Abstract
Our recently developed predoctoral training program in pharmacology and systems biology prepares students to become experts in systems-level models of disease that identify therapeutic targets and predict adverse effects or new uses of existing therapeutics. Multiple computational modeling modes are introduced throughout a curriculum that integrates basic cell and molecular sciences with the physiology and pathophysiology of disease states. Problem-based learning exercises enable students from different experimental and computational backgrounds to design experiments and interpret data quantitatively.
The need for and setting of the new predoctoral training program
New York City’s Mount Sinai School of Medicine has a tradition of interdisciplinary predoctoral training and a research continuum that encompasses basic, translational, and clinical investigations. This environment encourages PhD and MD/PhD students to become the agents of discovery and innovation as changes in biomedical research increase opportunities for clinical impact. Such a shifting research landscape accompanied burgeoning databases when genomics, transcriptomics, proteomics, and metabolomics emerged. The explosion of data necessitated use of computational approaches more broadly and deeply than ever before in biomedicine.1,2 The transformation of pharmacological sciences, in particular, into a more predictive science required experimental and computational models across scales, from cells to tissues, to organs, to organisms.3
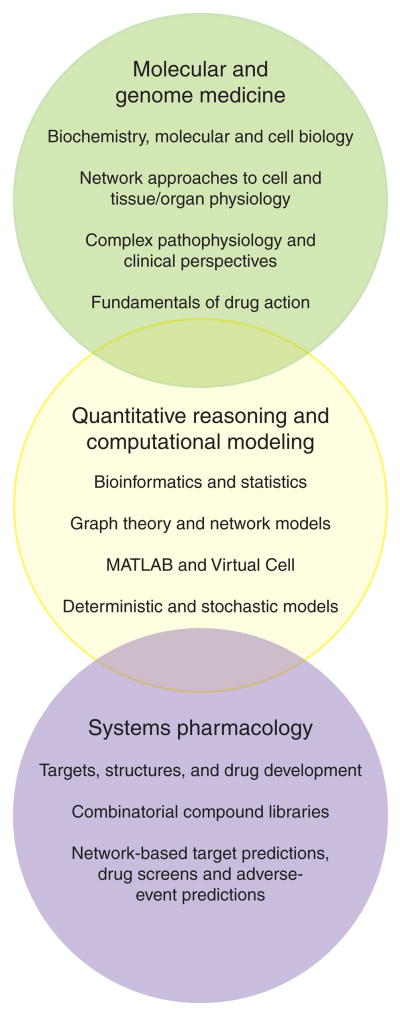
In this climate, a group of researcher– educators from several academic departments, catalyzed by the funding of a Systems Biology Center, have developed a new training program—Pharmacology and Systems Biology (PSB). PSB is un usual in the extent to which disease states, clinical data, diagnostics, and therapeutics are the context in which basic molecular/ cellular and physiological sciences are integrated with diverse systems and computational modeling approaches. Figure 1 is a diagrammatic representation of the interwoven domains of the PSB curriculum. In addition, we are committed to the principle that computational skills and quantitative reasoning should be developed across a PSB student group with different experimental and computational backgrounds. We expect all PSB students to learn to recognize opportunities to apply systems approaches to biomedical research problems and to collaborate effectively in doing so. A significant subset of PSB students is expected to become the developers of the new computational models and modeling approaches that lead to biomedical advances. The first presentation of PSB courses took place in the autumn of 2007. There are currently 29 PSB majors, 16 PhD students, and 13 MD/PhD students. Their undergraduate majors were as follows: biology/zoology/biomedicine/neurosciences, 16; engineering, 5; physics, 3; molecular biology/biochemistry, 2; mathematics, 1; chemistry, 1; social science, 1.
Figure 1.
The relationship between the integrated areas of the curriculum and the problems students will be prepared to approach. The top two domains are major features of the curriculum; the bottom domain illustrates areas of systems pharmacology in which Pharmacology and Systems Biology students will apply the skills acquired in the program.
Overall design of the PSB curriculum
The PSB curriculum is intertwined with general requirements of the graduate school (see Supplementary Table S1). The general requirements include research rotations, a course on responsible conduct of research, a statistics and quantitative reasoning course, and a schoolwide journal club (featuring articles coauthored and presented by students or postdocs) and seminar series. PSB students also participate in the well-attended Pharmacology Forum or another PSB-related journal club. In Pharmacology Forum, a student presents a recent research paper related to his or her research work followed by a research works-in-progress report. The faculty facilitator joins the subsequent discussion to pinpoint unresolved questions and controversies in the research area under discussion.
The PSB core curriculum is composed of three of four newly developed courses, depending on the track. The course load is comparable to that for other majors (see Supplementary Table S1 and the links to online PSB curriculum and course descriptions in Supplementary Information). The courses are (i) Systems Biomedicine: Molecules, Cells, and Networks, which presents molecular, cellular, and biochemical concepts and introduces modeling and network analysis in the context of disease states; (ii) Systems Biology: Computational Modeling, which promotes modeling skills using MATLAB (commercial software available at http://www.mathworks.com) and Virtual Cell (free software that can be used online at http://www.nrcam.uchc.edu) and prepares students for advanced computational and systems work; (iii) Quantitative Graduate Physiology; and (iv) Pharmacology. The Quantitative Graduate Physiology and Pharmacology courses have specific sections for graduate students that emphasize quantitative analyses, including systems and computational problem solving. These are integrated with sections of the medical school’s courses in these disciplines. Discussion and journal club sessions further amplify mechanistic and computational research approaches.
Two advanced elective courses are offered in PSB. The Advanced Cell Signaling course uses primary literature, including experiments and theoretical concepts that led to current knowledge, to develop a systems-level understanding of information flow through various cell signaling pathways and networks.4 The second course, Pharmacogenomics, analyzes differing drug effects in individuals that have their basis in polymorphisms in the human genome. Students explore the methods used to identify monogenic and polygenic clinical disorders, the central role of single-nucleotide polymorphisms, assays such as prognostic genotyping, diagnostic expression profiling, and genomic bioinformatics. Most of the remaining advanced course work uses the format of customized mini-courses. Groups of two to five students meet weekly with a faculty member for guided student presentations of research papers and critical-review papers in areas such as cell signaling networks and systems pharmacology, bioinformatics, network building and analysis, differential equation–based multiscale models, cardiac functions, and structural considerations in drug design.
Content and organization of the Systems Biomedicine and Systems Biology courses
The Systems Biomedicine course presents basic cell, biochemical, and molecular sciences in sufficient detail to provide appreciation of molecular mechanisms and regulatory complexity. This material is offered in a physiological and disease context that fosters appreciation of constraints of cell and tissue organization and differences in the functional anatomy and physiology of different organs. An introductory module introduces MATLAB as well as experimental methods and principles that will recur in the following four modules. Each of those four modules has a different disease as its focus, i.e., diabetes, cancer, renal disease, and drug abuse (Table 1). The disease contexts are chosen on the basis of the core basic science and pathophysiological principles they illustrate and the available faculty expertise in clinical, research, and computational activities related to the disease. Genetic, clinical, and epidemiological data are presented in each module. Current preventive and therapeutic interventions are noted, along with pipeline ideas for new therapeutics. Discussion topics include principles, methods, and challenges involved in clinical and epidemiological studies and in drug development. The importance of modeling systems at multiple scales emerges clearly as the students study disease states and the complexity of drug effects in diverse populations.5,6 Modeling approaches to complex data sets and networks are introduced in each module, along with computational problems that drive home insights into biological mechanisms.
Table 1.
Outline of the topics and principles presented in the Systems Biomedicine and Systems Biology courses in the Pharmacology and Systems Biology program
| Course | Module or topic | Subtopics and principles |
|---|---|---|
| Systems Biomedicine: Molecules, Cells, and Networks | Module | Representative concept areas |
| Module 1: General Mechanistic Principles and Introduction to MATLAB | Protein structure, nucleic acid structure Introduction to MATLAB enzyme kinetics (and MATLAB workshop) Receptor function and ligand binding, cell signaling Cell cycle Design of metabolic pathways Physiological homeostasis Genetics and pedigrees, transcription Epigenetics Omics: Genomics and proteomics, including pitfalls and principles of error assessment |
|
| Module 2: Diabetes | Protein processing Secretion and modulation of secretion Metabolic differentiation of different tissues and relationship to gene expression Modes of hormone-mediated signaling Protein processing, transporter-enzyme kinetics, organ crosstalk, ER stress and inflammation Learning from single-mutation-based diabetes SNPs and GWAS Epidemiology of diabetes/obesity Therapeutics and their mode of action, drug discovery |
|
| Module 3: Cancer | Signaling pathways and networks Growth control Cell cycle and mutations in the cell cycle (and MATLAB workshop) Cancer pathology Cancer genetics and genomics The lymphatic system and metastasis Chemotherapeutics and their pitfalls Mechanism-based new therapeutics Cancer epidemiology and statistical models |
|
| Module 4: Renal Disease | Anatomy of the kidney, its specialized cells Principles of filtration and uptake in the kidney Cell polarization and cytoskeleton Diseases of renal podocytes Systems approaches to understanding origins and progression of podocyte diseases vs. single-gene target approaches Channel and transporter structure–function; channelopathies, their systemic physiological effects, and therapeutics Personalized medicine for kidney disease and its systems basis |
|
| Module 5: Drug Abuse | Clinical perspective on drug abuse and its biological underpinnings Neurocircuitry of addiction Receptors and transporters in the relevant brain regions Neuroimaging of receptors Elements of synaptic structure and function Learning and memory Current and potential therapeutic approaches Systems modeling of addiction |
|
| Systems Biology: Computational Modeling | Topic | |
| Graph theory and networks | Representation of biological systems as graphs Tools for building metabolic and signaling networks Quantitative statistical analysis Identification of motifs |
|
| Statistical models for large data sets | Clustering Principal components analysis Partial least-squares regression |
|
| Deterministic models, biochemical signaling models | Representing reactions as systems of ODEs Models in MATLAB Stability analysis of dynamical systems, oscillatory cell cycle models, bistable (all-or-none) Signaling models: multicompartment ODE models Modeling spatial regulation: partial differential equations models |
|
| Physiological models | Action potential models, spatial propagation of electrical signals Calcium signaling models |
|
| Practical considerations in signaling models | Extracting parameters from the literature Estimating unknown parameters Estimating errors |
|
| Stochastic models | Waiting times Poisson probability distributions Gillespie’s algorithm Cell-to-cell variability |
ER, endoplasmic reticulum; GWAS, genome-wide association studies; ODE, ordinary differential equation; SNP, single-nucleotide polymorphism.
The Systems Biology course further builds computational skills through presentation of multiple modeling strategies at a more advanced introductory level. The instructors take a case-based approach to providing both conceptual and hands-on experience with the following contemporary techniques: graph theory and network analysis, statistical models and principal-component analysis, ordinary differential equation models, partial differential equation models, and stochastic models (Table 1). Problem sets require students both to implement models using MATLAB or other computational tools and to answer questions that require insight into the underlying biological process. Students are prepared to develop and perform simulations with simple models and to collaborate with experienced modelers to address more complex questions.
PSB progress report and plans
Students have been enthusiastic about the disease context of the PSB curriculum. They have provided invaluable suggestions on course refinements. Our experience shows that the integration of the elements in the introductory Systems Biomedicine course is challenging and depends on “integrators” who participate in the class and in online discussions. The integrators are the teaching assistants and module and course directors who are present throughout a specific module or throughout the whole course. They integrate material presented by faculty session leaders who have individual “domain expertise.” Integrators enter class discussions and engage students online, making connections between molecular/cellular mechanisms and physiological/clinical observations within and across modules. They emphasize recurring concepts as well as contrasts in mechanisms and therapeutic strategies in different organ system and disease contexts. They highlight areas that are yet to be addressed or for which there are conflicting data and send out new publications that impact those gaps. The Systems Biology course has also been well received and has successfully prepared students to pursue advanced systems/modeling studies and to initiate research projects that utilize these skills. One goal for the course is introduction of tiered problem sets that will work for students who have different levels of quantitative skills. Another goal is to emphasize modeling across scales. In both courses we will introduce workshop sessions in which students teach one another particular topics and introduce weekly problem-solving sessions for collaborative work by the students.
The course work prepares students for thesis research that covers a broad range of biological problems and approaches, both experimental and computational. Examples of research projects of current PSB students include the use of human protein–protein interaction networks to probe relationships between drug adverse events, drug targets, and genes involved in congenital diseases, related to long-QT syndrome. Te goals are to understand the basis whereby some drugs prolong the QT interval as a side effect and move toward predictive models for occurrence of such side effects. Another project involves multiscale hybrid models combining stochastic models of cell spreading with ordinary-differential-equation models to understand how integrin signaling networks control actin filament–driven cell spreading. Computational predictions are tested by experiments using total-internal-reflection fluorescence microscopy. A third project uses high-throughput RNA interference techniques to identify the signaling components involved in pathogen sensing and dendritic cell (DC) activation. The goal is to build a repository capturing information on the current level of understanding of signaling events in DCs to inform experiments that, together with further computational modeling, will describe and predict the response of human myeloid DCs to specific pathogens and their components.
Supplementary Material
Acknowledgments
The work of the authors on the training program described here was supported by training grant T32 GM-62754 for Integrated Training in Pharmacological Sciences (to T.A.K.) and Systems Biology Center grant P50 GM-071558 Systems Biology Center–New York (to R.I.) from the National Institute of General Medical Sciences of the National Institutes of Health. We also thank many colleagues for their participation in the original planning and the current execution of the program.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
CONFLICT OF INTEREST
The authors declared no conflict of interest.
References
- 1.Bialek W, Botstein D. Introductory science and mathematics education for 21st-century biologists. Science. 2004;303:788–790. doi: 10.1126/science.1095480. [DOI] [PubMed] [Google Scholar]
- 2.Morel NM, et al. Primer on medical genomics. Part XIV: Introduction to systems biology—a new approach to understanding disease and treatment. Mayo Clin Proc. 2004;79:651–658. doi: 10.4065/79.5.651. [DOI] [PubMed] [Google Scholar]
- 3.Preusch PC. What IS training in the pharmacological sciences? Mol Interv. 2002;2:270–275. doi: 10.1124/mi.2.5.270. [DOI] [PubMed] [Google Scholar]
- 4.Iyengar R, et al. Inquiry learning. Integrating content detail and critical reasoning by peer review. Science. 2008;319:1189–1190. doi: 10.1126/science.1149875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25:2466–2472. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wist AD, Berger SI, Iyengar R. Systems pharmacology and genome medicine: a future perspective. Genome Med. 2009;1:11. doi: 10.1186/gm11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.