Abstract
A scientific database can be a powerful tool for biologists in an era where large-scale genomic analysis, combined with smaller-scale scientific results, provides new insights into the roles of genes and their products in the cell. However, the collection and assimilation of data is, in itself, not enough to make a database useful. The data must be incorporated into the database and presented to the user in an intuitive and biologically significant manner. Most importantly, this presentation must be driven by the user’s point of view; that is, from a biological perspective. The success of a scientific database can therefore be measured by the response of its users – statistically, by usage numbers and, in a less quantifiable way, by its relationship with the community it serves and its ability to serve as a model for similar projects. Since its inception ten years ago, the Saccharomyces Genome Database (SGD) has seen a dramatic increase in its usage, has developed and maintained a positive working relationship with the yeast research community, and has served as a template for at least one other database. The success of SGD, as measured by these criteria, is due in large part to philosophies that have guided its mission and organisation since it was established in 1993. This paper aims to detail these philosophies and how they shape the organisation and presentation of the database.
Keywords: S. cerevisiae, database, genome-wide analysis, bioinformatics, yeast
INTRODUCTION
Saccharomyces cerevisiae, because it is a single–celled eukaryote with a relatively fast generation time, is a well-studied organism for which many types of scientific data exist. These include genetic and biochemical studies, a complete genomic sequence and, more recently, extensive data from genome-wide analyses. A little over ten years ago, even before the S. cerevisiae genome was sequenced to completion, it became clear that the marriage of database technology and biology could provide a mechanism for storing, organising and retrieving scientific information about this organism. The resulting database would provide convenient access to large amounts of data and, if organised correctly, would bring together different pieces of information in order to provide scientists with a larger view of the role of genes and their products in the cell. Furthermore, the solution to such puzzles in the model organism yeast might provide clues to the roles of related genes in other organisms.
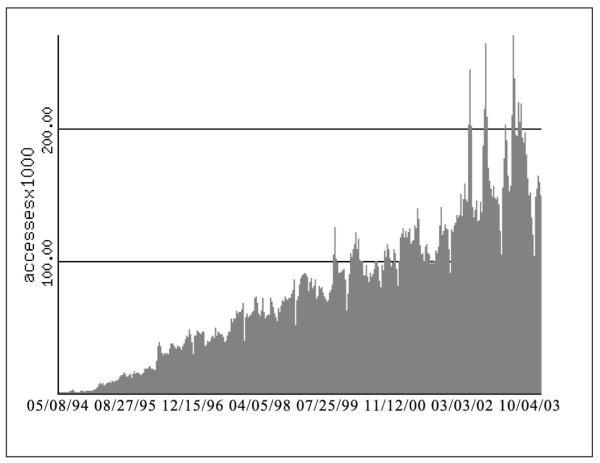
With these goals in mind, the Saccharomyces Genome Database (SGD)1 was established in 1993. It began as a set of spreadsheets, subsequently migrated to software that had originally been designed to house Caenorhaditis elegans data (ACEDB), 2 became available on the World-Wide Web in 1994, and is now built on top of a relational database. Since its inception, SGD has continued to grow in usage (Figure 1), currently averaging approximately 30,000 visits per week, and a total of 160,000 hits per week. Its success is quantified by its usage statistics,3 but can also be measured by the growth of the data it contains and the resources it provides. SGD has never entered a ‘maintenance’ mode, where the primary activity would be maintenance of existing data; rather, it has continually expanded in terms of its content, tools, interaction with outside communities and the expertise of its staff. SGD’s success can also be measured by the positive relationship it shares with the yeast community and others, as well as by the fact that it has served as a model for at least one emerging database, DictyBase,4 and several others that are in the planning stages.
Figure 1.
Usage of the Saccharomyces Genome Database website has increased steadily since its inception. This diagram covers the period from 8th May, 1994, to 4th October, 2003, and shows the number of requests per week for HTML pages, including those generated by cgi scripts accessing the database for information specific to a given gene. Image maps, redirections, requests from any computer in the Stanford.EDU domain, and personal WWW pages are not included in the usage statistics. Certain hosts have been excluded because they have indexed our site. The most up-to-date version of this graph is available online3
Since simply storing data is not enough to make a database useful, what is it that makes SGD a widely used and accepted database in the community it serves? In a general sense, this acceptance is due to a clarity of vision regarding the role and goals of SGD as a scientific database. This clarity of vision was present in SGD’s infancy and continues to guide strategies and daily decisions about the incorporation and presentation of data to the user. Specifically, SGD’s aim is to design and implement a resource that provides comprehensive annotated information about the Saccharomyces genome, with emphasis on the biology of the genes, their products and the interactions of these products in the cell. Thus, although SGD provides a genomic view of S. cerevisiae, its focus is on the biology of the cell’s components. Other important goals that drive SGD’s vision include broadening its relationship with the yeast and biomedical research communities, the incorporation of controlled, formalised vocabulary, the continual acquisition of new data and development of new resources, and the implementation of technical advances to the database. The achievement of these goals is made possible by the general principles that guide SGD, the daily decisions that drive the content, design and presentation of the database, and the working dynamics within SGD and with the community it serves. Each of these factors will be discussed below.
RESULTS
At the heart of SGD’s success as a scientific database lies a solid understanding of its scope and role. This understanding, in turn, is guided by two fundamental principles. These include the recognition that SGD exists as a service organisation, and the conviction that biology, as opposed to computer science, should drive the design, both of the user interfaces and of the underlying data storage. These principles also help guide the composition and dynamics of the SGD staff, which have contributed greatly to the character and success of the database.
SGD is a service organisation
The first guiding principle for SGD is the conviction that its primary function is service to the scientific community. Towards that end, SGD is committed to the free and open exchange of scientific data, to neutral presentation of all data, and to maintenance of a close, responsive relationship with the yeast research community. SGD’s funding as a National Research Resource attests to the fact that it is considered to provide continuing value to the community.
SGD is committed to the open exchange of scientific data
As a publicly funded database and service organisation, SGD is committed to providing free and open access to its data. Virtually all information is available for download, without restriction, from SGD’s ftp site.5 No limitations are placed on the use of these data, other than a request that SGD is cited as the source and that the data are not repackaged and sold. The funding sources of SGD mandate that it remain free to all users, both academic and commercial; it would be counter to SGD’s basic structure to require fees for access to these data. SGD’s location within the Department of Genetics6 in the Stanford University School of Medicine facilitates an open environment and helps SGD maintain an academic culture. The department is supportive of SGD’s efforts, and individual scientists at Stanford contribute both data and feedback to the organisation.
SGD takes a neutral position in the community
In order to serve the greater yeast research community, SGD curators strive to acquire and display data in a neutral, non-judgmental manner. A neutral position defines the database as a resource that represents and serves the entire community; in a sense, it allows the community to ‘own’ the database.
Neutrality necessitates that all data presented be derived from peer-reviewed publications; thus their validity is determined by the academic community rather than by SGD. Referencing all data in SGD helps ensure the quality and accountability. Historically, SGD has included some unreferenced data, primarily short descriptive phrases about genes and phenotypes. However, this policy has proven to be less than satisfactory, and all unreferenced information is currently being reviewed and associated with published sources. All new annotations and data in SGD cite published journal articles. For example, Gene Summary Paragraphs, composed by SGD curators based on their broad reading of the published literature, are referenced throughout with published journal articles. Similarly, all Gene Ontology (GO)7,8 annotations, describing the biological process, molecular function, or cellular component of a gene product, are associated with references. Currently, SGD’s GO annotations are supported by 4,400 different published references. Experimental data, such as microarray datasets or large-scale localisation studies, must also be published before being incorporated into SGD. In fact, SGD contains instances of published data that describe conflicting results for some gene products. The inclusion of conflicting results is a crucial aspect of this neutrality: rather than making judgments regarding conflicting information, SGD presents both sides to researchers and allows them to draw their own informed conclusions.
Another area in which SGD’s philosophy of maintaining neutrality becomes important is gene nomenclature. In 1994, shortly after SGD was created, the task of maintaining the S. cerevisiae Gene Name Registry,9 the complete list of all S. cerevisiae gene names, was transferred to SGD by Robert Mortimer, who had maintained the list for 30 years. Maintaining this list can sometimes present issues and conflicts that are difficult to resolve. To handle gene nomenclature in a consistent manner, SGD developed Gene Naming Guidelines that were reviewed and approved by the yeast community. Any changes or expansions to these guidelines are presented to the yeast research community for review and acceptance during key conferences. An important component of the Gene Naming Guidelines, and one that assists in neutrality, is that the yeast community itself is charged with the responsibility for naming yeast genes. Researchers studying a given gene use a web interface to propose a name that is then checked by SGD curators to ensure that it meets simple guidelines (such as uniqueness and correct formatting) before it is registered. Occasionally conflicts arise when two groups try to register different names for the same gene at roughly the same time, or when members of the yeast research community propose that an accepted gene name be changed. In these situations, the first priority is for the community to work together to establish consensus. SGD’s role is to foster communication between the various groups and to make sure that all interested parties are included in the discussion. SGD staff do not make judgments as to which of the competing gene names is more appropriate, but facilitate the process of reaching consensus within the community. In the fairly rare instance where consensus is not possible, the first published gene name becomes the standard name.
SGD has a close, responsive relationship with the yeast research community
Another essential requirement for a service organisation is the cultivation and maintenance of a strong relationship with the community it serves. SGD is dedicated to determining and responding to the needs of the scientific community. In addition, it relies heavily on the community to keep the information in the database accurate, current and relevant. Toward these ends, SGD invites community feedback and suggestions by providing several different forums for user commentary. In turn, the strong feedback SGD receives from the community is evidence that the community values the service SGD provides. This feedback includes correspondence from users, invitations to submit manuscripts and present at meetings, and the submission of large-scale and other data sets.
One forum that has consistently proven useful is daily e-mail communication with the yeast research community. SGD receives approximately 50 messages per week at its general ‘yeast-curator’ address, primarily through use of the ‘Send a Message to the SGD Curators’ link located at the bottom of most SGD web pages. SGD curators check this e-mail account daily, and reply personally and promptly to each message (typically within a single business day). In addition to questions about using the database, messages to SGD include corrections to existing data and suggestions for improvements, new resources, and new types of data. Reports of inaccuracies or errors encountered while using the database are viewed by SGD staff as opportunities to improve the database based on expert input from the community. Many user suggestions prompt improvements and expansions of SGD and are therefore considered invaluable. For example, the ‘Single page format’ for locus pages was developed in direct response to the community’s requests for an alternative display, preferred by some for viewing and printing locus information. In another example, ‘Webminer’,10,11 a microarray data search tool developed by Max Heiman at UCSF, was added as a database enhancement after suggestions were received via curator mail.
SGD staff members also meet directly with the community they serve by holding regular meetings with a scientific advisory board and by attending scientific meetings. Meetings with members of the advisory board, which includes both expert yeast researchers and bioinformatics specialists, are designed to assess the value of current SGD tools and to obtain guidance on future directions. By attending a wide variety of scientific meetings, SGD curators and programmers are able to meet a diverse group of researchers with differing interests and needs. At these meetings, SGD often presents computer demonstrations and posters as a way of both informing the community about its resources and speaking directly with users to better understand their needs. Computer demonstrations have proven to be particularly effective as they allow users to test resources and provide feedback directly to curators as they manoeuvre within the database.
The continual collection of usage statistics3 provides yet another form of feedback regarding how the database is used by members of the research community. These data include overall usage statistics as well as the number of hits for each of the top 30 web pages accessed, and thereby serve as an indicator of the most widely used resources (Figure 2). A relatively low number of hits may indicate that a particular resource is not well advertised, not easily found, difficult to use or of limited interest to the community. Analysing usage data also provides essential information about how people navigate the database. If users appear to be taking circuitous routes to get to a particular resource, it is often an indication that the resource is not easily found. Analysis of usage statistics guided the choice and ordering of menu items displayed in the navigation bar that appears at the top of most SGD pages, thereby providing links to the most commonly used resources from almost any location within the database.
Figure 2.

This pie chart breaks down requests during the week of 28th September, 2003, to 4th October, 2003, for HTML pages by document accessed, including those generated by cgi scripts accessing the database for information specific to a given gene. As is typical, requests for Locus pages make up nearly 30 per cent of all hits while requests for the home page itself make up about 10 per cent. The relative popularity of other tools varies from week to week. The individual slices in this pie chart show a representative sample of SGD’s most popular tools: 1. Locus pages (27.9 per cent), 2. Home page (10.5 per cent), 3. BLAST (7.0 per cent), 4. Sequence Retrieval (6.5 per cent), 5. ORF Map (6.0 per cent), 6. Gene/Sequence Resources (5.5 per cent), 7. Quick Search (4.4 per cent), 8. GO term pages (2.8 per cent), 9. Literature Guide (2.1 per cent), Other tools and pages (27.3 per cent). Access to weekly reports of SGD usage statistics is available via the main Saccharomyces Genome Database Usage Statistics page3
In addition to facilitating communication between itself and the yeast research community, SGD seeks to promote communication among researchers by serving as a place to store, organise and display their contact information. SGD colleague pages include ‘colleague’ data voluntarily supplied by our users, such as basic contact information, links to individual laboratory web pages, and text descriptions of users’ research interests. The ‘Yeast Laboratories Page’12 is a feature derived from colleague data that lists many of the research groups that study yeast. It can be searched using the name of a group’s Principal Investigator, the geographical location of the group, or key research interests of the group. SGD also posts information on upcoming scientific meetings and links to various community resources in its ‘Community Info’13 section.
SGD interacts with other community resources
In addition to its relationship with the yeast community, SGD also fosters its relationship with other databases and resources. SGD is a founding member of the GO Consortium,8 a group that is developing a controlled vocabulary that can be applied to all organisms. As such, SGD curators regularly participate in GO meetings with other database groups participating in the GO project. SGD has also begun to attend ‘biocurator’ meetings in which curators from several different databases meet to discuss current database issues and present new features. With respect to software, SGD is part of a joint effort with other databases called GMOD14 (Generic Model Organism Database), whose goal is to develop reusable components for creating new biological community databases. SGD’s involvement with each of these community groups allows it to share knowledge and results in new ideas and ways of thinking about database organisation and tools.
SGD has a biological emphasis
Because a database is physically a collection of computer software and hardware, it is easy to view it, and tempting to present it, as computer-centric. However, one of SGD’s founding principles is that the biology, rather than the computer technology, must drive the direction of the database. This principle is based on an understanding that the primary ‘consumers’ of the database are biologists whose interests lie largely in finding and analysing biological data in the easiest and most efficient manner possible, without understanding any aspect of the computer technology. Information is presented graphically using metaphors commonly used by biologists. This bio-centric approach is observed at all levels of SGD’s information management and is fundamental to the database’s information content, design, data displays, and tool and resource development.
Information content
As described in the previous section, SGD curators seek feedback from the community in order to tailor the information content to the needs of the yeast research community. Given the prolific rate of yeast research, this helps SGD prioritise the types of information to include and the level of detail it should provide. As such, SGD avoids large bottlenecks in curation by continually making decisions regarding quality versus quantity. In all cases, SGD seeks to provide the most comprehensive data in the most efficient manner. The policy of including only published data allows SGD to direct users to specific references when they seek more detail than is feasible to provide. In almost all cases, SGD designs its schema and interfaces to leave open the possibility of adding more detail in the future.
Much of the information included in SGD is mined from the literature by scientific curators. Published literature is tentatively associated with yeast genes by an automated script that searches the PubMed15 database weekly for mention of yeast genes or gene products. In these searches, papers whose title, abstract or MeSH terms contain the gene name, open reading frame (ORF) name or an alias name in addition to the text ‘Saccharomyces cerevisiae’ are identified and entered into the database. Curators then read the abstracts of these papers, and often the full text, in order to associate a given publication with appropriate genes and ‘Literature Guide Topics’. The Literature Guide is a resource that organises literature for a given gene according to various broad biological topics such as ‘Genetic interactions’, ‘Localisation’ and ‘Techniques and reagents’. The objective of this resource is to allow users to easily locate specific information regarding a given gene without time-intensive literature searches. While assigning publications to broad Literature Guide topics, scientific curators also look for information that can be used to update any of the other gene-specific fields in the database, including GO annotations and gene product descriptions. The process of curating the literature is understandably one that is continual. Recently, with an expanded staff, SGD has become close to reaching equilibrium between the number of papers it curates weekly compared with the new papers that enter the database.
As new genome-wide analysis techniques are developed and applied to S. cerevisiae, SGD has sought to incorporate the results of such large-scale analyses. The primary goal in incorporating these types of data is to make them accessible to both the traditional single-gene biologists and to bioinformatics researchers interested in global yeast studies. Toward that end, SGD typically makes large-scale results available for download on its ftp site, but also posts results for individual genes on the ‘Locus page’ (see below) for that gene. Occasionally SGD analyses data to provide a more comprehensive annotation of S. cerevisiae. For example, we recently integrated the results from three different genome-wide comparison studies16-18 to classify all S. cerevisiae ORFs as either ‘verified’, ‘uncharacterised’ or ‘dubious’. Although each group had published its own analysis, including the identification of ‘chance’ or ‘spurious’ ORFs, SGD curators collated the results from all groups and reanalysed them in the context of any additional literature or data from large-scale experiments, including genome-wide two-hybrid screens and localisation studies. The resulting ORF classifications are now included on SGD locus pages and in the files of gene information available for download.
More recently, as additional genomes are sequenced, the focus of SGD has broadened to include the goal of assimilating information from a variety of species. The database is being extended in this way because comparison with other organisms has great potential both for enhancing the understanding of S. cerevisiae and for extending the biological knowledge derived from the study of S. cerevisiae to other organisms.
Underlying database design (data storage and organisation)
In keeping with SGD’s philosophy, the underlying design of the database, however complex, is one that must promote the most logical organisation of the biological data from the perspective of the biologist. Good database design must anticipate the type of information a scientist would like to derive from the data and work backwards to create a schema that makes this possible. SGD’s table specifications are the result of close communication between a group comprising scientific programmers, database administrators, a systems administrator and scientific curators, with curators emphasising the scientific goals and the computing staff seeking to find the most efficient way, computationally, to achieve these goals. This involves the computing staff staying current with new hardware and technology; however, this new technology is used to enhance the biological goals rather than to become the centrepiece of the database design. Scientific curators understand database table structure and specifications, and are therefore able to work through suggestions put forth by the programming staff.
Accessing the data
Even if the underlying data in a database are well organised, they are still not of value unless they and the resources designed to assist in their analysis are organised on the website in a manner that is intuitive and allows users to ask biologically meaningful questions. In order to centralise information, tools and data are organised into broad categories of information at SGD, for instance ‘Homology & Comparisons’.19 In turn, each broad category has a contents page that lists the types of data and tools found in that category (Figure 3). These contents pages contain a common left-hand navigation menu that also allows users to view tools available in different categories. The primary motivation in designing tools is to allow biologists to easily find and analyse data in a way that can help establish relationships between gene products and functions. SGD has many different types of tools and continues to develop more in response to user feedback and new types of data.
Figure 3.
The SGD Homology and Comparisons page19 is found by clicking on ‘Homology & Comparisons’ located in the left-hand column on SGD’s Home page.1 The left-hand column of the Homology & Comparisons contents page, like other SGD contents pages and the SGD home page, provides a consistent navigation menu to move between the main categories of information to view the tools and resources available within each category. The main portion of the Homology & Comparisons index page lists all of the various tools and resources available within this category of information. At the top of the page, as on the Locus page (Figure 4), is the standard header and tool bar that appears on most SGD pages, providing easy and consistent navigation throughout the website
The organisation of data and resources described above provides a global view of SGD’s functionality. On another level, the organisation of specific biological data is also critical to allowing scientists to find detailed information about genes and their products. Such biological data are organised around genetic loci, such that each genetic feature is described on a single web page. In the case of individual genes or ORFs, this page provides links to literature, sequence, GO annotations, expression data, functional analysis studies and other types of information specific to the given locus (Figure 4). This is accomplished by a set of pull-down menus on the right-hand side of the page that allows users to go directly to information specific for the given locus. For instance, selecting ‘glucose limitation’ under the Functional Analysis menu on a Locus page takes the user directly to expression data for the locus from the selected study. This centralisation of biological data allows users to access information in an efficient and intuitive manner.
Figure 4.
The SGD Locus Page (the bottom of the page shown here is truncated for space considerations) is the central point for information about any gene in SGD. The left-hand column provides basic information about the specific gene, BNA1 in this example, including nomenclature (standard name, alias and systematic name), feature type, Gene Ontology (GO) annotations, biochemical pathways (where relevant), the sequence coordinates where the gene is located, and other basic information. The right-hand column contains a series of pull-down menus with links to related information and resources for this gene, including relevant literature, sequence analysis, protein information, localisation information, interactions and other resources. The help icon in the upper right corner links to a help page written specifically to explain the various features of the Locus Page and its organisation. A standard header and tool bar, which appears on most SGD pages, is found at the top of the page (also seen in Figure 3). At the top of the SGD Locus Page is the standard header and tool bar that appears on most SGD pages to provide consistent navigation to important tools from any location within SGD. The Quick Search box allows users to access the basic search for information in SGD from any page. The upper menu bar provides links to key help and search pages as well as a consistent link back to the home page. The lower menu bar provides links to some of SGD’s most popular tools, as determined by our usage statistics
Data display
In addition to a flexible schema design and a biologically meaningful organisation of the data, user-friendly interfaces are critical to providing the user with the most efficient means for accessing and interpreting biological data. Constructing clear, functional user interfaces has been and continues to be one of SGD’s greatest challenges. If a user interface is not intelligible, intuitive and designed with the user’s needs in mind, maximum value cannot be obtained from the underlying data. It is a time-intensive process that involves working through many version of an interface. Specific attributes of good interface design, such as simple presentation, clear organisation, intuitive use of links, logical paths of navigation and thorough help documentation are principles considered when designing new interfaces. All SGD curators play a role in the design process. The group dynamics at SGD are well suited to this methodology, and each member brings different ideas and visions to the discussion. The end result is a tool or display that has passed the inspection of several biological scientists.
Internal consistency is also important to the design of the user interface. Specifically, the consistent use of links, icons and colours promotes familiarity and ease of use. The resources mentioned above are examples of simple interfaces that provide logical links to related resources. Another example of simple, intuitive design is the header navigation bar present on most of SGD’s pages, as seen in Figures 3 and 4. This header contains two menu bars, one with links to assist the user in navigating basic pages within SGD’s web site (‘Help’, ‘Site Map’, ‘Full Search’ and ‘Home’), and the other with links to the most commonly used resources (eg ‘BLAST’20 and the ‘Virtual Library’21). It also contains a Quick Search feature designed so that users can readily search information from six popular fields, including ‘gene name’, ‘gene product’, ‘GO terms’ and ‘colleagues’ for any text entered. Additional search options are available from the ‘Full Search’ feature, which is accessible from a link in the header bar. This header navigation bar allows easy, consistent access to other tools and resources at SGD.
Examples of these design principles can be seen in two popular SGD resources, the Genomic View22 and the Chromosomal Features Map (Figure 5). The Genomic View displays the 16 chromosomes of S. cerevisiae graphically, giving the user an instantaneous overview of the genome (Figure 5a). Clicking at a desired spot along a chromosome and selecting a map type (‘chromosomal features’, ‘physical’ or ‘physical and genetic’) produces a more detailed map of that area. All three ‘detailed’ map options have similar layouts and navigation schemes. The chromosomal features map (Figure 5b) provides a graphical view of the genetic features along the chromosome. Clicking on a specific feature in any of the map displays takes the user to that genetic feature’s Locus or Feature page, which in turn displays a small, clickable version of the chromosomal features map. Users can also retrieve different views and resources for the same area via links at the bottom of the page. Even though the maps are relatively old interfaces, they have remained virtually unchanged and are still widely accessed, a testimony to their usability and design.
Figure 5.
(a) The Genomic View is a graphic display of all 16 chromosomes of S. cerevisiae. The horizontal bars represent the individual chromosomes. The length of each bar indicates the relative length of each chromosome, and the black dots denote the locations of centromeres. Each chromosome is also labelled with selected mapped genes to serve as positional markers. A variety of more detailed maps, such as the Chromosomal Features Map (b), can be accessed selecting a map type and clicking on a segment of a chromosome. (b) Chromosomal Features Map is a graphic display of the genetic features located on a specified region of a chromosome. The large horizontal bar at the top of the figure represents the chromosome, with the position of the centromere indicated with a black dot and the chromosomal coordinates indicated by the −103× scale bar, above. The position of the detailed section, displayed below, is indicated by a rectangle on the chromosome. Clicking and dragging this rectangle can be used to expand, narrow or move the view. The parallel lines indicate the two different complementary strands of DNA, and features encoded on either strand are labelled with rectangular boxes. Labelled features include centromeres, tRNAs, RNA genes, Ty transposons, TY LTR elements, rRNAs and snRNAs, with feature type indicated by colour (key not included on this figure)
Internal organisation of SGD
The guiding principles outlined above have influenced the composition of SGD’s staff, methods of communication, and the group’s organisation and dynamics. These aspects of SGD have all contributed significantly to its continued success as a scientific resource.
SGD staff
The composition of the staff is essential to SGD’s success. In keeping with the idea that SGD’s emphasis should be biological in perspective, its staff is composed almost entirely of scientists with biological backgrounds: all curators and most programmers have PhDs in various areas of biology. Scientific curators constitute the majority of the group. Like museum curators who collect art and display it to an interested public, SGD scientific curators use their knowledge of biology to capture and organise information for presentation to the user. The term ‘scientific curator’ is therefore descriptive in that it stresses the importance of the biological background of the curator, as well as the curator’s role in developing a product that will benefit the scientific community. Most of SGD’s curators have little or no bioinformatics experience upon joining SGD, since the focus is on hiring individuals who are biologists. Curators acquire bioinformatics skills largely at their own pace, as a result of working on the database. In a few cases, SGD curators have gone on to become database programmers, based on their own interest and initiative.
Because SGD is a community service project, its primary goal is to understand and meet the needs of the community. As such, the staff is composed of individuals who are driven by this common goal rather than by a desire to publish independently. A scientific curator position is therefore somewhat removed from the traditional scientific career path. However, there are many rewards in producing a database that is widely used by the yeast community, and this is reflected in the very low turnover rate of SGD’s staff. When individuals do move on they often pursue similar projects in other groups, for instance in Gene Ontology8or the Stanford Microarray Database.23
In addition to full-time scientific curators that work on site, SGD also employs part-time curators and remote curators. These curators all attend weekly curator meetings, either in person or via conference call. Remote curators also spend two weeks each year on site at SGD for working sessions, special meetings, and direct communication with other curators.
Methods of communication
SGD scientific curators are required to possess and further develop strong communication skills, using them both within SGD and in interactions with the outside community. Strong verbal communication skills allow curators to express opinions diplomatically in a group setting. Such discussions are essential to SGD’s daily decision-making process. Since SGD relies heavily on its interaction with the yeast community, both verbal and written communication skills are crucial to maintaining close relationships with the community. Diplomatic communication skills are often required to maintain SGD’s neutral position in the community, particularly when facilitating communication between researchers regarding gene name conflicts.
The physical layout of the workspace is designed to promote frequent interactions between scientific curators, programmers and the Principal Investigator. Most staff work in one of two open rooms without cubicles, with some curators and programmers working side by side. This allows the staff to discuss issues as they arise and to interact frequently and informally. In addition to direct communication, SGD relies heavily upon e-mail for internal communication and, to some extent, for decision-making. This ensures that remote curators can contribute to the conversation in real time. When complicated decisions or design issues arise, they are often postponed for discussion at weekly curator meetings. The SGD staff has found that these weekly meetings provide the ideal forum for discussing complex issues and reaching decisions by consensus.
Group organisation and dynamics
SGD’s internal organisation is somewhat unique, but contributes to the staff’s ability to work together in achieving the common goal of serving the scientific community. The SGD group is led by a Principal Investigator, but the organisation of the staff is essentially flat in structure. It is a team-based organisation, and the vast majority of decisions are made by consensus. In keeping with this, each scientific curator is trained in most of the various curator tasks. This equips each curator with background knowledge that allows him or her to contribute to group discussions on almost any issue that arises concerning the database. In order to make certain that responsibility exists for each of the various tasks, however, many of the curators have a defined area of specialisation. For instance, although all curators are responsible for adding GO annotations to the database, two curators take particular responsibility for some of the more detailed GO-specific tasks and for ensuring that GO annotations are assigned consistently. In this same spirit, there are two ‘Lead’ curators who are responsible for making certain that all tasks are organised effectively and completed efficiently. Nevertheless, virtually all decisions are made by consensus among a group of individuals familiar with all aspects of the database.
This joint understanding of various database tasks also allows the group flexibility. For instance, interested curators may take on programming tasks, thereby freeing the programmers for more complicated projects. It also encourages individuals to continually expand their skills and knowledge.
One important characteristic that allows SGD personnel to work successfully as a group is trust. All scientific curators have access to data files and are able to make modifications to them. As such, well-defined procedures exist for making updates in order to prevent simultaneous changes or changes that might affect an edit another curator is making. These procedures and others are described in help documentation written by all the curators.
Although the flat structure and consensus style of decision making often require extra time, SGD staff believe they are worthwhile. This method of operation enables SGD to consider a multitude of ideas, opinions and approaches, melding them into a carefully designed product.
CONCLUSION
Measuring by most standards, both tangible (user statistics, longevity, community feedback) and intangible (relationship with the community), the Saccharomyces Genome Database is a successful scientific resource. In this paper, the SGD group has reflected upon the reasons for this success, with the hope that relaying at least some aspects of SGD’s philosophy, emphasis and organisation might be useful in the development of other scientific databases.
Perhaps the most important reasons for SGD’s success are that its staff maintains a clear vision as to SGD’s identity as a service project and recognises the importance of designing the database from a biological perspective. The composition, internal organisation and working dynamics of the SGD group support this vision. It is striking that, even for a project as technologically intensive as a scientific database, the philosophy and culture of its human staff ultimately determine its level of achievement.
SGD looks forward to expanding its data content and increasing the features and resources it offers to the community. As more and more genomes are sequenced and larger-scale analyses are undertaken, the role of the biological database in assimilating and presenting information becomes increasingly important. The integration into SGD of large data sets that differ both in content and format presents a unique but important challenge. It requires synthesising many different types of information in a manner that will not only allow retrieval of the data but will also enhance their utility, with the goal being to offer researchers a way of making connections between data that would otherwise be very difficult to uncover. Novel schema, resource and interface design will be necessary to accomplish this goal. Integration of large data sets will also require SGD to draw on research both within the yeast community and among other communities in order to incorporate and link information that will be biologically relevant to its users. In this manner, SGD hopes to facilitate a more comprehensive view of the role of gene products and their interactions with one another in the cell.
Footnotes
The SGD group is a team of scientists who commit their full time and energy to the design, maintenance, and enhancement of the Saccharomyces Genome Database, a resource for scientific information about the model organism Saccharomyces cerevisiae (baker’s or budding yeast).
SGD promotes a broad biological understanding of S. cerevisiae
Several specific principles guide SGD
All annotations are referenced with published work
All information is free and available for download
Only published data is included
Daily user e-mail assists users and provides feedback and new ideas
SGD facilitates community consensus on S. cerevisiae gene names
Advisory board meetings and attendance at scientific meetings help keep SGD current
Usage statistics guide tool development and layout
Researcher contact information and laboratory descriptions are available
SGD contributes to joint projects
Published literature is the primary source for continuing annotation
Biology directs the information content
Information content is based on the needs of users
Database design anticipates user needs
The focus is on single genes, but large-scale genomic data is incorporated
Tools and resources are organised to allow easy discovery and use
As more fungal genomes are sequenced, the SGD information content is expanding
Information about a single gene or genetic focus is presented on a single web page
Interconnecting resources via hyperlinks allows easy navigation to all resources
Creating intuitive user interfaces and thorough help documentation are priorities
Design consistency in user interfaces promotes familiarity and ease of use
The typical staff member has an advanced degree in biology
Curators communicate directly and frequently with programmers
Staff communicate by email, weekly meetings, and phone and video conferencing
SGD is a team-based organisation
Curators learn all tasks and contribute to design decisions
Clear vision, commitment to biology and group culture all contribute to success
The increasing amount of genomic sequence and analysis data creates new challanges
References
- 1.SGD (Saccharomyces Genome Database) Stanford University; URL: http://www.yeastgenome.org [cited 9th October, 2003] [Google Scholar]
- 2.ACEDB Documentation . The Wellcome Trust Sanger Institute; URL: http://www.acedb.org/Documentation/[cited 9th October, 2003] [Google Scholar]
- 3.SGD Usage Statistics . Stanford University; URL: http://genome-www.stanford.edu/usage/sgd/[cited 9th October, 2003] [Google Scholar]
- 4.DictyBase URL: http://dictybase.org [cited 9th October, 2003]
- 5.SGD ftp site . Stanford University; URL: ftp://genome-ftp.stanford.edu/pub/yeast/[cited 9th October, 2003] [Google Scholar]
- 6.Department of Genetics. Stanford University . Stanford University; URL: http://genetics.stanford.edu/[cited 9th October, 2003] [Google Scholar]
- 7.The Gene Ontology Consortium Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gene Ontology Consortium . Gene Ontology Consortium; URL: http://www.geneontology.org [cited 9th October, 2003] [Google Scholar]
- 9.SGD Gene Registry Form . Stanford University; URL: http://db.yeastgenome.org/cgi-bin/SGD/registry/geneRegistry [cited 9th October, 2003] [Google Scholar]
- 10.Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webminer . Stanford University; URL: http://www.yeastgenome.org/webminer/[cited 9th October, 2003] [Google Scholar]
- 12.Yeast Labs Page . Stanford University; URL: http://www.yeastgenome.org/cache/yeastLabs.html [cited 9th October, 2003] [Google Scholar]
- 13.SGD Community Information . Stanford University; URL: http://www.yeastgenome.org/ComContents.shtml [cited 9th October, 2003] [Google Scholar]
- 14.GMOD . Cold Spring Harbor Laboratory; URL: http://www.gmod.org/[cited 9th December, 2003] [Google Scholar]
- 15.PubMed . NCBI; URL: http://www4.ncbi.nlm.nih.gov/PubMed/[cited 9th October, 2003] [Google Scholar]
- 16.Kellis M, Patterson N, Endrizzi M, et al. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423(6937):241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 17.Cliften P, Sudarsanam P, Desikan A, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301(5629):71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 18.Brachat S, Dietrich FS, Voegeli S, et al. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 2003;4(7):R45. doi: 10.1186/gb-2003-4-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homology and Comparisons Page . Stanford University; URL: http://www.yeastgenome.org/HCContents.shtml [cited 9th October, 2003] [Google Scholar]
- 20.Stanford University; Saccharomyces WU-BLAST2 Search. URL: http://seq.yeastgenome.org/cgi-bin/SGD/nph-blast2sgd [cited 9th October, 2003] [Google Scholar]
- 21.The World-Wide Web Virtual Library: Yeast . Stanford University; URL: http://www.yeastgenome.org/VL-yeast.html [cited 9th October, 2003] [Google Scholar]
- 22.S. cerevisiae Genomic View . Stanford University; URL: http://www.yeastgenome.org/MAP/GENOMICVIEW/GenomicView.shtml, [cited 9th October, 2003] [Google Scholar]
- 23.The Stanford Microarray Database . Stanford University; URL: http://genome-www5.stanford.edu// [cited 9th October, 2003] [Google Scholar]