ovo. Scx-GFP (28), Egr1−/− (29), Egr2−/−, or Egr2lacZ/+ (30) mouse embryos were collected after natural overnight matings. For staging, fertilization was considered to take place at midnight.
FACS of Mouse Limb Tendon Cells
Hind limbs and forelimbs from E11.5, E12.5, and E14.5 Scx-GFP embryos (28) were collected and dissociated with trypsin to obtain cell suspensions. Cell suspensions were subjected to FACS at room temperature using a MoFlo® XDP Flow Cytometer (Dako; with state laser, 488 nm) with Dako-Moflo Summit software or using a VantageTM Se option DiVa flow cytometer (laser, 488 nm). The GFP-positive fractions were collected in 2 mm PBS-EDTA supplemented with 20% of fetal calf serum.
RNA Isolation, Reverse Transcription, and Quantitative Real-time PCR
Total RNAs was extracted from FACS-sorted limb tendon cells or from tail tendons of E18.5 mutant and control mice using the RNeasy mini kit (Qiagen) and reverse-transcribed using the High Capacity Retrotranscription kit (Applied Biosystems) according to the manufacturer's instructions. Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems). Primer efficiencies were comprised between 95 and 103%, and for the primers that do not span introns, we performed control PCR minus RT to exclude genomic contamination of the cDNA samples. All primers were used at a concentration of 400 nm. Primer sequences for real-time PCR are listed in Table 1. The results were expressed as mRNA levels corrected for GAPDH levels in each sample. For mRNA level analyses of limb tendon cells, three independent RNA samples originating from three independent mouse litters for each embryonic stage were analyzed at least in triplicate. RNA quantity was extrapolated from the standard curve by the StepOnePlusTM real-time PCR software (Applied Biosystems). For mRNA level analyses in embryonic tail tendons, gene expression changes were quantified using the ΔΔCT method. The experiment was performed with eight independent samples for each genotype, and PCR was carried out in triplicate for each gene. Quantitative data shown as histograms are expressed as means and S.E. The fold changes between E11.5 and E12.5, between E12.5 and E14.5, between E11.5 and E14.5, or between wild-type and mutant mice stages were assessed for statistical significance by Student's t test. Asterisks in figures indicate the different p values (*, <0.05; **, <0.01; and ***, <0.001).
TABLE 1.
Primers used for quantitative real-time PCR
| Gene | Forward primer | Reverse primer | Accession no. |
|---|---|---|---|
| Col1a1 | 5′-tggagagagcatgaccgatg-3′ | 5′-GAGCCCTCGCTTCCGTACT-3′ | NM_007742 |
| Col1a2 | 5′-CCAGCGAAGAACTCATACAGC-3′ | 5′-GGACACCCCTTCTACGTTGT-3′ | NM_007743 |
| Col3a1 | 5′-CTAAAATTCTGCCACCCCGAA-3′ | 5′-AGGATCAACCCAGTATTCTCCACTC-3′ | NM_009930 |
| Col5a1 | 5′-CCTGGCATCAACTTGTCCGATGG-3′ | 5′-GTGGTCACTGCGGCTGAGGAACTTC-3′ | NM_015734 |
| Col12a1 | 5′-CCGTGTTGTGTATCGCCCT-3′ | 5′-CACCTTAGCAACCATCTGCCTC-3′ | NM_007730 |
| Col14a1 | 5′-GAGCAGAGACCACATTGGCC-3′ | 5′-CGTACAGCTCGAGGTCGGAA-3′ | NM_181277 |
| Egr1 | 5′-CAGCGCCTTCAATCCTCAAG-3′ | 5′-GCGATGTCAGAAAAGGACTCTGT-3′ | NM_007913 |
| Egr2 | 5′-AGGCCCCTTTGACCAGATG-3′ | 5′-GTCCGTGAGAAGGTGGGACA-3′ | NM_010118 |
| Scx | 5′-CCTTCTGCCTCAGCAACCAG-3′ | 5′-GGTCCAAAGTGGGGCTCTCCGTGACT-3′ | NM_198885.3 |
| Tnmd | 5′-AACACTTCTGGCCCGAGGTAT-3′ | 5′-AAGTGTGCTCCATGTCATAGGTTTT-3′ | NM_022322.2 |
| Mohawk | 5′-AGTAAAGACAGTCAAGCTGCCACTG-3′ | 5′-TCCTGGCCACTCTAGAAGCG3′ | NM_177595 |
| GAPDH | 5′-TTGTGGAAGGGCTCATGACC-3′ | 5′-TCTTCTGGGTGGCAGTGATG-3′ | NM_008084 |
FGF4 Bead Implantation
Human FGF4 recombinant protein was obtained from R&D Systems. Heparin beads were washed in PBS and soaked in 500 ng/ml of FGF4 for 1 h on ice. FGF4 or PBS beads were grafted into the right wings of normal embryos at E5. Four to 48 h after grafting, embryos were harvested and processed for in situ hybridization to tissue sections.
Somite and Neural Tube Electroporation
Embryos were co-electroporated in lateral somite at interlimb level at HH15 (31) or in the neural tube at limb level at HH13 (32) with pCAβ-GFP, a vector encoding the green fluorescent protein under the CMV-β-actin promoter and expression vectors encoding mouse Egr1 or Egr2 under the CMV-β-actin promoter (pCAβ-Egr1, pCAβ-Egr2). These expression vectors were used at 1 mg/ml (with the ratio 0.2 mg/ml pCAβ-GFP, 0.8 mg/ml pCAβ-Egr1/2). Embryos were allowed to develop for one or 2 days and processed for in situ hybridization to paraffin-embedded tissue sections or immunohistochemistry.
In Situ Hybridization, Immunohistochemistry, and Detection of β-Galactosidase Activity to Tissue Sections
Limbs from normal or manipulated chick embryos and limbs from mouse wild type or mutant embryos were fixed in Farnoy (60% (v/v) ethanol 100%, 30% (v/v) formaldehyde 40%, 10% (v/v) acetic acid) and processed for in situ hybridization to 8-μm wax tissue sections as described previously (17). The digoxigenin-labeled mRNA probes were used as described: mouse Egr1 (29), chick and mouse Egr2 (33), chick and mouse Scx (17), mouse Col1a1 (34), Col5a1 (35), and mouse Col12a1 (7). The probes for chick Egr1, Col1a1, Col3a1, Col5a1, Col12a1, and Col14a1 originate from the UMIST EST library (36). Differentiated muscle cells and nerves were detected on sections using the monoclonal antibodies MF20 or HNK1, respectively (Developmental Hybridoma Bank). The β-galactosidase activity was assessed on limb sections from E11.5 to E18.5 Egr2lacZ/+ mouse embryos as described (30). Chick collagen I protein was detected using polyclonal antibodies (Novotec). For type I collagen immunohistochemistry, chick embryos were fixed in 4% PFA and processed for cryostat sections.
Transmission Electronic Microscopy
The flexor digitorum longus tendons from hindlimbs of E18.5 wild type, Egr1−/−, or Egr2−/− mice were dissected and fixed in 2% glutaraldehyde, 2.5% paraformaldehyde in 0.2 m sodium cacodylate, pH 7.4. After post-fixation in 1% osmium tetraoxide for 1 h at room temperature, tendons were dehydrated in graded series of ethanol and embedded in Epoxy resin. Ultrathin sections were contrasted with uranyl acetate and lead citrate and examined with a Philips CM 120 electron microscope equipped with a GATAN ORIUS 200 2,000 × 2,000 digital camera. Two (Egr1) and three (Egr2) different mouse groups (including mutants and controls) were used for collagen fibril counting. A total of 50 section areas from each sample were analyzed. Measurements of collagen fibril number were performed using the ImageJ software. Quantitative data shown as histograms are expressed as means and S.E. The differences between control and mutant samples were assessed for statistical significance by Student's t test. Asterisks in figures indicate ***, p < 0.001.
Col1a1 Promoter Activity Analysis
The Egr1 and Egr2 expression plasmids (CMV-Egr1 and CMV-Egr2) were obtained by cloning the coding sequences for mouse Egr1 and Egr2 under the control of the CMV promoter. The pJ320 reporter construct contains a segment extending from −3.150 kb to +0.11 kb of the Col1a1 proximal promoter cloned upstream the lacZ gene in the pLacH plasmid (34). Transfection experiments were performed as previously (11) described. NIH/3T3 cells (ATCC-LGC Promochem) were transiently co-transfected with 900 ng of pJ320, 100 ng of control pGL3 vector (expressing the luciferase reporter gene under the control of the SV40 promoter) and various amounts of CMV-Egr1 or CMV-Egr2 expression vectors or of CMV-control. 48 h after transfection, cells were lysed, and the β-galactosidase and luciferase activities were assessed as described previously (11). All of the transfection experiments were done in triplicate and repeated at least three times. β-Galactosidase activities were normalized to luciferase activities to compensate for variations in transfection efficiency. Results are presented as fold activation of the relative β-galactosidase activities over the CMV control.
Chromatin Immunoprecipitation Assays
ChIP assays were performed as previously reported (37). Eighty hind limbs from E18.5 mouse embryos were homogenized using a mechanical disruption device (Lysing Matrix A, Fast Prep MP1, 3 × 30 s). 8 μg of each antibody, rabbit polyclonal anti-EGR1 antibody (C-19, Santa Cruz Biotechnology), anti-EGR2 antibody (Covance, Denver, PA, USA), or anti-acetylated histone H4 antibody (Upstate Biotechnology) as a positive control were used to immunoprecipitate 25 μg of sonicated chromatin. ChIP products were analyzed by PCR. Two pairs of primers were used to amplify fragments associated with tendon regulatory regions of the mouse Col1a1 proximal promoter, previously characterized (11, 34, 38). The reverse (5′-aaatgtagcaggtagggcac-3′) and forward (5′-atggcctttactccagacct-3′) primers were used to amplify a 230-base fragment (from −3.08 kb to −2.85 kb) of the Col1a1 promoter, flanking the TSE1 and TSE2 elements (11). The reverse (5′-cgtcaggaaagggtcatctg-3′) and forward (5′-aaaactagctcagggagggc-3′) primers were used to amplify a 330-base fragment (from −1.51 kb to −1.18 kb), included in the tendon-specific deletion region (1.537 kb to −0.22 kb).
RESULTS
Egr1 and Egr2 Expression Correlates with Increase of Tendon-associated Collagen Expression during Limb Tendon Differentiation
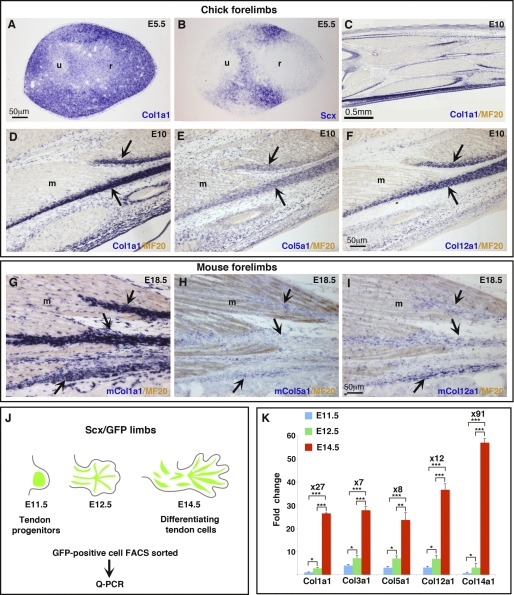
During vertebrate limb development, tendon cell differentiation is dependent on muscle and occurs in parallel to spatial arrangement of tendon primordia into individual tendons (reviewed in Ref. 4). The muscle-dependent phase of tendon formation has been established to initiate after E12.5 and E6 in mouse and chick limbs, respectively, based on the presence of Scx expression in E12.5 mouse and E6 chick muscleless limbs and its subsequent disappearance after these stages (8, 16, 17, 39). Before tendon differentiation and individualization, the main tendon collagen, Col1a1 was expressed ubiquitously in chick limb tissues (Fig. 1A), whereas the reference tendon marker Scx displayed an expression in dorsal and ventral limb regions (Fig. 1B). It is assumed that Scx early expression in dorsal and ventral limb regions labels tendon progenitors (7, 8, 40). When tendons are well individualized, Col1a1 expression is clearly enhanced in chick and mouse tendons, in addition to be expressed in other limb tissues such as muscle connective tissue cells, blood vessels, and dermis (Fig. 1, C, D, and G). Tendon-associated collagens, for example as Col5a1 and Col12a1, also label chick or mouse limb tendons (Fig. 1, E, F, H, and I). To analyze the expression levels of collagens and Egr genes specifically in tendon cells before and after the initiation of tendon differentiation process, we had to sort out tendon cells. We took advantage of the Scx-GFP mouse line (28). We isolated GFP-positive cells by FACS from limbs of Scx-GFP mouse line at different embryonic stages: E11.5 (tendon progenitors), E12.5 (transitory stage), and E14.5 (differentiating tendon cells) (Fig. 1J). Quantitative analysis of Col1a1 expression showed a dramatic 27-fold increase between E11.5 (progenitors) and E14.5 (tenocytes) (Fig. 1K), although a significant 2.7-fold increase of Col1a1 transcript levels was observed between E11.5 and E12.5 and a much greater increase of 10-fold increase occurred between E12.5 and E14.5 (Fig. 1K). This suggested that the initiation of Col1a1 increase was initiated between E11.5 and E12.5. The tendon-associated collagens, Col3a1, Col5a1, Col12a1, and Col14a1, followed similar expression patterns. They all displayed a massive increase of expression between E11.5 and E14.5 ranging from 7-fold for Col3a1 to 91-fold for Col14a1, and smaller but significant increases between E11.5 and E12.5 (Fig. 1K). This quantitative analysis of collagen expression in mouse limb tendon cells showed that tendon collagen expression increases dramatically between E11.5 and E14.5. The timing of the collagen expression increase is fully consistent with the timing of the muscle-dependent phase of tendon cell differentiation.
FIGURE 1.
Collagen expression during embryonic tendon development. A and B, adjacent transverse sections from E5.5 chick wings were hybridized with the digoxigenin-labeled antisense probes (blue) for Col1a1 (A) and Scx (B). Longitudinal sections from E10 chick forelimbs (C–F) and E18.5 mouse forelimbs (G–I) were hybridized with digoxigenin-labeled antisense probes (blue) for Col1a1 (C, D, and G), Col5a1 (E and H), and Col12a1 (F and I) and then incubated with the MF20 antibody (light brown) to visualize muscles. J, GFP+ cells were FACS-sorted from limbs of Scx/GFP transgenic mice at different embryonic stages E11.5, E12.5, and E14.5. K, quantitative real-time PCR (q-PCR) analyses of collagen mRNA levels in FACS-sorted tendon cells at different embryonic stages. Histograms, normalized for GAPDH, represent the means and S.E. of triplicate determination from three different samples of isolated tendon cells at each stage. Expression levels were normalized to that at E11.5. Fold changes between E11.5 and E14.5 are mentioned for each gene. Asterisks in the histograms indicate the different p values *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
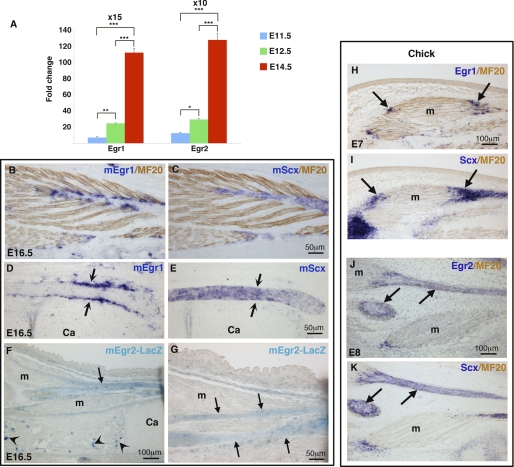
Quantitative analysis of Egr1 and Egr2 expression in tendon cells showed that Egr1 and Egr2 expression significantly increased between the stage of tendon progenitors (E11.5) and the stage of differentiating tendon cells (E14.5), 15-fold for Egr1 and 10-fold for Egr2 (Fig. 2A). These results indicated that Egr1 and Egr2 expression level followed that of collagens during tendon cell differentiation (Fig. 1, J and K, and 2A). In situ hybridization experiments confirmed that mEgr1 transcripts were first observed at E12.5 in Scx domains in forming tendons (supplemental Fig. S1, A and B). At E16.5, when tendons are fully individualized, mEgr1 transcripts display a restricted expression at the tendon attachment close to muscles (Fig. 2, B and C) (41) and delineate long tendons (Fig. 2, D and E). The mouse Egr2 probe failed to provide any significant signal in mouse limbs in our hands. We therefore analyzed Egr2 expression in Egr2lacZ/+ mice (30). lacZ (Egr2) was first observed in E14.5 limb tendons (data not shown) and displayed a general expression in all limb tendons at E16.5 (Fig. 2, F and G). In chick limbs, the onset of Egr1 and Egr2 expression was observed in differentiating tendons at E7 (Fig. 2, H–K, shown at E8 for Egr2). Similarly to the mouse limb situation, chick Egr2 was expressed in the whole tendon, whereas Egr1 displayed a more restricted expression at the tendon attachments close to muscles (Fig. 2, H–K). In addition, chick Egr1 transcripts were delineating long tendons in chick limbs (supplemental Fig. S2, A and B) as mEgr1 (Fig. 2, D and E). Egr1 displayed multiple other sites of expression in chick limbs and was also observed in subregions of axial tendons in E8 chick embryos (supplemental Fig. S2, C–H). It is worthy to note that Egr2 expression in limb tendons was transient, as its expression was lost at E14 in chick limb tendons and at E18 in EgrlacZ/+ mouse limb tendons (supplemental Fig. S3 and data not shown). No signal was detected using mouse Egr3 or Egr4 probes in tendon cells of E12.5 and E18.5 mouse limbs (supplemental Fig. S1).
FIGURE 2.
Expression of Egr1 and Egr2 in embryonic limb tendons. A, quantitative real-time PCR analyses of Egr1 and Egr2 mRNA levels in FACS-sorted tendon cells at different embryonic stages. Histograms, normalized for GAPDH mRNAs, represent the means and S.E. of triplicate determination from three different samples of isolated tendon cells at each stage. Expression levels were normalized to that at E11.5. Fold changes between E11.5 and E14.5 are mentioned for each gene. Asterisks in the histograms indicate the different p values *, p < 0.05; **, p < 0.01; and ***, p < 0.001. B–E, fore limbs of E16.5 mouse embryos were sectioned longitudinally. Adjacent sections were hybridized with the digoxigenin-labeled antisense probes (blue) for Egr1 (B and D) or Scx (C and E) and then incubated with the MF20 antibody (brown) to visualize muscles (B and C). Arrows in D and E point the Egr1 expression delineating the Scx expression domain in a long tendon. F and G, longitudinal sections of fore limbs from E16.5 Egr2lacZ/+ mouse embryos were stained with X-Gal for β-galactosidase activity. The arrows in F and G point to the lacZ staining in tendons. Arrowheads in F indicate Egr2 expression in bone. Chick wings of E7 (H and I), E8 (J and K) embryos were sectioned longitudinally. Adjacent sections were hybridized with the digoxigenin-labeled antisense probes (blue) for chick Egr1 (H) and Scx (I) or Egr2 (J) and Scx (K) and then incubated with the MF20 antibody (brown) to visualize muscles. Arrows in H and I indicate Egr1 and Scx expression in forming tendons. Arrows in J and K indicate Egr2 and Scx expression in forming tendons. m, muscle, Ca, cartilage.
Altogether, these expression data showed that Egr1 and Egr2 are expressed in embryonic tendons when they are individualized and that Egr expression is concomitant with the increase of collagen expression in differentiating tendon cells. We conclude that Egr1/2 expression is associated with embryonic limb tendon differentiation.
Expression of Egr1, Egr2, and Tendon Collagens Is Positively Regulated by FGF4
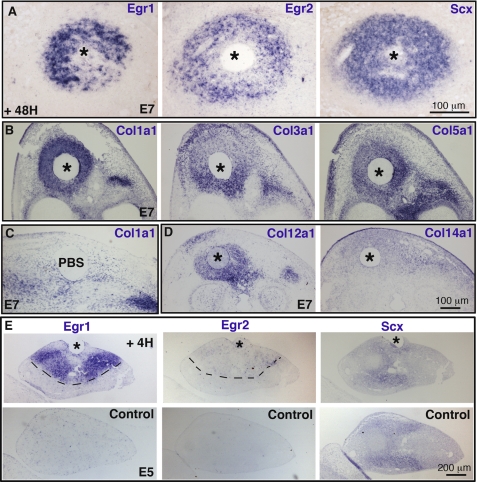
Chick tendon cell differentiation relies on a FGF muscle-derived signal (42–44). FGF4 expressed at the tips of limb and axial muscles close to tendons is a candidate for being a muscle signal involved in limb tendon differentiation (16, 43). To determine whether FGF4 could activate the expression of Egr genes, FGF4 beads were implanted into chick limbs before the onset of tendon differentiation (and of Egr1/2 endogenous expression), and Egr expression was analyzed by in situ hybridization. The expression of both Egr genes was activated by FGF4, 48 h after grafting as that of tendon reference marker, Scx (Fig. 3A). Application of FGF4 beads also induced the ectopic expression of the tendon-associated collagens, Col1a1, Col3a1, Col5a1, and Col12a1, and weakly induced the expression of Col14a1 (Fig. 3, B–D), strengthening the involvement of the FGF signal in tendon cell differentiation. We next determined the timing of induction of the Egr genes after FGF4 application and compared it with that of other tendon markers. Ectopic Egr transcripts could be detected as early as 4 h after FGF4 bead implantation, whereas Scx expression did not appear to change (Fig. 3E). The ectopic expression of Col1a1 and Col12a1 was observed 24 h after FGF4 application (Table 2). These results showed that Fgf4 activated the expression of a large variety of tendon differentiation markers and that induction of Egr expression was one of the earliest events, occurring before the induction of tendon-associated collagens.
FIGURE 3.
FGF4 activates the expression of Egr1, Egr2, and tendon-associated collagens. FGF4 (A, B, D, and E) or PBS (C) beads were implanted into the dorsal regions of chick wings of E5 embryos, and the embryos were fixed 48 h later (A–D) or 4 h later (E). Consecutive sections of the FGF4- or PBS-manipulated wings 48 h (A–D) or 4 h (E) after grafting and of the corresponding control left wings (E) were hybridized with Egr1 (A and E), Egr2 (A and E), Scx (A and E), Col1a1 (B and C), Col3a1 (B), Col5a1 (B), Col12a1 (D), and Col14a1 (D) probes. Adjacent sections are grouped accordingly. The asterisks indicate the positions of FGF4 beads in the grafted wings.
TABLE 2.
Activation of tendon gene expression by in situ hybridization following FGF4 bead implantation to chick limb buds
+, activation of ectopic expression; +/−, slight activation of ectopic expression; −, no detectable expression; n.d., not determined.
| Tendon markers | Time after FGF4 bead implantation |
||||
|---|---|---|---|---|---|
| 4 h | 6 h | 12 h | 24 h | 48 h | |
| Egr1 | + | + | + | + | + |
| Egr2 | +/− | + | + | + | + |
| Scx | +/− | + | + | + | + |
| Col1a1 | − | − | − | + | + |
| Col3a1 | n.d. | n.d. | n.d. | n.d. | + |
| Col5a1 | n.d. | n.d. | n.d. | n.d. | + |
| Col12a1 | n.d. | n.d. | − | + | + |
| Col14a1 | n.d. | n.d. | n.d. | n.d. | +/− |
Egr1 or Egr2 Is Sufficient for Tendon Marker Expression
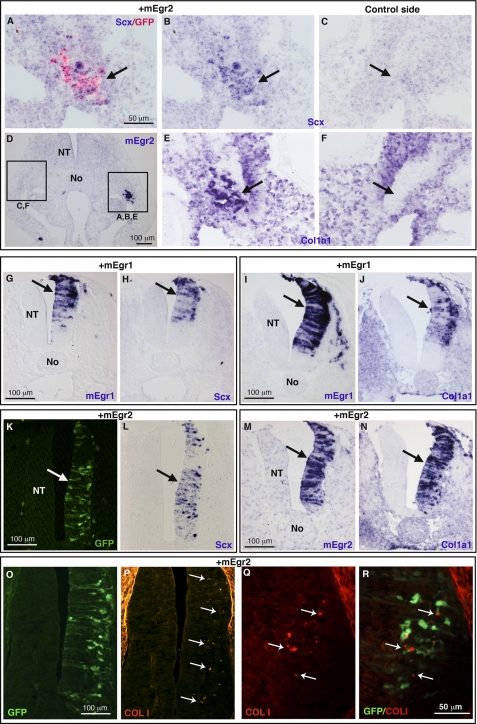
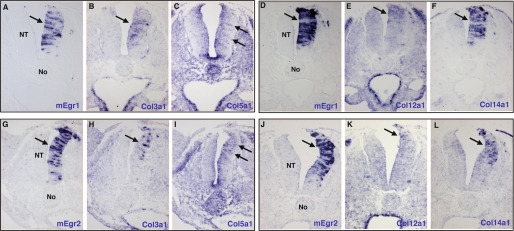
We next determined whether Egr genes were sufficient for tendon marker expression. Because Scx and Col1a1 displayed a wide expression in limbs before tendons are formed (Fig. 1, A and B), we tested the ability of Egr genes to activate tendon-specific targets, by overexpressing the Egr1 or Egr2 gene in another mesoderm-derived tissue, the muscle lineage, which is Scx- and Col1a1-negative. We targeted axial muscle cells using somite electroporation (31). Overexpression of mouse Egr1 or Egr2 gene in lateral somites induced ectopic expression of Scx and Col1a1 (shown for Egr2, Fig. 4, A–F, arrows, data not shown for Egr1). We next determined whether either Egr gene was sufficient for tendon gene expression in an unrelated embryonic tissue derived from the ectoderm, the neural tube. The developing neural tube does not express Scx or Col1a1 (supplemental Fig. S4, B and H) and can also be targeted by electroporation (32). Forced expression of a GFP control vector did not induce any tendon gene expression in neural tubes (supplemental Fig. S4). Forced expression of either mouse Egr1 or Egr2 induced ectopic expression of Scx and Col1a1 genes in the right sides of neural tubes (Fig. 4, G–N), the left sides served as internal controls. In addition, small patches of ectopic type I collagen protein was observed 48 h after mEgr2-forced expression (Fig. 4, O–R). The transience of gene misexpression with this technique precluded further analysis of collagen production in these experimental conditions. However, forced expression of mouse Egr1 or Egr2 activated the expression of the other tendon-associated collagens Col3a1, Col5a1, Col12a1, and Col14a1 24 h after electroporation (Fig. 5). These experiments showed that either Egr gene was sufficient for the expression of Scx and tendon-associated collagens. We conclude that either Egr gene has the ability to induce de novo expression of a large variety of tendon differentiation genes in ectopic contexts. This property was valid in both mesoderm- and ectoderm-derived tissues.
FIGURE 4.
Forced expression of mEgr1- or mEgr2-induced Scx and Col1a1 expression in mesoderm- and ectoderm-derived tissues. A–F, the hypaxial lips of dermomyotomes (mesoderm-derived) were electroporated with mEgr2 and GFP constructs in HH15 chick embryos. Ectopic gene expression was detected either with GFP protein (A) or mEgr2 transcripts (D), 24 h after electroporation. In regions where ectopic mEgr2 is observed, Scx and Col1a1 expression was up-regulated in lateral somites (B and E, arrows) compared with the equivalent somites of the non-electroporated sides (C and F, arrows). G–R, the neural tubes (ectoderm-derived) were electroporated with mEgr1 (G–J) or mEgr2 (K–R) constructs in HH13 chick embryos. mEgr1 or mEgr2 induced the ectopic expression of Scx (G, H, K, and L) and Col1a1 (I, J, M, N) in the right sides of neural tubes, 24 h after electroporation. O–R, 48 h after electroporation, spots of collagen I protein were observed (white arrows in P–R) adjacent to ectopic mEgr2 visualized by the GFP staining. No, notochord; NT, neural tube.
FIGURE 5.
Either Egr1 or Egr2 is sufficient for the expression of tendon-associated collagens in chick neural tubes. Adjacent transverse sections from different embryos electroporated with mEgr1 (A–F) or mEgr2 (G–L) were hybridized with probes for the ectopic gene mEgr1 (A and D) or mEgr2 (G and J) and the following tendon-associated collagens: Col3a1 (B and H), Col5a1 (C and I), Col12a1 (E and K), and Col14a1 (F and L). The expression of the tendon-associated collagens was up-regulated upon forced-expression of the mEgr1 and mEgr2 genes (arrows). Col5a1 (C) and Col12a1 (K) expression displayed a weak activation upon mEgr1- and mEgr2-forced expression, respectively. The adjacent sections corresponding to different embryos are grouped according to the embryos. No, notochord; NT, neural tube.
Effects of Egr1 and Egr2 Loss-of-function on Collagen Expression and Production
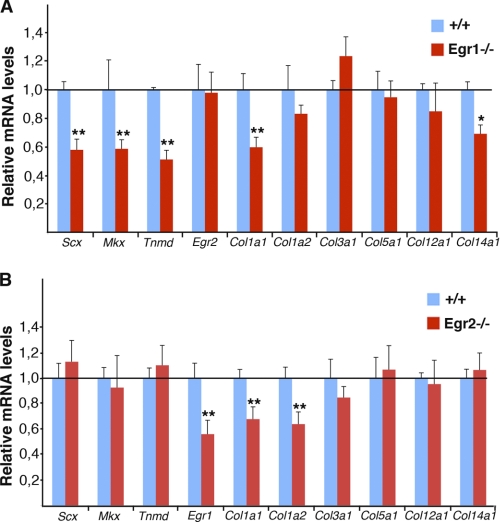
No tendon phenotype has been reported to date in Egr1 or Egr2 mutant mice. To analyze tendon marker expression in the absence of Egr1 or Egr2 activity, we examined the expression levels of various tendon markers from embryonic tail tendons by quantitative RT-PCR assays (Fig. 6). Loss of Egr1 activity led to a significant decrease of Col1a1 mRNA level in tail tendons compared with control tendons (Fig. 6A). In addition, the expression levels of the two DNA-binding factors, Scx and Mkx were also reduced in Egr1−/− mutant mice (Fig. 6A). In agreement with the fact that tenomodulin and Col14a1 are SCX target genes (7), the relative levels of tenomodulin and Col14a1 mRNAs were also decreased in Egr1−/− embryonic tendons (Fig. 6A). Egr2 loss-of-function led to a significant decrease of Col1a1 and Col1a2 mRNA levels, while not affecting other tendon markers (Fig. 6B). Egr1 expression was also reduced in Egr2−/− mutant mice (Fig. 6B). It has to be noted that these changes in mRNA levels observed by quantitative real time-PCR were not detectable by in situ hybridization to embryonic limb tendons from Egr1−/− or Egr2−/− mutant mice (data not shown), highlighting that in situ hybridization is not appropriate to detect quantitative changes in transcription. We conclude that the absence of Egr1 or Egr2 activity reduces the levels of Col1a1 transcription in embryonic tendons and that the expression levels of tendon markers are affected differentially in Egr1 and Egr2 embryonic defective tendons.
FIGURE 6.
Quantitative real-time PCR analyses of tendon gene expression in embryonic tendons of Egr1−/− and Egr2−/− mutant mice. Relative levels of tendon marker mRNAs in tail tendons from E18.5 Egr1−/− (A) and Egr2−/− (B) mutant mice compared with wild-type littermates. The following tendon markers have been analyzed: Scx, Mkx, tenomodulin, Egr1 (in Egr2−/−), Egr2 (in Egr1−/−), Col1a1, Col1a2, Col3a1, Col5a1, Col12a1, and Col14a1. mRNA levels of wild type and mutant mice were normalized to that of GAPDH in each experiment. The error bars represent S.E. The asterisks in the histograms indicate p values, *, p < 0.05; **, p < 0.01.
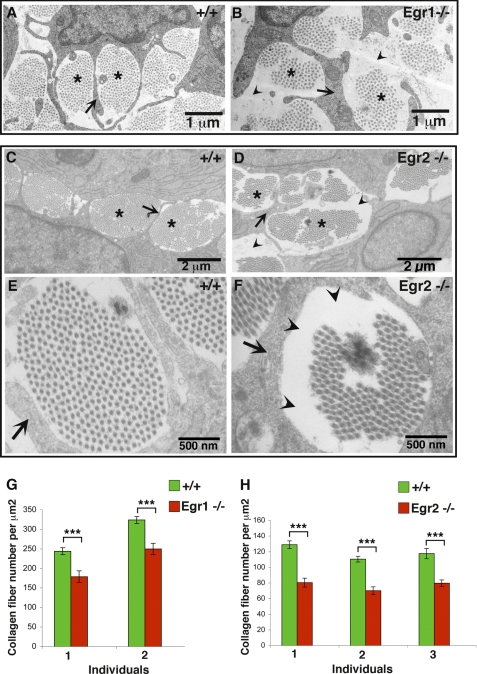
Because Col1a1 transcription was affected in each Egr mutant mice, we analyzed tendon structure in embryonic tendons from Egr1−/− or Egr2−/− mutant mice by transmission electronic microscopy (Fig. 7). Tendon collagen fibrils are orientated parallel to each other and can be observed on sections cut perpendicular to the tendon axis as bundles surrounded by cytoplasmic processes of tenocytes (Fig. 7, A, C, and E), (2). Bundles of collagen fibrils could be observed in E18.5 tendons depleted of Egr1 or Egr2 activity (Fig. 7, A–F). However, the amount of collagen fibrils in bundles was reduced leaving unoccupied spaces in mutant tendons compared with control tendons (Fig. 7, A–F). Measurements of the number of collagen fibrils per surface unit showed that a lack of Egr1 or Egr2 activity caused a 30 to 40% decrease in the number of collagen fibrils in E18.5 deficient tendons compared with control tendons (Fig. 7, G–H). Given that in E18.5 tendons, collagen fibrils are mainly type I collagen fibrils (2, 3), these results indicated a defect in type I collagen production in the absence of either Egr activity.
FIGURE 7.
Collagen fibril organization in Egr1- and Egr2-deficient tendons. Transmission electronic microscopy images of wild type (A, C, and E), Egr1−/− (B), or Egr2−/− (D and F) hind limb tendons at E18.5. Collagen fibrils are organized in bundles (asterisks in A–D) surrounded by cytoplasmic processes of tenocytes (arrows in A–F). E and F, show high magnification of a fibril bundle in control (E) and Egr2-defective tendon (F). Collagen fibrils are less abundant in mutants (B, D, and F) compared with control tendons (A, C, and E), leaving some unoccupied spaces (arrowheads in B, D, and F). G and H, the number of collagen fibrils per surface unit was analyzed in each sample from different mouse groups. The number of collagen fibril per surface unit diminished in the absence of Egr1 and Egr2 activity compared with control tendons. Histograms are expressed as means and S.E. Asterisks in the histograms indicate p values ***, p <0.001.
Because Col1a1 mRNA levels and type I collagen production were affected in embryonic tendons of Egr1−/− and Egr2−/− mutant mice, we analyzed Col1a1 expression in double Egr1−/−;Egr2−/− mutant mice. Despite infertility problems previously reported for Egr1−/− (29), we obtained double mutant embryos. However, we observed the presence of Col1a1 transcripts in Egr1−/−;Egr2−/− double mutant limbs by in situ hybridization (supplemental Fig. S5). This shows that the absence of both Egr1 and Egr2 activities is not enough to completely abolish Col1a1 expression, indicating that other factors compensate the absence of both Egr genes for Col1a1 transcription in tendons. We conclude that either Egr1 or Egr2 activity is required for complete Col1a1 transcription and normal type I collagen production in embryonic mouse tendons.
Egr1 or Egr2 Enhances Transcription of Mouse Col1a1 Proximal Promoter and Are Recruited to Tendon Regulatory Regions
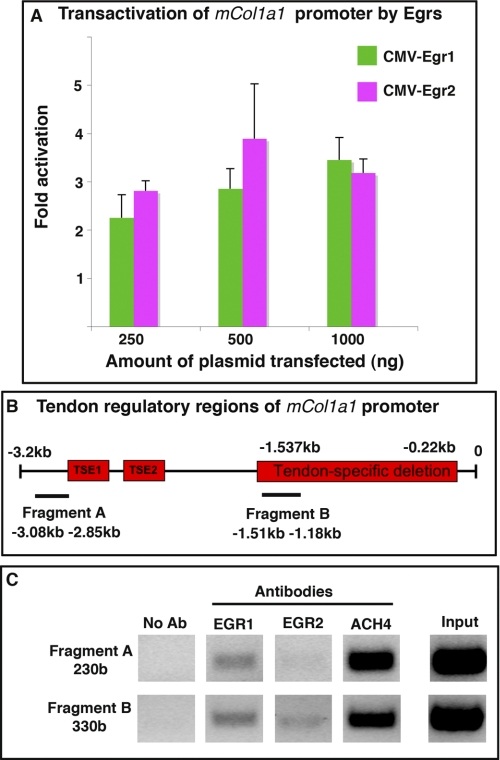
Given the diminution of Col1a1 RNAs and the reduction in the number of type I collagen fibrils in Egr-deficient mouse tendons (Figs. 6 and 7), and given the de novo activation of Col1a1 expression by Egr1/2 in chick gain-of-function experiments (Fig. 4), we analyzed the ability of EGR1 and EGR2 proteins to regulate Col1a1 transcription in tendons. We took advantage of the characterization of DNA regions driving Col1a1 expression in mouse tendons. A 3.2-kb segment of the mouse Col1a1 proximal promoter (pJ320) reconstitutes the Col1a1 endogenous expression in tendons and ligaments in transgenic mice (11, 34, 38). We observed that overexpression of EGR1 or EGR2 in NIH/3T3 cells increased the transcriptional activity of this region up to 3-fold (Fig. 8A). To determine whether the activation of the mouse Col1a1 proximal promoter by EGR proteins involves an interaction with the tendon regulatory regions, we performed ChIP experiments. The 3.2-kb segment of the mouse Col1a1 proximal promoter contains two tendon-specific elements, TSE1 and TSE2. These elements are required for the expression of Col1a1 in tenocytes and bind to SCX and NFATC proteins, respectively (11). However, to induce tendon-specific expression of Col1a1, TSE1 and TSE2 need to interact with downstream elements because the internal deletion of the region extending from −1.537 to −0.22 kb abolished the tendon expression of the reporter gene in transgenic mice, despite the presence of TSE1 and TSE2 (38). We chose two pairs of primers in order to target these two tendon regulatory regions. One primer set amplifies a 230-base fragment flanking the TSE1 and TSE2 elements (fragment A), and a second primer set amplifies a 330-base fragment (fragment B) included into the tendon-specific deletion region (−1.537 to −0.22 kb) (Fig. 8B). Chromatin was prepared from hind limbs and subjected to ChIP with EGR1 or EGR2 antibodies followed by PCR amplification. The DNA regions close to TSE1 and TSE1 (fragment A) were immunoprecipitated by EGR1, but not by EGR2, whereas the tendon-specific deletion region (fragment B) was immunoprecipitated by EGR1 and EGR2 (Fig. 8C). These results established that EGR1 and EGR2 are recruited differentially to tendon regulatory regions of the mouse Col1a1 proximal promoter in vivo. We conclude that EGR1 and EGR2 trans-activate the mouse Col1a1 proximal promoter and bind to tendon regulatory regions.
FIGURE 8.
Egr1 and Egr2 enhance the transcriptional activity of the mouse Col1a1 proximal promoter and are recruited to tendon regulatory regions. A, NIH/3T3 cells were transiently co-transfected with the reporter construct pJ320, containing a 3.2-kb region of the Col1a1 promoter including tendon-specific elements, cloned upstream of the lacZ gene, and with different amounts of CMV-Egr2, CMV-Egr1, or CMV control. Transfection of CMV-Egr1 or CMV-Egr2 had a trans-activating effect on pJ320. Results were expressed as mean and S.E. B, schematic representation of the 3.2-kb Col1a1 proximal promoter. The tendon-specific elements, TSE1 and TSE2, located around −2.8 and −2.3 kb and the region between −1.537 and −0.22 kb required for tendon specific expression of Col1a1 (named tendon-specific deletion) are represented by red boxes. Horizontal lines indicate the amplicon positions (not to scale) used for the ChIP assay. Fragment A is a 230-base region (−3.08 to −2.85 kb) flanking TSE1 and TSE2, and fragment B is a 330-base region (1.51 to −1.18 kb), included into the tendon-specific deletion (−1.537 to −0.22 kb). C, ChIP assays were performed on hind limbs from E18.5 mice with antibodies against EGR1, EGR2, or AcH4, (acetylated histone H4), as a positive control or without an antibody (no Ab) as a negative control. ChIP products were analyzed by PCR, to study the interaction of each EGR with tendon regulatory regions of mouse Col1a1 promoter. Primers targeting fragment A identify DNA regions immunoprecipitated by EGR1, but not by EGR2, whereas primers targeting fragment B identify DNA regions immunoprecipitated by EGR1 and EGR2.
DISCUSSION
EGRs Are DNA-binding Proteins Involved in Tendon Differentiation
To date, there are very few transcription factors identified as being involved in tendon formation. We have now identified EGR1 and EGR2 as other DNA-binding proteins involved in embryonic tendon formation. Egr expression is concomitant with tenocyte differentiation marked by the expression of various tendon collagens. In addition to be expressed at the time of increase of collagen expression in tendon cells, Egr genes are sufficient for their expression in ectopic situations. Both Egr genes are able to induce de novo expression of the tendon-associated collagens Col1a1, Col3a1, Col5a1, Col12a1, and Col14a1. Conversely, Col1a1 transcription is reduced in Egr1- and Egr2- deficient tendons. In addition to collagens, either Egr gene is sufficient for the expression of the reference tendon marker Scx, and the absence of Egr1 activity reduces the level of Scx expression in embryonic tendons. Given the timing of the endogenous expression of Scx (8) and Egr1/2 in tendon cells, Egr1/2 are obviously not involved in the initiation of Scx expression during development. Our data rather suggest that Egr1 maintains Scx expression in differentiated tendon cells. Besides Scx expression in tendon progenitors, Scx function has also been associated with tendon differentiation (7). Similarly to Egr-deficient tendons, the amount of collagen fibrils was also reduced and unoccupied spaces was observed by transmission electronic microscopy in embryonic tendons in the absence of Scx activity (7). However, the exact relationship between the DNA-binding proteins SCX and EGR during collagen fibrillogenesis remains to be clarified. TGFβ signaling has been shown to be crucial for mouse tendon formation as Scx expression is progressively lost in limb tendons from E12.5 in the absence of TGFβ signaling in mice. Moreover, TGFβ2 is able to induce ectopic Scx expression in mouse limb organ cultures and in mouse embryonic fibroblasts (40, 45). Because EGR1 has been shown to mediate the stimulation of collagen production elicited by TGFβ in fibrosis (46), it is possible that EGR proteins play similar role in embryonic tendon formation.
EGR Proteins Are Part of DNA-binding Protein Network Involved in Tendon-specific Expression of Col1a1
We have shown that either EGR1 or EGR2 is able to enhance the activity of the mouse proximal Col1a1 promoter and is recruited to tendon regulatory regions of this promoter. Moreover, the absence of Egr1 or Egr2 activity affects the levels of Col1a1 expression in embryonic mouse tendons, indicating that either Egr gene is involved in Col1a1 transcription. The presence of Col1a1 transcripts in embryonic tendons of double Egr1−/−, Egr2−/− mutant mice indicates that other factors act with EGR proteins to regulate Col1a1 transcription in tendons, reflecting a complex regulation of collagen transcription. Indeed, SCX has been shown to stimulate Col1a1 transcription by directly binding to a tendon specific element of the mouse Col1a1 promoter (11). SCX proteins have been shown to cooperate with the transcription factor NFATC4 in trans-activating the proximal promoter of Col1a1 in tendons through their binding to the tendon-specific elements, TSE1 and TSE2, respectively (11). Our data suggest that EGR proteins cooperate with SCX and NFATC4 to control Col1a1 transcription in tendons. Interestingly, NFAT proteins and EGR1 have been shown to cooperate to induce gene expression in various cell systems (47–49). Recently, it has been shown that Col1a1 transcription is also reduced in post-natal tendons of Mkx−/− mutant mice (12, 13), suggesting that the MKX homeobox protein may also participate in Col1a1 transcription in tendons.
Egr1 versus Egr2 in Tendon Differentiation
We have shown that either Egr gene is sufficient for de novo tendon marker expression, that either EGR activates the transcriptional activity of the proximal Col1a1 promoter and is recruited to tendon regulatory regions of this promoter and that the absence of either Egr gene leads to a decrease in Col1a1 mRNA levels and in the number of collagen fibrils in embryonic tendons. All of these data converge to a functional redundancy between Egr1 and Egr2 in tendon formation. However, Egr1 and Egr2 genes display differences in their expression pattern during tendon development, arguing against Egr redundancy. Egr2 transcripts displayed a general distribution indicating an expression in all limb tendon cells, whereas Egr1 transcripts display a restricted expression near muscles and delineating the long tendons. These data reveal the existence of different population of tendon cells. In addition, Egr2 expression in limb tendons is transient compared with the maintained Egr1 expression in limb tendons. Moreover, the mRNA levels of tendon markers are affected differentially in Egr1−/− and Egr2−/− mutant mice. As an example, the Scx transcript level is reduced in Egr1−/− mutant mice but unaltered in Egr2−/− mutant mice. Lastly, the recruitment of EGR1 and EGR2 proteins to the Col1a1 promoter appears to be different. EGR1 binds to two tendon regulatory regions, whereas EGR2 binds to only one region of the Col1a1 promoter. The differential roles of Egr1 and Egr2 genes in Col1a1 transcription and production in tendons remain to be further characterized.
Tendon Formation, Functional Analogy between Vertebrates and Drosophila
Despite a difference in embryological origin of Drosophila and vertebrate tendon cells (ectoderm versus mesoderm), we showed that homologue genes from the Egr family have similar function in tendon differentiation in both species. In Drosophila, Stripe is necessary and sufficient for epidermal tendon cell specification and differentiation (20–23). In vertebrates, we have shown that Egr genes are sufficient for the expression of tendon differentiation genes and required for complete collagen fibrillogenesis. However, there is a striking difference between their ability to induce tendon gene expression. In Drosophila, Stripe is able to induce tendon gene expression only in the ectoderm, (from which Drosophila tendon cells originate) and fails to induce any tendon marker expression in mesoderm (20, 22). In contrast, Egr genes are able to induce tendon gene expression in the ectoderm, in addition to doing so in mesoderm (from which vertebrate tendon cells originate). Egr1 and Egr2 are expressed too late for displaying a role in vertebrate tendon cell specification, suggesting a functional analogy between Egr genes and the StripeA isoform, which is involved in Drosophila tendon cell differentiation (23). We highlight similarities between Stripe and Egr genes regarding their capacities to be sufficient for tendon differentiation gene expression. However, the downstream genes of StripeA and Egr genes are probably different given the structural difference between Drosophila and vertebrate tendons. There is no clearly identified equivalent of Scx in Drosophila. Flies do not have fibrillar collagens (50), the main component of vertebrate tendons. Flies probably do not need the tensile strength provided by fibrillar collagens because of the existence of a chitinous exoskeleton. Our results establish a functional analogy between StripeA (Drosophila) and Egr (vertebrate) genes in tendon formation.
In conclusion, we have found that 1) the endogenous expression of Egr genes at the time of collagen increase during tendon differentiation, 2) Egr regulation by FGF4, 3) the Egr ability to induce de novo expression of Scx and tendon collagens, 4) the Egr requirement for normal collagen transcription and fibrillogenesis, and finally, 5) the EGR ability to activate and bind to the mouse Col1a1 proximal promoter are consistent with the notion that Egr genes are involved in tendon differentiation by regulating Col1a1 transcription.
Supplementary Material
Acknowledgments
We thank S. Dussurgey for expert technical assistance with FACS (Flow Cytometry Core Facility, IFR128 Bioscience Gerland, Lyon, France). Transmission electronic microscopy observations were performed at the “Centre Technique des Microstructures” (University Lyon 1, Villeurbanne, France).
This work was supported by the Association Française contre les Myopathies (AFM), Association pour la Recherche sur le Cancer (ARC), Agence Nationale de la Recherche (ANR), the sixth PCRDT through the MYORES network of excellence, the Rhône-Alpes region, and the Fondation pour la Recherche Médicale (FRM).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- E7
- embryonic day 7
- HH
- Hamburger and Hamilton.
REFERENCES
- 1. Banos C. C., Thomas A. H., Kuo C. K. (2008) Birth Defects Res. C. Embryo Today 84, 228–244 [DOI] [PubMed] [Google Scholar]
- 2. Zhang G., Young B. B., Ezura Y., Favata M., Soslowsky L. J., Chakravarti S., Birk D. E. (2005) J. Musculoskelet. Neuronal Interact. 5, 5–21 [PubMed] [Google Scholar]
- 3. Canty E. G., Kadler K. E. (2005) J. Cell Sci. 118, 1341–1353 [DOI] [PubMed] [Google Scholar]
- 4. Edom-Vovard F., Duprez D. (2004) Dev. Dyn. 229, 449–457 [DOI] [PubMed] [Google Scholar]
- 5. Ros M. A., Rivero F. B., Hinchliffe J. R., Hurle J. M. (1995) Anat. Embryol. 192, 483–496 [DOI] [PubMed] [Google Scholar]
- 6. Birk D. E., Mayne R. (1997) Eur. J. Cell Biol. 72, 352–361 [PubMed] [Google Scholar]
- 7. Murchison N. D., Price B. A., Conner D. A., Keene D. R., Olson E. N., Tabin C. J., Schweitzer R. (2007) Development 134, 2697–2708 [DOI] [PubMed] [Google Scholar]
- 8. Schweitzer R., Chyung J. H., Murtaugh L. C., Brent A. E., Rosen V., Olson E. N., Lassar A., Tabin C. J. (2001) Development 128, 3855–3866 [DOI] [PubMed] [Google Scholar]
- 9. Shukunami C., Takimoto A., Oro M., Hiraki Y. (2006) Dev. Biol. 298, 234–247 [DOI] [PubMed] [Google Scholar]
- 10. Docheva D., Hunziker E. B., Fässler R., Brandau O. (2005) Mol. Cell. Biol. 25, 699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Léjard V., Brideau G., Blais F., Salingcarnboriboon R., Wagner G., Roehrl M. H., Noda M., Duprez D., Houillier P., Rossert J. (2007) J. Biol. Chem. 282, 17665–17675 [DOI] [PubMed] [Google Scholar]
- 12. Ito Y., Toriuchi N., Yoshitaka T., Ueno-Kudoh H., Sato T., Yokoyama S., Nishida K., Akimoto T., Takahashi M., Miyaki S., Asahara H. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10538–10542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu W., Watson S. S., Lan Y., Keene D. R., Ovitt C. E., Liu H., Schweitzer R., Jiang R. (2010) Mol. Cell. Biol. 30, 4797–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kieny M., Chevallier A. (1979) J. Embryol. Exp. Morphol. 49, 153–165 [PubMed] [Google Scholar]
- 15. Kardon G. (1998) Development 125, 4019–4032 [DOI] [PubMed] [Google Scholar]
- 16. Edom-Vovard F., Schuler B., Bonnin M. A., Teillet M. A., Duprez D. (2002) Dev. Biol. 247, 351–366 [DOI] [PubMed] [Google Scholar]
- 17. Bonnin M. A., Laclef C., Blaise R., Eloy-Trinquet S., Relaix F., Maire P., Duprez D. (2005) Mech. Dev. 122, 573–585 [DOI] [PubMed] [Google Scholar]
- 18. Volk T. (1999) Trends Genet. 15, 448–453 [DOI] [PubMed] [Google Scholar]
- 19. Schweitzer R., Zelzer E., Volk T. (2010) Development 137, 2807–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Becker S., Pasca G., Strumpf D., Min L., Volk T. (1997) Development 124, 2615–2622 [DOI] [PubMed] [Google Scholar]
- 21. Frommer G., Vorbrüggen G., Pasca G., Jäckle H., Volk T. (1996) EMBO J. 15, 1642–1649 [PMC free article] [PubMed] [Google Scholar]
- 22. Vorbrüggen G., Jäckle H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8606–8611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Volohonsky G., Edenfeld G., Klämbt C., Volk T. (2007) Development 134, 347–356 [DOI] [PubMed] [Google Scholar]
- 24. Coulpier F., Decker L., Funalot B., Vallat J. M., Garcia-Bragado F., Charnay P., Topilko P. (2010) J. Neurosci. 30, 5958–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carter J. H., Tourtellotte W. G. (2007) J. Immunol. 178, 3038–3047 [DOI] [PubMed] [Google Scholar]
- 26. Eldredge L. C., Gao X. M., Quach D. H., Li L., Han X., Lomasney J., Tourtellotte W. G. (2008) Development 135, 2949–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamburger V., Hamilton H. L. (1992) Dev. Dyn. 195, 231–272 [DOI] [PubMed] [Google Scholar]
- 28. Pryce B. A., Brent A. E., Murchison N. D., Tabin C. J., Schweitzer R. (2007) Dev. Dyn. 236, 1677–1682 [DOI] [PubMed] [Google Scholar]
- 29. Topilko P., Schneider-Maunoury S., Levi G., Trembleau A., Gourdji D., Driancourt M. A., Rao C. V., Charnay P. (1998) Mol. Endocrinol. 12, 107–122 [DOI] [PubMed] [Google Scholar]
- 30. Schneider-Maunoury S., Topilko P., Seitandou T., Levi G., Cohen-Tannoudji M., Pournin S., Babinet C., Charnay P. (1993) Cell 75, 1199–1214 [DOI] [PubMed] [Google Scholar]
- 31. Bonnet A., Dai F., Brand-Saberi B., Duprez D. (2010) Mech. Dev. 127, 120–136 [DOI] [PubMed] [Google Scholar]
- 32. Delfini M. C., Duprez D. (2004) Development 131, 713–723 [DOI] [PubMed] [Google Scholar]
- 33. Giudicelli F., Taillebourg E., Charnay P., Gilardi-Hebenstreit P. (2001) Genes Dev. 15, 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rossert J., Eberspaecher H., de Crombrugghe B. (1995) J. Cell Biol. 129, 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roulet M., Ruggiero F., Karsenty G., LeGuellec D. (2007) Cell Tissue Res. 327, 323–332 [DOI] [PubMed] [Google Scholar]
- 36. Boardman P. E., Sanz-Ezquerro J., Overton I. M., Burt D. W., Bosch E., Fong W. T., Tickle C., Brown W. R., Wilson S. A., Hubbard S. J. (2002) Curr. Biol. 12, 1965–1969 [DOI] [PubMed] [Google Scholar]
- 37. Havis E., Anselme I., Schneider-Maunoury S. (2006) BioTechniques 40, 34, 36,, 38 passim [DOI] [PubMed] [Google Scholar]
- 38. Terraz C., Brideau G., Ronco P., Rossert J. (2002) J. Biol. Chem. 277, 19019–19026 [DOI] [PubMed] [Google Scholar]
- 39. Eloy-Trinquet S., Wang H., Edom-Vovard F., Duprez D. (2009) Dev. Dyn. 238, 1195–1206 [DOI] [PubMed] [Google Scholar]
- 40. Pryce B. A., Watson S. S., Murchison N. D., Staverosky J. A., Dünker N., Schweitzer R. (2009) Development 136, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McMahon A. P., Champion J. E., McMahon J. A., Sukhatme V. P. (1990) Development 108, 281–287 [DOI] [PubMed] [Google Scholar]
- 42. Brent A. E., Tabin C. J. (2004) Development 131, 3885–3896 [DOI] [PubMed] [Google Scholar]
- 43. Tozer S., Duprez D. (2005) Birth Defects Res. C. Embryo Today 75, 226–236 [DOI] [PubMed] [Google Scholar]
- 44. Brent A. E., Schweitzer R., Tabin C. J. (2003) Cell 113, 235–248 [DOI] [PubMed] [Google Scholar]
- 45. Lorda-Diez C. I., Montero J. A., Martinez-Cue C., Garcia-Porrero J. A., Hurle J. M. (2009) J. Biol. Chem. 284, 29988–29996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen S. J., Ning H., Ishida W., Sodin-Semrl S., Takagawa S., Mori Y., Varga J. (2006) J. Biol. Chem. 281, 21183–21197 [DOI] [PubMed] [Google Scholar]
- 47. Schabbauer G., Schweighofer B., Mechtcheriakova D., Lucerna M., Binder B. R., Hofer E. (2007) Thromb. Haemost. 97, 988–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alfonso-Jaume M. A., Mahimkar R., Lovett D. H. (2004) Biochem. J. 380, 735–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Decker E. L., Nehmann N., Kampen E., Eibel H., Zipfel P. F., Skerka C. (2003) Nucleic Acids Res. 31, 911–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hynes R. O., Zhao Q. (2000) J. Cell Biol. 150, F89–96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.