Abstract
Experiments were designed to evaluate changes in the transcriptome (mRNA levels) in the ovulatory, luteinizing follicle of rhesus monkeys, using a controlled ovulation model that permits analysis of the naturally selected, dominant follicle at specific intervals (0, 12, 24 and 36 h) after exposure to an ovulatory (exogenous hCG) stimulus during the menstrual cycle. Total RNA was prepared from individual follicles (n= 4–8/timepoint), with an aliquot used for microarray analysis (AffymetrixTM Rhesus Macaque Genome Array) and the remainder applied to quantitative real-time PCR (q-PCR) assays. The microarray data from individual samples distinctly clustered according to timepoints, and ovulated follicles displayed markedly different expression patterns from unruptured follicles at 36 h. Between timepoint comparisons revealed profound changes in mRNA expression profiles. The dynamic pattern of mRNA expression for steroidogenic enzymes (CYP17A, CYP19A, HSD3B2, HSD11B1 and HSD11B2), steroidogenic acute regulatory protein (StAR) and gonadotrophin receptors [LH/choriogonadotrophin receptor (LHCGR), FSH receptor (FSHR)] as determined by microarray analysis correlated precisely with those from blinded q-PCR assays. Patterns of mRNA expression for epidermal-growth-factor-like factors (amphiregulin, epiregulin) and processes [hyaluronan synthase 2 (HAS2), tumor necrosis factor alpha-induced protein 6 (TNFAIP6)] implicated in cumulus–oocyte maturation/expansion were also comparable between assays. Thus, several mRNAs displayed the expected expression pattern for purported theca (e.g. CYP17A), granulosa (CYP19A, FSHR), cumulus (HAS2, TNFAIP6) cell and surface epithelium (HSD11B)-related genes in the rodent/primate pre-ovulatory follicle. This database will be of great value in analyzing molecular and cellular pathways associated with periovulatory events in the primate follicle (e.g. follicle rupture, luteinization, inflammatory response and angiogenesis), and for identifying novel gene products controlling mammalian fertility.
Keywords: cumulus expansion, luteinization, oocyte maturation, ovulation, rhesus monkey
Introduction
Ovulation and luteinization of the mature follicle are essential processes for successful reproduction in mammals, including non-human primates and humans. It is well established that the midcycle rise in estradiol (E2) secreted from the maturing follicle elicits a surge in gonadotrophin hormones (LH and FSH) released from the anterior pituitary gland. The surge levels of gonadotrophins act on the maturing follicle to initiate a cascade of events leading to: (i) reinitiation of meiosis and cytoplasmic maturation in the enclosed oocyte, thereby producing a fertilizable oocyte, (ii) ovulation, thereby releasing the mature oocyte for potential fertilization in the oviduct and (iii) luteinization, thereby producing the corpus luteum (CL), which secretes a steroid hormone, progesterone, that is essential for the preparation of the uterus for implantation and maintenance of early pregnancy. Over the past several decades, there were numerous studies examining the morphologic and cellular changes that occur in the ovulatory follicle in various species, from laboratory rodent models to domestic animals and primates (Espey, 1978; Espey and Richards, 2006). In addition, biochemical approaches identified several key pathways, such as steroidogenesis (e.g. for progesterone synthesis), and inflammatory immune-type responses (e.g. cytokine production) in the cascade of events leading to ovulation or luteinization (Yoshimura and Wallach, 1987; Irianni and Hodgen, 1992; Espey, 1994; Richards et al., 1998). Recently, the activation of various proteases (e.g. A disintegrin and A metalloprotein with thrombospodin type 1 motif/ADAMTS proteases) (Ohnishi et al., 2005) and the production of angiogenic factors (e.g. vascular endothelial growth factor, VEGFA; angiopoietins) (Stouffer et al., 2001) was implicated in the tissue remodeling associated with follicle rupture and luteinization.
Although the above approaches contributed significantly to our understanding of periovulatory events in the follicle, previous investigators generally focused on a particular molecule or family of molecules (e.g. matrix metalloproteinases) (Chaffin and Stouffer, 1999; Curry and Smith, 2006) in the ovulatory cascade. To date, there have been few reports, mostly using non-primate models, attempting to examine gene expression on a large, comprehensive scale within the ovary or its regulation by gonadotrophin hormones, especially within the ovulatory, luteinizing follicle of the natural ovarian cycle (Leo et al., 2001; Liu et al., 2001; Chin et al., 2002; Jo et al., 2004; Friedmann et al., 2005; Agca et al., 2006; Hernandez-Gonzalez et al., 2006; Perlman et al., 2006).
Owing to species differences in reproductive processes among mammals, it would be ideal from a clinical perspective to study the cascade of periovulatory events during the natural menstrual cycle in a primate model. Recently, we developed a model wherein the final maturation and ovulatory timing of the dominant follicle can be controlled during the menstrual cycle in rhesus monkeys (Young et al., 2003). Control over the naturally selected dominant follicle was possible during the late follicular phase using an abbreviated (48 h) protocol of GnRH antagonist plus low-dose FSH + LH, followed by an ovulatory bolus of the LH-like hormone hCG. This controlled ovulation (COv) protocol offers a unique opportunity to experimentally analyze the dominant follicle at precise stages during the ovulatory process in primates. The current study was a collaboration between scientists at the Oregon National Primate Research Center (ONPRC), Portland, OR and Bayer Schering Pharma AG, Berlin, Germany to systematically evaluate the changes in the transcriptome (mRNA expression) that occur in the primate ovulatory follicle utilizing the COv protocol in rhesus monkeys. Levels and patterns of selected mRNAs were also quantified by real-time PCR (q-PCR) to further validate the data from macaque gene arrays.
Materials and Methods
Tissue collection during COv
The general care and housing of rhesus monkeys (Macaca mulatta) at ONPRC was described previously (Wolf et al., 1990). Animal protocols and experiments were approved by the Institutional Animal Care and Use Committee, and studies were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Menstrual cycles of adult female rhesus monkeys were monitored daily. Six days after the onset of menses, daily blood samples were collected by saphenous venipuncture. Serum was separated and assayed for concentrations of E2 and progesterone by specific electrochemoluminescent assays using a DPC Immulite 2000 (Diagnostic Products Corporation, Los Angeles, CA, USA) by the Endocrine Technology Laboratory at ONPRC (Young and Stouffer, 2004). When E2 levels reached 100–120 pg/ml, a GnRH antagonist (Acyline, 0.1 ml/kg; NICHD) plus recombinant (r)-human (h) FSH (NV Organon) and r-hLH (Merck Serono; 30 IU each) were administered at 16:00 that day (Day 1) and at 08:00 next day (Day 2). At 16:00 on Day 2, monkeys received either no treatment (timepoint 0) or 1000 IU r-hCG to initiate ovulatory events (Young et al., 2003). Daily blood samples were collected until the day of surgery. The ovary bearing the large dominant follicle was removed from anesthetized animals (Xu and Stouffer, 2005) by laparotomy at 0, 12, 24 and 36 h (unruptured follicle: 36−Ov, or ruptured follicle: 36 + Ov) post r-hCG treatment (n= 4–8/timepoint). The follicle was separated from the ovary, frozen in liquid nitrogen and stored at −80°C for later isolation of total RNA using Trizol (Invitrogen, Carlsbad, CA, USA) according to Invitrogen's standard protocol. Part of the RNA preparation from each follicle was sent to Bayer Schering Pharma AG (Berlin, Germany) for microarray assays, with the remainder used at ONPRC for verification of expression levels and patterns for selected genes by quantitative PCR.
Microarray assay
Affymetrix GeneChip® expression profiling experiments
A DNase I (Qiagen GmbH, Hilden, Germany) digestion step was performed to eliminate genomic DNA. The quality of the total RNA was checked for integrity with RNA LabChips on the Agilent Bioanalyzer 2100 (Agilent Technologies Inc., Santa Clara, CA, USA) and for concentration on the Peqlab NanoDrop (Peqlab Biotechnology, Erlangen, Germany). Total RNA (2 μg) was used to prepare biotinylated and fragmented cRNA following the instruction of the Affymetrix One-Cycle Target Labeling protocol, and individual samples were hybridized on the Affymetrix GeneChip®Rhesus Macaque Genome Array (Duan et al., 2007). The specifications of this gene chip are summarized at http://www.affymetrix.com/support/technical/datasheets/rhesus_datasheet.pdf. Chips were scanned using a GeneChip Scanner 3000 7G, and scanned images were extracted using the Affymetrix GCOS software. In total, 22 arrays were performed.
Expression analyses were performed using the Expressionist Pro 4.0 software (Genedata AG, Basel, Switzerland). The quality of the data files (CEL format) containing probe level expression data were checked and refined using the Expressionist Refiner software. Subsequently, refined CEL files were condensed with MAS5.0 and LOWESS normalized using all experiments as reference.
Unsupervised analysis
Principle component analysis (PCA) showing the relationships between individual samples was performed. Analysis of variance (ANOVA) was performed to detect differences in mRNA levels between the various timepoints, with the significance level set at false discovery rate (FDR) <0.01. The resulting gene list was clustered using the self-organizing map (SOM) method. All analyses were performed with the Expressionist Analyst Pro 4.0 software.
Supervised analysis
The data were subjected to a number of pair wise comparisons using the Expressionist Analyst Pro 4.0 software. Statistical analyses included pair wise comparisons between pretreatment samples (0 h) and samples following hCG exposure (12, 24 and 36 h). Probe sets were regarded to be hCG regulated if they were outside the triangular region in the Volcano plot [a plot of fold change (FC) versus t-test P-value)]with the corner values of an FC of five or higher and a t-test P < 0.000001.
Pathway analysis
The web-based GeneSifter& software (http://www.genesifter.net/web/) was used for gene ontological analysis (Bishop et al., 2009). Sets of significantly regulated genes (e.g. from SOM clustering analysis) were transferred to the MetaCore pathway analysis platform (GeneGo Inc., http://www.genego.com/) and overlaid to manually curated pathway maps.
Quantitative PCR
Reverse transcription was performed using 1 µg DNase (Promega, Madison, WI, USA)-treated RNA and Moloney murine leukemia virus reverse transcriptase (Promega) for 2 h at 37°C according to the supplier's protocol. The predicted M. mulatta gene sequences (NCBI database) were used to design TaqManTM probe and primer sets for the q-PCR assay using Primer Express software (Applied Biosystems, ABI, Foster City, CA, USA). Primers and probe sequences are listed in Table I. Primers were synthesized by Invitrogen (Carlsbad, CA, USA) and probes were synthesized by ABI. Q-PCR assays were performed using the TaqManTM PCR Core Reagent Kit with the ABI PRISM 7700 Sequence Detection System (ABI). To normalize the target signal, 18S rRNA was used as an active endogenous control in each well. Amplifications were conducted in a 10 µl final volume containing 250 nmol/l TaqManTM specific gene probe (labeled with the 5′ reporter dye 6-carboxyfluorescein and the 3′ MGBTM non-fluorescent quencher), 500 nmol/l forward and reverse primers, 250 nmol/l TaqManTM 18S probe (labeled with the 5′ reporter dye VIC), 80 nmol/l forward and reverse 18S primers, and 20 ng cDNA and 5 µl TaqManTM Universal PCR master mix containing ROX dye as a passive reference (Young and Stouffer, 2004). The PCRs were conducted in sealed 96-well optical plates with thermal cycler conditions of 2 min at 50°C, 10 min at 95°C and 50 cycles of 15 s at 95°C (DNA melting) and 1 min at 60°C (primer annealing/extension). The number of amplification cycles for the fluorescence to reach a determined threshold level (CT) was recorded for every unknown and an internal standard curve. The internal standard curve, used for relative mRNA quantification, was generated from five 10-fold dilutions of pooled macaque periovulatory samples (Young et al., 2002). CT values for unknown samples were used to extrapolate the amount of RNA equivalents from the internal standard curve. The RNA equivalent values were then divided by complimentary 18S rRNA equivalent values also derived from the same internal standard curve (Young and Stouffer, 2004).
Table I.
Primers and probes list.
| Gene name | Forward primer | Probe | Reverse primer |
|---|---|---|---|
| LHCGR | CAAATAAGCCTTCTCAGTCCACCTT | AGCTGTCCACATTGC | TAACACTCTGTGTAGCACGTCTTGTC |
| FSHR | GGTGATCCTAACTACCAGCCAATATAA | CACGGTCCCCAGGTT | TTCCAATGCAGAGATCAGCAA |
| StAR | GCACGGTCCCACTTGCAT | CGAAGACCAAACTTAC | AACCAC CCCTTGAGGTCGAT |
| HSD3B2 | AGGACGTCTCGGTCGTCATC | ACCGCCTGTATCATT | TACCTTTCACATTGACGTTCATGAT |
| HSD11B1 | ATGGGAGGACTAGACATGCTCATT | CCACATCACCAACACT | ATGCTTTTGCGCACATGGT |
| HSD11B2 | CACTGCCGATGCCCAATC | TCT GCG CCT CTC CAC T | AGGGTCTGTTTGGGCTCATG |
| CYP17A1 | CGTGGCCCCTATGCTCAT | CACAAGGCCAACGTT | TGATAACGTGTGTGCCCTTGTC |
| CYP19A | CCTCGCACCCAGATGAGACT | TCTTTACCCCCAGAAAC | GACCAGCCTTCTCTAGTGTTCCA |
| AREG | GGAATGGACCGCAATGACA | CGTGAACCATTTTC | CACTTCTTGAGGTAACCTCAAATCC |
| EREG | TGACATGAATGGCTACTGTTTGC | TGGACAGTGCATCTAC | CGGACACCAGTATAACCCACTTC |
| HSA2 | AGGGTCCCGGTGAGACAGAT | CGCAACACGTAACACA | TGCATGATGCAGATACTTTTGTTG |
| TNFAIP6 | TGGCGGCGTCTTTACAGATT | AGGCTTCCCAAATGA | AATGTGCCAGTAGCAGATTTGGT |
| KISS1 | CTTGGCAGCTACTGCTTTTCCT | TGCCACCCACTTTG | CAGAGGCCACCTTTTCTAATGG |
Statistical analyses
ANOVA followed by blocked Student–Newman–Keuls tests were performed to detect differences in mean mRNA levels between timepoints (Young and Stouffer, 2004), with the significance level set at P< 0.05, using the SigmaStat software package (SPSS Inc., Chicago, IL, USA).
Results
General characteristics of gene expression profiles
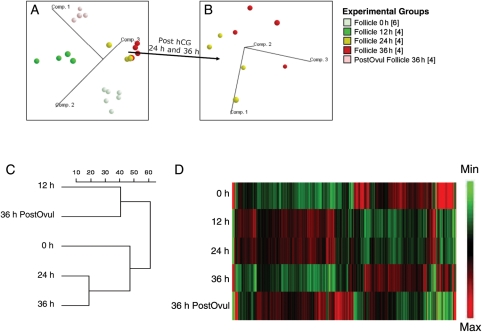
The initial simultaneous assessment of the gene expression values for individual follicles (see Materials and Methods) revealed uniform overall expression and distribution of transcripts within each timepoint, indicating expression profiles that were consistent with established standards for gene expression analyses (data not shown). The timepoints selected for collection of the macaque follicle (at 0, 12, 24 and 36 h of treatment) were estimated from previous studies in primates to cover the entire ovulatory phase (Fritz et al., 1992; Andersen et al., 1995). PCA revealed that samples from within the same timepoint after hCG treatment clustered together and clearly separated from each timepoint (Fig. 1A and B). PCA depicts the variance in gene expression profiles among samples. On the three-dimensional graphic, the distance between two-plotted spheres is proportional to the degree of similarity between the two groups' gene expression profiles, using all probe sets on the Affymetrix GeneChip Rhesus Macaque Genome Array. All sample groups formed distinct clusters. The clusters of 24 and 36 h (pre-ovulatory) samples are most similar but still separable as shown in Fig. 1B. The relatedness of gene expression between timepoints also revealed a similar gene expression patterns at 12 and 36 h post-ovulation (Fig. 1C). A heat map for all the probe sets whose intensity changed between 0 and 36 h post-ovulation (>2-FC; P< 0.001; ANOVA) is depicted at Fig. 1D. The entire database was submitted to NIH GEO (Submission number: GSE22776).
Figure 1.
PCA of RNA values from monkey follicles at different timepoints after hCG treatment defines relationships between individual samples. Samples are colored according to timepoints; the number of biological replicates is given in brackets. Each plotted ball represents the expression profile of an individual sample based on the projection of the data on the first three principal components, accounting for most of the variability in the data (labeled axes). The PCA is shown for all samples (A) and for the samples obtained 24 and 36 h post-hCG treatment (B). The dendrogram (C) illustrates the relatedness of the overall patterns of gene expression between the different timepoints with the scale bar representing an arbitrary measurement of Euclidean distance. (D) A gene expression heat map for all the probe sets from 0 to 36 h post-ovulation (>2-FC, P< 0.001).
Comparison of selected mRNA levels and patterns in the periovulatory follicle as estimated by q-PCR and microarray
Gonadotrophin receptors
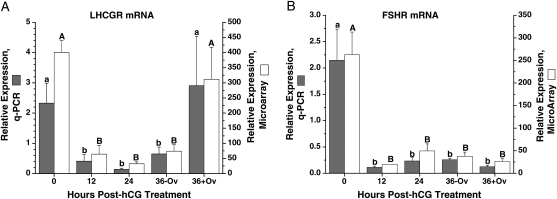
The levels and temporal patterns for mRNAs of two gonadotrophin receptors, LH/choriogonadotrophin receptor (LHCGR) and FSH receptor (FSHR), were analyzed. LHCGR mRNA levels, as quantified by q-PCR, were appreciable at 0 h, then significantly decreased at 12 h after hCG treatment (Fig. 2A). The levels remained low at 24 and 36 h post-hCG injection before ovulation, followed by a significant increase after ovulation. FSHR mRNA levels were also high at 0 h and decreased markedly at 12 h, but remained at low levels even after ovulation (Fig. 2B). The microarray results were identical to the q-PCR data.
Figure 2.
The relative expression of LH/choriogonadotrophin receptor (LHCGR) (A) and FSHR (B) mRNAs in the periovulatory follicle of the rhesus monkey. X-axis is the timepoint of follicle collection at 0 h (before hCG treatment), 12, 24 and 36 h (36−Ov, unruptured follicle; 36 + Ov, ruptured follicle) post-hCG treatment in a COv protocol (Young et al., 2003). Gray bars are the q-PCR quantification of mRNA levels normalized to 18S RNA. Open bars are the microarray evaluation of the relative gene expression. Different letters above the bars indicate a significant difference (P< 0.05) between the timepoints within one analysis (lower case, RT–PCR; upper case, microarray).
Steroid hormone biosynthesis
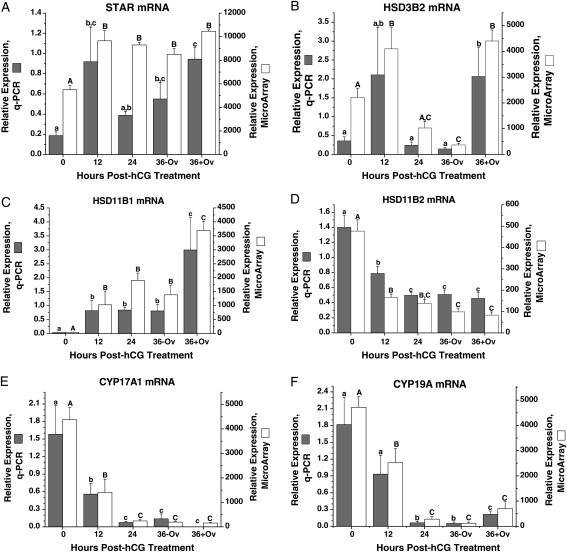
The mRNA levels and patterns of several proteins/enzymes involved in steroid hormone biosynthesis were also analyzed: steroidogenic acute regulatory protein (StAR), hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 (HSD3B2), hydroxysteroid (11-beta) dehydrogenase 1 (HSD11B1), hydroxysteroid (11-beta) dehydrogenase 2 (HSD11B2), cytochrome P450, family 17, subfamily A, polypeptide 1 (CYP17A1) and cytochrome P450, family 19, subfamily A, polypeptide 1 (CYP19A1). StAR mRNA (Fig. 3A) levels, as measured by q-PCR, were low before hCG treatment (0 h). After hCG administration, levels increased (P< 0.05) at 12 h, declined at 24 h and then returned to high levels by 36 h preovulation. HSD3B2 mRNA levels (Fig. 3B) were low at 0 h, tended to increase at 12 h, and then returned to low levels at 24 h. After ovulation, HSD3B2 mRNA levels increased (P< 0.05). The microarray data for StAR and HSD3B2 expression are similar to the q-PCR results.
Figure 3.
The relative expression of StAR (A), HSD3B2 (B), HSD11B1 (C), HSD11B2 (D), CYP17A1 (E) and CYP19A1 (F) mRNAs in the macaque periovulatory follicle. Refer to the legend of Fig. 2 for further details.
The level of HSD11B1 mRNA (Fig. 3C) was low but detectable at 0 h, then increased (P< 0.05) at 12 h post-hCG treatment followed by another increase after ovulation. In contrast, the expression pattern of HSD11B2 (Fig. 3D) was opposite to that of HSD11B1 (Fig. 3C); the highest levels of HDS11B2 mRNA were at 0 h, decreased (P< 0.05) at 12 h, followed another decrease at 24–36 h post-hCG treatment and remained low after ovulation. Both the HSD11B1 and HSD11B2 microarray data were identical to the q-PCR results.
The mRNA levels of CYP17A1 were highest at 0 h, then significantly decreased at 12 h after hCG treatment, followed by a further decrease to low but detectable levels by 24 h post-hCG that continued through ovulation (Fig. 3E). The pattern of CYP19A1 mRNA levels was comparable with that for CYP17A1, except levels increased modestly after ovulation (Fig. 3F). The microarray data for these two gene expressions were similar to the q-PCR results.
Cumulus–oocyte maturation
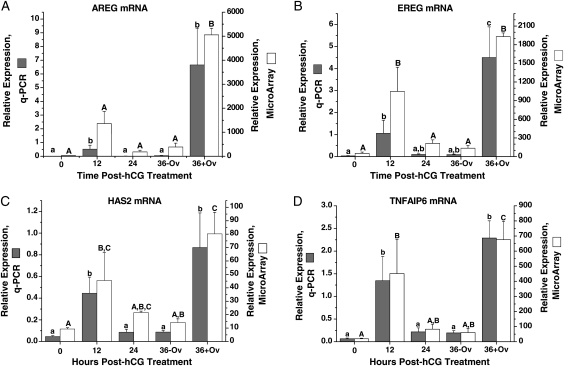
The mRNA expression patterns of three EGF-like factors (amphiregulin, AREG; epiregulin, EREG; and betacellulin, BTC), implicated in cumulus–oocyte events in the ovulatory follicle in rodents (Park et al., 2004; Conti et al., 2006) were analyzed (Fig. 4). AREG mRNA levels (Fig. 4A) were low but detectable at 0 h, increased at 12 h post-hCG, then declined at 24 h until ovulation. A significant secondary rise was observed at 36 h in the ruptured, but not unruptured, follicle. Similar to AREG, EREG mRNA levels (Fig. 4B) increased at 12 h, followed by an additional rise after ovulation. The microarray data for AREG and EREG are similar to their q-PCR results. In contrast, BTC mRNA levels were detectable but invariant between 0 and 36 h as measured by q-PCR or microarray (data not shown).
Figure 4.
The relative expression of AREG (A), EREG (B), HAS2 (C) and TNFAIP6 (D) mRNA in the macaque periovulatory follicle. Refer to the legend of Fig. 2 for further details.
The expression patterns of two genes, hyaluronan synthase 2 (HAS2) and tumor necrosis factor alpha-induced protein 6 (TNFAIP6), implicated in cumulus expansion in the rodent follicle (Kimura et al., 2002; Ochsner et al., 2003) were also analyzed. The level of HAS2 mRNA (Fig. 4C) was low at 0 h, followed by a significant increase at 12 h post-hCG treatment. Levels then decreased significantly to pretreatment levels at 24–36 h prior to follicle rupture. After ovulation, HAS2 mRNA levels again significantly increased. The biphasic pattern of TNFAIP6 mRNA levels (Fig. 4D) was similar to that for HAS2; peak levels occurred at 12 h in pre-ovulatory follicles and at 36 h after ovulation. The microarray data were similar to the q-PCR results.
Further analyses of expression patterns
ANOVA of the microarray data from each of the timepoints revealed 6947 significantly regulated probe sets (FDR < 0.01, data not shown). Preliminary analyses using profile distance search, identified probe sets that displayed expression patterns comparable with those of selected reference genes. For example, 154 probe sets displayed an expression profile that positively or negatively correlated (correlation coefficient of±0.83) with that of CYP17A1 (Fig. 3E). Moreover, 310 probe sets displayed an expression profile that correlated (correlation coefficient of±0.87) with that of AREG (Fig. 4A).
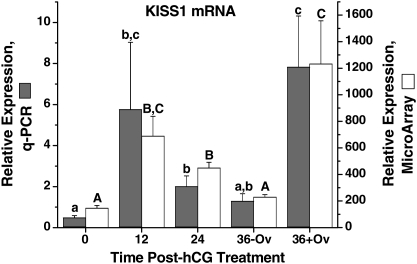
Review of the regulated probe sets identified other remarkable genes associated with periovulatory processes. The dynamic expression of KiSS-1 metastasis-suppressor (KISS1, kisspeptin), which directly governs the activation of GnRH neurons and hence hypothalamic-pituitary-gonadal function (Oakley et al., 2009), was evident in our microarray analysis of the macaque ovulatory follicle (Fig. 5). Q-PCR assay confirmed that KISS1 mRNA levels were low but detectable at 0 h, followed by a significant increase at 12 h post-hCG treatment, but declined at 36 h until follicle rupture. A second significant rise of KISS1 mRNA levels was at 36 h after ovulation.
Figure 5.
The relative expression of KISS1 mRNA in the macaque periovulatory follicle. Refer to the legend of Fig. 2 for further details.
Finally, gene ontology analysis of regulated probe sets revealed a number of cellular pathways or processes (Table II) altered during the periovulatory interval. Fifty-four significant gene ontology categories were identified, a number of which are involved in overlapping or similar cellular activities. In fact, five major themes accounted for the majority of the categories identified, including (i) cholesterol homeostasis, steroid synthesis and steroid metabolism; (ii) extracellular matrix synthesis, metabolism and binding; (iii) growth factors, growth factor receptors and growth factor signaling; (iv) immune function; as well as (v) prostaglandin synthesis and signaling.
Table II.
Ontologies of gene products significantly impacted by COva.
| Ontology | # genes presentb | Total # of genes in ontologyc | % of ontology representedd | z-score |
|---|---|---|---|---|
| Cytokine receptor activity | 14 | 39 | 35.9 | 5.89 |
| Steroid dehydrogenase activity, acting on the CH-OH group of donors | 9 | 19 | 47.4 | 5.86 |
| IgG binding | 5 | 7 | 71.4 | 5.78 |
| Oncostatin-M receptor activity | 3 | 3 | 100.0 | 5.52 |
| Steroid dehydrogenase activity | 9 | 21 | 42.9 | 5.44 |
| Immunoglobulin binding | 7 | 14 | 50.0 | 5.37 |
| Growth factor binding | 22 | 87 | 25.3 | 5.34 |
| Cytokine binding | 20 | 77 | 26.0 | 5.24 |
| Oxidoreductase activity, acting on the CH-CH group of donors | 7 | 15 | 46.7 | 5.11 |
| Receptor activity | 127 | 938 | 13.5 | 5.1 |
| Molecular transducer activity | 164 | 1283 | 12.8 | 5.06 |
| Signal transducer activity | 164 | 1283 | 12.8 | 5.06 |
| Proteoglycan binding | 6 | 12 | 50.0 | 4.97 |
| Pattern binding | 27 | 125 | 21.6 | 4.96 |
| Polysaccharide binding | 27 | 125 | 21.6 | 4.96 |
| Bile acid binding | 4 | 6 | 66.7 | 4.95 |
| Oxidoreductase activity, with oxidation of a pair of donors | 4 | 6 | 66.7 | 4.95 |
| Protein dimerization activity | 63 | 402 | 15.7 | 4.78 |
| Receptor binding | 90 | 636 | 14.2 | 4.7 |
| Lipopolysaccharide receptor activity | 3 | 4 | 75.0 | 4.62 |
| Prostaglandin E receptor activity | 3 | 4 | 75.0 | 4.62 |
| Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity | 3 | 4 | 75.0 | 4.62 |
| Heparan sulfate proteoglycan binding | 5 | 10 | 50.0 | 4.54 |
| MAP kinase phosphatase activity | 5 | 10 | 50.0 | 4.54 |
| MAP kinase tyrosine/serine/threonine phosphatase activity | 5 | 10 | 50.0 | 4.54 |
| Glycosaminoglycan binding | 24 | 114 | 21.1 | 4.53 |
| Ligand-dependent nuclear receptor activity | 12 | 46 | 26.1 | 4.07 |
| Interleukin-1 binding | 4 | 8 | 50.0 | 4.06 |
| Eicosanoid binding | 3 | 5 | 60.0 | 3.99 |
| Eicosatetraenoic acid binding | 3 | 5 | 60.0 | 3.99 |
| Interleukin-1 receptor activity | 3 | 5 | 60.0 | 3.99 |
| Transforming growth factor beta receptor activity, type I | 3 | 5 | 60.0 | 3.99 |
| Heparin binding | 18 | 85 | 21.2 | 3.95 |
| Insulin-like growth factor binding | 7 | 21 | 33.3 | 3.91 |
| Lipopolysaccharide binding | 4 | 9 | 44.4 | 3.72 |
| Transmembrane receptor activity | 80 | 617 | 13.0 | 3.56 |
| Protein binding | 634 | 6459 | 9.8 | 3.54 |
| Outward rectifier potassium channel activity | 3 | 6 | 50.0 | 3.52 |
| Oxidoreductase activity, with incorporation or reduction of molecular oxygen | 18 | 94 | 19.1 | 3.46 |
| Monocarboxylic acid binding | 10 | 41 | 24.4 | 3.46 |
| Pattern recognition receptor activity | 4 | 10 | 40.0 | 3.43 |
| Glutathione transferase activity | 5 | 15 | 33.3 | 3.3 |
| Transmembrane receptor protein phosphatase activity | 5 | 15 | 33.3 | 3.3 |
| Carbohydrate binding | 39 | 266 | 14.7 | 3.28 |
| Protein complex binding | 26 | 159 | 16.4 | 3.28 |
| Insulin-like growth factor receptor binding | 4 | 11 | 36.4 | 3.18 |
| Cofactor binding | 31 | 203 | 15.3 | 3.17 |
| Extracellular matrix binding | 6 | 21 | 28.6 | 3.14 |
| Hedgehog receptor activity | 3 | 7 | 42.9 | 3.14 |
| High-density lipoprotein binding | 3 | 7 | 42.9 | 3.14 |
| Molybdenum ion binding | 3 | 7 | 42.9 | 3.14 |
| Prostaglandin receptor activity | 3 | 7 | 42.9 | 3.14 |
| Hyaluronic acid binding | 5 | 16 | 31.3 | 3.12 |
| Protein homodimerization activity | 36 | 251 | 14.3 | 3.01 |
az-score >3.0 as determined by GeneSifter software analysis and >3 transcripts per ontogeny.
bIndicates the number of differentially expressed genes present in the periovulatory array database that belong to the indicated ontology.
cThe total number of genes assigned to the indicated ontology.
dDenotes the percentage of genes in the rhesus macaque ovulatory follicle that comprise each ontology.
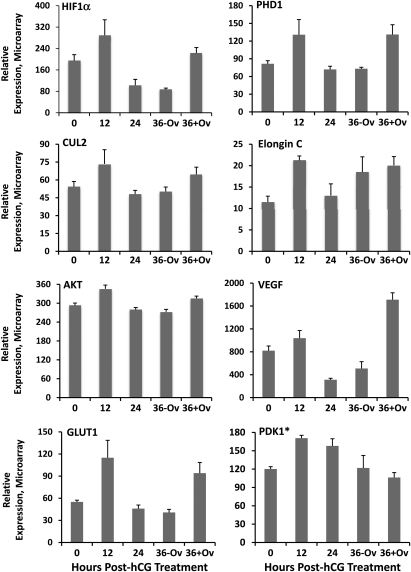
To further identify those genes with a similar pattern of expression involved in specific cellular processes, the pathway analysis software MetaCore™ was utilized. The periovulatory mRNA expression data were analyzed for significant (P< 0.05, ANOVA) overlapping or conserved cellular activities based on the software's curated database of protein–protein interactions. One pathway of note possessing numerous genes with a parallel pattern of expression included the hypoxia/hypoxia-inducible factor (HIF)-1 system. While all of the components involved in HIF1α degradation, stabilization, transcription, as well as key target genes were found to be abundantly expressed in the ovulatory follicle (Fig. 6 and Supplementary data, Fig. S1), a large subset of these genes exhibited a similar pattern of expression (Fig. 6) that included higher mRNA levels in 12 h pre-ovulatory follicles and at 36 h after ovulation relative to follicles isolated prior to hCG injection (0 h). In contrast, the level of these same mRNAs in unruptured follicles collected 24 and 36 h post-hCG were similar or lower than those observed before hCG administration (0 h).
Figure 6.
Genes of the HIF1α system that exhibit a similar pattern of differential expression in the rhesus macaque ovulatory follicle. MetaCore pathway analysis identified genes involved in regulating HIF1α stability, transcription, as well as those that are downstream HIF1α targets as being coordinately and significantly (P< 0.05 by ANOVA) regulated through the rhesus macaque periovulatory interval. The asterisk (*) indicates PDK1 was included in this category due to a significant increase in expression at 12 h after hCG, but does not exhibit a secondary increase in the ruptured 36 h follicle as is the case for the other genes. See Fig. 7 for definition of gene symbols and placement in HIF1α signaling and degradation pathways.
Discussion
This study details the changes in the transcriptome (mRNA levels) occurring within the primate periovulatory follicle at specific timepoints immediately prior (time 0) to and after (12, 24 and 36 h) the administration of an ovulatory hCG bolus. This laboratory group and others recently performed gene analyses on the granulosa cell and cumulus–oocyte compartments in human and non-human primate follicles generated by controlled ovarian stimulation (COS) protocols. These protocols take advantage of the fact that considerable material is available following multiple antral follicle development at precise timepoints after the ovulatory hCG bolus, but it is recognized that COS cell preparations arise from heterogeneous populations of follicles that differ in health, development and ability to ovulate or produce a mature oocyte capable of fertilization (Laufer et al., 1984; Goldman et al., 1993; Chaffin and Stouffer, 2000). Therefore, the current study utilized a COv protocol (Young et al., 2003) that permits the natural selection and maturation of the dominant, pre-ovulatory follicle within the menstrual cycle. In this protocol, pituitary gonadotrophin control of the follicle is acutely suppressed, thereby preventing a spontaneous midcycle gonadotrophin surge, which is replaced by an exogenous bolus of hCG. Thus, the single, pre-ovulatory follicle of the cycle can be evaluated at intervals up to and immediately after the follicle rupture.
In a prior study, we determined that ovulation occurred in the COv protocol at the earliest timepoint examined, i.e. 48 h after hCG administration. In the current study, 50% of the follicles ruptured within 36 h of hCG treatment. We, therefore, divided follicles collected at this timepoint into two categories: non-ovulated versus ovulated follicles. Based on evidence, including laparoscopic evaluation, that primate follicles begin to ovulate within 36 h of onset of the LH/hCG surge (Rawson and Dukelow, 1973; Weick et al., 1973), we propose that follicles collected at this timepoint are immediately (within a few hours) before or after timely ovulation. Since animals selected for COv protocols (≥120 pg E/ml; low dose, exogenous LH and FSH for 24–36 h prior to the hCG bolus) routinely ovulate (Young et al., 2003), it is unlikely that the unruptured follicles are degenerating so late in the ovulatory process. Nevertheless, caveats of the current study include: (i) since the pharmacokinetics of hCG levels after a bolus injection do not mimic the pattern of LH secretion during the midcycle gonadotrophin surge, the observed patterns of gene expression may differ somewhat from those in the periovulatory follicle of the spontaneous menstrual cycle, and (ii) since the levels of mRNAs detected in the entire follicle reflect their abundance and changes in various tissue compartments, the values may be dominated by activity in the mural granulosa or theca (when compared with oocyte–cumulus or ovarian surface epithelial) compartments. Thus, patterns of mRNA expression may not reflect accurately the changes occurring in each follicular compartment. Nevertheless, these data provide a genome-wide analysis of the transcripts in the primate pre/periovulatory follicle that are regulated directly or indirectly by a gonadotrophin (LH/CG) surge, and provide a database for comparison with results from primates following COS protocols, from other species, and from specific follicular compartments (Leo et al., 2001; Liu et al., 2001; Chin et al., 2002; Jo et al., 2004; Friedmann et al., 2005; Agca et al., 2006; Hernandez-Gonzalez et al., 2006; Perlman et al., 2006).
The current microarray analysis confirms recent reports that multiple mRNAs from various gene ontologies are up- or down-regulated in the mature follicle after exposure to an ovulatory, luteinizing bolus of gonadotrophin. Based on a 2006 review article by Espey and Richards (2006), less than 100 gene products were identified as specifically involved in ovulation-related processes, prior to the genomic revolution. In the past few years, investigators using limited or genome-wide DNA microarrays have expanded the database to include 100–1000 s of genes whose mRNA levels change in response to exposure to an ovulatory/surge level of LH or CG. In some cases, previous sampling was limited to two timepoints before versus after LH/CG exposure, but other attempts included three or more timepoints during the periovulatory interval, for analyzing whole ovaries or follicle cell preparations from rodents (Jo et al., 2004; Carletti and Christenson, 2009), or in the current study, whole pre/periovulatory follicles from macaques. Multiple timepoints permit more detailed analyses of the patterns of gene activity, and provided evidence for biphasic patterns of mRNA expression that suggest several, temporally dependent roles or activities during the cascade of events culminating in ovulation. Such patterns may be crucial to understanding the role(s) of gene products in the interwoven processes of cumulus–oocyte maturation, luteinization and ovulation that occur within the primate periovulatory follicle, as well as discerning the direct actions of LH/CG from those that are mediated by local factors produced or suppressed by gonadotrophin action.
The Affymetrix™ rhesus genome array is a recently developed research tool which, based on multi-center comparisons of RNA data from different macaque tissues, provides reproducible and valid information on levels of gene products (Duan et al., 2007). Nevertheless, our experimental design included selective verification of the general pattern and quantification of mRNA levels within the same tissue samples using q-PCR. The gene products chosen for comparison of microarray and q-PCR results encompassed a number of follicular activities. Our goal was not only to validate the microarray database, but to extrapolate examples based on the current literature from primarily non-primate species to gene activities and functions anticipated in the primate follicle. As noted in combination with our completed studies (Bogan et al., 2008; Bishop et al., 2009), the relative levels and general patterns of mRNA determined by q-PCR were typically comparable with those determined by microarray, with a few (e.g. StAR) exceptions.
Changes in mRNA levels for gonadotrophin receptors (LHCGR, FSHR) and steroidogenic enzymes were consistent with reported processes resulting in the conversion of the estrogen-secreting mature follicle into the primarily progesterone-secreting CL. According to the 2 cell-two gonadotrophin model for estrogen production, which appears relevant to the primate pre-ovulatory follicle (Sasano et al., 1989; Sanders and Stouffer, 1997), appreciable mRNA levels for FSHR, LHCGR, CYP17A1 (catalyzing androgen production) and CYP19A (converting androgen to estrogen) were expected and observed in follicles prior to hCG administration (0 h). The more rapid decline in mRNA levels for the gonadotrophin receptors (near baseline by 12 h post-hCG), compared with CYP17A1 and CYP19A, (baseline by 24 h post-hCG), may reflect receptor down-regulation (Menon et al., 2004; Hirsh et al., 2005) as well as luteal differentiation. Notably, the return of high mRNA levels for LHCGR, but not FSHR, after ovulation (36 h + Ov), is consistent with evidence that the developing CL in primates is a target tissue for the luteotropic LH, but not FSH (Eyster and Stouffer, 1985). The rapid (12 h post-hCG) increase in StAR and HSD3B2 mRNAs supports evidence that functional luteinization occurs within this time interval in the primate periovulatory follicle; progesterone levels in follicular fluid and both StAR and HSD3B2 mRNA levels in mural granulosa cells increase over 10-fold by 12 h post-hCG in COS protocols (Chaffin et al., 1999, 2000). Interestingly, biphasic patterns for StAR and HSD3B2 mRNA expression and enzyme activity were noted in these earlier studies (Chaffin et al., 2000, 1999). Although we could not rule out artifact (due to increasing numbers of degenerating, anovulatory follicles) in the COS protocols, the current results strongly suggest that this biphasic response in the progestogenic pathway (LDLR and CYP11A1 as well as StAR and HSD3B2) in luteinizing cells (Chaffin et al., 2000) also occurs during the ovulatory cascade in the dominant follicle of the menstrual cycle. The data are consistent with the concept that early progesterone production is important, at least locally in the follicle, for progesterone receptor-mediated actions promoting cell health, luteinization and ovulation (Hibbert et al., 1996; Peluso, 2003; Bridges et al., 2006), whereas the later, second increase after ovulation reflects further development of the early CL and progesterone production for hormonal as well as local actions (Stouffer, 2006).
Notably, the mRNA levels for two other steroidogenic enzymes, HSD11B1 and HSD11B2, displayed opposite patterns after hCG administration, with the former increasing and the latter decreasing markedly by 12 h post-hCG. Moreover, HSD11B1 expression increased further after ovulation. The HSD11B enzymes control glucocorticoid (primarily cortisol in primates) levels with the B1 form promoting cortisol production from cortisone, whereas the B2 form promotes the reverse step to form cortisone (Rae and Hillier, 2005). The HSD11B isoforms are expressed in several ovarian cell types of various species (see, Lewicka et al., 2003) including primates, and it is hypothesized that the ratio of their expression and products (cortisol : cortisone) has a role in controlling cell proliferation and local inflammatory responses (Rae and Hillier, 2005). Our results extend the findings of Fru et al. (2006) of the opposite expression patterns for HSD11B1 and 2 in macaque granulosa cells from COS follicles following hCG administration, and suggest an important role for cortisol during and after ovulation in the luteinizing follicle. Fru et al. (2006) also reported that hCG treatment promoted the expression of CYP2IA2, which with HSD11B1 and 2 could promote mineralocorticoid synthesis and action in the macaque periovulatory follicle. Further studies are warranted to evaluate the synthesis, regulation and roles of local gluco- and mineralo-corticoids in periovulatory events, and possibly oocyte quality (Lewicka et al., 2003).
In addition to the above gene products purportedly expressed within granulosa or theca cells of the follicle wall and/or the ovarian surface epithelium (HSD11), several others were selected that reportedly play key roles in rodents during cumulus–oocyte maturation and expansion (Richards, 2005). Notably, mRNAs for two EGF-like growth factors, AREG and EREG (but not BTC), appeared to increase in the macaque follicle within 12 h post-hCG. The data agree with Fru et al.'s (2007) report on AREG and EREG expression within 3–12 h post-hCG in macaque granulosa cells from COS follicles, and are consistent with the possibility that hCG-induced EGFs play a role in meiotic resumption and cumulus expansion in primate, as well as rodent (Conti et al., 2006), pre-ovulatory follicles. However, the decline in AREG and EREG expression prior to ovulation (24–36 h post-hCG) and renewed expression post ovulation, also suggests these growth factors have other undefined roles after cumulus–oocyte extrusion—presumably during luteal development. A remarkably similar mRNA pattern was noted for two critical components that are induced to produce extracellular matrix during cumulus expansion in rodents (Richards, 2005): HAS-2, the enzyme controlling hyaluronan synthesis; and TNFAIP6, a secreted protein that forms a stable complex with serum-derived interα trypsin inhibitor heavy-chain proteins and hyaluronan to stabilize the expanded matrix. Thus, key components of this process are temporally expressed and may play a critical role in cumulus expansion and oocyte quality (McKenzie et al., 2004) in primates. However, as for the EGFs, the second peak in HAS-2/TNFAIP6 expression after ovulation suggests additional roles after cumulus–oocyte extrusion, during development and function of the CL.
Further analyses of the transcriptome should yield valuable information on here-to-fore unrecognized or underappreciated pathways associated with the cascade of events within the primate periovulatory follicle that are important for cumulus–oocyte maturation, rupture of the follicle and release of the fertilizable oocyte, and conversion of the ruptured follicle wall into the newly forming CL. Inspection of the database can provide some remarkable observations, such as the discovery that mRNA levels for the KISS gene are dynamically expressed in the macaque follicle following the hCG bolus. A peptide product of KISS1 was originally isolated based on its suppression of tumor metastasis and named ‘metastin’, but studies since the mid-2000s identified its role controlling gonadotrophin secretion through its direct actions on hypothalamic GnRH neurons (Smith et al., 2006). Now termed kisspeptin, it is recognized as a key signal in the neural control of the onset (Tena-Sempere, 2006) and cyclicity (Caraty et al., 2007) of reproduction. Its transient expression in the pre-ovulatory follicle and subsequent re-expression in the ruptured follicle during luteal development in the rat (Castellano et al., 2006; Gaytan et al., 2009) and monkey (current study) supports potential ovarian actions of kisspeptin, perhaps in controlling tissue remodeling, angiogenesis and vascular flow (Mead et al., 2007), or a local GnRH regulatory system.
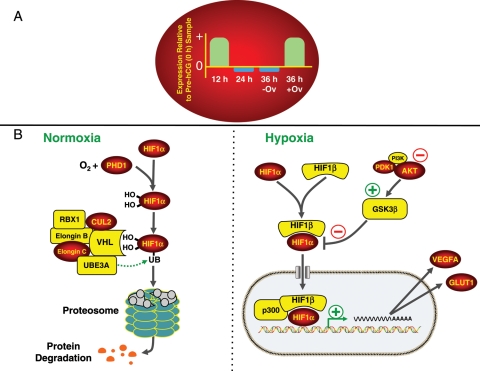
Currently, sophisticated software programs are being employed to analyze and discern different expression patterns, plus gene ontologies and cell pathways whose components are dynamically expressed during the periovulatory interval. Studying clusters of gene products with similar expression patterns (e.g. the profile of AREG, Fig. 4A) may aid in unraveling the processes associated with temporally distinct events, such as cumulus–oocyte maturation, follicle rupture and luteal development. Likewise, pathway analyses may identify key processes that are critical for follicle, oocyte and subsequent embryonic quality (Wiener-Megnazi et al., 2004). For example, recent evidence suggests that hypoxia-inducible factors in the pre-ovulatory follicle are critical for ovulation in mice (Kim et al., 2009), and that LH/CG regulates HIF1α mRNA expression in human luteinized granulosa cells (van den Driesche et al., 2008). The transcription factor HIF1α is regulated by oxygen concentrations, mainly via posttranslational modifications that determine its rate of degradation. Under normal atmospheric oxygen concentrations, two HIF1α prolines are hydroxylated (amino acid residues 564 and 462) by the oxygen-sensitive enzyme prolyl hydroxylase 1, which results in the interaction of HIF1α with an E3 ubiquitin-ligase complex containing von Hippel–Lindau protein and other accessory proteins (Nakayama, 2009; Webb et al., 2009). This, in turn, leads to the ubiquitinylation and degradation of HIF1α protein by the 26S proteasome. In a hypoxic environment, however, unmodified HIF1α is stabilized through its pairing with HIF1β (also known as aryl hydrocarbon receptor nuclear translocator, or ARNT), allowing for the translocation of the heterodimer into the nucleus and the increased expression of several target genes including VEGF and glucose transporter-1 (GLUT1). The effects of hypoxia on HIF1α activation and subsequent induction of target gene expression can be further fine tuned through several intracellular signaling components, including the AKT/ glycogen synthase kinase 3β pathway (Mottet et al., 2003). Taken together, the pathway analyses, as summarized in Fig. 7, revealed a period of heightened HIF1α regulation of target gene expression (e.g. VEGF and GLUT1) in the cells of the primate follicle 12 h post-hCG and following rupture.
Figure 7.
Dynamics of expression of hypoxia-related genes. The conserved pattern of gene expression is indicated in the red oval whereby a significant increase (P< 0.05) in mRNA levels was noted in follicles obtained 12 h post-hCG and following rupture (36 h post-hCG; 36 + Ov) relative to the pre-hCG (0 h) follicles. In contrast, similar or reduced mRNA levels of these same genes were observed in the unruptured follicles isolated 24 and 36 h (36−Ov) following hCG injection relative to pre-hCG samples. Columns for each timepoint represent a general change in expression (up and down) relative to the pre-hCG samples and not the FC in mRNA levels. (B) An overview of the mechanisms responsible for regulating HIF-1α degradation or stabilization/transcription that occurs under normoxic and hypoxic conditions, respectively. All genes denoted with a red oval exhibited a pattern of expression similar to the pattern shown in (A). All genes in yellow denote abundant but unchanging mRNA levels in follicles isolated throughout the periovulatory interval. Abbreviations include: v-akt murine thymoma viral oncogene homolog 1, AKT1, cullin 2, CUL2; glucose transporter 1, GLUT1; glycogen synthase kinase 3β, GSK3β; hypoxia-inducible factor 1α, HIF1α; hypoxia-inducible factor 1β, HIF1β; 3-phosphoinositide dependent protein kinase 1, PDK; phosphoinositide-3-kinase, PI3K; prolyl hydroxylase 1, PHD1; ring-box 1, RBX1; ubiquitin protein ligase E3A, UBE3A; vascular endothelial growth factor, VEGF; von Hippel–Lindau tumor suppressor, VHL.
In conclusion, genes whose mRNA levels varied during the interval from just prior to exposure to an ovulatory gonadotrophin (hCG) stimulus to after follicle rupture were identified in the dominant follicle of the rhesus monkey during the menstrual cycle. The patterns of mRNA expression for selected genes, as generated from macaque genome arrays, were typically verified by blinded q-PCR assays. The patterns of mRNA expression for selected genes confirmed and expanded information on purported theca (e.g. CYP17A, AREG), granulosa (CYP19A, FSHR), cumulus (HAS2, TNFAIP6) and surface epithelial (HSD11B)—associated gene products in the rodent/primate pre-ovulatory follicle. Moreover, the subsequent re-expression of several gene products (e.g. AREG, HAS2 and TNFAIP6) in the ruptured follicle suggests additional roles during development of the CL. This database will be of great value in identifying novel or unappreciated genes involved in molecular and cellular pathways leading to cumulus–oocyte maturation, ovulation, and luteal development. In addition, the transcriptome provides the basis for considering their translation and the role of their protein products in periovulatory events.
Authors’ roles
F.X. performed the majority of the experiments, collected all the samples, analyzed all data, supervised q-PCR experiments and drafted the manuscript. R.L.S. was the laboratory director, designed the project with input from B.L. at Schering, reviewed progress and data sets and coordinated review of the paper. J.M., G.L., M.P., T.W. and B.L. performed the microarrays at Schering, analyzed the gene expression profiles and made Fig. 1A and B. J.D.H. reviewed the q-PCR results, compiled Figs 6 and 7 and the Supplementary data, and assisted in manuscript preparation. J.W.W. performed q-PCR of HSD11B1, HSD11B2, CYP17A1 and CYP19A. A.B. performed q-PCR of LHCGR and FSHR. M.T. performed q-PCR of AREG and EREG. M.S. performed q-PCR of KISS1.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Funding
This work was supported by Schering Female AMPPA; National Institutes of Health (R01 HD20869, R01 HD42000, U54 CDRC HD55744, T32DK007674, RR000163); M.J. Murdock Charitable Trust.
Acknowledgements
A special thanks to the dedicated animal care technicians, and members of the Surgery Unit, Division of Animal Services, for their assistance in the animal protocols and tissue procurement. The expert services provided by the Hormone Technology (Assay) Core, Molecular & Cellular Biology Core at ONPRC are gratefully acknowledged.
Conflict of interest: none declared.
References
- Agca C, Ries JE, Kolath SJ, Kim JH, Forrester LJ, Antoniou E, Whitworth KM, Mathialagan N, Springer GK, Prather RS, et al. Luteinization of porcine preovulatory follicles leads to systematic changes in follicular gene expression. Reproduction. 2006;132:133–145. doi: 10.1530/rep.1.01163. [DOI] [PubMed] [Google Scholar]
- Andersen AG, Als-Nielsen B, Hornnes PJ, Franch Andersen L. Time interval from human chorionic gonadotrophin (HCG) injection to follicular rupture. Hum Reprod. 1995;10:3202–3205. doi: 10.1093/oxfordjournals.humrep.a135888. [DOI] [PubMed] [Google Scholar]
- Bishop CV, Hennebold JD, Stouffer RL. The effects of luteinizing hormone ablation/replacement versus steroid ablation/replacement on gene expression in the primate corpus luteum. Mol Hum Reprod. 2009;15:181–193. doi: 10.1093/molehr/gap005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan RL, Murphy MJ, Stouffer RL, Hennebold JD. Systematic determination of differential gene expression in the primate corpus luteum during the luteal phase of the menstrual cycle. Mol Endocrinol. 2008;22:1260–1273. doi: 10.1210/me.2007-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges PJ, Komar CM, Fortune JE. Gonadotrophin-induced expression of messenger ribonucleic acid for cyclooxygenase-2 and production of prostaglandins E and F2alpha in bovine preovulatory follicles are regulated by the progesterone receptor. Endocrinology. 2006;147:4713–4722. doi: 10.1210/en.2005-1575. [DOI] [PubMed] [Google Scholar]
- Caraty A, Smith JT, Lomet D, Ben Said S, Morrissey A, Cognie J, Doughton B, Baril G, Briant C, Clarke IJ. Kisspeptin synchronizes preovulatory surges in cyclical ewes and causes ovulation in seasonally acyclic ewes. Endocrinology. 2007;148:5258–5267. doi: 10.1210/en.2007-0554. [DOI] [PubMed] [Google Scholar]
- Carletti M, Christenson L. Rapid effects of luteinizing hormone on gene expression in the mural granulosa cells of mouse periovulatory follicles. Reproduction. 2009;137:843–855. doi: 10.1530/REP-08-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Gaytan M, Roa J, Vigo E, Navarro VM, Bellido C, Dieguez C, Aguilar E, Sanchez-Criado JE, Pellicer A, et al. Expression of KiSS-1 in rat ovary: putative local regulator of ovulation? Endocrinology. 2006;147:4852–4862. doi: 10.1210/en.2006-0117. [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Stouffer RL. Expression of matrix metalloproteinases and their tissue inhibitor messenger ribonucleic acids in macaque periovulatory granulosa cells: time course and steroid regulation. Biol Reprod. 1999;61:14–21. doi: 10.1095/biolreprod61.1.14. [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Stouffer RL. Role of gonadotrophins and progesterone in the regulation of morphological remodelling and atresia in the monkey peri-ovulatory follicle. Hum Reprod. 2000;15:2489–2495. doi: 10.1093/humrep/15.12.2489. [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Hess DL, Stouffer RL. Dynamics of periovulatory steroidogenesis in the rhesus monkey follicle after ovarian stimulation. Hum Reprod. 1999;14:642–649. doi: 10.1093/humrep/14.3.642. [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Dissen GA, Stouffer RL. Hormonal regulation of steroidogenic enzyme expression in granulosa cells during the peri-ovulatory interval in monkeys. Mol Hum Reprod. 2000;6:11–18. doi: 10.1093/molehr/6.1.11. [DOI] [PubMed] [Google Scholar]
- Chin KV, Seifer DB, Feng B, Lin Y, Shih WC. DNA microarray analysis of the expression profiles of luteinized granulosa cells as a function of ovarian reserve. Fertil Steril. 2002;77:1214–1218. doi: 10.1016/s0015-0282(02)03114-x. [DOI] [PubMed] [Google Scholar]
- Conti M, Hsieh M, Park JY, Su YQ. Role of the epidermal growth factor network in ovarian follicles. Mol Endocrinol. 2006;20:715–723. doi: 10.1210/me.2005-0185. [DOI] [PubMed] [Google Scholar]
- Curry TE, Jr, Smith MF. Impact of extracellular matrix remodeling on ovulation and the folliculo-luteal transition. Semin Reprod Med. 2006;24:228–241. doi: 10.1055/s-2006-948552. [DOI] [PubMed] [Google Scholar]
- Duan F, Spindel ER, Li YH, Norgren RB., Jr. Intercenter reliability and validity of the rhesus macaque GeneChip. BMC Genomics. 2007;8:61. doi: 10.1186/1471-2164-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espey LL. Ovulation. New York: Plenum Press; 1978. [Google Scholar]
- Espey LL. Current status of the hypothesis that mammalian ovulation is comparable to an inflammatory reaction. Biol Reprod. 1994;50:233–238. doi: 10.1095/biolreprod50.2.233. [DOI] [PubMed] [Google Scholar]
- Espey LL, Richards JS. Ovulation. 3rd edn. New York: Elsevier Academic Press; 2006. [Google Scholar]
- Eyster KM, Stouffer RL. Adenylate cyclase in the corpus luteum of the rhesus monkey. II. Sensitivity to nucleotides, gonadotropins, catecholamines, and nonhormonal activators. Endocrinology. 1985;116:1552–1558. doi: 10.1210/endo-116-4-1552. [DOI] [PubMed] [Google Scholar]
- Friedmann S, Dantes A, Amsterdam A. Ovarian transcriptomes as a tool for a global approach of genes modulated by gonadotropic hormones in human ovarian granulosa cells. Endocrine. 2005;26:259–265. doi: 10.1385/endo:26:3:259. [DOI] [PubMed] [Google Scholar]
- Fritz MA, McLachlan RI, Cohen NL, Dahl KD, Bremner WJ, Soules MR. Onset and characteristics of the midcycle surge in bioactive and immunoactive luteinizing hormone secretion in normal women: influence of physiological variations in periovulatory ovarian steroid hormone secretion. J Clin Endocrinol Metab. 1992;75:489–493. doi: 10.1210/jcem.75.2.1639949. [DOI] [PubMed] [Google Scholar]
- Fru KN, VandeVoort CA, Chaffin CL. Mineralocorticoid synthesis during the periovulatory interval in macaques. Biol Reprod. 2006;75:568–574. doi: 10.1095/biolreprod.106.053470. [DOI] [PubMed] [Google Scholar]
- Fru KN, Cherian-Shaw M, Puttabyatappa M, VandeVoort CA, Chaffin CL. Regulation of granulosa cell proliferation and EGF-like ligands during the periovulatory interval in monkeys. Hum Reprod. 2007;22:1247–1252. doi: 10.1093/humrep/del519. [DOI] [PubMed] [Google Scholar]
- Gaytan F, Gaytan M, Castellano JM, Romero M, Roa J, Aparicio B, Garrido N, Sanchez-Criado JE, Millar RP, Pellicer A, et al. KiSS-1 in the mammalian ovary: distribution of kisspeptin in human and marmoset and alterations in KiSS-1 mRNA levels in a rat model of ovulatory dysfunction. Am J Physiol Endocrinol Metab. 2009;296:E520–E531. doi: 10.1152/ajpendo.90895.2008. [DOI] [PubMed] [Google Scholar]
- Goldman S, Dirnfeld M, Gonen Y, Koifman M, Lissak A, Abramovici H. Different morphology and proliferative ability of cumulus and granulosa cells originating from cystic follicles aspirated from stimulated in vitro fertilization patients. Fertil Steril. 1993;59:601–605. doi: 10.1016/s0015-0282(16)55807-5. [DOI] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol Endocrinol. 2006;20:1300–1321. doi: 10.1210/me.2005-0420. [DOI] [PubMed] [Google Scholar]
- Hibbert ML, Stouffer RL, Wolf DP, Zelinski-Wooten MB. Midcycle administration of a progesterone synthesis inhibitor prevents ovulation in primates. Proc Natl Acad Sci USA. 1996;93:1897–1901. doi: 10.1073/pnas.93.5.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh L, Ben-Ami I, Freimann S, Dantes A, Tajima K, Kotsuji F, Amsterdam A. Desensitization to gonadotropic hormones: a model system for the regulation of a G-protein-coupled receptor with 7-transmembrane spanning regions. Biochem Biophys Res Commun. 2005;326:1–6. doi: 10.1016/j.bbrc.2004.10.168. [DOI] [PubMed] [Google Scholar]
- Irianni F, Hodgen GD. Mechanism of ovulation. Endocrinol Metab Clin North Am. 1992;21:19–38. [PubMed] [Google Scholar]
- Jo M, Gieske MC, Payne CE, Wheeler-Price SE, Gieske JB, Ignatius IV, Curry TE, Jr, Ko C. Development and application of a rat ovarian gene expression database. Endocrinology. 2004;145:5384–5396. doi: 10.1210/en.2004-0407. [DOI] [PubMed] [Google Scholar]
- Kim J, Bagchi IC, Bagchi MK. Signaling by hypoxia-inducible factors is critical for ovulation in mice. Endocrinology. 2009;150:3392–3400. doi: 10.1210/en.2008-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Konno Y, Miyoshi K, Matsumoto H, Sato E. Expression of hyaluronan synthases and CD44 messenger RNAs in porcine cumulus-oocyte complexes during in vitro maturation. Biol Reprod. 2002;66:707–717. doi: 10.1095/biolreprod66.3.707. [DOI] [PubMed] [Google Scholar]
- Laufer N, Tarlatzis BC, DeCherney AH, Masters JT, Haseltine FP, MacLusky N, Naftolin F. Asynchrony between human cumulus-corona cell complex and oocyte maturation after human menopausal gonadotropin treatment for in vitro fertilization. Fertil Steril. 1984;42:366–372. doi: 10.1016/s0015-0282(16)48074-x. [DOI] [PubMed] [Google Scholar]
- Leo CP, Pisarska MD, Hsueh AJ. DNA array analysis of changes in preovulatory gene expression in the rat ovary. Biol Reprod. 2001;65:269–276. doi: 10.1095/biolreprod65.1.269. [DOI] [PubMed] [Google Scholar]
- Lewicka S, von Hagens C, Hettinger U, Grunwald K, Vecsei P, Runnebaum B, Rabe T. Cortisol and cortisone in human follicular fluid and serum and the outcome of IVF treatment. Hum Reprod. 2003;18:1613–1617. doi: 10.1093/humrep/deg352. [DOI] [PubMed] [Google Scholar]
- Liu HC, He Z, Rosenwaks Z. Application of complementary DNA microarray (DNA chip) technology in the study of gene expression profiles during folliculogenesis. Fertil Steril. 2001;75:947–955. doi: 10.1016/s0015-0282(01)01706-x. [DOI] [PubMed] [Google Scholar]
- McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, Amato P, Matzuk MM. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 2004;19:2869–2874. doi: 10.1093/humrep/deh535. [DOI] [PubMed] [Google Scholar]
- Mead EJ, Maguire JJ, Kuc RE, Davenport AP. Kisspeptins are novel potent vasoconstrictors in humans, with a discrete localization of their receptor, G protein-coupled receptor 54, to atherosclerosis-prone vessels. Endocrinology. 2007;148:140–147. doi: 10.1210/en.2006-0818. [DOI] [PubMed] [Google Scholar]
- Menon KM, Munshi UM, Clouser CL, Nair AK. Regulation of luteinizing hormone/human chorionic gonadotropin receptor expression: a perspective. Biol Reprod. 2004;70:861–866. doi: 10.1095/biolreprod.103.024471. [DOI] [PubMed] [Google Scholar]
- Mottet D, Dumont V, Deccache Y, Demazy C, Ninane N, Raes M, Michiels C. Regulation of hypoxia-inducible factor-1alpha protein level during hypoxic conditions by the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3beta pathway in HepG2 cells. J Biol Chem. 2003;278:31277–31285. doi: 10.1074/jbc.M300763200. [DOI] [PubMed] [Google Scholar]
- Nakayama K. Cellular signal transduction of the hypoxia response. J Biochem. 2009;146:757–765. doi: 10.1093/jb/mvp167. [DOI] [PubMed] [Google Scholar]
- Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30:713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner SA, Russell DL, Day AJ, Breyer RM, Richards JS. Decreased expression of tumor necrosis factor-alpha-stimulated gene 6 in cumulus cells of the cyclooxygenase-2 and EP2 null mice. Endocrinology. 2003;144:1008–1019. doi: 10.1210/en.2002-220435. [DOI] [PubMed] [Google Scholar]
- Ohnishi J, Ohnishi E, Shibuya H, Takahashi T. Functions for proteinases in the ovulatory process. Biochim Biophys Acta. 2005;1751:95–109. doi: 10.1016/j.bbapap.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Park JY, Su YQ, Ariga M, Law E, Jin SL, Conti M. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science. 2004;303:682–684. doi: 10.1126/science.1092463. [DOI] [PubMed] [Google Scholar]
- Peluso JJ. Progesterone as a regulator of granulosa cell viability. J Steroid Biochem Mol Biol. 2003;85:167–173. doi: 10.1016/s0960-0760(03)00192-4. [DOI] [PubMed] [Google Scholar]
- Perlman S, Bouquin T, van den Hazel B, Jensen TH, Schambye HT, Knudsen S, Okkels JS. Transcriptome analysis of FSH and FSH variant stimulation in granulosa cells from IVM patients reveals novel regulated genes. Mol Hum Reprod. 2006;12:135–144. doi: 10.1093/molehr/gah247. [DOI] [PubMed] [Google Scholar]
- Rae MT, Hillier SG. Steroid signalling in the ovarian surface epithelium. Trends Endocrinol Metab. 2005;16:327–333. doi: 10.1016/j.tem.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Rawson JM, Dukelow WR. Observation of ovulation in Macaca fascicularis. J Reprod Fertil. 1973;34:187–190. doi: 10.1530/jrf.0.0340187. [DOI] [PubMed] [Google Scholar]
- Richards JS. Ovulation: new factors that prepare the oocyte for fertilization. Mol Cell Endocrinol. 2005;234:75–79. doi: 10.1016/j.mce.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Richards JS, Russell DL, Robker RL, Dajee M, Alliston TN. Molecular mechanisms of ovulation and luteinization. Mol Cell Endocrinol. 1998;145:47–54. doi: 10.1016/s0303-7207(98)00168-3. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Stouffer RL. Localization of steroidogenic enzymes in macaque luteal tissue during the menstrual cycle and simulated early pregnancy: immunohistochemical evidence supporting the two-cell model for estrogen production in the primate corpus luteum. Biol Reprod. 1997;56:1077–1087. doi: 10.1095/biolreprod56.5.1077. [DOI] [PubMed] [Google Scholar]
- Sasano H, Okamoto M, Mason JI, Simpson ER, Mendelson CR, Sasano N, Silverberg SG. Immunolocalization of aromatase, 17 alpha-hydroxylase and side-chain-cleavage cytochromes P-450 in the human ovary. J Reprod Fertil. 1989;85:163–169. doi: 10.1530/jrf.0.0850163. [DOI] [PubMed] [Google Scholar]
- Smith JT, Clifton DK, Steiner RA. Regulation of the neuroendocrine reproductive axis by kisspeptin-GPR54 signaling. Reproduction. 2006;131:623–630. doi: 10.1530/rep.1.00368. [DOI] [PubMed] [Google Scholar]
- Stouffer RL. Sturcture, Function, and Regulation of the Corpus Luteum. 3rd edn. Elsevier Academic Press; 2006. [Google Scholar]
- Stouffer RL, Martinez-Chequer JC, Molskness TA, Xu F, Hazzard TM. Regulation and action of angiogenic factors in the primate ovary. Arch Med Res. 2001;32:567–575. doi: 10.1016/s0188-4409(01)00323-x. [DOI] [PubMed] [Google Scholar]
- Tena-Sempere M. KiSS-1 and reproduction: focus on its role in the metabolic regulation of fertility. Neuroendocrinology. 2006;83:275–281. doi: 10.1159/000095549. [DOI] [PubMed] [Google Scholar]
- van den Driesche S, Myers M, Gay E, Thong KJ, Duncan WC. HCG up-regulates hypoxia inducible factor-1 alpha in luteinized granulosa cells: implications for the hormonal regulation of vascular endothelial growth factor A in the human corpus luteum. Mol Hum Reprod. 2008;14:455–464. doi: 10.1093/molehr/gan040. [DOI] [PubMed] [Google Scholar]
- Webb JD, Coleman ML, Pugh CW. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell Mol Life Sci. 2009;66:3539–3554. doi: 10.1007/s00018-009-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick RF, Dierschke DJ, Karsch FJ, Butler WR, Hotchkiss J, Knobil E. Periovulatory time courses of circulating gonadotropic and ovarian hormones in the rhesus monkey. Endocrinology. 1973;93:1140–1147. doi: 10.1210/endo-93-5-1140. [DOI] [PubMed] [Google Scholar]
- Wiener-Megnazi Z, Vardi L, Lissak A, Shnizer S, Reznick AZ, Ishai D, Lahav-Baratz S, Shiloh H, Koifman M, Dirnfeld M. Oxidative stress indices in follicular fluid as measured by the thermochemiluminescence assay correlate with outcome parameters in in vitro fertilization. Fertil Steril. 2004;82(Suppl 3):1171–1176. doi: 10.1016/j.fertnstert.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Wolf DP, Thomson JA, Zelinski-Wooten MB, Stouffer RL. In vitro fertilization-embryo transfer in nonhuman primates: the technique and its applications. Mol Reprod Dev. 1990;27:261–280. doi: 10.1002/mrd.1080270313. [DOI] [PubMed] [Google Scholar]
- Xu F, Stouffer RL. Local delivery of angiopoietin-2 into the preovulatory follicle terminates the menstrual cycle in rhesus monkeys. Biol Reprod. 2005;72:1352–1358. doi: 10.1095/biolreprod.104.037143. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Wallach EE. Studies of the mechanism(s) of mammalian ovulation. Fertil Steril. 1987;47:22–34. [PubMed] [Google Scholar]
- Young KA, Stouffer RL. Gonadotropin and steroid regulation of matrix metalloproteinases and their endogenous tissue inhibitors in the developed corpus luteum of the rhesus monkey during the menstrual cycle. Biol Reprod. 2004;70:244–252. doi: 10.1095/biolreprod.103.022053. [DOI] [PubMed] [Google Scholar]
- Young KA, Hennebold JD, Stouffer RL. Dynamic expression of mRNAs and proteins for matrix metalloproteinases and their tissue inhibitors in the primate corpus luteum during the menstrual cycle. Mol Hum Reprod. 2002;8:833–840. doi: 10.1093/molehr/8.9.833. [DOI] [PubMed] [Google Scholar]
- Young KA, Chaffin CL, Molskness TA, Stouffer RL. Controlled ovulation of the dominant follicle: a critical role for LH in the late follicular phase of the menstrual cycle. Hum Reprod. 2003;18:2257–2263. doi: 10.1093/humrep/deg467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.