Abstract
Summary
GO::TermFinder comprises a set of object-oriented Perl modules for accessing Gene Ontology (GO) information and evaluating and visualizing the collective annotation of a list of genes to GO terms. It can be used to draw conclusions from microarray and other biological data, calculating the statistical significance of each annotation. GO::TermFinder can be used on any system on which Perl can be run, either as a command line application, in single or batch mode, or as a web-based CGI script.
Availability
The full source code and documentation for GO::TermFinder are freely available from http://search.cpan.org/dist/GO-TermFinder/
Contact
sherlock@genome.stanford.edu
INTRODUCTION: MOTIVATION AND DESIGN
The amount of data that can be produced by experimental platforms such as microarrays can be overwhelming. A typical microarray experiment can generate many lists of genes, each containing dozens or hundreds of genes of interest. The challenge to the biologist is to determine whether there is a common theme to those genes, which will help in interpretation of the experiment. The Gene Ontology (GO) Consortium (Ashburner et al., 2000) provides controlled vocabularies, which model Biological Process, Molecular Function and Cellular Component, that are structured into directed acyclic graphs (DAGs). Gene products may be annotated to one or more GO nodes, and because of the structure of GO, a gene annotated to a given node is thus also annotated to all ancestral nodes (parent, grandparent, etc.) of that specific node.
THE APPLICATION PROGRAMMING INTERFACE (API)
GO:TermFinder comprises an extensible set of object-oriented Perl modules that can be used to determine the significance of a GO annotation to a list of genes, and to access GO information and annotation information through a well-documented API. The software defines two abstract classes, OntologyProvider and AnnotationProvider, which provide APIs for handling ontology and annotation information, respectively. Also provided are concrete implementations of both of these abstract classes that parse the annotation and ontology files provided by the GO Consortium (www.geneontology.org), as shown in Figure 1. This software design defines a plug-in architecture, which allows the creation of alternate concrete subclasses, which for instance might read from a database instead of flat files.
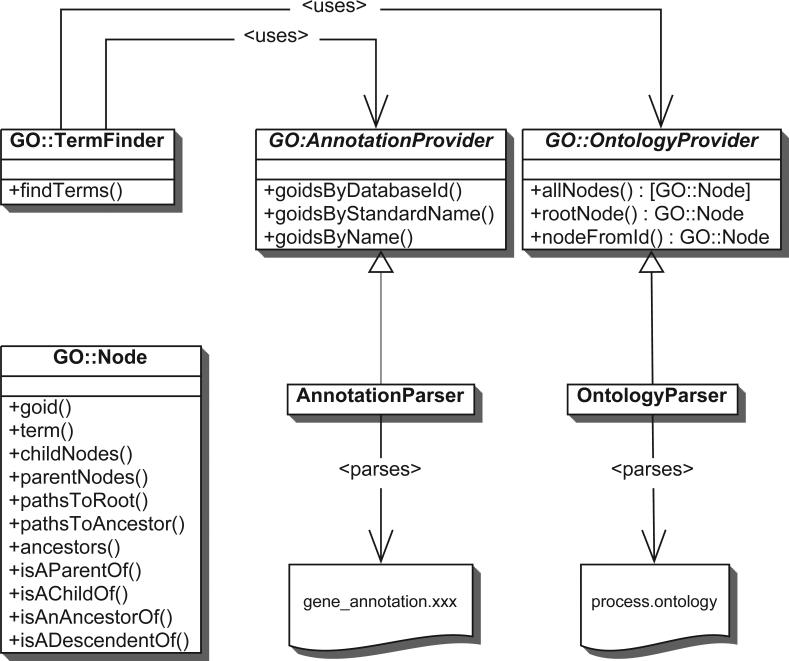
Fig. 1.
Simplified UML diagram of the architecture of GO::TermFinder and associated modules. Public methods defined by the abstract base class, GO::OntologyProvider, which are implemented by concrete subclasses, such as the GO::OntologyProvider::OntologyParser class that we have written, return either a single GO::Node, or an array of GO::Node instances. A subset of the public interface to GO::Node is shown, illustrating the various methods that exist to query the attributes of a GO::Node, as well as to traverse the GO structure.
CALCULATION OF STATISTICAL SIGNIFICANCE
To determine whether any GO terms annotate a specified list of genes at a frequency greater than that would be expected by chance, GO::TermFinder calculates a P-value using the hypergeometric distribution:
In this equation, N is the total number of genes in the background distribution, M is the number of genes within that distribution that are annotated (either directly or indirectly) to the node of interest, n is the size of the list of genes of interest and k is the number of genes within that list which are annotated to the node. The background distribution by default is all the genes within a given annotation file, though the software also allows a user-defined background distribution, such that biases in the sampling population (e.g. the genes represented on a microarray) can be accounted for correctly. The hypergeometric distribution is sampling without replacement—for instance, consider a bag with 500 red and 500 green beads. If 20 beads were selected randomly, and beads were not replaced after each selection, and 17 were green, we would use the hypergeometric distribution to calculate the P-value as the probability of picking 17, or more, green beads from 20, given that there are 500 of each in the background distribution.
MULTIPLE HYPOTHESIS CORRECTION
In a statistical experiment, a P-value is considered significant if it is less than that experiment's chosen alpha value. The alpha value specifies the accepted level of certainty at which a result is considered statistically significant when it is in fact merely the result of random chance. For example, in an experiment using an alpha value of 0.05, there isa1in20 that any given true ‘null’ test would seem significant just by chance. When multiple hypotheses are tested, each hypothesis has a probability of being falsely determined to be significant. If 10 hypotheses are tested and the alpha level is 0.05, then the chance of finding at least one apparently significant difference due to random chance equals 0.4 (which is 1 − 0.9510).
Correction for multiple hypotheses attempts to maintain the probability of falsely finding any significant hypothesis at the alpha value. The most common multiple hypothesis correction method used is the Bonferroni correction, whereby the alpha value is simply divided by the number of tests, and the overall chance of finding any false positive remains the same as in a single hypothesis experiment. The Bonferroni correction assumes that the tests are independent, and is usually considered a conservative adjustment (Sokal and Rohlf, 1995). In our case, the hypotheses (GO nodes) are not independent, because the nodes themselves are structured in a DAG and it is thus not clear whether a Bonferroni adjustment would be appropriate. To determine whether the Bonferroni correction is appropriate for multiple hypothesis correction, we implemented a simulation-based correction within GO::TermFinder. For each simulation, the same number of genes as were provided in the real data were picked randomly from the list of genes that define the background distribution, and P-values were calculated as normal. Adjusted P-values for the real data were calculated for each node as the fraction of 1000 null-hypothesis simulations having any node with a P-value as good or better than the P-value for that node, where the null hypothesis states that a randomly chosen list of genes should not be significantly annotated by any GO nodes. Examining the output of simulations, to determine a correction factor that would need to be applied to uncorrected P-values, and comparing it to the Bonferroni adjusted P-values, we determined that the Bonferroni adjustment is in fact somewhat liberal, rather than conservative. Both simulation and Bonferroni are provided as options for multiple hypothesis correction, though while the simulation based analysis is the most accurate, it also takes three orders of magnitude longer to run, as 1000 independent simulations are needed.
FALSE DISCOVERY RATES
A concern with classical multiple hypothesis correction is that it aims to control the probability of making even a single type I error (a false positive) within the tested family of hypotheses. This can be overly restrictive, and result in lots of false negatives instead. An alternative methodology for multiple hypothesis testing is to calculate the false discovery rate (FDR), which is the expected proportion of true null hypotheses rejected out of the total number of null hypotheses rejected (Benjamini and Hochberg, 1995), i.e. it is the proportion of hypotheses deemed to be significant, that are not actually significant. Based on 50 simulations, GO::TermFinder calculates the FDR for each hypothesis from the real data as the average number of nodes per simulation that have a P-value as good or better than the real node's P-value, divided by the number of nodes in the real data that have a P-value as good or better than that P-value. Comparison of P-values corrected by simulation versus the FDR (Table 1) shows the conservative nature of classic multiple hypothesis testing. Using a cutoff of 5% false discovery in this example results in 27 hypotheses being chosen as significant with an FDR of 1.63%, and less than 1 expected false positive. However, the P-value at that level is 0.137, higher than that would be typically used as a cutoff. Using a P-value cutoff of 0.05 would result in picking 22 hypotheses as significant, suggesting that using the corrected P-value is likely to result in more false negatives.
Table 1.
Comparison of Bonferroni corrected P-values, simulation corrected P-values and FDR for the 28 most significant GO nodes.
| GO term | Rank | FDR (%) | Expected false positives | Uncorrected P-value | Bonferroni corrected P-value | Simulation corrected P-value | Simulation/Bonferroni |
|---|---|---|---|---|---|---|---|
| Invasive growth (sensu Saccharomyces) | 1 | 0 | 0 | 1.93E–09 | 1.35343E–07 | 0.0001 | N/A |
| Negative regulation of transcription by carbon catabolites | 2 | 0 | 0 | 1.25E–08 | 8.737E–07 | 0.0001 | N/A |
| Negative regulation of transcription by glucose | 3 | 0 | 0 | 1.25E–08 | 8.737E–07 | 0.0001 | N/A |
| Regulation of transcription by carbon catabolites | 4 | 0 | 0 | 1.25E–08 | 8.737E–07 | 0.0001 | N/A |
| Regulation of transcription by glucose | 5 | 0 | 0 | 1.25E–08 | 8.737E–07 | 0.0001 | N/A |
| Protein-vacuolar targeting | 6 | 0 | 0 | 2.36E–07 | 1.652E–05 | 0.0001 | N/A |
| Growth pattern | 7 | 0 | 0 | 4.45E–07 | 3.117E–05 | 0.0001 | N/A |
| Filamentous growth | 8 | 0 | 0 | 4.45E–07 | 3.117E–05 | 0.0001 | N/A |
| Protein processing | 9 | 0 | 0 | 4.97E–07 | 3.476E–05 | 0.0001 | N/A |
| Growth | 10 | 0 | 0 | 5.16E–07 | 3.609E–05 | 0.0001 | N/A |
| Cell differentiation | 11 | 0 | 0 | 4.11E–05 | 2.874E-03 | 0.0062 | 2.157 |
| Sporulation | 12 | 0 | 0 | 4.11E–05 | 2.874E-03 | 0.0062 | 2.157 |
| Cellular morphogenesis | 13 | 0 | 0 | 4.35E–05 | 3.047E-03 | 0.0065 | 2.133 |
| Morphogenesis | 14 | 0 | 0 | 4.35E–05 | 3.047E-03 | 0.0065 | 2.133 |
| Development | 15 | 0 | 0 | 5.86E–05 | 4.100E-03 | 0.0115 | 2.805 |
| Negative regulation of transcription, DNA-dependent | 16 | 0.125 | 0.02 | 1.38E-04 | 9.627E-03 | 0.0237 | 2.462 |
| Negative regulation of transcription | 17 | 0.118 | 0.02 | 1.47E-04 | 0.0103 | 0.0238 | 2.309 |
| Protein targeting | 18 | 0.111 | 0.02 | 1.60E-04 | 0.0112 | 0.0293 | 2.618 |
| Cellular physiological process | 19 | 0.105 | 0.02 | 2.20E-04 | 0.0154 | 0.0352 | 2.286 |
| Intracellular protein transport | 20 | 0.100 | 0.02 | 2.29E-04 | 0.0160 | 0.0359 | 2.237 |
| Protein transport | 21 | 0.095 | 0.02 | 2.62E-04 | 0.0183 | 0.0416 | 2.271 |
| Cellular process | 22 | 0.091 | 0.02 | 3.00E-04 | 0.0210 | 0.0429 | 2.043 |
| Intracellular transport | 23 | 0.087 | 0.02 | 4.74E-04 | 0.0332 | 0.0635 | 1.914 |
| Sporulation (sensu Saccharomyces) | 24 | 0.083 | 0.02 | 5.67E-04 | 0.0397 | 0.074 | 1.865 |
| Sporulation (sensu Fungi) | 25 | 0.320 | 0.08 | 7.27E-04 | 0.0509 | 0.0905 | 1.779 |
| Cell growth and/or maintenance | 26 | 0.615 | 0.16 | 8.85E-04 | 0.0619 | 0.1057 | 1.706 |
| Protein-membrane targeting | 27 | 1.630 | 0.44 | 1.41E-03 | 0.0989 | 0.1373 | 1.389 |
| Meiosis | 28 | 5.786 | 1.62 | 2.94E-03 | 0.2055 | 0.5511 | 2.681 |
For a group of genes that show sensitivity to 1 M NaCl and 10 μM nystatin [(Giaever et al., 2002); SNF7 STP22 VPS28 SNF8 VPS36 VPS25 YGR122W RIM20 RIM21 RIM8 RIM101 DFG16 RIM9 YGL046W RIM13 YNR029]. Note that the Bonferroni correction is up to 2.8-fold less conservative than the simulation method that controls the Family Wise Error Rate. N/A, not applicable—cases where no P-values better than that node's P-value were seen in simulations.
VISUALIZATION OF RESULTS
The GO::TermFinder set of libraries includes a module, GO::View, for visualizing the output of an analysis of a set of genes for enriched GO terms. The module is configurable such that both nodes in the output, and genes annotated to those nodes can be linked to URLs, with their identifiers embedded in those URLs. Additionally, the colors of the nodes themselves are based on the calculated P-values, so that attention is drawn to the most significant nodes. Thus, the output data can be easily and intuitively viewed and explored in a web browser, as shown in Figure 2, which was generated from the ‘methionine cluster’, which is discussed below.
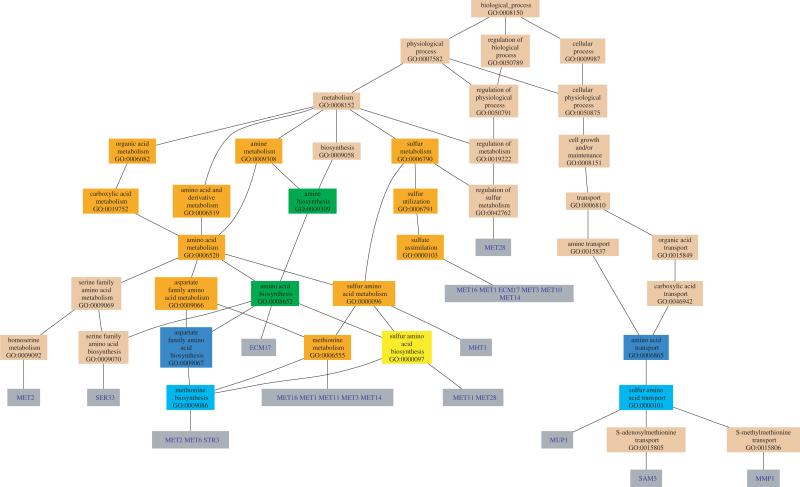
Fig. 2.
Visualizing output from GO::TermFinder. GO graph layout that includes the significant GO nodes annotated by the ‘methioine cluster’, which contains ICY2, MET11, MXR1, SAM3, MET28, STR3, MMP1, MET1, SER33, MHT1, MET14, MET16, MET3, MET10, ECM17, MET2, MUP1 and MET6. The color of the nodes is an indication of their Bonferroni corrected P-value (orange <= 1e-10; yellow 1e-10 to 1e-8; green 1e-8 to 1e-6; cyan 1e-6 to 1e-4; blue 1e-4 to 1e-2; tan > 0.01).
EXAMPLE OF A GO::TERMFINDER ANALYSIS OF MICROARRAY DATA
Spellman et al. (1998) characterized the yeast cell cycle using microarrays. The authors called one set of coherently regulated genes the ‘methionine cluster’ because it contained many genes whose name begins with ‘MET’ (Figure 4b in that paper, containing ICY2, MET11, MXR1, SAM3, MET28, STR3, MMP1, MET1, SER33, MHT1, MET14, MET16, MET3, MET10, ECM17, MET2, MUP1, MET6—note that seven of the genes in this cluster have been named since that study). GO::TermFinder identifies (using the Biological Process ontology, and the SGD-provided gene associations file on May 3, 2004) many GO nodes with significant Bonferroni-adjusted P-values for this list. The top three nodes are: sulfur metabolism (2.75e−21), sulfur amino acid metabolism (1.5e−19) and methionine metabolism (3.39e−16). While the initial naming of the cluster as the methionine cluster was close to the mark, GO::TermFinder is more informative, and provides a robust statistical basis on which to draw conclusions about observations from microarray data. In all, there are 23 GO terms that are selected as significant using a 5% FDR as the cut-off, and at that level, the number of expected false positives would be less than 1, with an FDR of 2.17%.
INCLUDED TOOLS
GO::TermFinder includes within its distribution a number of useful tools for enabling users to use the functionality provided within the libraries. Two batch processing tools exist, that allow the analysis of any number of files, each of which contain a list of genes. One of these simply produces text output, while the other generates html pages with browsable representations of the GO, as depicted in Figure 2. Additionally, there are some simple tools for retrieving GO information, such as the parents, children or ancestors of a particular node. Although potentially useful in their own right, these tools are also useful examples for programmers wanting to implement their own client scripts of these libraries.
COMPARABLE SOFTWARE
While this work was in progress, many similar tools have become available. These include FunSpec (Robinson et al., 2002), Onto-Express (Draghici et al., 2003a,b; Khatri et al., 2002) and FatiGO (Al-Shahrour et al., 2004) which are web applications; GoMiner (Zeeberg et al., 03), a Java application; GeneMerge (Castillo Davis and Hartl, 2003), which is a standalone Perl script whose source is available; and FuncAssociate (Berriz et al., 2003), which is a web application, comprising Perl and C code that are available for download. Each of these supports the calculation of P-values for annotations for a given set of genes, with various multiple hypothesis correction strategies, and some have support for FDR calculation. Although there is a significant overlap in functionality, GO::TermFinder has some unique attributes not found in these other tools. First, the source code for GO::TermFinder is fully and freely available under a very permissive Open Source license (the MIT license), which is not the case for any of the above tools. Those whose source code is available (GeneMerge and FuncAssociate) are available under more restrictive licenses. Second, GO::TermFinder is modular so it is easy to incorporate into other applications or analysis pipelines, or to improve or modify its behavior—additionally, the MIT license asserts no ownership on any improvements made to the software. Third, GO::TermFinder defines an API for accessing GO and Annotation information, which is well documented, and can easily be used by Perl programmers. Fourth, GO::TermFinder can create browsable, visual presentations of the significant nodes, making it very easy for biologists to interpret their data. Finally, GO::TermFinder includes a number of tools, as described above. Thus despite the considerable overlap with other tools, we believe GO::TermFinder to be a significant contribution.
DISCUSSION
The ability to determine rapidly significant GO annotations for a list of genes, generated by any means, is a powerful tool in a biologist's arsenal in these days of genomic scale biology. GO::TermFinder is flexible, extensible and easy to reuse and incorporate into analysis pipelines. In future, we will write data adaptors to the CHADO schema, which is being designed as a generic model organism database (www.gmod.org). This will enable new databases to incorporate the GO::TermFinder functionality without additional coding.
ACKNOWLEDGEMENTS
The authors thank all the members of the Stanford Microarray Database and the Saccharomyces Genome Database for stimulating discussions, and Rob Tibshirani and Trevor Hastie for statistical discussions. This work was supported by a grant from the US National Institutes of Health to G.S. (1R01HG002732).
REFERENCES
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL. GeneMerge–post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003a;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. Global functional profiling of gene expression. Genomics. 2003b;81:98–104. doi: 10.1016/s0888-7543(02)00021-6. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Grigull J, Mohammad N, Hughes TR. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: the priniciples and Practice of Statistics in Biological Research. 3rd ed. Freeman; New York: 1995. [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]