Abstract
In view of its critical role in influenza A virus (IAV) tropism and pathogenesis, we evaluated the receptor binding properties of HA proteins of the closely related swine and new pandemic human IAVs. We generated recombinant soluble trimeric H1 ectodomains of several IAVs and analyzed their sialic acid binding properties using fetuin-binding and glycan array analysis. The results show that closely related swine and new pandemic H1 proteins differ dramatically in their ability to bind these receptors. Although new pandemic H1 protein exhibited hardly any binding, swine H1 bound efficiently to a number of α2–6-linked sialyl glycans. The responsible amino acids were identified by analyzing chimeric H1 proteins and by performing systematic site-directed mutagenesis of swine and new pandemic human H1 proteins. The difference was found to map to residues at positions 200 and 227. Although substitution of either residue significantly affected the binding phenotype, substitution of both was found to act synergistically and reverse the phenotype almost completely. Modeling of the T200A and E227A substitutions into the crystal structure of the new pandemic human H1 protein revealed the loss of potential hydrogen bond formation with Gln191, which is part of the 190-loop of the receptor binding site, and with the penultimate galactose, respectively. Thus, a residue not belonging to the receptor binding site may affect the interaction of HA with its receptor. Interestingly, whereas alanine at position 200 is found in most new pandemic human viruses, the residue at position 227 in these viruses is invariably a glutamic acid.
Keywords: Glycosylation, Mutant, Receptors, Viral Protein, Virus, Glycan Array, Hemagglutinin, Influenza A Virus, Sialic Acid
Introduction
All influenza A virus (IAV)2 pandemics known so far originated from avian or swine IAV strains that managed to cross the species barrier to humans and acquire the capacity of human-to-human transmission. The most recent example of this was the new H1N1 swine origin IAV that emerged in 2009 and rapidly spread around the world (1, 2). The specificity of the interaction of HA with sialic acid (SIA), the cellular receptor, largely explains the host range of IAVs (3). Thus, viruses that infect humans bind preferentially to SIA linked to the penultimate galactose in an α2–6 configuration, whereas avian viruses prefer binding to SIA with α2–3 linkages (4). However, the adaptations in HA required for swine IAVs to become infectious for humans and to establish themselves in the human population are much less characterized.
The HA receptor binding site (RBS) is formed by three structural elements at the tip of the HA molecule, an α-helix composed by residues 190–198 (the 190-helix) and two loop structures formed by residues 133–138 (the 130-loop) and 220–229 (the 220-loop). Four conserved residues, comprising Tyr98, Trp153, His183, and Tyr195, form the base of the RBS (5). The amino acid residues in the RBS that are critical for the recognition of either avian or human receptors have been well characterized (4, 6, 7). For H1, glutamic acid and glycine residues at positions 190 and 225, respectively, result in binding to avian SIA receptors, whereas H1 proteins that carry aspartic acid residues at these positions interact with human SIA receptors (8–11). For H2 and H3, mutations of glutamine and glycine residues at positions 226 and 228 to leucine and serine, respectively, correlate with a shift from avian to human receptor specificity (6). The same mutations also allow binding of H5 to human SIA receptors (11).
In addition to birds, pigs also serve as a reservoir of new human IAVs. As pigs carry cell surface receptors for both avian and human IAVs, they may act as intermediate hosts or mixing vessels for the generation of IAV reassortants that can be transmitted to humans (12, 13), as was recently demonstrated by the new H1N1 swine origin IAV (1). The swine origin H1N1 virus is a reassortant with at least three parents. Six of the segments, including the one coding for HA, are closest in sequence to those of H1N2 “triple-reassortant” IAVs isolated from pigs in North America. The remaining two segments (for neuraminidase (NA) and M1) are from different Eurasian “avian-like” viruses of pigs (14). Not much, however, is known about the adaptations required for swine IAVs to overcome barriers of cross-species transmission. Although mutations in the IAV polymerase can enable such transmissions (15, 16), efficient spread in the human population is very likely to also involve adaptive mutations in HA. These may involve changes in the binding preference of HA, from α2–3- to α2–6-sialyl glycans, as well as, for swine viruses already having an α2–6 preference, more subtle changes affecting the avidity and/or specificity with which the diverse spectrum of different α2–6-linked SIA receptors are recognized (17, 18).
In view of the important role of HA in host range tropism and pathogenesis, we decided to study and compare the receptor binding properties of the H1 proteins of closely related swine and new pandemic swine origin IAVs. To this end, we generated recombinant soluble trimeric H1 ectodomains of several IAVs and analyzed their sialic acid binding properties using fetuin solid phase and hemagglutination assays and glycan array analysis. The results show that closely related swine and new pandemic swine origin H1 proteins differ dramatically in their ability to bind these receptors. Although hardly any binding could be observed for the new pandemic swine origin H1, the swine H1 protein bound efficiently to several α2–6 SIA-containing glycans. The amino acid residues responsible for the difference in receptor binding were identified by analyzing chimeric H1 proteins as well as by performing systematic site-directed mutagenesis of swine and new pandemic swine origin H1 proteins. Molecular modeling of the responsible mutations (T200A and E227A) demonstrated the loss of two potential hydrogen bond interactions with Gln191 in the 190-helix and with the penultimate galactose, respectively, thereby providing a rationale for the observed differences in H1-receptor binding.
EXPERIMENTAL PROCEDURES
Genes, Expression Vectors, Protein Expression, and Purification
Codon optimized H1-encoding cDNAs (Genscript) of A/Cal/04/09 (GenBankTM accession no. FJ966082; referred to as swine origin H1), A/Swine/Ohio/01 (GenBankTM accession no. AF455675; referred to as swine H1), A/Kentucky/07 (GenBankTM accession no. CY028163; referred to as seasonal H1), A/Puerto Rico/8/34 (GenBankTM accession no. NP_040980.1; referred to as PR8 H1), and A/Duck/NZL/76 (GenBankTM accession no. ABB20429.1; referred to as duck H1) were cloned into pCD5 expression as described previously (19). The swine origin IAV A/Cal/04/09 replicates efficiently in cell culture and has been shown to replicate in and transmit among guinea pigs with similar efficiency to that of a seasonal H3N2 influenza virus (20). Site-directed mutagenesis of the H1-encoding sequences was performed with the QuikChange Site-directed Mutagenesis kit (Stratagene). Chimeric HA expression plasmids were generated by conventional cloning using SgRAI, AgeI, NheI, and PacI restriction enzymes. The HA proteins were expressed in HEK293S GnT1(−) cells and purified from the cell culture supernatants as described previously.
HA Receptor Binding Assays
Binding of HA to fetuin was assessed using a fetuin solid phase binding assay as described previously (19, 21). Briefly, purified, soluble trimeric H1 was precomplexed with HRP-linked anti-Strep tag mouse antibody and with HRP-linked anti-mouse IgG (4:2:1 molar ratio) prior to incubation of limiting dilutions on the fetuin-coated (1 μg/ml fetuin per well) 96-well Nunc MaxiSorp plates. HA binding was subsequently detected using tetramethylbenzidine substrate (BioFX) in an ELISA reader (EL-808 (BioTEK)), reading the OD at 450 nm. Hemagglutination assays were performed with 0.5% chicken or human erythrocytes using HA precomplexed as described previously (19) at a starting concentration of 10 μg/ml.
Glycan Array Analyses
Glycan array analysis of the HA proteins was performed by the Core H of the Consortium for Functional Glycomics as described previously (19, 22).
Modeling
Three-dimensional crystal structures of swine origin H1 (Protein Data Bank code 3LZG (23)) and of the H1 protein from A/Swine/Iowa/30 (Protein Data Bank code 1RV0, in binary complex with an α2–6-linked SIA trisaccharide (24)) were downloaded from the Protein Data Bank. Substitutions T200A and E227A were modeled on the Cal/04/09 structure using SWISS-MODEL (25). Subsequent energy minimizations were not necessary as inspection of the modeled structure by GROMOS revealed no unfavorable energy interactions. Next, the Cα-backbone atoms of residues lining the receptor binding site of Swine/Iowa/30 were superpositioned with the corresponding atoms of the modeled Cal/04/09 (mutant) H1 protein. The root mean square deviation of the superpositioned atoms was smaller than 1.02 Å, allowing the SIA receptor to be copied from the Swine/Iowa/30 structure into the Cal/04/09 structures. Molecular interactions were further examined using the Swiss-Pdb Viewer (26).
RESULTS
H1 Proteins of Swine and New Pandemic IAVs Differ in Their Receptor Binding Properties
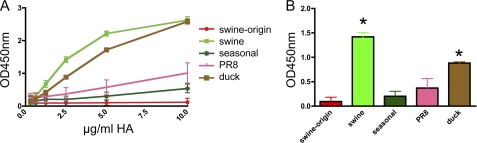
In a previous study (19), we showed that recombinant soluble trimeric HA proteins produced in mammalian cells are excellent tools for performing HA receptor binding studies. Here, we exploited this approach to study and compare receptor binding properties of the H1 proteins of a new pandemic swine origin H1N1 virus (swine origin H1) and of a triple-reassortant swine H1N2 virus (swine H1). The HA gene segment of the swine origin H1N1 virus is closest in sequence to those of the swine H1N2 triple-reassortant IAVs, with the particular swine H1 protein used in this study being the first hit in a protein blast search. The selected swine origin and swine H1 proteins differ only at 26 amino acid residues in their ectodomains. As controls, soluble trimeric H1 proteins of a human seasonal H1N1 virus (seasonal H1), of IAV Duck/NZL/76/H1N3 (duck H1) and of IAV Puerto Rico/8/34/H1N1 (PR8 H1) were taken along. An alignment of these different H1 proteins is shown in supplemental Fig. S1. The purified proteins were first tested for their ability to bind fetuin (Fig. 1). Fetuin is a blood glycoprotein with mono-, bi-, and triantennary glycans containing α2–3 and α2–6 SIA in a 2:1 ratio (27–29). Binding to fetuin by swine origin H1 was hardly detectable. In comparison, swine and duck H1 exhibited strong binding to fetuin, whereas seasonal and PR8 H1 displayed intermediate binding efficiency. The different binding properties of the swine and swine origin H1 proteins were confirmed by hemagglutination assays using either chicken or human erythrocytes (supplemental Fig. S2).
FIGURE 1.
Fetuin binding of recombinant soluble trimeric H1 proteins. A, limiting dilutions of recombinant soluble H1 trimers, complexed with HRP-conjugated antibodies, were applied in the fetuin binding assay. B, bar graph of HA-fetuin binding at a HA concentration of 2.5 μg/ml. S.D. are indicated. Asterisks indicate H1 binding significantly different from that of swine origin H1 (p < 0.05; Student's t test).
Identification of Responsible Residues
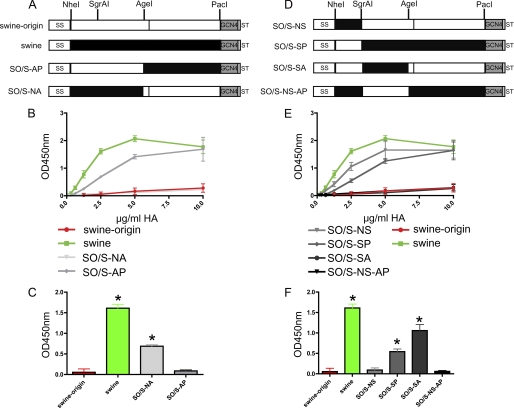
To identify the residues responsible for their different SIA-binding properties, a set of chimeric HA proteins was generated. In the first two, the HA1 domains were exchanged (Fig. 2A). Clearly, only the chimeric protein with the HA1 domain derived from the swine H1 (SO/S-NA) was able to efficiently bind to fetuin (Fig. 2, B and C); the inverse chimeric protein (SO/S-AP) did not exhibit binding just like the swine origin H1 protein. Additional chimeric proteins (Fig. 2D) were generated to narrow down the HA1 region responsible for the receptor binding difference. Chimeric proteins carrying the amino terminus of the swine or the swine origin H1 protein (SO/S-NS and SO/S-SP proteins, respectively) were not affected in their (in)ability to bind fetuin (Fig. 2, E and F). In contrast, exchanging the C-terminal HA1 region clearly affected the fetuin-binding capacity of the resulting H1 proteins (Fig. 2, E and F). Thus, the chimeric protein, in which this region was derived from the swine H1 (SO/S-SA), demonstrated efficient fetuin binding, whereas the opposite was observed for the reciprocal chimera (SO/S-NS-AP) (Fig. 2, E and F).
FIGURE 2.
Fetuin binding of chimeric H1 proteins. A and D, schematic representation of the soluble trimeric swine origin and swine H1 proteins and chimers thereof. The HA ectodomains are preceded by a signal sequence (SS) and contain a GCN4 trimerization motif (GCN4) and the Strep tag II (ST) at their carboxyl terminus. The positions in the HA protein corresponding with the sequences recognized by the restriction enzymes (NheI, SgrAI, AgeI, and PacI) used to generate the chimeric proteins are indicated. White and black boxes correspond with swine origin and swine H1 protein sequences, respectively. B and E, limiting dilutions of recombinant soluble H1 trimers, complexed with HRP-conjugated antibodies, were applied in the fetuin binding assay. C and F, bar graphs of HA-fetuin binding at a HA concentration of 2.5 μg/ml. S.D. are indicated. Asterisks indicate H1 binding significantly different from that of swine origin H1 (p < 0.05; Student's t test).
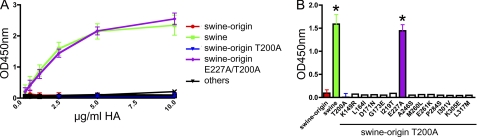
The results show that the region between residues 143 and 332 of the H1 protein (H3 numbering) is responsible for the observed difference in the receptor binding between swine and swine origin H1 proteins. Within this region, the two proteins differ at 14 positions (supplemental Fig. S1). To find out which amino acids are responsible for the binding difference, we changed these residues one by one, in the background of the swine origin H1 protein, to those of the swine H1 protein. All HA proteins containing a single amino acid mutation were expressed, purified, and tested for fetuin binding. Only one of the mutated swine origin H1 proteins, the one carrying the E227A mutation, showed a significant increase in fetuin binding (Fig. 3, A and B). The fetuin binding of this mutant protein was, however, less than that of the wild type swine H1 protein, indicating that additional residues must play a role.
FIGURE 3.
Fetuin binding of swine origin H1 proteins carrying single amino acid substitutions. A, limiting dilutions of swine origin H1 proteins carrying single amino acid substitutions, complexed with HRP-conjugated antibodies, were applied in the fetuin binding assay. Wild type swine origin and swine H1 proteins were taken along as controls. B, bar graph of HA-fetuin binding at a HA concentration of 2.5 μg/ml. S.D. are indicated. Asterisks indicate H1 binding significantly different from that of swine origin H1 (p < 0.05; Student's t test).
To identify any additional residues important for the more efficient SIA receptor binding of the swine H1 protein, the same mutations were made now in the background of the swine origin E227A mutant protein. The subsequent fetuin binding assay showed that, with one exception, all double mutant proteins displayed a similar intermediate SIA binding as the swine origin E227A H1 protein, though some small differences were sometimes observed (Fig. 4, A and B). The exception was the swine origin T200A/E227A H1 protein, which bound to fetuin with similar efficiency as the swine H1 protein. Thus, while changing the Thr at position 200 in the swine origin H1 protein did not have an apparent effect on fetuin binding (Fig. 3), in combination with the E227A mutation, its consequences were significant (Fig. 4). Similar results were obtained when these HA proteins were tested in the hemagglutination assay (supplemental Fig. S2).
FIGURE 4.
Fetuin binding of swine origin H1 proteins carrying amino acid substitutions in addition to E227A. A, limiting dilutions of swine origin H1 proteins carrying amino acid substitutions in addition E227A, complexed with HRP-conjugated antibodies, were applied in the fetuin binding assay. Wild type swine origin and swine H1 proteins as well as the swine origin E227A H1 protein were taken along as controls. B, bar graph of HA-fetuin binding at a HA concentration of 2.5 μg/ml. S.D. are indicated. Asterisks indicate H1 binding significantly different from that of swine origin E227A H1 p < 0.05; Student's t test).
Next, we analyzed whether the T200A mutation also had a synergistic effect on fetuin binding in combination with other mutations than the E227A substitution. Therefore, the same mutations were made again, now in the background of the swine origin T200A H1 protein. However, none of the mutant HA proteins containing two amino acid substitutions displayed increased fetuin binding with the exception of the swine origin T200A/E227A HA protein (Fig. 5, A and B). These results demonstrate, that the positive effect of the T200A mutation on fetuin binding is only observed in combination with the E227A substitution.
FIGURE 5.
Fetuin binding of swine origin H1 proteins carrying amino acid substitutions in addition to T200A. A, limiting dilutions of swine origin H1 proteins carrying amino acid substitutions in addition T200A, complexed with HRP-conjugated antibodies, were applied in the fetuin binding assay. Wild type swine origin and swine H1 proteins as well as the swine origin T200A H1 protein were taken along as controls. B, bar graph of HA-fetuin binding at a HA concentration of 2.5 μg/ml. S.D. are indicated. Asterisks indicate H1 binding significantly different from that of swine origin T200A H1 (p < 0.05; Student's t test).
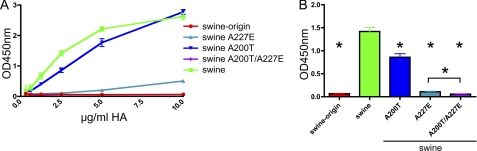
To confirm the importance of the Ala residues found at positions 200 and 227 in the swine protein for strong fetuin binding of HA, these amino acids were changed to the residues found in the swine origin H1 protein, both separately and combined. The fetuin binding assay revealed that, although the effect of the A200T substitution was small, the A227E substitution had a much larger negative effect on SIA binding (Fig. 6). Again, the combination of both mutations had the most drastic effect, resulting in the same nondetectable fetuin binding as the swine origin H1 protein. Thus, by introducing two substitutions in the swine origin (T200A and E227A) or in the swine (A200T and A227E) H1 proteins, HA proteins are obtained with fetuin-binding properties that mirror each other. We conclude that the amino acid differences at these two positions are largely responsible for the different SIA binding properties of the swine and swine origin H1 proteins, at least as assessed by the fetuin-binding and hemagglutination assays.
FIGURE 6.
Fetuin binding of swine H1 proteins carrying single or double amino acid substitutions. A, limiting dilutions of swine H1 proteins carrying single or double amino acid substitutions, complexed with HRP-conjugated antibodies, were applied in the fetuin binding assay. Wild type swine origin and swine H1 proteins were taken along as controls. B, bar graph of HA-fetuin binding at a HA concentration of 2.5 μg/ml. S.D. are indicated. Asterisks indicate H1 binding significantly different from that of swine H1. The significant difference between the swine A227E and the swine A200T/A227E H1 proteins is also indicated (p < 0.05; Student's t test).
Glycan Array Analysis
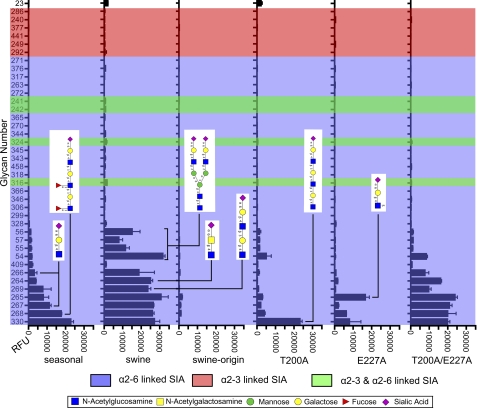
For a detailed study of the SIA binding properties of the swine origin H1 proteins in comparison with the closely related swine and the seasonal H1 proteins, we subjected our soluble trimeric HA preparations to glycan array analysis in collaboration with the Consortium for Functional Glycomics. None of the proteins bound to α2–3-linked SIA-containing glycans (Fig. 7 and supplemental Table), as might be expected from the presence of residues Asp190 and Asp225 in all proteins (8–10). Remarkably, the seasonal and the swine H1 proteins and the swine origin T200A/E227A double mutant H1 all bound most efficiently to the same set of 12 glycans, albeit with differences in their relative binding avidity. For example, only the swine H1 bound with high avidity to bi-antennary N-linked type glycans 54–57 (glycan numbering corresponds with that of supplemental Table, whereas their structures are shown in Fig. 7) containing two α2–6-linked SIAs. These N-linked type glycans have been demonstrated to occur in human upper respiratory epithelium (30) and primary swine respiratory epithelial cells (31). Note that all other bi-antennary glycans were not bound by any of the H1 proteins. Consistent with the fetuin-binding and hemagglutination assays, the swine origin HA protein displayed hardly any binding to the glycan array.
FIGURE 7.
Glycan array analysis of (mutant) H1 proteins. Seasonal, swine, swine origin, swine origin T200A (T200A), swine origin E227A (E227A), and swine origin T200A/E227A (T200A/E227A) H1 proteins were subjected to glycan array analysis. Glycan numbers are given on the y axis and are sorted according to the binding profile of the HA protein of the seasonal H1 protein. α2–6-Linked, α2–3-linked, and α2–3- and α2–6-linked sialyl glycans are indicated blue, red, and green, respectively. Several glycan structures (generated with GlycoWorkbench (39)) are shown. The complete glycan array results as well as the identity of all glycans are shown in supplemental Table.
Introduction of the T200A mutation into the swine origin H1 protein resulted in modest binding to the same set of 12 glycan species, with the exception of the long, three lactosamine repeat-containing glycan 330, which was bound much more efficiently. However, when this glycan was fucosylated (glycan 268), this increased binding was less apparent. Importantly, linear lactosamine repeat-containing glycans have been detected in swine and human respiratory epithelial cells (30, 31). Introduction of the E227A mutation into the swine origin HA protein also resulted in more efficient binding to the three lactosamine repeat-containing glycans 268 and 330, as well as to a sulfated single lactosamine repeat-containing glycan 265. This increased binding was not apparent for similar nonsulfated glycans 266–267. Combining the T200A and E227A mutations had a synergistic effect and resulted in a HA protein with a similar binding profile as the swine H1 protein, although the biantennary glycans 54–57 were less efficiently bound by the double mutant H1 protein.
Modeling of T200A and E227A Substitutions
We modeled the T200A and E227A substitutions into the crystal structure of the swine origin H1 protein (23) and examined how changes in the molecular interactions could explain the altered binding with human SIA receptors. An α2–6-linked SIA trisaccharide receptor was fitted into the modeled structure by superposition with the H1 protein from A/Swine/Iowa/30 in binary complex with α2–6-linked SIA trisaccharide (24). Fig. 8 shows the relevant part of the RBS of the swine origin H1 and of the double mutant. The view of the RBS is such that the potential hydrogen bond interactions in the swine origin H1 between Thr200 and Gln191 and between Glu227 and penultimate galactose (GlcNAc not shown) are visible. Substitution of the residues at position 200 and 227 by alanine resulted in the loss of these potential hydrogen bonds. It is conceivable that the loss of a direct interaction of the receptor with position 227 in the RBS of the double mutant can affect the dynamic interactions between the receptor and the RBS, thereby resulting in altered, in this case increased, binding of particular glycans. Position 200, which is located on the surface, outside of the RBS, does not directly interact with the receptor. However, in the wild type swine origin H1, a potential hydrogen bond between Thr200 and Gln191 occurs, which is lost in the double mutant. Gln191 is a highly conserved amino acid in the 190-helix, of which amino acids Asp190 and Ser193 have been shown to interact directly with the receptor. Any potential hydrogen bond with this helix may obviously influence its exact orientation and thereby have an indirect effect on receptor binding.
FIGURE 8.
Structural model of the swine origin H1 RBS in complex with a α2–6-linked sialyl glycan. A, structural model of the swine origin H1 RBS in complex with a α2–6-linked sialyl glycan. Key amino acids are indicated and shown in a stick representation (gray, carbon; red, oxygen; blue, nitrogen). The α2–6-linked SIA is shown in red, and penultimate galactose is shown in blue. Potential hydrogen bond interactions are shown by dotted lines. B, substitution of the residues at positions 200 and 227 resulted in the loss of potential hydrogen bond interactions with Gln191 and with galactose. The red dotted circles highlight the presence or absence of the potential hydrogen bond interactions.
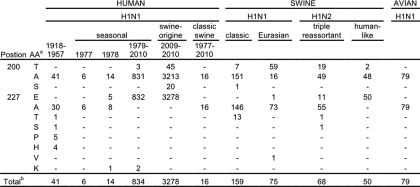
Amino Acid Distribution at Positions 200 and 227
We analyzed the amino acid distribution at positions 200 and 227 of all H1 proteins of IAVs of human, swine, and avian origin (Table 1). The ancestral H1 from the 1918 pandemic strain contains Ala200 and Ala227. Ala200 is (almost) completely conserved in avian (100%) and human seasonal H1N1 (99.7%) viruses. Swine H1N1 and H1N2 viruses display a more diverse pattern. Ala200 is dominant in H1 from classical swine H1N1 (95%) human-like H1 from H1N2 (96%) viruses. However, Thr200 is dominant in Eurasian swine H1N1 viruses (79%) and occurs frequently in triple-reassortant H1N2 viruses (28%). Remarkably, Thr200 is only present in 1.4% of pandemic 2009 H1N1 viruses. The frequency of its occurrence remained at a low level since the start of the pandemic (data not shown).
TABLE 1.
Frequency of residues at positions 200 and 227 in H1
a Amino acids are indicated by single letters.
b Total number of unique full-length sequences are shown. Only for new pandemic H1N1 all full-length sequences were counted-indicates absence of sequences with particular residues.
The H1 proteins derived from the swine origin new pandemic H1N1 virus invariably contain E227 (Table 1). The segment encoding this protein is derived from the H1N2 triple-reassortant influenza virus isolated from pigs in North America. The H1 proteins of these viruses contain either Glu (17%) or Ala (83%) at position 227 (Table 1). The triple-reassortant virus, in turn, obtained its HA-encoding segment from the classical swine H1N1 virus. Remarkably, most HA proteins of these latter viruses contain Ala at position 227 (91%), whereas Glu has never been detected.
DISCUSSION
Transmission of IAVs from birds and swine to humans is posing a serious global health threat. Although the molecular determinants governing the switch from avian to human receptor specificity have been elucidated for several HAs (4, 6, 32), much less is known about the HA requirements for pig to human transmission and subsequent efficient spread in the population. As a first step to address this point, we analyzed the differences in the receptor binding properties of the H1 proteins of closely related swine and new pandemic swine origin IAVs. Although the swine H1 protein efficiently bound to several α2–6-linked SIA-containing glycans, the swine origin H1 protein displayed only very poor binding to these glycans. Although the two proteins differ at 26 amino acid residues, their different receptor binding properties were found to be largely determined by only two residues, located at positions 200 and 227. Substitution of both these residues was shown to have a synergistic effect. Importantly, reciprocal results were obtained when the residues at these positions were mutated in the context of the swine H1 protein. These findings provide new insights into H1 receptor interactions and show that residues 227 and 200, which are located within and outside of the RBS, respectively, can affect HA receptor binding properties.
In a previous study, in which glycan arrays were probed with whole viruses, dual receptor specificity was reported for new pandemic swine origin H1N1 viruses (33). However, our recombinant trimeric swine origin H1 preparations exclusively bound to α2–6- and not to α2–3-linked sialyl glycans, in agreement with other studies using recombinant new pandemic swine origin H1 trimers (34, 35). However, recombinant H1 proteins of several new pandemic swine origin H1N1 viruses (35), which all contain Ala200, appeared to display more efficient binding to the glycan array than our recombinant H1 protein with the T200A substitution. These differences may be attributed to the H1 proteins differing at other residues but also to the different expression systems used. Although the HA proteins used by Yang and corkers were produced in insect cells (35), which contain paucimannose N-glycans, our H1 proteins were generated in HEK293S GnTI(−) cells, which produce proteins with high mannose N-glycans. We recently demonstrated that recombinant soluble HA proteins with paucimannose N-linked glycans display more promiscuous receptor binding when compared with proteins that carry high mannose or desialylated complex N-glycans (19).
In contrast to the swine H1 protein, the swine origin H1 protein displayed only very modest receptor binding, for which the residues at positions 200 and 227 were shown to be largely responsible. Yet, we observed highly similar receptor specificities for these various H1 proteins. Close inspection of the glycan array data (Fig. 7 and supplemental Table) revealed that selecting the 12 strongest binding glycans for each (mutant) H1 protein yielded almost completely overlapping sets. From these results, we conclude that the amino acids at positions 200 and 227 mainly contribute to the receptor binding affinity.
Our observations demonstrate that the identity of the residue at position 200, which is not part of RBS, can affect the interaction of HA with its receptor. Modeling of the T200A substitution into the structure of the swine origin H1 protein (23) revealed the loss of a potential hydrogen bond with residue Gln191 in the 190-loop, thereby probably affecting the interaction of H1 with SIA-containing glycans. Other studies already hinted at the putative involvement of residue 200 in H1 receptor binding. Characterization of zanamivir-selected drug-resistant H1N1 viruses not only revealed a massive deletion in the region encoding the NA active center but also an A200T mutation in HA (36), in agreement with the notion that HA mutations resulting in weaker cell attachment often arise during in vitro selection with NA inhibitors (37). Furthermore, passaging in naïve mice of high avidity binding PR8 mutant viruses (H1N1), which had been selected in mice immunized with influenza vaccine, selected for additional HA substitutions, including an A200T substitution, that resulted in decreased cell binding (38).
Substituting the residue at position 227, which is located within the RBS, had a much larger effect on receptor binding of H1 than substitution of the residue at position 200. Maines and co-workers (34) suggested that the presence of Glu227 in combination with Ile219 in the new pandemic H1 protein would disrupt optimal contacts with α2–6-sialylated glycans. In this study, we demonstrate that the E227A substitution indeed results in increased receptor binding regardless, however, of the identity of the 219 residue. In agreement with our results, also for the H1 protein of PR8, substitution of the alanine at position 227, this time by a threonine residue, correlated with decreased cell binding (38). According to our model (Fig. 8), the E227A substitution disrupts the potential hydrogen bond interaction with the penultimate galactose of the sialyl glycan. Loss of a hydrogen bond may affect the dynamic interactions between receptor and RBS and thereby change receptor binding affinity. However, the synergistic effect of substituting the residues at positions 200 and 227 is not easily explained on the basis of a static model.
In contrast to the T200A substitution, which is found in most new pandemic swine origin H1N1 viruses, the identity of residue 227 in the HA protein of all swine origin H1N1 isolates is invariably a glutamic acid. The T200A substitution may provide a selective advantage to the swine origin H1N1 virus, by its subtle effect on receptor binding (this work) and/or by modifying antigenicity (38). However, it appears that the larger increase in receptor binding resulting from mutation of Glu227 is not compatible with spread of the swine origin H1N1 virus in the human population. It will be of interest to determine the biological consequences of these mutations in H1 and to establish their relationship to the efficient propagation of H1N1 viruses in humans.
Supplementary Material
Acknowledgments
We thank the Core H of the Consortium for Functional Glycomics for glycan array analyzes, with special thanks to David F. Smith and Jamie Heimburg-Molinaro.
This work was supported by the program “Impulse Veterinary Avian Influenza Research” in the Netherlands.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1 and S2.
- IAV
- influenza A virus
- RBS
- receptor binding site
- SIA
- sialic acid
- NA
- neuraminidase.
REFERENCES
- 1. Dawood F. S., Jain S., Finelli L., Shaw M. W., Lindstrom S., Garten R. J., Gubareva L. V., Xu X., Bridges C. B., Uyeki T. M. (2009) N. Engl. J. Med. 360, 2605–2615 [DOI] [PubMed] [Google Scholar]
- 2. Ilyushina N. A., Kim J. K., Negovetich N. J., Choi Y. K., Lang V., Bovin N. V., Forrest H. L., Song M. S., Pascua P. N., Kim C. J., Webster R. G., Webby R. J. (2010) Emerg. Infect. Dis. 16, 314–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taubenberger J. K., Kash J. C. (2010) Cell Host Microbe 7, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Connor R. J., Kawaoka Y., Webster R. G., Paulson J. C. (1994) Virology 205, 17–23 [DOI] [PubMed] [Google Scholar]
- 5. Skehel J. J., Wiley D. C. (2000) Annu. Rev. Biochem. 69, 531–569 [DOI] [PubMed] [Google Scholar]
- 6. Matrosovich M., Tuzikov A., Bovin N., Gambaryan A., Klimov A., Castrucci M. R., Donatelli I., Kawaoka Y. (2000) J. Virol. 74, 8502–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naeve C. W., Hinshaw V. S., Webster R. G. (1984) J. Virol. 51, 567–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glaser L., Stevens J., Zamarin D., Wilson I. A., García-Sastre A., Tumpey T. M., Basler C. F., Taubenberger J. K., Palese P. (2005) J. Virol. 79, 11533–11536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tumpey T. M., Maines T. R., Van Hoeven N., Glaser L., Solórzano A., Pappas C., Cox N. J., Swayne D. E., Palese P., Katz J. M., García-Sastre A. (2007) Science 315, 655–659 [DOI] [PubMed] [Google Scholar]
- 10. Stevens J., Blixt O., Glaser L., Taubenberger J. K., Palese P., Paulson J. C., Wilson I. A. (2006) J. Mol. Biol. 355, 1143–1155 [DOI] [PubMed] [Google Scholar]
- 11. Stevens J., Blixt O., Tumpey T. M., Taubenberger J. K., Paulson J. C., Wilson I. A. (2006) Science 312, 404–410 [DOI] [PubMed] [Google Scholar]
- 12. Ma W., Kahn R. E., Richt J. A. (2008) J. Mol. Genet. Med. 3, 158–166 [PMC free article] [PubMed] [Google Scholar]
- 13. Ito T., Couceiro J. N., Kelm S., Baum L. G., Krauss S., Castrucci M. R., Donatelli I., Kida H., Paulson J. C., Webster R. G., Kawaoka Y. (1998) J. Virol. 72, 7367–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith G. J., Vijaykrishna D., Bahl J., Lycett S. J., Worobey M., Pybus O. G., Ma S. K., Cheung C. L., Raghwani J., Bhatt S., Peiris J. S., Guan Y., Rambaut A. (2009) Nature 459, 1122–1125 [DOI] [PubMed] [Google Scholar]
- 15. Mehle A., Doudna J. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21312–21316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neumann G., Kawaoka Y. (2006) Emerg. Infect. Dis. 12, 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gambaryan A., Yamnikova S., Lvov D., Tuzikov A., Chinarev A., Pazynina G., Webster R., Matrosovich M., Bovin N. (2005) Virology 334, 276–283 [DOI] [PubMed] [Google Scholar]
- 18. Nicholls J. M., Chan R. W., Russell R. J., Air G. M., Peiris J. S. (2008) Trends Microbiol. 16, 149–157 [DOI] [PubMed] [Google Scholar]
- 19. de Vries R. P., de Vries E., Bosch B. J., de Groot R. J., Rottier P. J., de Haan C. A. (2010) Virology 403, 17–25 [DOI] [PubMed] [Google Scholar]
- 20. Steel J., Staeheli P., Mubareka S., García-Sastre A., Palese P., Lowen A. C. (2010) J. Virol. 84, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cornelissen L. A., de Vries R. P., de Boer-Luijtze E. A., Rigter A., Rottier P. J., de Haan C. A. (2010) PLoS One 5, e10645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu R., Ekiert D. C., Krause J. C., Hai R., Crowe J. E., Jr., Wilson I. A. (2010) Science 328, 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gamblin S. J., Haire L. F., Russell R. J., Stevens D. J., Xiao B., Ha Y., Vasisht N., Steinhauer D. A., Daniels R. S., Elliot A., Wiley D. C., Skehel J. J. (2004) Science 303, 1838–1842 [DOI] [PubMed] [Google Scholar]
- 25. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 27. Rogers G. N., Daniels R. S., Skehel J. J., Wiley D. C., Wang X. F., Higa H. H., Paulson J. C. (1985) J. Biol. Chem. 260, 7362–7367 [PubMed] [Google Scholar]
- 28. Baenziger J. U., Fiete D. (1979) J. Biol. Chem. 254, 789–795 [PubMed] [Google Scholar]
- 29. Spiro R. G., Bhoyroo V. D. (1974) J. Biol. Chem. 249, 5704–5717 [PubMed] [Google Scholar]
- 30. Chandrasekaran A., Srinivasan A., Raman R., Viswanathan K., Raguram S., Tumpey T. M., Sasisekharan V., Sasisekharan R. (2008) Nat. Biotechnol. 26, 107–113 [DOI] [PubMed] [Google Scholar]
- 31. Bateman A. C., Karamanska R., Busch M. G., Dell A., Olsen C. W., Haslam S. M. (2010) J. Biol. Chem. 285, 34016–34026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bateman A. C., Busch M. G., Karasin A. I., Bovin N., Olsen C. W. (2008) J. Virol. 82, 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Childs R. A., Palma A. S., Wharton S., Matrosovich T., Liu Y., Chai W., Campanero-Rhodes M. A., Zhang Y., Eickmann M., Kiso M., Hay A., Matrosovich M., Feizi T. (2009) Nat. Biotechnol. 27, 797–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maines T. R., Jayaraman A., Belser J. A., Wadford D. A., Pappas C., Zeng H., Gustin K. M., Pearce M. B., Viswanathan K., Shriver Z. H., Raman R., Cox N. J., Sasisekharan R., Katz J. M., Tumpey T. M. (2009) Science 325, 484–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang H., Carney P., Stevens J. (2010) PLoS Curr. 22, 2, RRN1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baz M., Abed Y., Boivin G. (2007) Antiviral Res. 74, 159–162 [DOI] [PubMed] [Google Scholar]
- 37. McKimm-Breschkin J. L. (2000) Antiviral Res. 47, 1–17 [DOI] [PubMed] [Google Scholar]
- 38. Hensley S. E., Das S. R., Bailey A. L., Schmidt L. M., Hickman H. D., Jayaraman A., Viswanathan K., Raman R., Sasisekharan R., Bennink J. R., Yewdell J. W. (2009) Science 326, 734–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bohne-Lang A., von der Lieth C. W. (2005) Nucleic Acids Res. 33, W214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.