Abstract
This review focuses on the reaction mechanism of enzymes that use B12 and tetrahydrofolate (THF) to catalyze methyl group transfers. It also covers the related reactions that use B12 and tetrahydromethanopterin (THMPT), which is a THF analog used by archaea. In the past decade, our understanding of the mechanisms of these enzymes has increased greatly because the crystal structures for three classes of B12-dependent methyltransferases have become available and because biophysical and kinetic studies have elucidated the intermediates involved in catalysis. These steps include binding of the cofactors and substrates, activation of the methyl donors and acceptors, the methyl transfer reaction itself, and product dissociation. Activation of the methyl donor in one class of methyltransferases is achieved by an unexpected proton transfer mechanism. The cobalt (Co) ion within the B12 macrocycle must be in the Co(I) oxidation state to serve as a nucleophile in the methyl transfer reaction. Recent studies have uncovered important principles that control how this highly reducing active state of B12 is generated and maintained.
I. Introduction to Methyltransferases and Their Cofactors
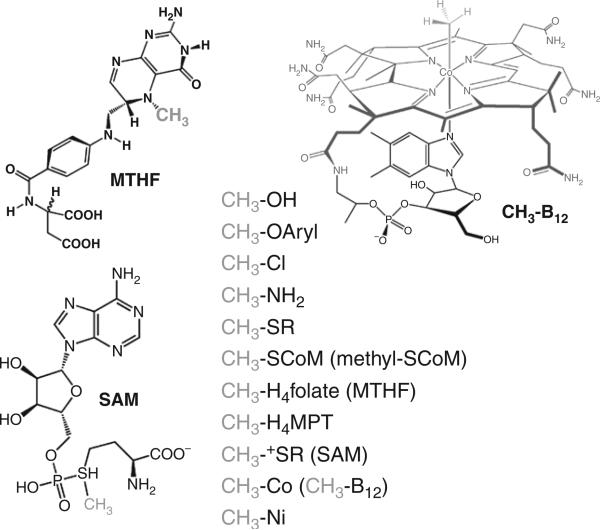
An organic chemist wishing to insert a methyl group into a compound might use methyltriflate or diazomethane. In biology, the methyl donors are much less explosive. Figure 10.1 shows some of the simple methyl donors that are found in nature like methanol, methylamines, methanethiols as well as the cofactors that are involved in methylation reactions like methyltetrahydrofolate (MTHF), S-adenosyl-L-methionine (SAM), and methyl-B12. A methyltransferase catalyzes the transfer of a methyl group from one of the donors shown in Fig. 10.1, like MTHF or SAM, to an acceptor like homocysteine or the N6 group on adenine in DNA (Fig. 10.2).
Figure 10.1.
Methyl donors in biology.
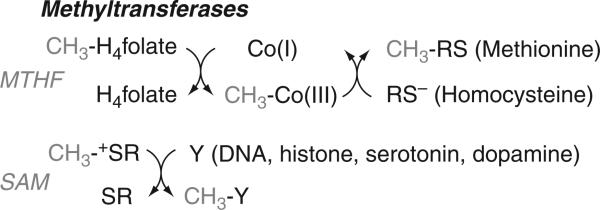
Figure 10.2.
Two types of methyltransferases.
Two cofactors figure prominently in methyltransferase chemistry: vitamin B12 (or cobalamin) and tetrahydrofolate (THF). The history of vitamin B12 dates back to its description as the antipernicious anemia factor (Minot and Murphy, 1926; Whipple and Robscheit-Robbins, 1925). Cobalamin was isolated by Smith and Folkers in 1948 (Rickes et al., 1948; Smith, 1948) and its structure was crystallized (Rickes et al., 1948) and its structure was determined in 1956 (Hodgkin et al., 1956). Like heme, F430, and chlorophyll, vitamin B12 is a tetrapyrrolic cofactor with a central Co atom coordinated by the four equatorial pyrrole nitrogen ligands (Fig. 10.1). Extending from one of the pyrrole rings is a propanolamine-linked group that can serve as the lower axial ligand to Co. The lower ligand is dimethylbenzimidazole in cobalamins, while the upper axial ligand is a cyano-, methyl-, or 5′-deoxyadenosyl- group in vitamin B12,methylcobalamin (MeCbl1), and adenosylcobalamin (AdoCbl) or coenzyme B12, respectively. The biological role of MeCbl as an essential coenzyme for a methyltransferase (Guest et al., 1962) was revealed a few years after a role for AdoCbl as the coenzyme for glutamate mutase was discovered (Barker et al., 1958).
Tetrahydrofolate (THF) is the reduced form of the vitamin folic acid, which was first recognized as the compound in brewer's yeast that can reverse anemia. Folic acid was isolated in a highly purified form from 4 tons of spinach leaves by Esmond Snell, Herschel Mitchell, and Roger Williams (Mitchell et al., 1941). The vitamin was crystallized soon after (Pfiffner et al., 1945; Stokstad, 1943) and synthesized in 1946 by Lederle Laboratories (Angier et al., 1946). Besides serving as a cofactor in methyltransferase reactions, THF is the major one-carbon carrier in cells and is needed for protein and DNA synthesis and is important in nitrogen metabolism. Its essentiality in such key metabolic processes makes THF metabolism a key target for anticancer and antimicrobial drugs (M.P. Costi, S. Ferrari, 2001).
There are two classes of methyltransferases, which differ in their use of an activated versus an unactivated methyl group donor. One class of methyltransferases, exemplified by methionine synthase, use an unactivated methyl donor and an intermediate methyl carrier, cobalamin, which in the Co(I) oxidation state is an extremely potent nucleophile that can react with the methyl group of MTHF to generate an intermediate organometallic MeCbl intermediate. The methyl group is then transferred from MeCbl to the final acceptor, which for methionine synthase is homocysteine, generating methionine. The other class of methyltransferases uses an activated methyl donor in the form of SAM to directly methylate the N6 of adenine or the amino groups of lysine or arginine in histones, and so on. This review focuses on the first class of methyltransferases that utilize THF and B12.
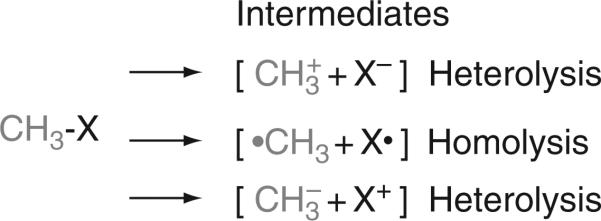
In all methyltransferase-catalyzed reactions, as shown in Fig. 10.3, transfer of the methyl group involves cleavage of a methyl-X bond, where “X” can be one of a variety of functional groups (N, S, Cl, O, etc.). Heterolysis can lead to two products: a methyl cation or a methyl anion. On the contrary, homolysis leads to a methyl radical. For methyl group transfers, heterolysis is most common, leading to the formation of a methyl carbocation equivalent.
Figure 10.3.
Three ways to cleave a methyl-X bond.
II. Three Component Systems Required for B12/THF-Dependent Methyltransferases
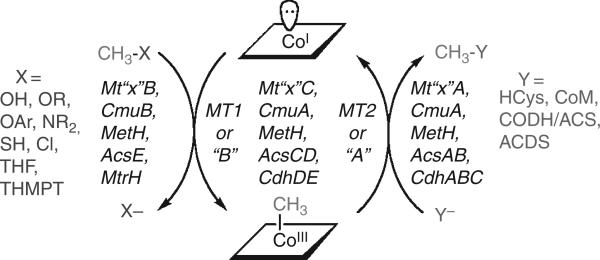
All B12-dependent methyltransferases contain three components, as indicated in Fig. 10.4 and Table 10.1. In these systems, the “B” component binds the methyl donor, a “C” component binds B12, and an “A” component binds the methyl group acceptor. An alternative nomenclature in use describes the B component as MT1 and the A component as MT2. Recent literature references for each methyltransferase system are given in Table 10.1. In each case, the methyl group is unactivated, that is, it is a secondary alcohol, an amine (primary, secondary, or tertiary), or a thiolate. The required electrophilic activation is accomplished by the “B” component, as described in more detail below. The “C” component supplies the supernuclophilic Co(I) state of B12, which acts as an intermediary methyl group acceptor that interacts with the “B” component to accept the methyl group from the substrate, forming MeCbl. The C Component then interacts with the A component to transfer the methyl group to the ultimate methyl group acceptor.
Figure 10.4.
B12-dependent methyltransferases.
Table 10.1.
B12-dependent methyltransferases
| Methyl donor | Methyltransferase components | Biological system | Ultimate methyl acceptor | Reference |
|---|---|---|---|---|
| MTHF | MetH | Methionine Synthesis | Homocysteine | (Matthews, 2001) |
| CH3NH | MtmABC | Methanogenesis | Coenzyme M | (Krzycki, 2004) |
| (CH3)2N | MtbABC | Methanogenesis | Coenzyme M | (Soares et al., 2005) |
| (CH3)3N | MttABC | Methanogenesis | Coenzyme M | (Soares et al., 2005) |
| CH3SH | MtsAB | Methanogenesis | Coenzyme M | (Tallant et al., 2001) |
| MTHMPT | MtrA-H | Methanogenesis | CoM | (Gottschalk and Thauer, 2001) |
| CH3OH | MtaABC | Methanogenesis | Coenzyme M | (Hagemeier et al., 2006) |
| MTHF | AcsABCDE | Acetogenesis | THF, CODH/ACS | (Doukov et al., 2007) |
| MTHF | MtaABC | Acetogenesis | THF | (Das et al., 2007) |
| CH3OAr | MtvABC | Acetogenesis | THF | (Naidu and Ragsdale, 2001) |
| CH3Cl | CmuAB | Dehalorespiration | THF | (Studer et al., 2001) |
III. Biological Systems Impacted by B12 and Folate-Dependent Methyltransferases
A. Methionine biosynthesis
B12-dependent methyltransferases play an important role in microbial and eukaryotic metabolism. Biosynthesis of methionine in most microbes and eukaryotes depends upon the B12- and THF-dependent enzyme, methionine synthase (MetH). Since methionine is the precursor of SAM, methionine synthase supports the many methyltransferases involved in methylation of DNA, proteins, and neurotransmittors (Fig. 10.2). An elevated level of homocysteine is linked to a number of pathological states, including premature heart disease and neural tube defects and methionine synthase plays an important role in controlling homocysteine homeostasis (Col et al., 2007).
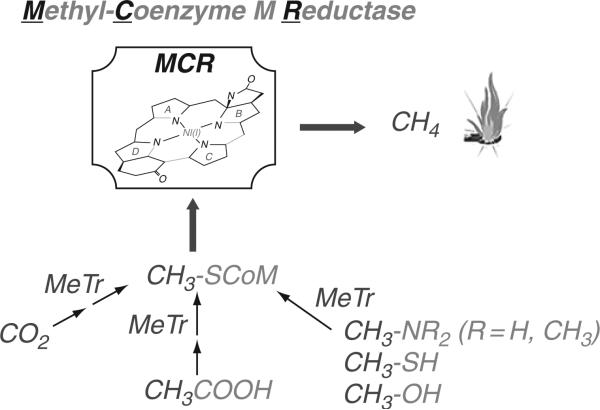
B. Methanogenesis
As shown in Fig. 10.5, growth of methanogens requires the conversion of substrates (CO2, methylamines, methylthiols, and acetate) to methyl- SCoM. The nickel containing enzyme methyl-SCoM reductase then catalyzes the conversion of methyl-SCoM to methane. Table 10.1 lists six B12-dependent methyltransferases that are important in growth of methanogenic archaea. Most of these methyltransferases catalyze the transfer of a methyl group from the methyl donor (Methyl-X) to CoM, thus providing methyl-SCoM needed for methane formation. These methanogenic methyltransferases use B12 (or an analog with a slight modification in the benzimidazole component) bound to the “C” component. Growth on acetate or H2/CO2 involves the generation of methyltetrahydromethanopterin (MTHMPT), which is converted to methyl-SCoM by the Mtr system. As described below, this is interesting membrane-bound system links the methyl transfer reaction to generation of a sodium ion gradient, which is utilized to make ATP (Gottschalk and Thauer, 2001).
Figure 10.5.
Methanogenesis.
C. Methanogenic methyltransferases and the 22nd amino acid
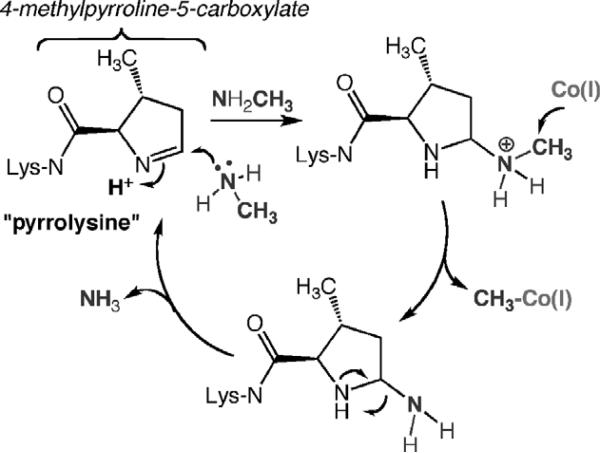
Study of the methanogenic methyltransferases has uncovered the 22nd amino acid, pyrrolysine, which is encoded by a UAG stop codon. This system is reminiscent of selenocysteine, encoded by the UGA codon. For pyrrolysine synthesis, there is an enzymatic system, encoded by the pylBCD genes to synthesize pyrrolysine, a dedicated pyrrolysyl-tRNA synthetase (encoded by pylS), which aminoacylates a specific amber-decoding tRNA (encoded by the pylT gene) with pyrrolysine (Blight et al., 2004; Longstaff et al., 2007). Thus, like the other 20 natural amino acids, pyrrolysine is co-translationally placed into the nascent polypeptide chain. The role of pyrrolysine in the methyltransferase reaction is discussed below.
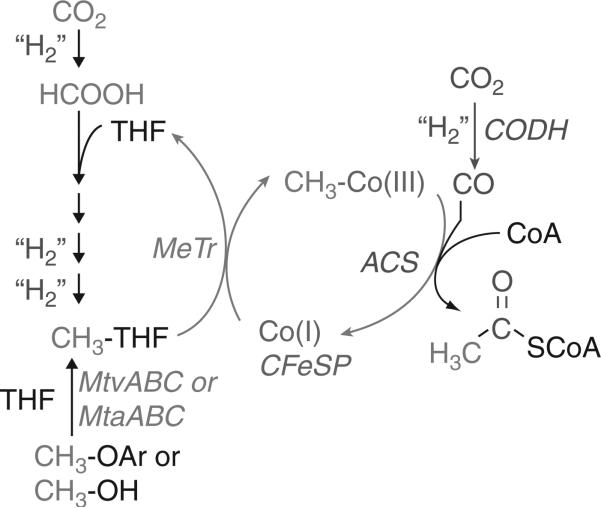
D. Acetogenesis
A role for B12 in anaerobic CO2 fixation was described in ~1965 (Ljungdahl et al., 1966; Poston et al., 1964). There are two B12-dependent methyltransferases involved in the Wood-Ljungdahl pathway of CO2 fixation (Fig. 10.6). The first, MTHF: corrinoid iron-sulfur protein (CFeSP) methyltransferase is encoded by the acsE gene and catalyzes transfer of the methyl group from MTHF to the Co(I)-B12 site in the CFeSP, which is encoded by the acsCD genes. In the subsequent reaction, the methylated CFeSP transfers the methyl group to a NiFeS cluster in acetyl-CoA synthase (ACS); thus, this is a metal (Co) to metal (Ni) methyl group transfer between two proteins. ACS then catalyzes the condensation of its Ni-bound methyl group with CO (generated by CO dehydrogenase, CODH) and coenzyme A to generate acetyl-CoA. The series of organometallic intermediates is one of the novel features of the Wood-Ljungdahl pathway.
Figure 10.6.
Methyltransferases in acetogenesis.
Two other methyltransferase systems that are shown in Fig. 10.6 couple to the Wood-Ljungdahl pathway. The MtvABC system transfers the methyl group of vanillate or other methoxylated aromatics to THF to generate MTHF (Engelmann et al., 2001; Naidu and Ragsdale, 2001), while MtaABC catalyzes the synthesis of MTHF from the methyl group of methanol and THF (Das et al., 2007). MtvB was shown to catalyze methyl transfer from the aromatic phenylmethyl ether to the corrinoid component of MtvC and MtvA catalyzed the MtvC-dependent methylation of THF (Naidu and Ragsdale, 2001). The sequence homology between the Mtv and Mta systems suggests that the A, B, and C components in the two systems share similar functions (Das et al., 2007).
E. Other metabolic systems in which B12- and folate-dependent methyltransferases play a key role
Growth of Sphingomonas paucimobilis SYK-6 on lignin-derived biaryls and monomers requires the ligM gene, which is homologous to mtvB, to catalyze the transfer of the methyl group from vanillate to THF. Since the ligM gene in this organism is located in the same gene cluster as two genes encoding THF-dependent enzymes, it is likely that lignin degradation is linked directly to one-carbon metabolism (Abe et al., 2005). However, unlike the methyltransferase reactions described in the previous section, these remain to be characterized.
IV. Structure and Function of B12 in Methyltransferases
A. Binding of B12 to the enzymes
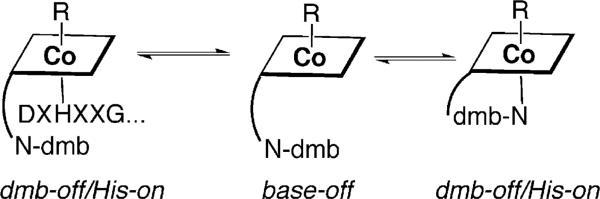
Three modes of B12 binding have been described: “dmb-on,” “dmb-off ”/“his-on,” and “base-off,” which refers to whether or not a lower axial ligand is coordinated to Co (Fig. 10.7). Thus, “dmb-on” indicates that the dimethylbenzimidazole group, which is appended to one of the tetrapyrrole rings, is ligated to Co, while “his-on” refers to the state in which the dmb ligand is replaced by a His residue donated by the protein. “Dmb-on” and “dmb-off ” have often been referred to as “base-off ” and “base-on”; however, this is inaccurate because the His ligand also acts as a base. It is more accurate to only refer to the “base-off” binding mode when there is no nitrogenous base ligand. Such a coordination mode was first revealed by spectroscopic studies of the CFeSP in the Wood-Ljungdahl pathway (Ragsdale et al., 1987) and recently confirmed in the crystal structure (Svetlitchnaia et al., 2006). In most cases, unambiguous definition of the ligation state, that is, “dmb-on”, “dmb-off”, “base-off ”, was first revealed by spectroscopic studies. For example, a “dmb-off” state was revealed for several proteins by electron paramagnetic resonance (EPR) spectroscopy (Ragsdale et al., 1987) (Stupperich, 1990, #220) several years before a crystal structure revealed a “his-on”“dmb-off ” binding mode in the B12-binding domain of methionine synthase (Drennan et al., 1994). EPR spectroscopic studies have been successful in revealing the ligation mode in a number of other enzymes (Abend et al., 1998, #6281; Yamanishi et al., 1998, #2310; Lawrence et al., 1999, #4157; Abend et al., 1999, #3897; Ke et al., 1999, #6277). The EPR spectrum of the Co(II) state of B12 is particularly diagnostic of the axial ligation state because the unpaired electron in the orbital exhibits strong interactions with the axial ligand. If this ligand is a nitrogen atom, with a nuclear spin (I) of 1, each of the eight hyperfine lines (due to splitting of the resonance by interactions with the Co nucleus with I = 7/2) in the EPR spectrum exhibit a three-line splitting. The “base-off ” mode of B12 binding is characterized by the presence of singlets instead of triplets at each of the eight resonant positions, as was observed in the EPR spectrum of the CFeSP (Ragsdale et al., 1987). The “his-on” mode is clearly indicated by adding 15N-His to the growth medium, which replaces the natural abundance histidine (mostly 14N) in enzymes. Since 15N has a nuclear spin of ½, doublet instead of triplet superhyperfine lines are observed at each of the eight resonant positions in the EPR spectrum. Observation of the triplet spectrum in B12 proteins labeled with 15N-His suggests a “dmb-on” binding mode.
Figure 10.7.
Three modes of B12 binding.
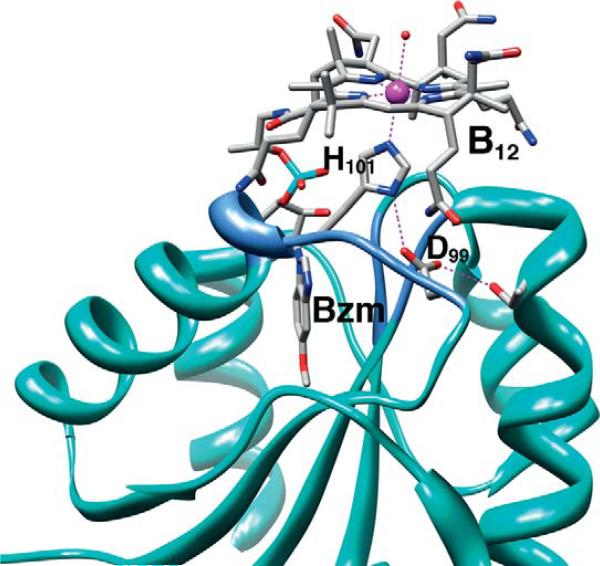
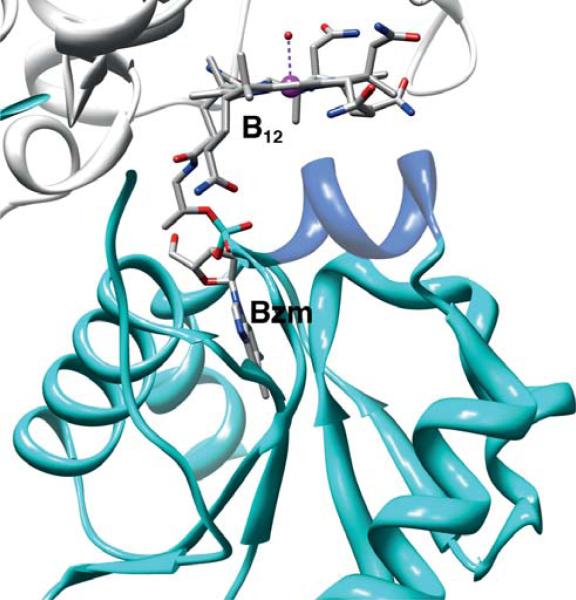
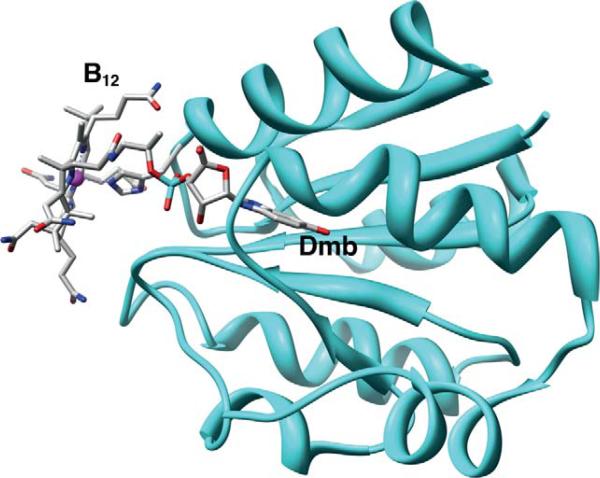
Crystallographic studies of methyltransferases have revealed the elegant molecular details of how proteins bind B12. The crystal structures of the cobalamin-binding component of several methyltransferases are available, including the B12-binding domain of methionine synthase in several states (Bandarian et al., 2002, 2003; Drennan et al., 1994), MtaC (Das et al., 2007 ) (Fig. 10.8) and the CFeSP (AcsCD) from M. thermoacetica (Svetlitchnaia et al., 2006) (Fig. 10.9), and the MtaBC complex from Methanosarcina barkeri (Hagemeier et al., 2006). In all of these methyltransferase structures, cobalamin is bound within a Rossman α/β fold (Fig. 10.10). The “C” components that have a “dmb-off/his-on” mode of B12 binding share a sequence motif DXHXXGX41SXLX26–28GG in which the His is the lower axial ligand. The Asp, His, and Ser residues in this sequence are referred to as the catalytic triad and facilitate formation of the “base-off ” conformation by protonating the His ligand, (Ludwig and Matthews, 1997). In the M. thermoacetica MtaC, the Asp and His are present in the catalytic triad (Fig. 10.6); however, it appears that a Thr residue replaces Ser. The dmb side chain is deeply embedded and is responsible for much of the binding energy that tethers the cobalamin to the protein. A “dmb-on” structure is not available in the methyltransferase class, which is somewhat surprising since several AdoCbl-dependent isomerases share the “dmb-on” binding mode, including diol dehydratase (Abend et al., 1998; Shibata et al., 1999; Yamanishi et al., 1998), ribonucleotide reductase (Lawrence et al., 1999; Sintchak et al., 2002), and ethanolamine ammonia lyase (Abend et al., 1999; Ke et al., 1999).
Figure 10.8.
B12-binding site in M. thermoacetica MtaC (Das et al., 2007). The region containing the conserved DXH motif (see text) is shown in cornflower blue. Generated from PDB ID# 1Y80 using Chimera.
Figure 10.9.
B12-binding site in the M. thermoacetica CFeSP (Svetlitchnaia et al., 2006 ). The region containing the hydrophobic helix (see text) is shown in cornflower blue. Generated using Chimera from PDB ID code 2H9A.
Figure 10.10.
B12-binding site in M. barkeri MtaC focusing on the Rossman domain involved in ligating the Dmb moiety, generated from PDB ID code 2I2X, using Chimera.
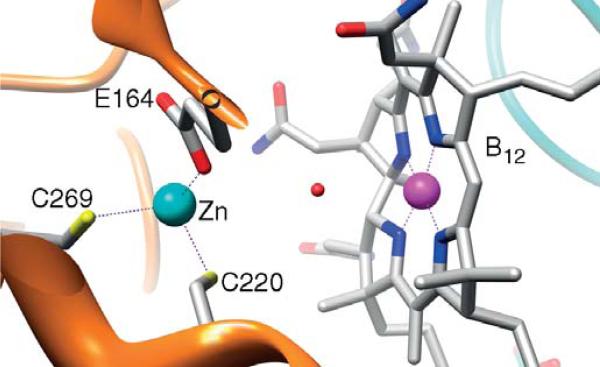
Most of the methyltransferases share the “dmb-off/his-on” binding mode, while the CFeSP involved in transferring the methyl group to the ACS component in the Wood-Ljungdahl pathway is in the “base-off ” state, as revealed by spectroscopic studies (Ragsdale et al., 1987) and confirmed by crystallographic studies (Svetlitchnaia et al., 2006) of the CFeSP (Fig. 10.9). A related protein in the methanogenic acetyl-CoA decarbonylase synthase complex also is in the “base-off ” state (Jablonski et al., 1993). Proteins that bind B12 in the “base-off ” state lack the DXH . . . signature sequence. Instead, they contain a relatively hydrophobic helix below the plane of the cobalamin (SVLTAWAA) (Fig. 10.9). A coordinating water molecule in the upper axial position is replaced by a methyl group during catalysis. The EPR spectrum of the CFeSP in H2 17O exhibits 17O-induced hyperfine broadening, providing conclusive demonstration that H2O coordinates to the metal center in one of the open axial positions (Stich et al., 2006).
B. Generation and maintenance of the active Co(I) state of B12
Cobalt in B12 can exist in the (I), (II), and (III) states. Cobalt cycles between the Co(I) and methyl-Co(III) states during catalysis. In the Co(I) state, Co has a d8 configuration. In B12 and related corrinoids, Co(I) is a supernucleophile (Schrauzer and Deutsch, 1969; Schrauzer et al., 1968) and is weakly basic, with a pKa below 1 for the Co(I)-H complex (Tackett et al., 1963). Protein-bound Co(I) is also highly reducing with a standard reduction potential for the Co(II)/(I) couple below −500 mV (Banerjee et al., 1990b). These properties make Co(I) fairly unstable and subject to inactivation. For example, in cobalamin-independent methionine synthase, the Co(I) center undergoes oxidative inactivation to the 2+ state once in every 100–2,000 turnovers (Drummond et al., 1993; Fujii et al., 1977). The Co(II)/Co(I) reduction potential of the CFeSP-bound corrinoid in the anaerobic microbial system (AcsA-E), is −504 mV (Harder et al., 1989), and the Co(I) intermediate is oxidized to the catalytically inactive Co(II) state once every 100 turnovers (Menon and Ragsdale, 1999).
Once oxidative inactivation occurs, the B12-dependent methyltransferases require reductive activation to reenter the catalytic cycle. This difficult reduction of the inactive Co(II) to the Co(I) state is accomplished by different systems. In cobalamin-independent methionine synthase, the unfavorable one-electron reduction is coupled to the highly exergonic demethylation of SAM, forming MeCbl (Banerjee et al., 1990a). In E. coli, flavodoxin is the electron donor, while, in humans, the donor is methionine synthase reductase (Olteanu and Banerjee, 2001). The reductive methylation reaction is necessary for reactivation of the Co(II) state because the quinone/ hydroquinone and hydroquinone/semiquinone couples of the E. coli flavodoxin have a significantly more positive redox potential (~ −250 mV and −450 mV, respectively (Vetter and Knappe, 1971)), than the Co(II)/(I) couple of methionine synthase (−526 mV) (Banerjee et al., 1990c) (Olteanu, 2004, #6302). Before reductive activation, the His ligand dissociates from the Co center to generate the “base-off ” conformation. The rationale for generating the “base-off ” state is that the nitrogen ligand would donate electron density to the Co center, making the reduction more difficult. In methionine synthase, binding of flavodoxin, the redox partner responsible for reductive activation, leads to dissociation of the His ligand (Hoover et al., 1997). Removal of the dmb ligand also is an intermediate step in the electrochemical reduction of Co(II) to the Co(I) state of B12 in solution (Lexa and Savéant, 1976). Thus, methyltransferases appears to have evolved a mechanism to facilitate the reductive activation that can be understood based on the principles of inorganic chemistry and electrochemistry.
While SAM-dependent reductive methylation is used to return Co to the catalytic cycle in methionine synthase, in the methanol- (Daas et al., 1996b) and dimethylamine- (Wassenaar et al., 1998) methyltransferases, an ATP-dependent activating protein is involved. ATP-dependent activation also appears to be required for the aromatic O-demethylase from some acetogens (Kaufmann et al., 1998).
In the M. thermoaceticum MTHF:CFeSP methyltransferase involved in the Wood-Ljungdahl pathway, the inactive Co(II) state of the CFeSP is already “base-off ” in the resting enzyme, which represents a “ready” state for electron transfer to form a four-coordinate Co(I) state (Harder et al., 1989; Ragsdale et al., 1987; Stich et al., 2006; Wirt et al., 1993, 1995). The direct electron donor is the [4Fe-4S] cluster of the AcsC subunit of the CFeSP (Menon and Ragsdale, 1999). This cluster has a reduction potential of −523 mV (Harder et al., 1989), which is nearly isopotential with the Co(II)/Co(I) couple of the CFeSP-bound B12 and can accept electrons from a low-potential ferredoxin or directly from enzymatic systems that couple to ferredoxin, including CO/CODH, H2/hydrogenase, or pyruvate/pyruvate ferredoxin oxidoreductase (Menon and Ragsdale, 1999). These low potential electron donors have a reduction potential similar to that of the Co(II)/(I) couple, which probably explains why this system does not require coupling to ATP or reductive methylation as in the systems described above.
C. The importance of the “dmb-off”/“dmb-on” equilibrium
Kinetic and thermodynamic studies of alkylated corrinoids have shown that the nature of the lower ligand strongly influences the rate and mode of Co–C bond breaking (Hogenkamp et al., 1965; Kräutler, 1987; Pratt, 1999). Since most of the methyltransferase “C” components contain a Co-N-His bond replacing the Co-N-dmb ligation in the free cofactor, enzymatic (Dorweiler et al., 2003) and model (Fasching et al., 2000) studies have been conducted to determine if this ligand switch conferred a special mechanistic advantage. On the basis of studies of methyl transfer from MTHF to Co(I) in methionine synthase, it was concluded that the replacement of 5,6-dimethylbenzimidazole by imidazole has little effect on the kinetics of the methyl transfer reaction. This is in accord with studies of B12 models by Kräutler, which confirm the expectation that imidazole and benzimidazole are comparable as axial ligands (Fasching et al., 2000). However, it was noted that the imidazole nitrogen in imidazolylcobamide allows pH-dependent control of reactivity by protonation/deprotonation of the axial ligand, since the pKa of the coordinating nitrogens in imidazole and benzimidazole are 7.0 (Datta and Grzybowski, 1966) and 5.5 (Catalan et al., 1983; Lane and Quinlan, 1960), respectively. Accordingly, the pKa of the imidazole ligand is 1.4 pH units higher than that of 5,6-dimethylbenzimidazole (pKa = 2.9) in the methyl- Co(III) states (Fasching et al., 2000).
Complete removal of the axial donor ligand to Co is expected to markedly affect the methyl transfer reaction. The presence of an N-donor ligand in the lower axial ligand position inhibits heterolytic cleavage, whereas O-donor ligands, such as water, do not exhibit this inhibition. For example, MeCbl reacted with homocysteinethiolate at least 1500-fold more slowly than methylcobinamide (Norris and Pratt, 1996). This axial ligand effect appears to be recapitulated in the M. thermoacetica CFeSP in which transfer of the methyl group from free methyl-cobinamide (lacking the lower axial dimethylbenzimidazole ligand) to the nickel center in ACS occurs at a rate ~103 times faster than that with free MeCbl (Seravalli et al., 2001). Furthermore, in methyltransferases with a “dmb-off/ his-on” conformation, such as methionine synthase, the His ligand to the Co ion is removed to facilitate reduction of the inactive Co(II) enzyme. A similar axial His ligand dissociation from the Co(II) center has been observed upon binding of the ATP-dependent activating protein to the methanogenic MtaABC system from M. barkeri (Daas et al., 1996a). A “catalytic triad” consisting of His759, Asp757, and Ser810 in methionine synthase, which is conserved among the “dmb-off/his-on” proteins, is involved in removal of this axial ligand by protonating the N-atom that ligates to Co, as illustrated by proton uptake associated with reduction of Co(II) to Co(I) (Hoover et al., 1997). The role of ligand dissociation on methionine synthase takes on special significance since this coordination state change is accompanied by a global conformational change in the protein (Hoover et al., 1997). There was only a minor affect of the axial ligation on the rates of methyl transfer between exogenous cobalamin cofactor and folate bound to the N-terminal domains of methionine synthase (residues 2–649) (Dorweiler et al., 2003). In fact, based on the minor effect of the axial ligand on the methyl transfer from MTHF to Co(I) in methionine synthase, it was concluded that the primary role of the ligand triad is to control the conformational equilibria during catalysis, rather than to control axial ligation (Dorweiler et al., 2003). These major conformational changes are an exciting hallmark of the catalytic cycle of methionine synthase.
Other effects of the protein, besides the N-donor ligand, on the methyl transfer reaction are observed in methyltransferases. In the CFeSP, the Co–OH2 bond is lengthened by ~0.2 A, which is proposed to further enhance reduction of the Co(II) species (Stich et al., 2006). Evidence for this enhancement is suggested by the 100-fold faster methylation of ACS by the methylated CFeSP than by free MeCbi+. This rate enhancement is likely due to a combination of favorable protein–protein interactions between ACS and CFeSP and enzyme-induced elongation of the lower axial ligand bond (Stich et al., 2006). Lengthening the Co–OH2 bond in the methyl-Co(III) state would stabilize the unoccupied Co orbital, increasing mixing of this orbital with the corrin-based occupied frontier orbitals, effectively making the Co center more “Co(I)-like,” which would facilitate heterolytic Co–C bond cleavage. Furthermore, partially dissociating the lower water ligand would discourage homolytic bond cleavage given the strong preference of Co(II)corrinoids to retain an axial ligand.
V. Activation of the Methyl Group Donors
A. Binding of the methyl group donor to the Mt“x”B (MTII) component
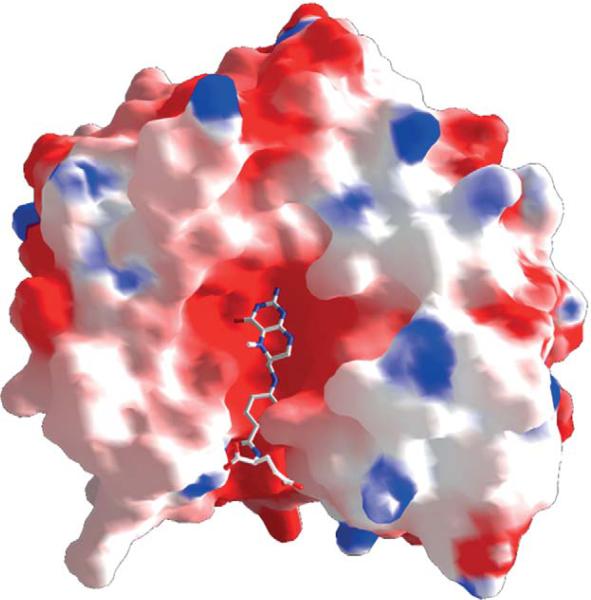
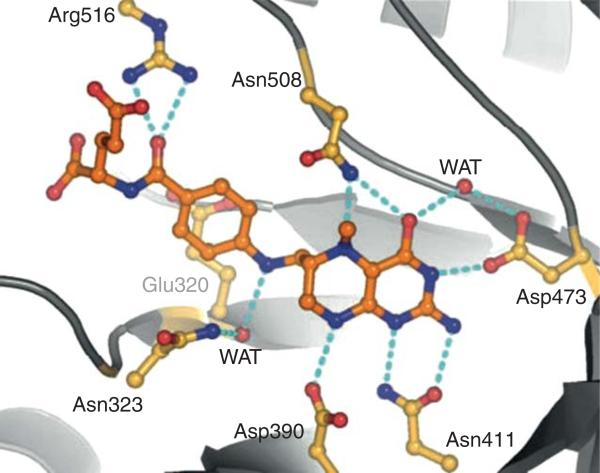
The substrates that donate the methyl groups in biology vary in size from methanol (32 Da) to MTHF (443 Da for the monoglutamate derivative). Thus, the size of the substrate-binding site must vary markedly among different methyltransferases. The electronic properties also differ, since the methyl group is bonded to a hydroxy, phenoxy, thiol, or amine (primary, secondary, or tertiary) group. Regardless, in all methyltransferases, the domain/protein that binds the methyl group donor folds into an α/β triosephosphate isomerase (TIM) barrel structure, as observed in the crystal structures of the methanol-binding protein MtaB (Hagemeier et al., 2006), the homocysteine and MTH-binding domains of methionine synthase (Evans et al., 2004), the methylamine-binding protein MtmB (Hao et al., 2002), the MTHF:CFeSP methyltransferase (Doukov et al., 2000, 2007), and even the MTHF-binding domain of the corrinoid-independent methionine synthase (MetE) (Pejchal and Ludwig, 2005). In all of these proteins, the methyl donor binds within the cavity formed by the TIM barrel, as shown in the electrostatic surface rendering of the structure of the MTHF:CFeSP methyltransferase (Fig. 10.11). The red surface at the MTHF-binding site indicates the negative charge, which complements positively charges on the substrate. MTHF (or THF) is strongly cemented within this cavity (Kd<10 μM) by hydrogen bonds that are conserved between MTHF: CFeSP methyltransferase and methionine synthase (Fig. 10.12). The MTHF-binding site also resembles the binding site for the related pterin substrate (dihydropteroate) in dihydropteroate synthase (Achari et al., 1997; Hampele et al., 1997), with conserved residues D75, N96, and D160, forming a “pterin hook” (Doukov et al., 2000).
Figure 10.11.
The methyltetrahydrofolate (MTHF)-binding site in the MTHF:CFeSP methyltransferase (AcsE).
Figure 10.12.
Methyltetrahydrofolate (MTHF)-binding site in MetH. From Evans et al. (2004).
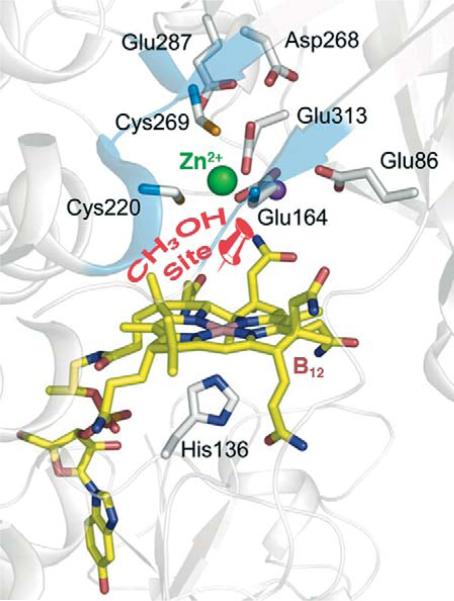
Based on the structure of the MtaBC complex and on biochemical studies, it was proposed that methanol binds between a Zn ion in the TIM barrel of MtaB and the cobalamin site in MtaC (Fig. 10.13) (Evans et al., 2004). In the case of the methylamine methyltransferase (MtmB), a novel amino acid, pyrrolysine, is located within the cavity of the TIM barrel (Fig. 10.14) (Hao et al., 2002). Biochemical and crystallographic results indicate that the methylamine substrate forms a covalent complex with pyrrolysine as part of the activation mechanism (Fig. 10.15), as discussed in more detail below.
Figure 10.13.
Proposed methanol-binding site between the Co and Zn ions (pinpoint) in MtaBC. Modified from Hagemeier et al. (2006). Generated from PDB ID code 2I2X using Chimera.
Figure 10.14.
Pyrrolysine (stick diagram) in the channel formed by the triosephosphate isomerase (TIM) barrel of MtmB. Generated from 1NTH (Hao et al., 2002) using Chimera.
Figure 10.15.
Proposed role of pyrrolysine in activating methylamines. Modified from Hao et al. (2002 ).
B. General acid catalysis, Lewis acid catalysis, and covalent catalysis to accomplish electrophilic activation of the methyl group donor
Among the methyltransferases, a similar principle underlies the activation of the methyl group that will be transferred: to donate positive charge to the heteroatom attached to the methyl group, leading to electrophilic activation of the methyl group. There is a solid chemical basis for this mechanism. Quaternary amines, with a full positive charge on nitrogen, do not require activation, as shown by studies of methyl transfer from a variety of quaternary ammonium salts to Co(I)-cobaloxime (Hilhorst et al., 1994), trimethylphenylammonium cation to cob(I)alamin (Pratt et al., 1994), and dimethylaniline at low pH to Co(I)-cobyrinate (Zheng et al., 1999). In addition, methyl transfer is catalyzed by Lewis acids, such as Zn(II) (Wedemeyer-Exl et al., 1999). Among the various methyltransferases, different mechanisms are used to accomplish electrophilic activation of the methyl group, including general acid catalysis in which proton transfer occurs to the N5 group on the pterin of MTHF, Lewis acid catalysis involving binding of methanol to a Zn(II) site, and covalent catalysis in which an adduct is formed between the methyl donor and a pyrrolysine residue.
In the methyltransferases that transfer the methyl group of MTHF to cobalamin, protonation of the N5 group of MTHF leads to electrophilic activation of the methyl group (Fig. 10.16). This mechanism has been most thoroughly studied in methionine synthase and the M. thermoacetica MTHF: CFeSP methyltransferase. Proton uptake has been measured with pH indicators, by transient kinetics, and nuclear magnetic resonance (NMR) (Seravalli et al., 1999; Smith and Matthews, 2000). The protonation step also has been studied by following the pH dependencies of the steady state and transient reaction kinetics of the MTHF:CFeSP methyltransferase (Zhao et al., 1995) and methionine synthase (Matthews, 2001) and by studies of variants that are compromised in acid–base catalysis (Doukov et al., 2007). Thus, in both methionine synthase and MTHF:CFeSP methyltransferase, the rate of reaction of CH3-H4folate with the protein-bound or exogenous cobalamin increases as the pH is lowered (with a pKa of 5.6–6.0) and the reverse reactions exhibit the opposite pH-rate profile (Matthews, 2001; Zhao et al., 1995). Furthermore, binding of CH3-H4folate to MTHF:CFeSP methyltransferase is coupled to proton uptake from solution with a pH profile similar to that of the methyl transfer reaction (Seravalli et al., 1999), suggesting protonation in the binary complex. Surprisingly, when a similar experiment was performed with the MetH(2–649) fragment, proton release rather than proton uptake is observed (Smith and Matthews, 2000). Thus, it is clear that general acid catalysis facilitates the methyl transfer reaction, but whether this occurs in the binary or ternary complex, or perhaps in the transition state for the methyl transfer reaction remains undecided. Perhaps methionine synthase and MTHF:CFeSP methyltransferase differ in this aspect of catalysis.
Figure 10.16.
Proton transfer networks without an obvious proton donor. From Doukov et al. (2007).
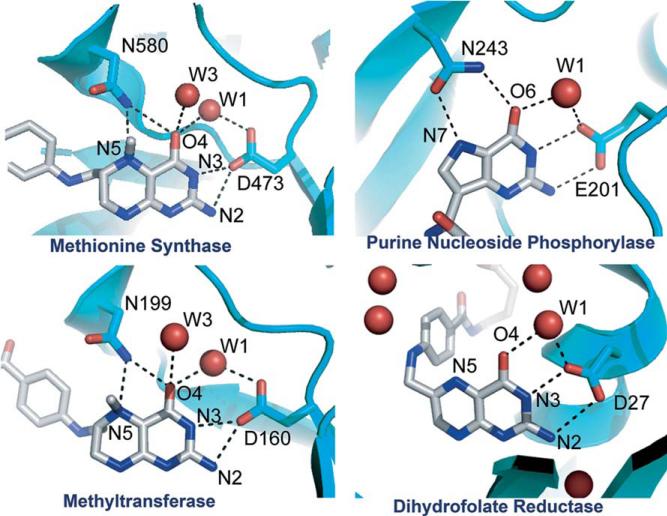
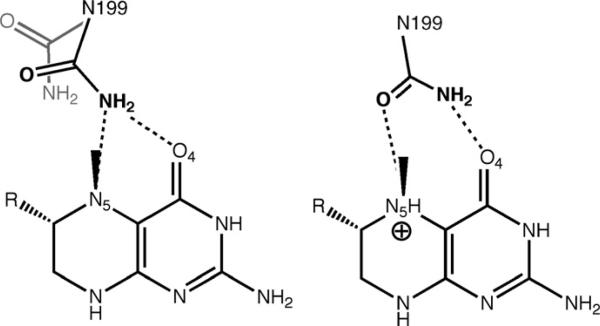
The crystal structure of the MTHF:CFeSP methyltransferase revealed no obvious proton donor within H-bonding distance of the N5 position of CH3-H4 folate and the only amino acid located near enough to N5 to participate in H-bonding is the side chain of Asn199 (Fig. 10.16) (Doukov et al., 2007). Although Asn is not chemically suitable to be a proton donor, it is conserved in all methyltransferases. The recent crystal structures of the binary complexes of CH3-H4folate bound to methionine synthase (Evans et al., 2004) and the MTHF:CFeSP methyltransferase (Doukov et al., 2007) reveal very similar environments around N5 of the pterin (Fig. 10.16). An important role for the Asn residue is indicated by its movement from a distant position to within H-bonding distance of the N5 atom upon CH3-H4folate binding, where it becomes part of an extended H-bonding network that includes several water molecules that are also conserved in the methionine synthase structure.
The lack of a discernable proton transfer pathway is seen in a number of enzymes, including methionine synthase (Evans et al., 2004), dihydrofolate reductase (Rod and Brooks, 2003), and purine nucleoside phosphorylase (Fedorov et al., 2001). Figure 10.16 shows the similarity in the H-bonding patterns among these proteins. Given the provocative location of Asn199 in MTHF:CFeSP methyltransferase and its potential role in transition state stabilization in the transmethylation reaction, an Asn199 variant was prepared by site-directed mutagenesis and the properties of this variant were compared to those of the wild-type protein by kinetic and structural studies to evaluate the contribution of this residue to catalysis (Doukov et al., 2007). These experiments are consistent with the involvement of an extended H-bonding network in proton transfer to N5 of the folate that includes Asn199, a conserved Asp (Asp160), and a water molecule. This situation is reminiscent of purine nucleoside phosphorylase, which involves protonation of the purine N7 in the transition state and is accomplished by an extended H-bonding network that includes water molecules, a Glu residue, and an Asn residue (Kicska et al., 2002). Similarly, in MTHF: CFeSP methyltransferase, an N199A variant exhibits only ~20-fold weakened affinity for CH3-H4folate, but a much more marked 20,000–40,000-fold effect on catalysis, suggesting that Asn199 plays an important role in stabilizing a transition state or high-energy intermediate for methyl transfer (Doukov et al., 2007). Thus, we speculate that the conformation of Asn in the transition state for methyl transfer resembles that of the Asn243 in the phosphorylase in which the carboxamide oxygen would accept the H-bond from N5-H, and the carboxamide nitrogen would donate a H-bond to O4 (Fig. 10.17). This dual H-bonding function could rationalize the placement of Asn at this key position in the methyltransferases.
Figure 10.17.
Left: Asn199 in the apo and MeTr-bound states. Right: Proposed transition state for proton transfer. Modified from Doukov et al. (2007).
In the methyl group transfer from methanol to CoM, catalyzed by the methanogenic MtaB, activation by protonation of the hydroxyl group is not feasible since the pKa of CH3OH2+ is −1.5 (Olah, 1993). Biochemical experiments provided strong evidence that activation involves Lewis acid catalysis by a Zn ion, which donates positive charge to the oxygen of the hydroxyl group of the substrate (Sauer and Thauer, 1997). The biochemical studies are complemented by model studies, which demonstrate Zn(II)-catalyzed methylation of cob(I)alamin by methanol (Schnyder et al., 1998). The catalytic metal-binding site of MtaB is deep within a funnel at the C-terminus, where the Zn(II) is ligated by two sulfurs from Cys residues and one carboxylate O from Glu (Fig. 10.18) (Hagemeier et al., 2006). Zn and the Co in the cobalamin are only 7.7 Å apart. It was proposed that the Lewis acid activation of methanol occurs by coordination of the hydroxyl group of methanol to the empty fourth coordination site at Zn(II). This binding mode is similar to that observed in alcohol dehydrogenase and carbonic anhydrase, both of which use Zn-based Lewis acid catalysis for substrate activation. There are other charged residues in the second coordination sphere that may also provide H-bonds and a suitable electrostatic environment for the electrophilic activation of the methyl group, facilitating attack on the methyl group by the Co center.
Figure 10.18.
Zn site in M. barkeri MtaB (gold) where methanol is proposed to be activated (Hagemeier et al., 2006). The MtaC subunit is shown in cyan. Generated using Chimera from PDB# 2I2X.
Figure 10.15 summarizes the proposed mechanism of covalent catalysis by pyrrolysine in activation of the methyl group of mono-, di-, and trimethylamines (Hao et al., 2002). Recent structures of MtmB in the presence of hydroxylamine and N-methyl-hydroxylamine demonstrate the adduct between the amine and C-2 of pyrrolysine (Hao et al., 2004). Thus, as shown in Fig. 10.15, nucleophilic attack of the methylamine substrate on the imino group of pyrrolysine generates a substituted methyl ammonium adduct at C-2. The positive charge on nitrogen is expected to lead to electrophilic activation of the methyl group, facilitating attack by Co(I), which would generate methyl-Co(III) and leave a covalent amine adduct on pyrrolysine. Proton transfer associated with elimination of the amine as ammonia would regenerate pyrrolysine for the next round of catalysis.
Thus, there are at least three ways that enzymes activate the methyl donor in methyltransferases: general acid catalyzed protonation of the N5 of pterins in MTHF (and probably in MTHMPT) through a H-bonding network, Lewis acid catalysis using a Zn active site near the cobalamin, and covalent catalysis using a novel amino acid.
VI. Activation of the Methyl Group Acceptors: Zn Thiolates and NiFeS Clusters
The methyl group acceptors listed in Table 10.1 are amines (THF, THMPT), thiols (CoM or homocysteine), and metal ions (a NiFeS cluster). Transfer of the methyl group to THF or THMPT is simply the reverse of the methylation of cobalamin by MTHF or MTHMPT and will not be discussed here, where the focus will be on methylation of homocysteine by methionine synthase, of CoM by several of the “A” components, or of the Ni center in the A-Cluster of ACS.
A. Methylation of thiol acceptors
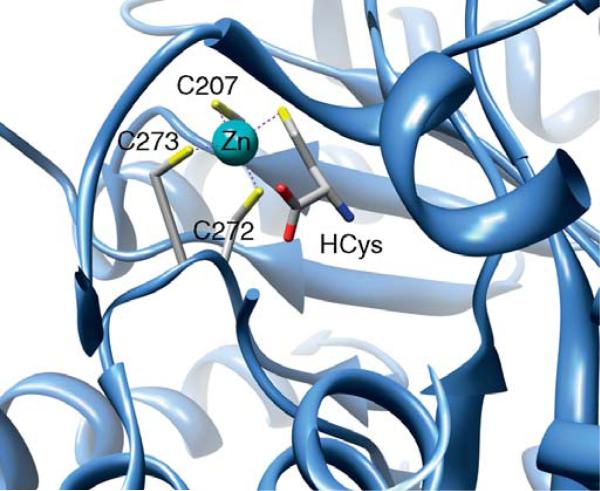
A general theme for enzymes catalyzing alkyl transfers to thiols is that they possess a catalytic Zn site, which is considered to be important in enhancing the nucleophilicity of the thiol at neutral pH (Matthews and Goulding, 1997). The role of Zn appears to be activation of the thiol group by decreasing its pKa value, since a proton is released upon binding of homocysteine to methionine synthase (Goulding and Matthews, 1997). Although the nucleophilicity of a Zn-bound thiol is less than that of a free thiolate, the pKa of the free thiol of CoM or homocysteine is too high to allow significant amounts of the thiolate to be present, and the Zn-bound thiolate is much more nucleophilic than a thiol. Thus, metal ion-catalyzed activation poises homocysteine or CoM for nucleophilic attack on an intermediate methyl donor, such as MeCbl.
A subset of these methyltransferases, including methionine synthase, MtaA, MtbA, and MtsA share a Cys-X-His-Xn-Cys motif, where Zn coordinates to the His and two Cys residues (Gencic et al., 2001; Krüer et al., 2002; Tallant et al., 2001; Zhou et al., 1999). In methioine synthase, the ligation of homocysteine directly to a Zn site was shown by XAS studies using selenohomocysteine (Peariso et al., 2001). The crystal structure of the homocysteine domain of methionine synthase reveals an (αβ)8 TIM barrel, like that of the MTHF-binding domain. Although they exhibit little sequence identify, both cobalamin-dependent (MetH) and cobalamin–independent (MetE) methionine synthases contain Zn-binding sites. The Zn site in cobalamin-independent methionine synthase contains two Cys and one His ligands, while in cobalamin-dependent methionine synthase (Fig 10.19), it consists of three Cys ligands ((Evans et al., 2004; Peariso et al., 1998, 2001) and references therein). Homocysteine binds similarly to a (Cys)3Zn site within the TIM barrel in betaine–homocysteine methyltransferase (BHMT) (Evans et al., 2002) .
Figure 10.19.
Homocysteine ligated to Zn within the triosephosphate isomerase (TIM) barrel of the N-terminal domain of MetH. Generated from PDB ID# 1Q8A using Chimera.
The “A” components of a variety of methanogenic methyltransferases also contain catalytic Zn sites that are responsible for binding CoM or homocysteine, including the MtaA component of the methanol:CoM methyltransferase system, which is responsible for methyl transfer from methyl-Co(III) on MtaC to CoM (Gencic et al., 2001). MtsA (Tallant et al., 2001) and MtbA (Krüer et al., 2002), which are MeCbl:CoM methyltransferases. Other Zn enzymes that bind thiols during their catalytic mechanism include the E.coli Ada protein (Myers et al., 1994; Wilker and Lippard, 1997), S-methylmethionine:homocysteine methyltransferase (Thanbichler et al., 1999), epoxyalkane:coenzyme M transferase (Allen et al., 1999; Ensign and Allen, 2003), and protein farnesyl transferase (Huang et al., 1997; Strickland et al., 1998).
B. Methylation of the NiFeS cluster of ACS
Acetyl-CoA Synthase (ACS), encoded by the acsB gene in M. thermoacetica, is part of a complex that catalyzes the conversion of CO2, CoA, and a methyl group to acetyl-CoA, as shown on the right hand side of Fig. 10.6. Several reviews that focus on the structure, function, and mechanism of ACS are available (Brunold, 2004; Drennan et al., 2004; Lindahl, 2004; Ragsdale, 2006; Riordan, 2004). The other component of this complex is CODH, which is encoded by the acsA gene, and catalyzes the reduction of CO2 to CO. CODH and ACS contain internal channels that interlink to form a 70 Å channel that sequesters CO and facilitates its delivery to the ACS active site (Tan et al., 2005; Doukov et al., 2008). This channel has been identified biochemically (Maynard and Lindahl, 1999; Seravalli and Ragsdale, 2000) and by X-ray crystallography (Darnault et al., 2003; Doukov et al., 2002, 2008).
The catalytic strategy of ACS is to use a Ni active site, the A-Cluster, to form organometallic intermediates. The A-Cluster consists of a [4Fe-4S] cluster bridged to a binuclear NiNi center, which contains a Ni site (Nip) that is thiolate bridged to another Ni ion in a thiolato- and carboxamidotype N2S2 coordination environment (Darnault et al., 2003; Doukov et al., 2002; Ragsdale et al., 1985; Svetlitchnyi et al., 2004). Although the details are under discussion, the basic mechanism of ACS involves metal-centered catalysis that includes the following bioorganometallic intermediates: methyl-Ni, Ni-CO, and acetyl-Ni. The mechanism involves transfer of a methyl group from the MeCbl state of the CFeSP to a Ni center, which also binds CO and CoA; then, ACS catalyzes condensation of the C–C and C–S bonds to form acetyl-CoA (Ragsdale and Wood, 1985). The methyl transfer reaction could occur by either a radical or an SN2-type nucleophilic mechanism. Model studies of the reaction between methyl-Co3+ (CH3-Co3+ dimethylglyoximate) and a Ni1+ macrocycle provide precedent for a methyl radical transfer (Ram and Riordan, 1995; Ram et al., 1997). A radical methyl transfer would require homolysis of the CH3–Co bond of the methylated CFeSP, which Martin and Finke pointed out was not favorable because reduction of CH3-Co3+ requires redox potentials (< − 1 V) that are too low for physiological electron donors (Martin and Finke, 1990). Rapid kinetic studies and stereochemical studies using a chiral methyl donor also indicate that the transmethylation reaction involves an SN2-type nucleophilic attack of Ni on the methyl group of the methylated CFeSP (CH3–Co3+) to generate methyl-Ni and Co1+(Lebertz et al., 1987; Menon and Ragsdale, 1998, 1999). Thus, it is likely that the metal-to-metal methyl transfer reaction is similar to the other B12-dependent methyltransferase reactions described above. In fact, kinetic studies indicate that the Ni (I) site on ACS is as strong a nucleophile as is the Co1+ site in the CFeSP (Tan et al., 2003).
ACKNOWLEDGMENTS
I thank NIH (GM39451) for supporting the work in my laboratory on methyltransferases. I thank Ruma Banerjee, Joe Krzycki, Vadim Gladyshev, and Rick Finke for their comments on parts of the manuscript.
Footnotes
Publisher's Disclaimer: This chapter was originally published in the book Vitamins and Hormones: Folic Acid and Folates - Vol. 79. The copy attached is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non-commercial research and educational use. This includes without limitation use in instruction at your institution, distribution to specific colleagues, and providing a copy to your institution.s administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution.s website or repository, are prohibited. For exceptions, permission may be sought for such use through Elsevier's permissions site at: http://www.elsevier.com/locate/permissionusematerial
REFERENCES
- Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 2005;187:2030–2037. doi: 10.1128/JB.187.6.2030-2037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abend A, Bandarian V, Nitsche R, Stupperich E, Retey J, Reed GH. Ethanolamine ammonia-lyase has a “base-on” binding mode for coenzyme B-12. Arch. Biochem. Biophys. 1999;370:138–141. doi: 10.1006/abbi.1999.1382. [DOI] [PubMed] [Google Scholar]
- Abend A, Nitsche R, Bandarian V, Stupperich E, Retey J. Dioldehydrase Binds Coenzyme B12 in the “Base-On” Mode: ESR Investigations on Cob(II)alamin. Angew. Chem. Int. Ed. 1998;37:625–627. doi: 10.1002/(SICI)1521-3773(19980316)37:5<625::AID-ANIE625>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Achari A, Somers DO, Champness JN, Bryant PK, Rosemond J, Stammers DK. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- Allen JR, Clark DD, Krum JG, Ensign SA. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc. Natl. Acad. Sci. USA. 1999;96:8432–8437. doi: 10.1073/pnas.96.15.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angier RB, et al. The Structure and Synthesis of the Liver L. casei Factor. Science. 1946;103:667–669. [PubMed] [Google Scholar]
- Bandarian V, Ludwig ML, Matthews RG. Factors modulating conformational equilibria in large modular proteins: a case study with cobalamin-dependent methionine synthase. Proc. Natl. Acad. Sci. USA. 2003;100:8156–8163. doi: 10.1073/pnas.1133218100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandarian V, Pattridge KA, Lennon BW, Huddler DP, Matthews RG, Ludwig ML. Domain alternation switches B(12)-dependent methionine synthase to the activation conformation. Nat. Struct. Biol. 2002;9:53–56. doi: 10.1038/nsb738. [DOI] [PubMed] [Google Scholar]
- Banerjee R, Frasca V, Ballou DP, Matthews RG. Participation of Cob(I) alamin in the Reaction Catalyzed by Methionine Synthase from Escherichia coli: A Steady-State and Rapid Reaction Kinetic Analysis. Biochem. 1990a;29:11101–11109. doi: 10.1021/bi00502a013. [DOI] [PubMed] [Google Scholar]
- Banerjee RV, Frasca V, Ballou DP, Matthews RG. Participation of cob(I) alamin in the reaction catalyzed by methionine synthase from Escherichia coli:A steady-state and rapid reaction kinetic analysis. Biochemistry. 1990b;29:11101–11109. doi: 10.1021/bi00502a013. [DOI] [PubMed] [Google Scholar]
- Banerjee RV, Harder SR, Ragsdale SW, Matthews RG. Mechanism of reductive activation of cobalamin-dependent methionine synthase: an electron para-magnetic resonance spectroelectrochemical study. Biochem. 1990c;29:1129–1135. doi: 10.1021/bi00457a005. [DOI] [PubMed] [Google Scholar]
- Barker HA, Weissbach H, Smyth RD. A coenzyme containing pseudo-vitamin B12. Proceedings of the National Academy of Sciences, USA. 1958;44:1093–1097. doi: 10.1073/pnas.44.11.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, Krzycki JA. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- Brunold TC. Spectroscopic and Computational Insights into the Geometric and Electronic Properties of the A Cluster of Acetyl-Coenzyme A Synthase. J. Biol. Inorg. Chem. 2004;9:533–541. doi: 10.1007/s00775-004-0566-8. [DOI] [PubMed] [Google Scholar]
- Catalan J, Elguero J, Flammang R, Maquestiau A. On the relative basicities of imidazole and benzimidazole. Angew. Chem. Suppl. 1983;4:411–418. [Google Scholar]
- Col B, Oltean S, Banerjee R. Translational regulation of human methionine synthase by upstream open reading frames. Biochim. Biophys. Acta. 2007;1769:532–540. doi: 10.1016/j.bbaexp.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costi MP, Ferrari S. Update on antifolate drugs targets. Curr Drug Targets. 2001;2:135–166. doi: 10.2174/1389450013348669. [DOI] [PubMed] [Google Scholar]
- Daas PJH, Hagen WR, Keltjens JT, Vanderdrift C, Vogels GD. Activation mechanism of Methanol:5- hydroxybenzimidazolylcobamide methyltransferase from Methanosarcina barkeri. J. Biol. Chem. 1996a;271:22346–22351. doi: 10.1074/jbc.271.37.22346. [DOI] [PubMed] [Google Scholar]
- Daas PJH, Wassenaar RW, Willemsen P, Theunissen RJ, Keltjens JT, Vanderdrift C, Vogels GD. Purification and properties of an enzyme involved in the ATP-dependent activation of the methanol:2- mercaptoethanesulfonic acid methyltransferase reaction in Methanosarcina barkeri. J. Biol. Chem. 1996b;271:22339–22345. doi: 10.1074/jbc.271.37.22339. [DOI] [PubMed] [Google Scholar]
- Darnault C, Volbeda A, Kim EJ, Legrand P, Vernede X, Lindahl PA, Fontecilla-Camps JC. Ni-Zn-[Fe(4)-S(4)] and Ni-Ni-[Fe(4)-S(4)] clusters in closed and open alpha subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase. Nat. Struct. Biol. 2003;10:271–279. doi: 10.1038/nsb912. [DOI] [PubMed] [Google Scholar]
- Das A, et al. Characterization of a corrinoid protein involved in the C1 metabolism of strict anaerobic bacterium Moorella thermoacetica. Proteins. 2007;67:167–176. doi: 10.1002/prot.21094. [DOI] [PubMed] [Google Scholar]
- Datta SP, Grzybowski AK. The acid dissociation constant of the imidazolium ion. J. Chem. Soc. (B) 1966:136–140. [Google Scholar]
- Dorweiler JS, Finke RG, Matthews RG. Cobalamin-dependent methionine synthase: probing the role of the axial base in catalysis of methyl transfer between methyltetrahydrofolate and exogenous cob(I)alamin or cob(I)inamide. Biochemistry. 2003;42:14653–14662. doi: 10.1021/bi035525t. [DOI] [PubMed] [Google Scholar]
- Doukov T, Seravalli J, Stezowski J, Ragsdale SW. Crystal structure of a methyltetrahydrofolate and corrinoid dependent methyltransferase. Structure. 2000;8:817–830. doi: 10.1016/s0969-2126(00)00172-6. [DOI] [PubMed] [Google Scholar]
- Doukov TI, Blasiak LC, Seravalli J, Ragsdale SW, Drennan CL. Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Biochemistry. 2008;47:3474–3483. doi: 10.1021/bi702386t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doukov TI, Hemmi H, Drennan CL, Ragsdale SW. Structural And Kinetic Evidence For An Extended Hydrogen Bonding Network In Catalysis Of Methyl Group Transfer: Role Of An Active Site Asparagine Residue In Activation Of Methyl Transfer By Methyltransferases. Journal of Biological Chemistry. 2007;282:6609–6618. doi: 10.1074/jbc.M609828200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doukov TI, Iverson T, Seravalli J, Ragsdale SW, Drennan CL. A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Science. 2002;298:567–572. doi: 10.1126/science.1075843. [DOI] [PubMed] [Google Scholar]
- Drennan CL, Doukov TI, Ragsdale SW. The Metalloclusters of Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase: A Story in Pictures. J. Biol. Inorg. Chem. 2004;9:511–515. doi: 10.1007/s00775-004-0563-y. [DOI] [PubMed] [Google Scholar]
- Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. How a protein binds B12: A 3.0Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- Drummond JT, Huang S, Blumenthal RM, Matthews RG. Assignment of enzymatic function to specific protein regions of cobalamin-dependent methionine synthase from Escherichia coli. Biochem. 1993;32:9290–9295. doi: 10.1021/bi00087a005. [DOI] [PubMed] [Google Scholar]
- Engelmann T, Kaufmann F, Diekert G. Isolation and characterization of a veratrol:corrinoid protein methyl transferase from Acetobacterium dehalogenans. Arch. Microbiol. 2001;175:376–383. doi: 10.1007/s002030100275. [DOI] [PubMed] [Google Scholar]
- Ensign SA, Allen JR. Aliphatic epoxide carboxylation. Annu. Rev. Biochem. 2003;72:55–76. doi: 10.1146/annurev.biochem.72.121801.161820. [DOI] [PubMed] [Google Scholar]
- Evans JC, Huddler DP, Hilgers MT, Romanchuk G, Matthews RG, Ludwig ML. Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase. Proc. Natl. Acad. Sci. USA. 2004;101:3729–3736. doi: 10.1073/pnas.0308082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JC, Huddler DP, Jiracek J, Castro C, Millian NS, Garrow TA, Ludwig ML. Betaine-homocysteine methyltransferase: zinc in a distorted barrel. Structure. 2002;10:1159–1171. doi: 10.1016/s0969-2126(02)00796-7. [DOI] [PubMed] [Google Scholar]
- Fasching M, Schmidt W, Krautler B, Stupperich E, Schmidt A, Kratky C. Co alpha-(1H-imidazolyl)-Co beta-methylcob(III)amide: Model for protein-bound corrinoid cofactors. Helv. Chim. Acta. 2000;83:2295–2316. [Google Scholar]
- Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, Almo SC. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- Fujii K, Galivan JH, Huennekens FM. Activation of methionine synthase: further characterization of flavoprotein system. Arch. Biochem. Biophys. 1977;178:662–670. doi: 10.1016/0003-9861(77)90238-7. [DOI] [PubMed] [Google Scholar]
- Gencic S, LeClerc GM, Gorlatova N, Peariso K, Penner-Hahn JE, Grahame DA. Zinc-thiolate intermediate in catalysis of methyl group transfer in Methanosarcina barkeri. Biochemistry. 2001;40:13068–13078. doi: 10.1021/bi0112917. [DOI] [PubMed] [Google Scholar]
- Gottschalk G, Thauer RK. The Na(+)-translocating methyltransferase complex from methanogenic archaea. Biochim. Biophys. Acta. 2001;1505:28–36. doi: 10.1016/s0005-2728(00)00274-7. [DOI] [PubMed] [Google Scholar]
- Goulding CW, Matthews RG. Cobalamin-dependent methionine synthase from Escherichia coli: involvement of zinc in homocysteine activation. Biochemistry. 1997;36:15749–15757. doi: 10.1021/bi971988l. [DOI] [PubMed] [Google Scholar]
- Guest JR, Friedman S, Woods DD, Smith EL. A methyl analog of cobamide coenzyme in relation to methionine synthesis by bacteria. Nature. 1962;195:340–342. doi: 10.1038/195340a0. [DOI] [PubMed] [Google Scholar]
- Hagemeier CH, Krer M, Thauer RK, Warkentin E, Ermler U. Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex. Proc. Natl. Acad. Sci. USA. 2006;103:18917–18922. doi: 10.1073/pnas.0603650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampele IC, D'Arcy A, Dale GE, Kostrewa D, Nielsen J, Oefner C, Page MG, Schonfeld HJ, Stuber D, Then RL. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J. Mol. Biol. 1997;268:21–30. doi: 10.1006/jmbi.1997.0944. [DOI] [PubMed] [Google Scholar]
- Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- Hao B, Zhao G, Kang PT, Soares JA, Ferguson TK, Gallucci J, Krzycki JA, Chan MK. Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid. Chem. Biol. 2004;11:1317–1324. doi: 10.1016/j.chembiol.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Harder SA, Lu W-P, Feinberg BF, Ragsdale SW. Spectroelectrochemical studies of the corrinoid/iron-sulfur protein from Clostridium thermoaceticum. Biochemistry. 1989;28:9080–9087. doi: 10.1021/bi00449a019. [DOI] [PubMed] [Google Scholar]
- Hilhorst E, Iskander AS, Chen TBRA, Pandit U. Alkyl transfer from quaternary ammonium salts to cobalt(I): model for the cobalamin-dependent methionine synthase reaction. Tetrahedron. 1994;50:8863–8870. [Google Scholar]
- Hodgkin DC, Kamper J, Mackay M, Pickworth J, Trueblood KN, White JG. Structure of vitamin B12. Nature. 1956;178:64–66. doi: 10.1038/178064a0. [DOI] [PubMed] [Google Scholar]
- Hogenkamp HP, Rush JE, Swenson CA. Observations on the organo-metallic bond of the corrinoid coenzymes. J. Biol. Chem. 1965;240:3641–3644. [PubMed] [Google Scholar]
- Hoover DM, Jarrett JT, Sands RH, Dunham WR, Ludwig ML, Matthews RG. Interaction of Escherichia coli cobalamin-dependent methionine synthase and its physiological partner flavodoxin: Binding of flavodoxin leads to axial ligand dissociation from the cobalamin cofactor. Biochemistry. 1997;36:127–138. doi: 10.1021/bi961693s. [DOI] [PubMed] [Google Scholar]
- Huang CC, Casey PJ, Fierke CA. Evidence for a catalytic role of zinc in protein farnesyltransferase. Spectroscopy of Co2+-farnesyltransferase indicates metal coordination of the substrate thiolate. J. Biol. Chem. 1997;272:20–23. doi: 10.1074/jbc.272.1.20. [DOI] [PubMed] [Google Scholar]
- Jablonski PE, Lu WP, Ragsdale SW, Ferry JG. Characterization of the metal centers of the corrinoid/iron-sulfur component of the CO dehydrogenase enzyme complex from Methanosarcina thermophila by EPR spectroscopy and spectroelectrochemistry. J. Biol. Chem. 1993;268:325–329. [PubMed] [Google Scholar]
- Kaufmann F, Wohlfarth G, Diekert G. O-demethylase from Acetobacterium dehalogenans--substrate specificity and function of the participating proteins. Eur. J. Biochem. 1998;253:706–711. doi: 10.1046/j.1432-1327.1998.2530706.x. [DOI] [PubMed] [Google Scholar]
- Ke SC, Torrent M, Museav DG, Morokuma K, Warncke K. Identification of dimethylbenzimidazole axial coordination and characterization of (14)N super-hyperfine and nuclear quadrupole coupling in Cob(II)alamin bound to ethanolamine deaminase in a catalytically-engaged substrate radical-Cobalt(II) biradical state. Biochemistry. 1999;38:12681–12689. doi: 10.1021/bi983067w. [DOI] [PubMed] [Google Scholar]
- Kicska GA, Tyler PC, Evans GB, Furneaux RH, Shi W, Fedorov A, Lewandowicz A, Cahill SM, Almo SC, Schramm VL. Atomic dissection of the hydrogen bond network for transition-state analogue binding to purine nucleoside phosphorylase. Biochemistry. 2002;41:14489–14498. doi: 10.1021/bi026636f. [DOI] [PubMed] [Google Scholar]
- Kräutler B. Thermodynamic trans-effects of the nucleotide base in the B12 coenzymes. Helvetica Chimica Acta. 1987;70:1268–1278. [Google Scholar]
- Krüer M, Haumann M, Meyer-Klaucke W, Thauer RK, Dau H. The role of zinc in the methylation of the coenzyme M thiol group in methanol:coenzyme M methyltransferase from Methanosarcina barkeri. Eur. J. Biochem. 2002;269:2117–2123. doi: 10.1046/j.1432-1033.2002.02860.x. [DOI] [PubMed] [Google Scholar]
- Krzycki JA. Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 2004;8:484–491. doi: 10.1016/j.cbpa.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Lane TJ, Quinlan KP. Metal binding of the benzimidazoles. J. Am. Chem. Soc. 1960;82:2994–2997. [Google Scholar]
- Lawrence CC, Gerfen GJ, Samano V, Nitsche R, Robins MJ, Retey J, Stubbe J. Binding of Cob(II)alamin to the adenosylcobalamin-dependent ribo-nucleotide reductase from Lactobacillus leichmannii. Identification of dimethylbenzimidazole as the axial ligand. J. Biol. Chem. 1999;274:7039–7042. doi: 10.1074/jbc.274.11.7039. [DOI] [PubMed] [Google Scholar]
- Lebertz H, Simon H, Courtney LF, Benkovic SJ, Zydowsky LD, Lee K, Floss HG. Stereochemistry of acetic acid formation from 5-methyl-tetrahydrofolate by Clostridium thermoaceticum. J. Am. Chem. Soc. 1987;109:3173–3174. [Google Scholar]
- Lexa D, Savéant J-M. Electrochemistry of vitamin B12. I. Role of the base-on/base-off reaction in the oxidoreduction mechanism of the B12r-B12s system. J. Am. Chem. Soc. 1976;98:2652–2658. doi: 10.1021/ja00425a039. [DOI] [PubMed] [Google Scholar]
- Lindahl PA. Acetyl-Coenzyme A Synthase: The Case for a Nip0-Based Mechanism of Catalysis. J. Biol. Inorg. Chem. 2004;9:516–524. doi: 10.1007/s00775-004-0564-x. [DOI] [PubMed] [Google Scholar]
- Ljungdahl L, Irion E, Wood HG. Role of corrinoids in the total synthesis of acetate from CO2 by Clostridium thermoaceticum. Fed. Proc. 1966;25:1642–1648. [PubMed] [Google Scholar]
- Longstaff DG, Larue RC, Faust JE, Mahapatra A, Zhang L, Green-Church KB, Krzycki JA. A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine. Proc. Natl. Acad. Sci. USA. 2007;104:1021–1026. doi: 10.1073/pnas.0610294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig ML, Matthews RG. Structure-based perspectives on B-12-dependent enzymes. Annu. Rev. Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- Martin BD, Finke RG. Co-C homolysis and bond dissociation energy studies of biological alkylcobalamins: methylcobalamin, including a ≥1015 Co-CH3 homolysis rate enhancement at 25 °C following one-electron reduction. J. Am. Chem. Soc. 1990;112:2419–2420. [Google Scholar]
- Matthews RG. Cobalamin-dependent methyltransferases. Acc. Chem. Res. 2001;34:681–689. doi: 10.1021/ar0000051. [DOI] [PubMed] [Google Scholar]
- Matthews RG, Goulding CW. Enzyme-catalyzed methyl transfers to thiols: the role of zinc. Curr. Opin. Chem. Biol. 1997;1:332–339. doi: 10.1016/s1367-5931(97)80070-1. [DOI] [PubMed] [Google Scholar]
- Maynard EL, Lindahl PA. Evidence of a molecular tunnel connecting the active sites for CO2 reduction and acetyl-CoA synthesis in acetyl-CoA synthase from Clostridium thermoaceticum. Journal of the American Chemical Society. 1999;121:9221–9222. [Google Scholar]
- Menon S, Ragsdale SW. Role of the [4Fe-4S] cluster in reductive activation of the cobalt center of the corrinoid iron-sulfur protein from Clostridium thermoaceticum during acetyl-CoA synthesis. Biochemistry. 1998;37:5689–5698. doi: 10.1021/bi9727996. [DOI] [PubMed] [Google Scholar]
- Menon S, Ragsdale SW. The role of an iron-sulfur cluster in an enzymatic methylation reaction: methylation of CO dehydrogenase/acetyl-CoA synthase by the methylated corrinoid iron-sulfur protein. J. Biol. Chem. 1999;274:11513–11518. doi: 10.1074/jbc.274.17.11513. [DOI] [PubMed] [Google Scholar]
- Minot GR, Murphy WP. Treatment of pernicious anaemia by a special diet. J. Am. Med. Assn. 1926;87:470–476. [PMC free article] [PubMed] [Google Scholar]
- Mitchell HK, Snell EE, Williams RJ. The concentration of “folic acid”. J. Am. Chem. Soc. 1941;63:2284. [Google Scholar]
- Myers LC, Cushing TD, Wagner G, Verdine GL. Metal-coordination sphere in the methylated Ada protein-DNA co-complex. Chem. Biol. 1994;1:91–97. doi: 10.1016/1074-5521(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Naidu D, Ragsdale SW. Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica. J. Bacteriol. 2001;183:3276–3281. doi: 10.1128/JB.183.11.3276-3281.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris PR, Pratt JM. Methyl transfer reactions of protein-free Co corrinoids. BioFactors. 1996;5:240–241. [Google Scholar]
- Olah GA. Superelectrophiles. Angew. Chem. Int. Ed. 1993;32:767–788. [Google Scholar]
- Olteanu H, Banerjee R. Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J. Biol. Chem. 2001;276:35558–35563. doi: 10.1074/jbc.M103707200. [DOI] [PubMed] [Google Scholar]
- Olteanu H, Wolthers KR, Munro AW, Scrutton NS, Banerjee R. Kinetic and thermodynamic characterization of the common polymorphic variants of human methionine synthase reductase. Biochemistry. 2004;43:1988–1997. doi: 10.1021/bi035910i. [DOI] [PubMed] [Google Scholar]
- Peariso K, Goulding CW, Huang S, Matthews RG, Penner-Hahn JE. Characterization of the zinc binding site in methionine synthase enzymes of Escherichia coli: The role of zinc in the methylation of homocysteine. J. Am. Chem. Soc. 1998;120:8410–8416. [Google Scholar]
- Peariso K, Zhou ZS, Smith AE, Matthews RG, Penner-Hahn JE. Characterization of the zinc sites in cobalamin-independent and cobalamin-dependent methionine synthase using zinc and selenium X-ray absorption spectroscopy. Biochemistry. 2001;40:987–993. doi: 10.1021/bi001711c. [DOI] [PubMed] [Google Scholar]
- Pejchal R, Ludwig ML. Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication. PLoS Biol. 2005;3:e31. doi: 10.1371/journal.pbio.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfiffner JJ, Calkins DG, O'Dell BL, Bloom ES, Brown RA, Campbell CJ, Bird OD. Isolation of an antianemia factor (vitamin Bc conjugate) in crystalline form from yeast. Science. 1945;102:228–230. doi: 10.1126/science.102.2644.228. [DOI] [PubMed] [Google Scholar]
- Poston JM, Kuratomi K, Stadtman ER. Methyl-vitamin B12 as a source of methyl groups for the synthesis of acetate by cell-free extracts of Clostridium thermoaceticum. Ann. N.Y. Acad. Sci. 1964;112:804–806. doi: 10.1111/j.1749-6632.1964.tb45057.x. [DOI] [PubMed] [Google Scholar]
- Pratt JM. The roles of Co, corrin, and protein: I. Co-ligand bonding and the trans ligand effect. In: Banerjee R, editor. Chemistry and biochemistry of B12. Vol. 1. John Wiley and Sons; New York: 1999. pp. 73–112. [Google Scholar]
- Pratt JM, Norris PR, Hamza SA, Bolton R. Methyl Transfer from Nitrogen to Cobalt: Model for the B12-Dependent Methyl Transfer Enzymes. J. Chem. Soc., Chem. Commun. 1994:1333–1334. [Google Scholar]
- Ragsdale SW. Metals and their scaffolds to promote difficult enzymatic reactions. Chem. Rev. 2006;106:3317–3337. doi: 10.1021/cr0503153. [DOI] [PubMed] [Google Scholar]
- Ragsdale SW, Lindahl PA, Münck E. Mössbauer, EPR, and optical studies of the corrinoid/Fe-S protein involved in the synthesis of acetyl-CoA by Clostridium thermoaceticum. J. Biol. Chem. 1987;262:14289–14297. [PubMed] [Google Scholar]
- Ragsdale SW, Wood HG. Acetate biosynthesis by acetogenic bacteria: Evidence that carbon monoxide dehydrogenase is the condensing enzyme that catalyzes the final steps of the synthesis. J. Biol. Chem. 1985;260:3970–3977. [PubMed] [Google Scholar]
- Ragsdale SW, Wood HG, Antholine WE. Evidence that an iron-nickel-carbon complex is formed by reaction of CO with the CO dehydrogenase from Clostridium thermoaceticum. Proc. Natl. Acad. Sci. USA. 1985;82:6811–6814. doi: 10.1073/pnas.82.20.6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram MS, Riordan CG. Methyl transfer from a cobalt complex to Ni(tmc) (+) yielding Ni(tmc)Me(+): A model for methylcobalamin alkylation of CO dehydrogenase. J. Am. Chem. Soc. 1995;117:2365–2366. [Google Scholar]
- Ram MS, Riordan CG, Yap GPA, Liable-Sands L, Rheingold AL, Marchaj A, Norton JR. Kinetics and mechanism of alkyl transfer from organocobalt (III) to nickel(I): Implications for the synthesis of acetyl coenzyme A by CO dehydrogenase. J. Am. Chem. Soc. 1997;119:1648–1655. [Google Scholar]
- Rickes EL, Brink NG, Koniuszy FR, Wood TR, Folkers K. Crystalline Vitamin B12. Science. 1948;107:396–397. doi: 10.1126/science.107.2781.396. [DOI] [PubMed] [Google Scholar]
- Riordan C. Synthetic Chemistry and Chemical Precedents for Understanding the Structure and Function of Acetyl Coenzyme A Synthase. J. Biol. Inorg. Chem. 2004;9:542–549. doi: 10.1007/s00775-004-0567-7. [DOI] [PubMed] [Google Scholar]
- Rod TH, Brooks CL., 3rd. How dihydrofolate reductase facilitates protonation of dihydrofolate. J. Am. Chem. Soc. 2003;125:8718–8719. doi: 10.1021/ja035272r. [DOI] [PubMed] [Google Scholar]
- Sauer K, Thauer RK. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri-Zinc dependence and thermodynamics of the methanol:cob(I) alamin methyltransferase reaction. Eur. J. Biochem. 1997;249:280–285. doi: 10.1111/j.1432-1033.1997.t01-1-00280.x. [DOI] [PubMed] [Google Scholar]
- Schnyder A, Darbre T, Keese R. Methyl transfer from methanol to Co-cobyrinate: a medel for the coenzyme B12 dependent methyltransferase. Angew. Chem. Int. Ed. Engl. 1998;37:1283–1285. doi: 10.1002/(SICI)1521-3773(19980518)37:9<1283::AID-ANIE1283>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Schrauzer GN, Deutsch E, Windgassen RJ. The nucleophilicity of vitamin B12s. J. Am. Chem. Soc. 1968;90:2441–2442. doi: 10.1021/ja01011a054. [DOI] [PubMed] [Google Scholar]
- Schrauzer GN, Deutsch EJ. Reactions of cobalt(I) supernucleophiles.The alkylation of vitamin B12s, cobaloximes (I), and related compounds. J. Am. Chem. Soc. 1969;91:3341–3350. doi: 10.1021/ja01040a041. [DOI] [PubMed] [Google Scholar]
- Seravalli J, Brown KL, Ragsdale SW. Acetyl-Coenzyme A Synthesis From Unnatural Methylated Corrinoids: Requirement for “Base-Off” Coordination at Cobalt. J. Am. Chem. Soc. 2001;123:1786–1787. doi: 10.1021/ja005709k. [DOI] [PubMed] [Google Scholar]
- Seravalli J, Ragsdale SW. Channeling of Carbon Monoxide during Anaerobic Carbon Dioxide Fixation. Biochemistry. 2000;39:1274–1277. doi: 10.1021/bi991812e. [DOI] [PubMed] [Google Scholar]
- Seravalli J, Shoemaker RK, Sudbeck MJ, Ragsdale SW. Binding of (6R, S)-methyltetrahydrofolate to methyltransferase from Clostridium thermoaceticum: Role of protonation of methyltetrahydrofolate in the mechanism of methyl transfer. Biochemistry. 1999;38:5736–5745. doi: 10.1021/bi9824745. [DOI] [PubMed] [Google Scholar]
- Shibata N, Masuda J, Tobimatsu T, Toraya T, Suto K, Morimoto Y, Yasuoka N. A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase. Structure. 1999;7:997–1008. doi: 10.1016/s0969-2126(99)80126-9. [DOI] [PubMed] [Google Scholar]
- Sintchak MD, Arjara G, Kellogg BA, Stubbe J, Drennan CL. The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer. Nat. Struct. Biol. 2002;9:293–300. doi: 10.1038/nsb774. [DOI] [PubMed] [Google Scholar]
- Smith AE, Matthews RG. Protonation state of methyltetrahydrofolate in a binary complex with cobalamin-dependent methionine synthase. Biochemistry. 2000;39:13880–13890. doi: 10.1021/bi001431x. [DOI] [PubMed] [Google Scholar]
- Smith EL. Purification of anti-pernicious anæmia factors from liver. Nature. 1948;161:638–639. doi: 10.1038/161638a0. [DOI] [PubMed] [Google Scholar]
- Soares JA, Zhang L, Pitsch RL, Kleinholz NM, Jones RB, Wolff JJ, Amster J, Green-Church KB, Krzycki JA. The residue mass of L-pyrrolysine in three distinct methylamine methyltransferases. J. Biol. Chem. 2005;280:36962–36969. doi: 10.1074/jbc.M506402200. [DOI] [PubMed] [Google Scholar]
- Stich TA, Seravalli J, Venkateshrao S, Spiro TG, Ragsdale SW, Brunold TC. Spectroscopic Studies of the Corrinoid/Iron-Sulfur Protein from Moorella thermoacetica. Journal of the American Chemical Society. 2006;128:5010–5020. doi: 10.1021/ja054690o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokstad ELR. Some properties of a growth factor for Lactobacillus casei. J. Biol. Chem. 1943;149:573–574. [Google Scholar]
- Strickland CL, Windsor WT, Syto R, Wang L, Bond R, Wu Z, Schwartz J, Le HV, Beese LS, Weber PC. Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue. Biochemistry. 1998;37:16601–16611. doi: 10.1021/bi981197z. [DOI] [PubMed] [Google Scholar]
- Studer A, Stupperich E, Vuilleumier S, Leisinger T. Chloromethane: tetrahydrofolate methyl transfer by two proteins from Methylobacterium chloromethanicum strain CM4. Eur. J. Biochem. 2001;268:2931–2938. doi: 10.1046/j.1432-1327.2001.02182.x. [DOI] [PubMed] [Google Scholar]
- Stupperich E, Eisinger JJ, Albracht SPJ. Evidence for a super-reduced cobamide as the major corrinoid fraction in vivo and a histidine residue as a cobalt ligand of the p-cresolyl cobamide in the acetogenic bacterium Sporomusa ovata. Eur. J. Biochem. 1990;193:105–109. doi: 10.1111/j.1432-1033.1990.tb19310.x. [DOI] [PubMed] [Google Scholar]
- Svetlitchnaia T, Svetlitchnyi V, Meyer O, Dobbek H. Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis. Proc. Natl. Acad. Sci. USA. 2006;103:14331–14336. doi: 10.1073/pnas.0601420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlitchnyi V, Dobbek H, Meyer-Klaucke W, Meins T, Thiele B, Romer P, Huber R, Meyer O. A functional Ni-Ni-[4Fe-4S] cluster in the monomeric acetyl-CoA synthase from carboxydothermus hydrogenoformans. Proc. Natl. Acad. Sci. USA. 2004;101:446–451. doi: 10.1073/pnas.0304262101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett SL, Collat JW, Abbott JC. The formation of hydridocobalamin and its stability in aqueous solution. Biochemistry. 1963;2:919–923. doi: 10.1021/bi00905a004. [DOI] [PubMed] [Google Scholar]
- Tallant TC, Paul L, Krzycki JA. The MtsA subunit of the methylthiol: coenzyme M methyltransferase of Methanosarcina barkeri catalyses both half-reactions of corrinoid-dependent dimethylsulfide: coenzyme M methyl transfer. J. Biol. Chem. 2001;276:4485–4493. doi: 10.1074/jbc.M007514200. [DOI] [PubMed] [Google Scholar]
- Tan X, Loke HK, Fitch S, Lindahl PA. The tunnel of acetyl-coenzyme a synthase/carbon monoxide dehydrogenase regulates delivery of CO to the active site. J. Am. Chem. Soc. 2005;127:5833–5839. doi: 10.1021/ja043701v. [DOI] [PubMed] [Google Scholar]
- Tan X, Sewell C, Yang Q, Lindahl PA. Reduction and Methyl Transfer Kinetics of the alpha Subunit from Acetyl Coenzyme A Synthase. J. Am. Chem. Soc. 2003;125:318–319. doi: 10.1021/ja028442t. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Neuhierl B, Bock A. S-methylmethionine metabolism in Escherichia coli. J. Bacteriol. 1999;181:662–665. doi: 10.1128/jb.181.2.662-665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter H, Jr, Knappe J. Flavodoxin and ferredoxin of Escherichia coli. Hoppe. Seylers. Z. Physiol. Chem. 1971;352:433–446. doi: 10.1515/bchm2.1971.352.1.433. [DOI] [PubMed] [Google Scholar]
- Wassenaar RW, Keltjens JT, van der Drift C. Activation and reaction kinetics of the dimethylamine/coenzyme M methyltransfer in Methanosarcina barkeri strain Fusaro. Eur. J. Biochem. 1998;258:597–602. doi: 10.1046/j.1432-1327.1998.2580597.x. [DOI] [PubMed] [Google Scholar]
- Wedemeyer-Exl C, Darbre T, Keese R. A model for the cobalamin-dependent methionine synthase. Helvetica Chimica Acta. 1999;82:1173–1184. [Google Scholar]
- Whipple GH, Robscheit-Robbins FS. Blood regeneration in severe anemia. II. Favorable influence of liver, heart and skeletal muscle in diet. Am. J. Physiol. 1925;72:408–418. [Google Scholar]
- Wilker JJ, Lippard SJ. Alkyl transfer to metal thiolates: Kinetics, active species identification, and relevance to the DNA methyl phosphotriester repair center of Escherichia coli ada. Inorg. Chem. 1997;36:969–978. doi: 10.1021/ic961082o. [DOI] [PubMed] [Google Scholar]
- Wirt MD, Kumar M, Ragsdale SW, Chance MR. X-ray absorption spectroscopy of the corrinoid/iron sulfur protein involved in acetyl-CoA synthesis by Clostridium thermoaceticum. J. Am. Chem. Soc. 1993;115:2146–2150. [Google Scholar]
- Wirt MD, Wu J-J, Scheuring EM, Kumar M, Ragsdale SW, Chance MR. Structural and electronic factors in heterolytic cleavage: Formation of the Co(I) intermediate in the corrinoid/iron-sulfur protein from Clostridium thermoaceticum. Biochemistry. 1995;34:5269–5273. doi: 10.1021/bi00015a042. [DOI] [PubMed] [Google Scholar]
- Yamanishi M, Yamada S, Muguruma H, Murakami Y, Tobimatsu T, Ishida A, Yamauchi J, Toraya T. Evidence for axial coordination of 5,6-dimethylbenzimidazole to the cobalt atom of adenosylcobalamin bound to diol dehydratase. Biochemistry. 1998;37:4799–4803. doi: 10.1021/bi972572a. [DOI] [PubMed] [Google Scholar]
- Zhao S, Roberts DL, Ragsdale SW. Mechanistic studies of the methyltransferase from clostridium thermoaceticum: origin of the pH dependence of the methyl group transfer from methyltetrahydrofolate to the corrinoid/iron-sulfur protein. Biochemistry. 1995;34:15075–15083. doi: 10.1021/bi00046a013. [DOI] [PubMed] [Google Scholar]
- Zheng D, Darbre T, Keese R. Methanol and dimethylaniline as methylating agents of Co(I) corrinoids under acidic conditions. J. Inorg. Biochem. 1999;73:273–275. [Google Scholar]
- Zhou ZHS, Peariso K, PennerHahn JE, Matthews RG. Identification of the zinc ligands in cobalamin- independent methionine synthase (MetE) from Escherichia coli. Biochemistry Usa. 1999;38:15915–15926. doi: 10.1021/bi992062b. [DOI] [PubMed] [Google Scholar]