Abstract
Specific and efficient recognition of import cargoes is essential to ensure nucleocytoplasmic transport. To this end, the prototypical karyopherin importin β associates with import cargoes directly or, more commonly, through import adaptors, such as importin α and snurportin. Adaptor proteins bind the nuclear localization sequence (NLS) of import cargoes while recruiting importin β via an N-terminal importin β binding (IBB) domain. The use of adaptors greatly expands and amplifies the repertoire of cellular cargoes that importin β can efficiently import into the cell nucleus and allows for fine regulation of nuclear import. Accordingly, the IBB-domain is a dedicated NLS, unique to adaptor proteins that functions as a molecular liaison between importin β and import cargoes. This review provides an overview of the molecular role played by the IBB-domain in orchestrating nucleocytoplasmic transport. Recent work has determined that the IBB-domain has specialized functions at every step of the import and export pathway. Unexpectedly, this stretch of ∼40 amino acids plays an essential role in regulating processes such as formation of the import complex, docking and translocation through the nuclear pore complex (NPC), release of import cargoes into the cell nucleus and finally recycling of import adaptors and importin β into the cytoplasm. Thus, the IBB-domain is a master regulator of nucleocytoplasmic transport, whose complex molecular function is only recently beginning to emerge.
Keywords: IBB, Importin β binding domain; NLS, Nuclear Localization Sequence; NPC, Nuclear Pore Complex; Nuclear Import; Karyopherins; importin β; importin α; Ran
1. Introduction
1.1 The IBB-domain is a specialized NLS found in transport adaptors
In 1984 Daniel Kalderon et al. reported the discovery of a molecular signal responsible for the nuclear localization of the SV40 large T-antigen [1, 2]. This sequence, hence named nuclear localization sequence (NLS), was characterized by two biochemical signatures: a small size of ∼7 residues and a highly basic sequence, enriched in lysines and arginines. These two criteria, together with the availability of entire genome sequences made possible the identification of many other NLSs, with monopartite and bipartite organization of basic amino acids as those found in SV40 large T-antigen [1, 2] and nucleoplasmin [3], respectively. Over the past twenty years, the universe of basic transport sequences has been greatly expanded leading to the definition of classical and, more recently, non-classical NLSs [4]. While classical NLSs are restricted to SV40 large T-antigen [1, 2] and nucleoplasmin [3] -like sequences that are imported by a heterodimer of importin β/α, non-classical NLSs can be more complex in sequence, length and amino acid composition, as well as may require recognition of the cargoes tertiary structure [5]. Of particular interest to this review is the importin β binding (IBB) domain found in adaptor proteins importin α [6], snurportin [7] and XRIPα [8]. The IBB-domain functions as a specialized NLS found only in import adaptor proteins that specifically evolved to transport cargoes into the cell nucleus together with importin β. Like classical NLSs, the IBB-domain is highly enriched in basic amino acids and, when fused to an exogenous protein, confers Ran-dependent nuclear accumulation. The IBB-domain possesses three defining characteristics, which sets it apart from classical NLSs. First, it adopts a folded conformation in complex with the receptor importin β, characterized by an N-terminal 310 helix connected by a short loop (5–7 residues) to a ∼30 residue long C-terminal straight α-helix [9, 10]. The folded conformation of IBB is very different from the structure of short classical NLSs bound to importin α, or longer sequences such as the M9 [11, 12] and PTHrP-NLS [13] in complex with importin β-like receptors, which lack secondary structure elements and adopt an extended conformation (Fig. 1). Second, all IBB-domains characterized thus far are located at the amino terminus of adaptor proteins with the N-terminal 310 helix within 25 residues from the adaptor N-terminus (Fig. 2A). Third, IBB-domains bind the import receptor importin β with nanomolar affinity and this interaction is competed off by the small GTPase Ran, in a GTP bound conformation. Taken together, these three defining properties render the IBB-domain both a molecular tether between the import receptor and cargoes, as well as a powerful regulator of nucleocytoplasmic transport. This review will touch upon several aspects of this important network, with emphasis on the regulatory role of the IBB-domain in nuclear transport and recycling of import factors to the cytoplasm.
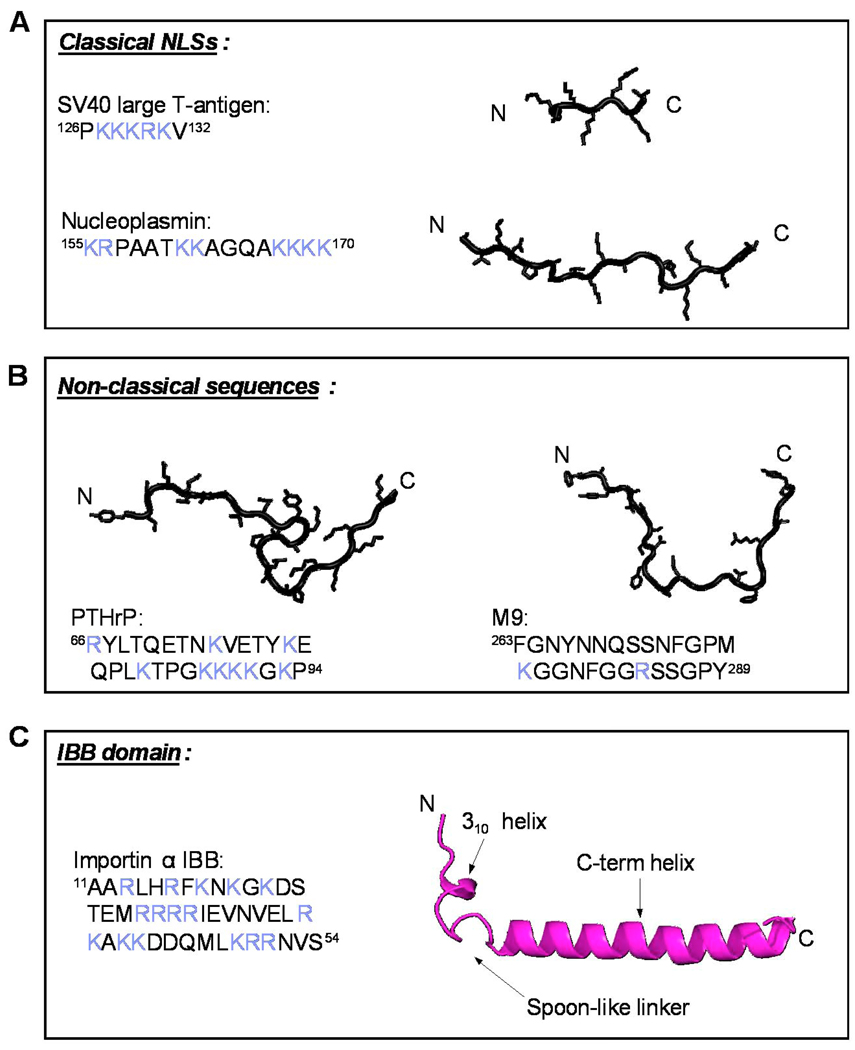
Fig. 1. Diversification of NLSs three-dimensional structure and amino acid sequence.
All structures are taken from the crystallographic complexes of the NLS bound to their respective receptor/adaptor. In the amino acid sequence, basic amino acids are colored in blue. (A) Classical monopartite and bipartite NLS found in SV40 T-large antigen (pdb 1EJL) and nucleoplasmin (pdb 1EJY) (top and bottom, respectively) that bind importin α. (B) Extended non-classical NLSs recognized directly by β-karyopherins. In the left panel is the PTHrP-NLS (res. 67–94) (pdb 1M5N) that binds importin β N-terminal HEAT repeats 1–11 [13]. In the right panel is the M9-NLS of hnRNP A1, which binds transportin (pdb 2H4M) [12]. (C) Ribbon diagram of importin α IBB-domain (pdb 1QGK). This ∼40 amino acid domain contains an N-terminal 310 helix connected by a loop of variable length to a long C-terminal α-helix.
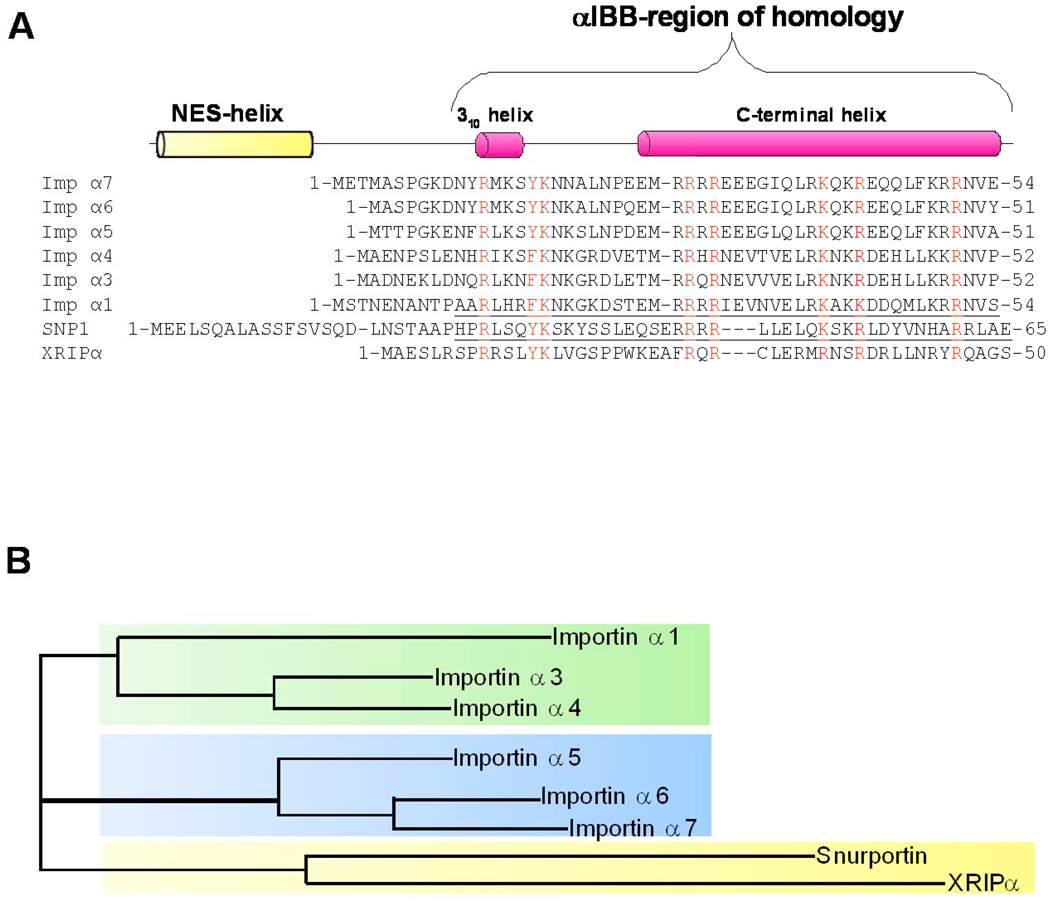
Fig. 2. Evolution and conservation of IBB-domains in adaptor proteins.
(A) Sequence alignment of IBB-domains found in six human isoforms of importin α (αl, α3, α4, α5, α6, α7), human snurportin (SNP1), and human XRIPα. A schematic representation of the IBB-secondary structure is drawn above the sequence alignment. In the alignment, highly conserved basic residues are colored in red. The underlined sequences refer to the IBB-domains that were crystallized in complex with importin β, namely importin αl IBB-domain (res. 11–54) [9] and snurportin IBB-domain (res. 25–65) [10]. Notice that the N-terminal extension comprising the NES [56] is conserved only in sIBB, and in other snurportin homologues [10], but not in αIBBs and xXRIPα. (B) Phylogenetic tree of human IBB-domain sequences shown in panel (A). In the tree, the branch lengths are proportional to the predicted evolutionary time between sequences. Three subfamilies of IBBs (shaded in green, blue and yellow) are identified. Both sequence alignment and phylogenetic tree were generated using the program ClustalW [118].
1.2. Design Principles of Nucleocytoplasmic Transport
Transport across the nuclear envelope is an active, receptor-mediated process, which is essential for normal cell function (reviewed in [14–21]). The Nuclear Pore Complex (NPC) is the only gateway between the cytoplasm and the nucleus. It is a large proteinaceous structure (∼60 MDa) that perforates the nuclear envelope and facilitates nucleocytoplasmic transport (reviewed in [22–26]). The NPC has octagonal symmetry around the central channel formed by eight spokes with the channel sitting perpendicular to the plane of the nuclear envelope. For every spoke there is a filament that freely reaches 35nm into the cytoplasm. On the nucleoplasmic side, eight fibrils emanating from each of the NPC spokes merge together to form the nuclear basket. Overall, import and export complexes moving between the cytoplasm and nucleoplasm must travel well over 100 nm through the NPC to complete a round of translocation. In higher eukaryotes the NPC is composed of approximately thirty proteins, termed nucleoporins (nups), which are found in multiple copies. A subset of nups is highly enriched in Phenylalanine-Glycine repeats (FG-nups), which account for ∼50% of the NPC mass [15]. FG-regions of nups adopt a natively unfolded conformation that is thought to fill the pore and make it impermeable to cargoes larger than ∼40kDa [27, 28]. In contrast, smaller cargoes (M.W. < 40kDa), ions, and metabolites can freely enter the cell nucleus by diffusion though the NPC.
Proteins responsible for recognizing transport cargoes are part of a super family of transport factors known as β-karyopherins (also termed importins or exportins), which share an N-terminal RanGTP-binding domain [29, 30]. There are 20 known β-karyopherins in humans [31], and importin β is undoubtedly the best-characterized member of this family. In the classical nuclear import pathway [32], cytoplasmic cargoes bearing a classical NLS are first recognized by importin α, which associates with importin β via its N-terminal importin β binding (IBB) domain. The ternary import complex importin β/ α/NLS translocates through the NPC in a process that requires interaction of importin β with FG-nups [27, 28, 33, 34]. As a result of the active nature of nuclear transport, translocation of cargoes can occur against a concentration gradient. The small GTPase Ran is the source of both energy and directionality of transport. Ran exists in different nucleotide bound conformational states across the nuclear envelope where nuclear Ran is predominately GTP-bound; the cytoplasmic pool of Ran is mainly GDP-bound. This differential compartmentalization, which is maintained in part by the sequestrating of RanGEF (Ran Guanine nucleotide Exchange Factor, also known as RCC1) and RanGAP (GTPase-Activating Protein) into the nucleus and cytoplasm, respectively [35, 36], permits importins and exportins to load and unload their cargoes in the appropriate compartment. Whereas RanGTP is responsible for the dissociation of import complexes, it is required for the formation of export complexes [37–39]. Upon entry into the nucleus importin β encounters RanGTP, which it binds to and in turn releases the importin α/NLS complex. Dissociation of the NLS-bearing cargo from importin α requires the IBB-domain.
2. Diversification and evolution of IBB-domains
Most β-karyopherins bind directly to their cargoes in the absence of adaptors. In contrast, importin β binds import cargoes primarily in complex with adaptor proteins. An IBB-domain has been identified and functionally characterized in three proteins, all active as nuclear import adaptors, namely importin α, snurportin and XRIPα. Since these adaptor proteins cannot translocate through the NPC on their own, the IBB-domain is the critical functional liaison between import adaptors and importin β that allows nuclear import to occur. Separating the cargo binding domain (importin α, snurportin and XRIPα) from the nucleoporin and Ran binding domain (importin β) allows for specialization and regulation of import, a feature that has likely appeared during evolution of higher eukaryotes.
The first identified protein containing an IBB, importin α, is the general import adaptor of cargoes bearing a classical NLS [40]. We will refer to this IBB-domain as αIBB (Fig. 1C, and Fig. 2A). In yeast, there is only one gene encoding importin α, while 6 isoforms of importin α exist in humans (Fig. 2A), which fall into three phylogenetically distinct groups, the α1s, α2s and α3s [40]. Most likely, an ancestral animal importin α1(s) gave rise to the animal α2s and α3s. As a result, the gene encoding importin α1 is found in all eukaryotes, including animals, fungi and plants, while importin α2 and α3 genes are unique to metazoan animals. This suggests that the functional diversification of importin α1 occurred throughout evolution together with the evolution of multicellular animals, presumably to perform more complex cell and tissue specific functions during development and differentiation [40]. Although importin α1 is usually considered the universal adaptor involved in nuclear import, other importin α paralogs also share the ability to bind and import classical NLS cargoes [41]. Interestingly, while the importin α family exhibits a broad degree of functional redundancy, certain non-classical cargoes are very specific for certain animal isoforms. For instance, importin α5 (also known as NPI-1 [42]) is involved in the nuclear import of dimeric phosphorylated STAT1 [43], and influenza virus polymerase PB2 [44], or importin α3 (Quip-1) mediates translocation of NF-κB p50/p65 heterodimer into the nucleus [45] .
Importin α and importin β are likely evolutionarily related proteins [46, 47], made up of similar helical repeats, known as armadillo (ARM) and HEAT, respectively [48]. Both in importin α and β, arrays of ARM and HEAT repeats stack together to generate a concave and a convex surface, vital for the function of these import factors [49–52]. Importin α is predominantly rigid due to the presence of 10 stacked ARM repeats, each formed by three α-helices [40]. In contrast, β-karyopherins contain approximately 20 copies of HEAT repeats [52], a two helices motif, which adopts a hairpin-like structure with the outer α-helix, helix A, having a slightly bent conformation. Evolutionarily, it is possible that importin α arose concurrently with importin β from an α-independent import receptor [47]. This agrees well with the observation that HEAT repeats, found in both prokaryotes and eukaryotes, arose before ARM repeats (found in eukaryotes only) [53, 54]. Sequence alignment of all human importin αs shows a variable degree of sequence conservation between the C-terminal ARM-core and the N-terminal IBB-domain. While the ARM core presents a much greater variability (sequence identity of ∼33%), the IBB-domain is much more conserved among the different importin α isoforms (Fig. 2A–B). Finally, the IBB-domain of importin α1, and likely all αIBBs, binds importin β with exceptionally high affinity (Kd ∼2 nM) and dissociates from the receptor with slow a off-rate (for αIBB koff = 2.07e−4 ± 0.3e−4 s−1) [10].
A second well-characterized IBB-domain is found in snurportin (referred herein as sIBB), the import adaptor for ribosomal U snRNPs [7], a large class of ribonucleoproteins complexes critical for RNA splicing (reviewed in [55]). Like importin α, snurportin has two functional domains: an N-terminal IBB-domain spanning residues 1–65 and a large C-terminal m3G cap binding domain [7]. Whereas there is no structural similarity between the importin α and snurportin C-terminal domains, sIBB and αIBB are highly similar both in amino acid sequence (Fig. 2A) and three-dimensional structure [10]. Interestingly, the sIBB-domain is more complex than the αIBB and contains two distinct moieties [10]. Region 25–65 of the sIBB-domain closely resembles the αIBB, with which it shares all basic residues critical for importin β binding (Fig. 2A). The only noticeable difference between these two regions is a three amino acid gap between residues 46–47 of sIBB (Fig. 2A). We will refer to region 25–65 of the sIBB-domain as the αIBB-region of homology. Whereas different isoforms of importin α have poor sequence conservation upstream of the IBB-domain, residues 1–24 are well conserved in all snurportins [10]. This region contains a functional Nuclear Export Sequence (NES) between residues 1–17 [56] and overall, the region comprising residues 1–24 of sIBB shares 42% sequence identity and 79% similarity to a region of Nup153 spanning residues 1011–1035 [10]. Although the functional significance of this observation is unclear, it is interesting that the same region of Nup153 binds importin β with high affinity, likely via interactions with FG-repeats [46]. Together, both determinants in sIBB function synergistically to confer low nanomolar binding affinity for importin β (Kd ∼2 nM) [10].
The third and last known IBB-domain was identified in XRIPα (referred herein as xIBB) [8] (Fig. 2A and B), the specific adaptor protein of replication protein A (RPA), a protein involved in DNA metabolism (reviewed in [57]). It is not clear why this replication factor uses a specialized import adaptor rather than importin α, to gain access to the cell nucleus. XRIPα presents three domains, an N-terminal IBB domain, a central acidic region, and a C-terminal Zn-finger motif [8]. A trimeric complex of importin β/XRIPα/RPA could be immunoprecipitated from Xenopus egg extracts using an affinity-purified anti-XRIPα antibody, suggesting a high affinity interaction of the adaptor/cargo complex for importin β [8]. The specificity of importin β and XRIPα for RPA was also confirmed by nuclear import assays in digitonin permeabilized cells, where RPA is efficiently imported in the nucleus by XRIPα and importin β in a Ran- and energy-dependent manner [8]. Although the structure of the xIBB-domain is not known, attentive analysis of the XRIPα N-terminal domain meets all the criteria of an IBB. It is confined to the N-terminal 50 residues of the protein, and contains basic conserved residues both in the N-terminal 310 helix and C-terminal IBB-α-helix (Fig. 2A). Finally, xIBB binds importin β in a RanGTP-regulated manner, which is essential to ensure nuclear import of RPA [8].
3. Role of the IBB-domain in import complex assembly
3.1. Autoinhibition of NLS-cargo binding
The cytoplasm is the site of recognition and interaction of NLS-containing cargoes with adaptors. Since the IBB-domain of adaptor proteins is also an efficient NLS, a regulatory mechanism must be in place to ensure importin β imports only cargo-loaded adaptors and thus prevent futile import of empty importin β/α heterodimers. Key to this regulatory mechanism in importin α, is the IBB-domain. In 1999 Bostjan Kobe demonstrated crystallographically that the IBB-domain of mammalian importin α1 occupies a large part of the NLS binding grove in the absence of NLS cargo, effectively acting as an internal NLS [58]. This autoinhibition acts as a regulatory switch for NLS binding. When importin α is not bound to an NLS-cargo, its IBB is not exposed to importin β, thereby preventing wasteful import of empty adaptor [58]. Likewise, autoinhibition functions to reduce the affinity of importin α for NLS when importin β is not available. A construct of importin α that lacks the IBB-domain binds a classical NLS-peptide with approximately 10 nM affinity whereas the full-length importin α affinity is only in the micromolar range [59], suggesting that the IBB-domain inhibits the binding of NLS-cargo. This intramolecular interaction is stabilized by a network of basic contacts that resemble the binding mode of a classical NLS to importin α (Fig. 3A). In the latter, the basic side chains of an NLS occupy a shallow groove within the ARM repeats 1–4 of importin α. Five points of contact between NLS and importin α (usually referred as positions P1-P5, Fig. 3B) have been identified from the analysis of several importin α/NLS complexes [60–65]. Together, structural and mutational data have demonstrated the crucial role played by the NLS basic lysine at the P2 site, which contacts the Trp/Asn pair between importin α1 ARM repeats 3 and 4 [58], [62, 66] (Fig. 3B). Consistent with the idea of an intramolecular NLS, the IBB-domain contains several basic amino acids that mimic the side chains found at position P1-P4 of classical NLSs. For instance, mutation of the IBB basic patch 49KRR51 to 49AAA51 dramatically diminishes IBB’s autoinhibitory capability and increases the binding affinity of NLS peptides to importin α [67]. Further mutational analysis revealed that K49 in the IBB is the most critical residue in the autoinhibitory sequence, equivalent to the critical P2 site seen for classical NLSs [67] (Fig. 3B). This site is so specific for lysine, that replacing this position in the IBB to arginine results in a noticeable drop in the binding affinity for importin α [67]. IBB-residues 50RRNV53 downstream of the critical K49 bind at the P3-P6 sites of importin α. Like in classical NLSs, these residues are less conserved among importin α isoforms (Fig. 2A) and may be dispensable for binding and autoinhibition. Another interesting structural observation is that the main autoinhibitory amino acids are not the same residues responsible for binding to importin β, which are found upstream of the autoinhibitory moiety in the IBB [68]. Therefore it is possible that importin β can “peel off” the IBB from its autoinhibitory state by binding to the exposed residues not directly contacting importin α. Because the αIBB binds to the NLS binding groove of importin α with micromolar affinity [58, 69], and importin β with nanomolar affinity [10], it is unlikely that once bound to importin β, the IBB would dissociate to re-occupy the NLS binding pocket. Therefore, formation of the trimeric import complex (importin β/α/NLS-cargo) is likely to occur cooperatively in the cytoplasm (Fig. 3C), when both NLS-cargo and importin β are simultaneously present to synergistically compete off the α IBB-domain for binding to the P2 site. The switch in αIBB from the autoinhibitory mode to an importin β-competent mode signals that the trimeric import complex is assembled and ready for transport.
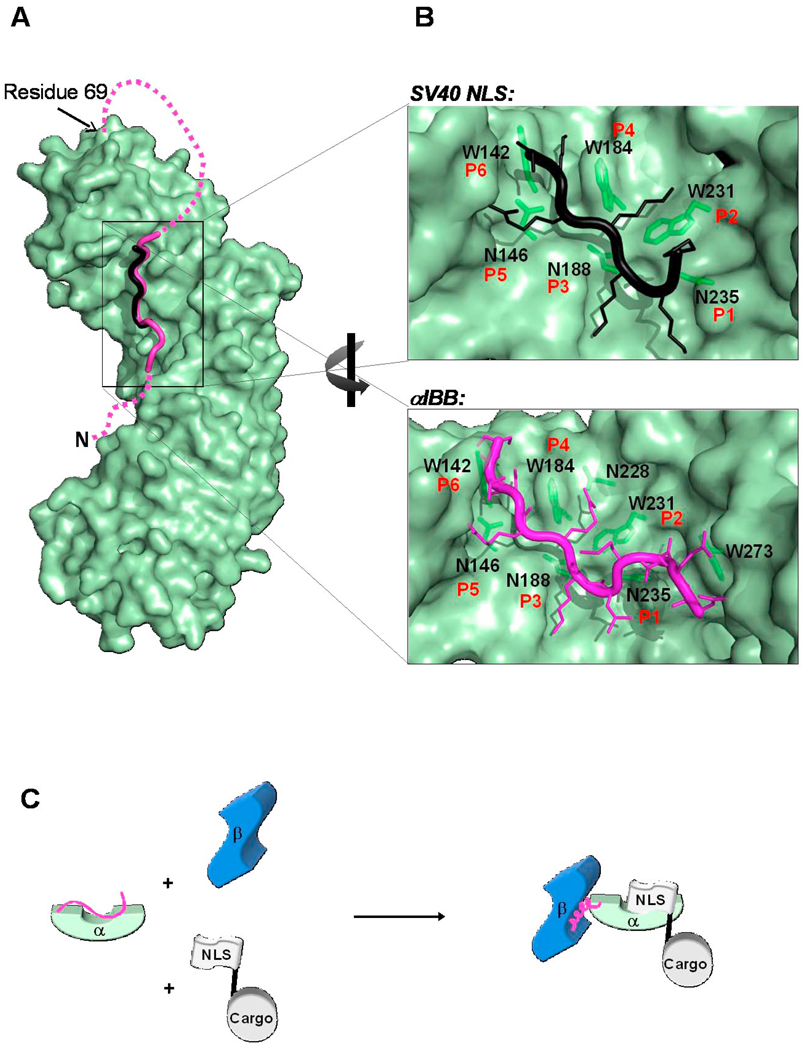
Fig. 3. The autoinhibited conformation of importin α IBB-domain.
(A) Surface representation of the mammalian importin α (in green) with the αIBB-domain (in magenta) in an autoinhibited conformation (pdb 1IAL). The residues that connect the autoinhibitory IBB-sequence with the ARM repeat core, as well as those amino acids N-terminal of the bound peptide were not seen in the crystal structure and therefore are modeled as a magenta dashed line. The structure of the SV40 NLS (in black) is also docked to importin α. The NLS and the IBB occupy the same position on importin α. (B) Blowup view of the binding site for NLS (top) and IBB (bottom) in importin α. Key interactions between the NLS/IBB and importin α at sites P1-P6 are shown. Importin α Trp and Asn residues contacting basic NLS/IBB side chains are shown as green sticks. (C) Cartoon schematic depicting how the IBB functions to regulate formation of the trimeric import complex of importin β/α/NLS when all three factors are present.
Interestingly, the αIBB residues involved in autoinhibition as well as the Trp/Asn that form the main NLS binding pocket are conserved across all importin α isoforms (Fig. 2A). However, the IBB-mediated autoinhibition of cargo binding is not a general feature of all NLSs. For instance, STAT1, a non-classical phosphorylation-dependent import cargo that uses the isoform importin α5, binds full length and ΔIBB-importin α5 with equal affinity [70]. This suggests the IBB-domain of importin α5 does not compete intramolecularly with activated STAT1. Thus, the autoinhibitory role of αIBB is likely specific to the recognition of classical NLS by importin α and cannot be generalized to all cargoes imported through importin αs. Additionally, autoinhibitory residues critically important in αIBB are not found in the sIBB-domain (Fig. 2A). Even at a 100 molar excess, the sIBB is unable to compete off the m3G cap from the C-terminal region of snurportin [71]. The cap-binding domain of snurportin recognizes only the 5’-2,2,7-terminal trimethylguanosine cap structure rather than the mono-methylated cap [71]. Because the hypermethylation of the cap only occurs when the RNA/core proteins are completely and properly assembled, the overall specificity of the snurportin cap-binding domain for its cargo, as opposed to the sIBB-domain, is thought to function as a checkpoint to ensure proper import of functional U-snRNPs.
3.2. The importin β bound conformation of IBB-domain
The extent of importin α, snurportin and XRIPα interaction with importin β is limited to the IBB domain. Although the IBB-domain is small compared to importin β (∼40 versus 876 residues), binding of the IBB-domain has vast structural and functional effects on importin β. In solution, unliganded importin β is highly flexible and adopts an elongated conformation [72, 73], highly susceptible to proteolysis [74]. In complex with the IBB-domain, the protein undergoes dramatic conformational changes and folds into a compact, nearly globular structure, with a diameter of only 85 Å [9, 10]. The structure of importin β bound to αIBB and sIBB was determined crystallographically in several crystal forms (Fig. 4A–C) [9, 10, 75]. These structures revealed that electrostatic interactions make up the majority of contacts between IBBs and importin β. As previously pointed out, importin β contains arrays of HEAT repeats, which generate an internal acidic surface consisting of B-helices and an outer, rather hydrophobic surface formed by A-helices. The IBB-domain, which is composed of almost 40% basic residues and is highly positively charged at physiological pH, occupies two thirds of the internal binding surface of importin β (HEATs 7–19). The αIBB C-terminal α-helix is intimately associated with importin β HEAT repeats 12–19. The αIBB α-helix is 41 Å in length and is partially amphipathic, with its six acidic residues positioned on the opposite face of the surface that contacts importin β. Electrostatic contacts dominate the interaction of importin β with αIBB α-helix, with a total of 13 salt bridges, mainly involving highly conserved aspartate and glutamate residues of importin β and basic IBB-side chains [9]. Overall, it was hypothesized that importin β exerts a protective function with respect to the IBB-domain and other basic substrates that are highly susceptible to proteolytic degradation and aggregate easily with cytoplasmic polyanions, such as RNA. This property is shared by other β-karyopherins that may act as intracellular chaperones for exposed basic domains, just as heat shock proteins function as chaperones for exposed hydrophobic patches [76].
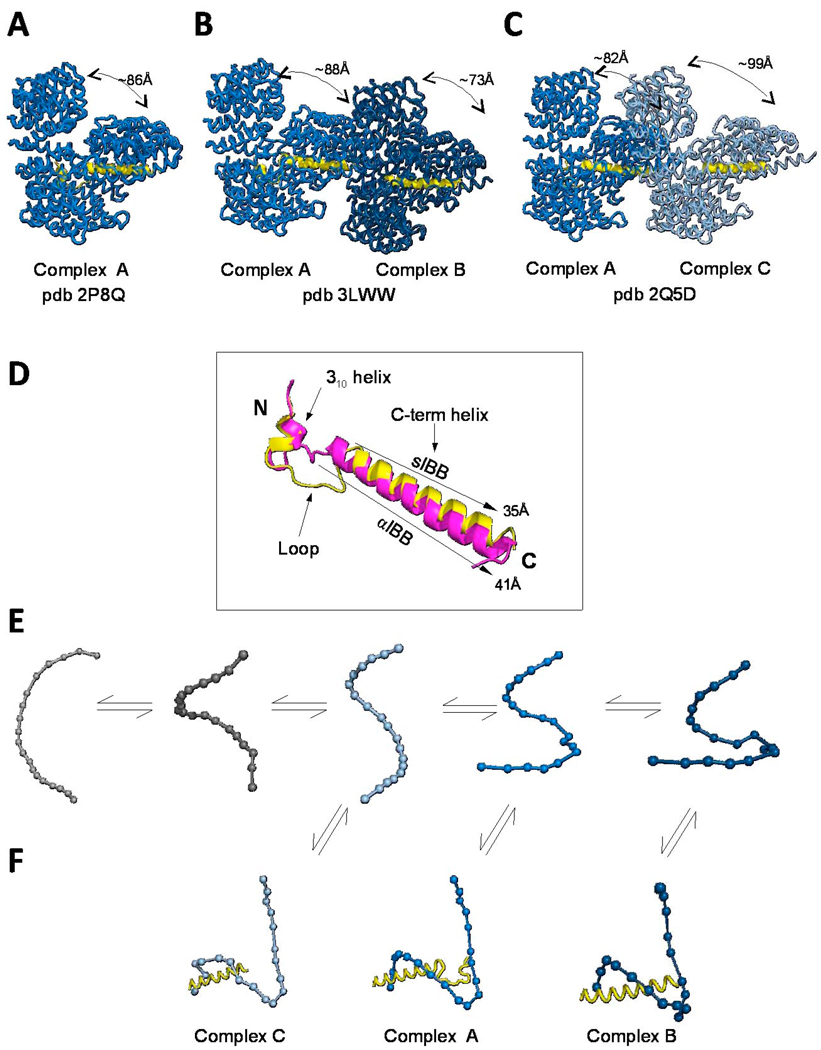
Fig. 4. Structure and recognition of IBB-domains by importin β.
Structural views of importin β bound to the sIBB (res 25–65) in three distinct crystal forms. (A) 2.35Å resolution structure (pdb 2P8Q) of importin β bound to sIBB (res. 25–65) containing one complex in the asymmetric unit. This structure represents the strained conformation of the protein. (B) 3.15Å resolution structure (pdb 3LWW) with two importin β/sIBB complexes (indicated as A–B) in the same asymmetric unit. Complex A is identical to the high resolution structure in panel (A). In contrast, complex B presents an ultra-strained conformation of importin β (C) 3.2Å resolution structure (pdb 2Q5D) with two importin β/sIBB (res. 25–65) complexes (indicated as A–C) in the same asymmetric unit. Complex A is equivalent to importin β/sIBB complex A in panel (A) and (B), while complex C has a dramatically less strained conformation of importin β. All structures in (A–C) are aligned using importin β in complex A as reference. In all complexes, the sIBB is colored in yellow and the distance between N- and C-terminal HEAT repeats of importin β is indicated. (D) Ribbon diagram of sIBB superimposed onto the αIBB domain (in yellow and magenta, respectively). Both IBBs were crystallized in complex with full length human importin β (pdb 2P8Q and 1QGK, respectively). The sIBB α-helix is ∼5 Å shorter than the αIBB C-terminal α-helix due to a three amino acid gap in its amino acid sequence between residues 46–47 (Fig. 2A). (E) Population selection model proposed for the recognition of sIBB by importin β. Putative conformers of importin β are shown as beads-on-string, where each HEAT repeat is represented by a sphere. This view emphasizes the different degree of intramolecular opening in different importin β conformers. Grey molecules are conformations of importin β not observed crystallographically, but likely seen by small angle X-ray scattering [72]. (F) Stabilization of different conformers by the sIBB-domain is thought to populate importin β conformers, which can be crystallized and hence trapped in a crystal lattice [75].
The importin β/sIBB complex has also been solved crystallographically both with full-length importin β and a construct of importin β lacking the first 127 amino acids [10, 77]. In the structure of full-length importin β bound to sIBB (res. 25–65) is very similar to that of importin β/αIBB complex (Fig. 4A–C) [10]. sIBB adopts a similar fold as αIBB, with an N-terminal 310 helix, followed by a loop, and terminated with the long C-terminal α-helix. There are three main differences between the two IBBs. First, the C-terminal α-helix of the sIBB is 6 Å shorter than the αIBB α-helix (Fig. 4D), due to the three amino acid gap in sIBB (as compared to αIBB) between residues 46–47 (Fig. 2A). As a result, structural superimposition of sIBB and αIBB reveals that the polybasic stretch 43Arg-Arg-Arg-Arg 46 conserved in both IBBs is shifted down in sIBB, as compared to the αIBB (27Arg-Arg-Arg-Arg 31), and only two of the four arginines contact importin β directly. Second, although the tertiary structure of importin β bound to either αIBB or sIBB is essentially identical, the sIBB tilts 5 degrees toward the concave surface of importin β compared to the αIBB-domain [10]. And thirdly, the linker region between the sIBB N-terminal moiety and its C-terminal α-helix is much longer than in αIBB [10].
The α- and sIBB-bound conformation of importin β seen crystallographically provides an atomic view of the strained conformation of importin β, which is likely adopted when the import complex crosses the NPC. This structural state differs greatly from the elongated and flexible solenoid-like structure of unliganded importin β seen in solution by small angle X-ray scattering (SAXS) [72, 73]. Likewise, while the unliganded importin β is highly susceptible to proteolysis, the IBB-bound conformation is extraordinarily resistant to protease digestion [74]. Therefore, the IBB-domain is a specific modulator of importin β structure and flexibility. Thermodynamically, the highly strained conformation of importin β bound to IBB-domains [49, 52] can be thought of as a tertiary structure where backbone and side chain conformations do not fall in local energy minima. The charge complementarity between the acidic concave surface of importin β and the highly positive IBB-domain provides the energetic contribution to overcome the energetic cost of distorting importin β structure. The enthalpic gain owed to surface complementarity outweighs the entropic cost of being ‘strained’, and makes the overall free energy of binding negative. In analogy with the structure of the export factor Cse1p bound to Kap60p and RanGTP [78], the strained conformation of importin β bound to IBB-domains can be described as a “spring-loaded” solenoid, which although trapped in a closed conformation has propensity to release the cargo and return to a “ground level” conformation. This function requires the small GTPase Ran.
3.3. Models for the recognition of IBB-domains by importin β
Efficient and specific recognition of cargoes and adaptor/cargo complexes is essential to ensure nucleocytoplasmic transport. Two models have been proposed to describe how β-karyopherins recognize their cargoes [79–81] and could be used to interpret the molecular recognition of IBB-domains by importin β. According to the induced fit hypothesis, importin β changes its structure upon binding to the IBB-domain to adopt a conformation that is more complementary to the IBB-domain. This induced recognition results in a large energy release, thereby locking importin β structure in a strained conformation that is sufficiently stable to be populated in solution. In contrast, according to the population selection model [75, 79–81], importin β exists in solution as an ensemble of conformers that freely inter-exchange at equilibrium, due to the low free energy of interconversion. A ligand like the IBB-domain would interact with one or more importin β “conformers”, thus shifting the equilibrium of the entire population. Although the induced fit model has been widely accepted for many decades, recent experimental evidences support the population selection model. This model is energetically less costly than the classical induced fit. Since structural conformers of a flexible molecule like importin β can freely interconvert in solution, the activation barrier for binding to an IBB-domain is usually smaller than that predicted by the induced fit model. In the latter, large conformational rearrangements necessary to drive the importin β/IBB complex to a final state would require large activation energies. Conformational selection has been shown to play an important role in enzyme catalysis [82] and protein association reactions involving binding to small molecule ligands, other proteins, DNA and RNA [79].
Two new crystal forms of importin β bound to the sIBB-domain (Fig. 4B and C) have recently provided clues to distinguish between the induced fit and population selection models [75]. In the first crystal form two importin β/sIBB (res. 25–65) complexes (complex A and B, respectively) were trapped in the asymmetric unit in drastically different conformations. In complex A (Fig. 4B), importin β is bound to the sIBB-domain (res. 25–65) in a closed conformation that is nearly identical to the high resolution structure of the importin β/sIBB complex previously described (r.m.s.d. 1.32Å) (Fig. 4A) [10]. Unexpectedly, in complex B in the same asymmetric unit (Fig. 4B), importin β presents an ultrastrained conformation with N- and C-termini interacting with each other via an extensive network of intramolecular contacts. The structure of importin β in complex B is so strained that the distance between the protein N- and C-termini is only ∼73Å, as compared to ∼88Å in complex A (Fig. 4B). As a result of the highly strained conformation of importin β backbone in complex B, the electron density corresponding to sIBB residues 25–38 is not observed. The extensive network of intramolecular contacts within N- and C termini of importin β in complex B compensates for the reduced binding surface with the sIBB-domain, making complex A and B sufficiently populated at equilibrium to be crystallized. In a second crystal form, an opposite scenario was observed [10]. While one importin β/sIBB complex is identical to complex A, the other importin β in the asymmetric unit adopts a highly unstrained conformation (complex C), with N- and C-termini spaced ∼99Å away from each other (Fig. 4C). Overall, these independently determined crystallographic structures of the importin β/sIBB complex provide three structural snapshots of importin β conformers bound to sIBB (Fig. 4A–C). If the importin β/sIBB complex was the final product of an “induced fit recognition” like in the classical Fischer "lock and key" analogy, only one stably populated complex in the same crystal lattice would be expected. Instead, the presence of two drastically different populations of importin β/sIBB trapped in the same crystallographic asymmetric unit supports the population selection model. Accordingly, it has been hypothesized that in solution importin β exists as an ensemble of different conformers characterized by a different curvature and relative position of the N- and C-termini (Fig. 4E). Selection of a given population by the sIBB-domain is hypothesized to shift the equilibrium among these conformers to formation of a tight complex that becomes sufficiently populated in solution to be crystallized [75] (Fig. 4F). Two molecular determinants have been hypothesized to influence the selection of importin β conformers: first, the enthalpic complementarity between acidic importin β and the basic sIBB-domain, which provides the energy necessary to strain the importin β solenoid structure. As shown crystallographically [10, 75], this can be achieved either by extensive intermolecular contacts between importin β and sIBB, or by extensive intramolecular contacts between importin β N- and C-termini. Second, the IBB-domain secondary structure, or simply its propensity to fold in solution, governs the mode of recognition by importin β. It was demonstrated using circular dichroism spectroscopy that the sIBB-domain, and not its counterpart αIBB, adopts a helical structure in solution [69, 75]. This folded structure could serve as a template to select importin β conformers, thereby shifting the equilibrium of the importin β population to a defined number of conformers that yield high enthalpic complementarity.
4. Role of IBB in translocation of the import complex through the NPC
Import complexes moving through the NPC interact with FG-repeats of nucleoporins (Fig. 5). Although the exact molecular mechanism of passage through the NPC remains unknown, several models for translocation of large macromolecules above the diffusion limit (MW >40kDa) have been proposed (reviewed in [14–20]). While the import complex moves through the NPC, importin β is the only component of the import complex responsible for binding to nucleoporins and has been shown to have a stochastic movement in the NPC channel [83]. Paradoxically, the import cargo does not ‘see’ the NPC, although its size and shape can certainly influence the energetics and kinetics of transport [84]. The crystal structure of importin β bound to an FxFG fragment of a nucleoporin revealed that FG-repeats bind the convex surface of importin β, on the opposite surface as adaptor and Ran [33]. This compartmentalization of binding surfaces allows importin β to interact simultaneously with import cargoes while trafficking through the pore. Given the pivotal role of IBB-domains in modulating importin β structure and plasticity, it is not surprising that the IBB-domain is also involved in cross-talk with FG-nups. Interestingly, recent work [85] has defined a new function of the IBB-domain in modulating the movement of import complexes through the NPC. It is well documented that nuclear import of the importin β/α/NLS-cargo complex requires energy for translocation [86–88]. In permeabilized cells, the classical import complex is conveniently recapitulated by importin β bound to a chimera of αIBB-fused to GFP. In a nuclear import assay in digitonin permeabilized HeLa cells [89], the importin β/αIBB-GFP complex localizes at the nuclear envelope (NE) when GTP is depleted from the cells [27]. This represents a stalled import complex that is bound to the NPC. However, under identical conditions, an import complex consisting of importin β and a chimera of sIBB-fused to GFP (sIBB-GFP) is able to translocate efficiently into the nucleus with significantly reduced energy requirement [85, 90]. Lott et al. determined that the different karyophilic properties of sIBB in permeabilized cells, as compared to αIBB are caused by the reduced affinity of importin β bound to sIBB for the central channel nucleoporin Nup62 [85] (Fig. 5). Whereas importin β bound to αIBB has nanomolar affinity for Nup62, when complexed with sIBB, the affinity for this nucleoporin drops to ∼1.5 µM [27]. Further analysis revealed that differences in the IBB amino acid sequences and the length of the IBB C-terminal α-helix are the two main determinants in sIBB regulating importin β avidity for Nup62. A three amino acid insertion in sIBB C-terminal α-helix that extends the helix to mimic that of the αIBB, restores importin β high affinity binding for Nup62. Furthermore, electron microscopic (EM) analysis of import reactions in Xenopus oocytes microinjected with import complexes coupled to colloidal gold revealed that the persistence of the importin β/sIBB import complex at the NPC is dramatically different to that of the importin β/αIBB complex. When bound to sIBB, importin β is seen at the cytoplasmic and nuclear sides of the NPC, but is completely absent from the central channel. This is not the case for the importin β/αIBB import complex, which is evenly distributed among the cytoplasmic fibrils, central channel, and nuclear basket [85]. Together this data supports the idea that the nature of the IBB-domain can modulate the importin β pathway through the NPC by altering the affinity of importin β for FG-nucleoporins. In agreement with this data, Rollenhagen et al. also showed that sIBB causes importin β to have preferential binding of certain nucleoporins as compared to the αIBB-import complex [91]. Nuclear import of U1 snRNPs requires the presence of the cytoplasmic Nup214 in order to translocate through the NPC. However, when binding to this nucleoporin was specifically blocked with antibodies, nuclear import of U1 snRNPs was inhibited, while the classical import pathway still persisted. The differences between importin α and snurportin-mediated import was also seen on the nuclear side of the NPC, where the importin β/αIBB import complex remains associated with the nuclear basket longer than the importin β/sIBB complex [91].
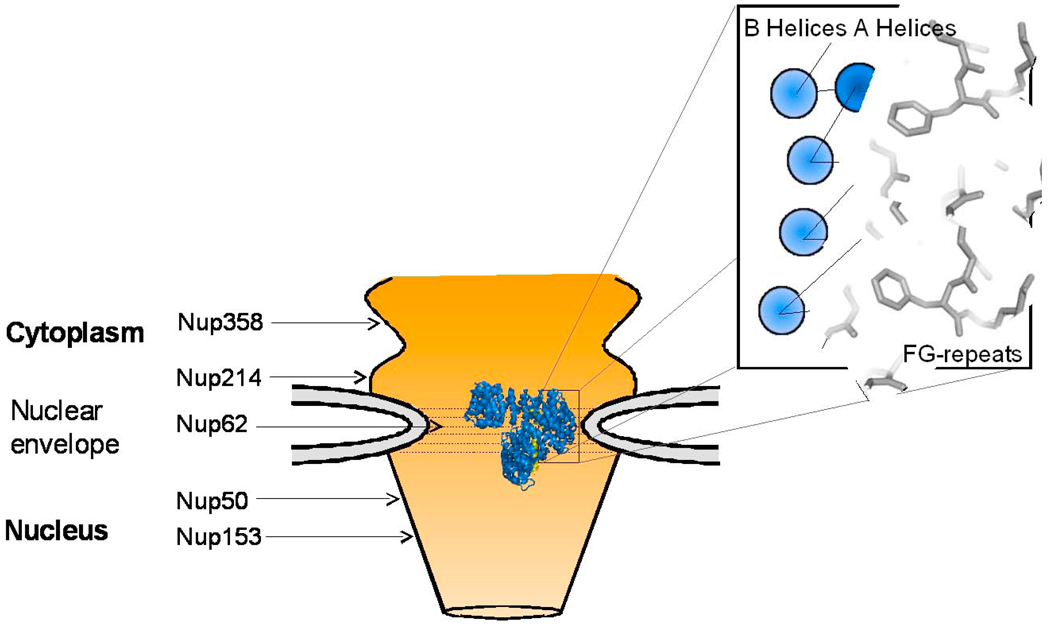
Fig. 5. Role of IBB-domain in translocation of import complexes through the NPC.
Structure of the importin β/sIBB complex (colored in blue and yellow, respectively) at the central channel of the NPC, which is formed primarily by Nup62 complex [119]. This schematic is not to scale: the structure of importin β is greatly magnified with respect to the NPC central channel. Key FG-nucleoporins are indicated respective to their location in the NPC. In the blowup is cartoon of a region of the importin β HEAT repeats bound to FG-motifs. HEAT repeats are represented as circles, with the outer A helices in dark blue, and the concave B helices as light blue. B helices are responsible for binding to the IBB-domain while the convex surface interacts with the nucleoporins. Phenylalanine side chains emanating from FG-nups are modeled interacting with the groove in between A helices of importin β.
The differential energy requirement of different import complexes during translocation may reflect a fundamental aspect of the NPC biology. For instance, it is possible that through its interaction with a given IBB, importin β binding affinity for the NPC can be modulated not only at the central channel, but also at the cytoplasmic filaments and possibly even at the nuclear basket. However, if the IBB-domain controls importin β avidity for the NPC, it is unclear how this may affect the kinetics of import complex movement through the NPC. Unfortunately, the current models proposed for translocation of cargoes through the NPC do not allow predicting for whether a reduced affinity of the import complex for FG-nups would translate as faster or slower movement through the NPC. In summary, the IBB-domain is a key regulator of importin β flexibility, which functionally results in a differential affinity of the import complex for FG-nucleoporins.
5. The IBB is important for recycling of adaptor proteins and importin β to the cytoplasm
5.1. Role of IBB-domain in recycling of importin α
Upon entry into the nucleus, the import complex encounters RanGTP, which binds importin β with higher affinity compared to adaptor proteins [92]. As seen in the crystal structure of Kap95p (the yeast homologue of importin β) bound to RanGTP, the small GTPase makes contacts with two regions of importin β [93], located between HEAT repeats 1–4 and 12–15 [93]. Given the extended nature of this binding interaction and the high affinity of importin β for the IBB-domain, Lee et al. hypothesized that RanGTP dissociates the importin β/IBB complex in a series of steps, which change the overall helicoidal pitch of importin β. Superimposition of the structure of Kap95p in complex with RanGTP to importin β bound to αIBB reveals a 20 Å displacement in the C-terminus of Kap95p, directly caused by RanGTP binding [93]. Thus, RanGTP and αIBB occupy partially overlapping binding sites on importin β [94] and RanGTP may act allosterically on importin β to trigger a conformational change that renders the protein incapable of binding to importin α [93].
Once importin β is released from the trimeric complex, the NLS-bearing cargo needs to dissociate from importin α to be released in the cell nucleus. This likely occurs through a series of defined binding events, involving the IBB-domain, nucleoporins, the export factor CAS, and RanGTP (Fig. 6). Spontaneous dissociation of the NLS/importin α complex is too slow to account for the rapid import rates observed [69, 95]. Recent work has suggested that the NPC may act as a catalyst to facilitate dissociation of import complexes. Nup50, an FG-nucleoporin localized to the nuclear basket of the NPC [96], has been shown to increase the rate of import complex disassembly [97] by directly binding to importin α The crystal structure of ΔIBB-importin α bound to a fragment of Nup50 spanning residues 1–109 revealed two distinct nup binding sites on importin α [95]. The first, and the higher affinity, is located on the outer surface of the importin α ARM repeats 9 and 10, which binds residues 24–46 of Nup50. Alanine mutations of the basic region on Nup50 completely abolished binding to importin α, suggesting electrostatic interactions are essential to stabilize this region of contact [98]. Binding of Nup50 to the C-terminus of importin α positions Nup50 residues 1–15 near the NLS binding groove of importin α. This second site of interaction has a weaker affinity compared to the first region, but with a high local concentration set up by the initial binding step, it is likely sufficiently strong to actively compete off the NLS from importin α [95] (Fig. 6). Other nucleoporins might share this role and overlap in this function [96]. For instance, it was found that another nuclear basket nucleoporin, Nup153, could also accelerate disassembly of the classical nuclear import complex in vitro [97]. Thus, the view of the NPC as a catalyst that increases the rate of import complex disassembly is gaining quick support in the literature [95, 97, 99].
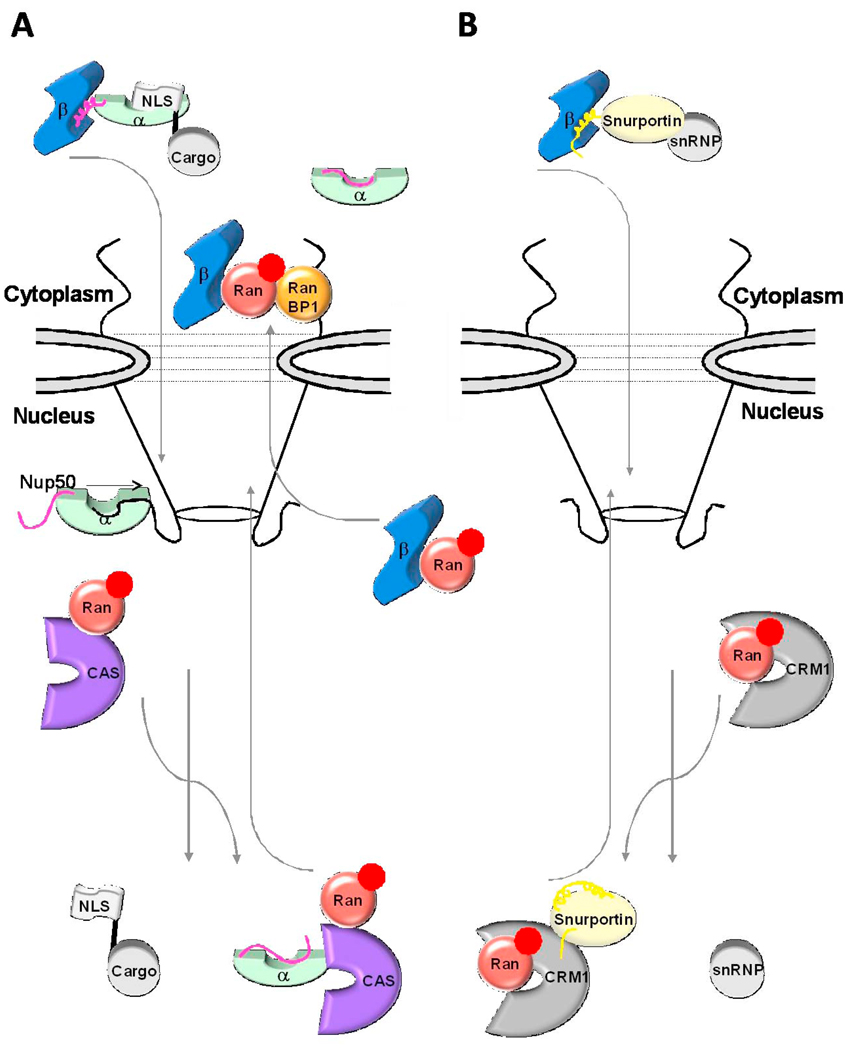
Fig. 6. Role of IBB-domain in recycling of adaptors and importin β to the cytoplasm.
(A) Cartoon diagram of the importin β/α/NLS-cargo complex (in blue, green, and grey, respectively) assembled in the cytoplasm. Upon nuclear entry RanGTP (in red) displaces importin β, and the nucleoplasmic nucleoporins Nup50 (black line) binds importin α NLS-groove and displaces the NLS-cargo. The export complex of CAS and RanGTP (shown in purple and red, respectively) displaces Nup50 from importin α and with the αIBB locked in an autoinhibited position an export-competent forms to be recycled to the cytoplasm. (B) An analogous scenario for the importin β/snurportin/snRNP import complex (in blue, yellow and grey, respectively). Upon nuclear entry the N-terminal seventeen amino acids of the sIBB binds to the export factor CRM1 (shown in silver). Colored in dark yellow is the remainder of the sIBB not involved in interactions with CRM1, shown as two surface exposed helices separated by a kinked loop.
At this point in the import pathway Nup50 is bound to an NLS-free importin α quite strongly, however if Nup50 remained tightly bound to importin α it would slow down the overall rate of nuclear import by not allowing for efficient recycling of the adaptor. The process of removing Nup50 from importin α involves both the CAS/RanGTP export complex and the IBB-domain (Fig. 6). The autoinhibitory signal of the IBB is necessary, as importin α lacking the IBB-domain is unable to cycle through multiple transport events [68], but not sufficient for importin α reshuttling to the cytoplasm. Efficient displacement from the NPC requires the concomitant presence of the export receptor CAS bound to RanGTP. The trimeric export complex formed by importin α/CAS/RanGTP is highly specific and forms cooperatively in the cell nucleus [100–102]. CAS belongs to the importin β superfamily of transport receptors and like other export factors requires RanGTP for complex formation. The importin α/CAS/RanGTP export complex requires all three binding partners to form and is specific for NLS-free importin α [100–102]. This is essential to avoid the wasteful return of import cargoes to the cytoplasm before proper unloading in the nucleus.
Attentive examination of the importin α/CAS/RanGTP structure [78] reveals that the IBB-domain is the critical regulatory signal controlling assembly of a productive export complex. In the trimeric complex, Cse1p (the yeast homologue of CAS) wraps around RanGTP and the C-terminus of Kap60p (yeast homologue of importin α) (Fig. 7A). Cse1p folds into a superhelix of 20 stacked HEAT repeats, with a long loop in HEAT 19 that folds back onto itself to contact both RanGTP and Kap60p. The structure of RanGTP in the trimeric complex is almost identical to that seen in the import complex bound to Kap95p [93] (r.m.s.d. 0.4 Å) as does the ARM repeats in Kap60p with the autoinhibited importin α structure (r.m.s.d. 1.4 Å) [78]. This suggests that there is no allosteric affects of Csep1 on Kap60p to change its conformation. Instead, it is the αIBB-domain that folds back to signal dissociation of Nup50. The αIBB is locked into its export-competent conformation by four discrete binding regions located throughout its length (Fig. 7B and C) [78]. The first region spans amino acids 12–18 of IBB and interacts with Cse1p HEAT repeats 2–4. The second region-comprises IBB residues 33–36 and folds back to bind to the minor NLS binding site. The third region (amino acids 40–45) reaches up to bind the outer surface of Cse1p between HEAT repeats 5–7. Finally, residues 54–58 of the IBB loop back down to interact with the major NLS binding site. Thus, the αIBB-domain alternate binding between CAS and the NLS binding groove signals to CAS that a cargo-free importin α is ready to be recycled into the cytoplasm.
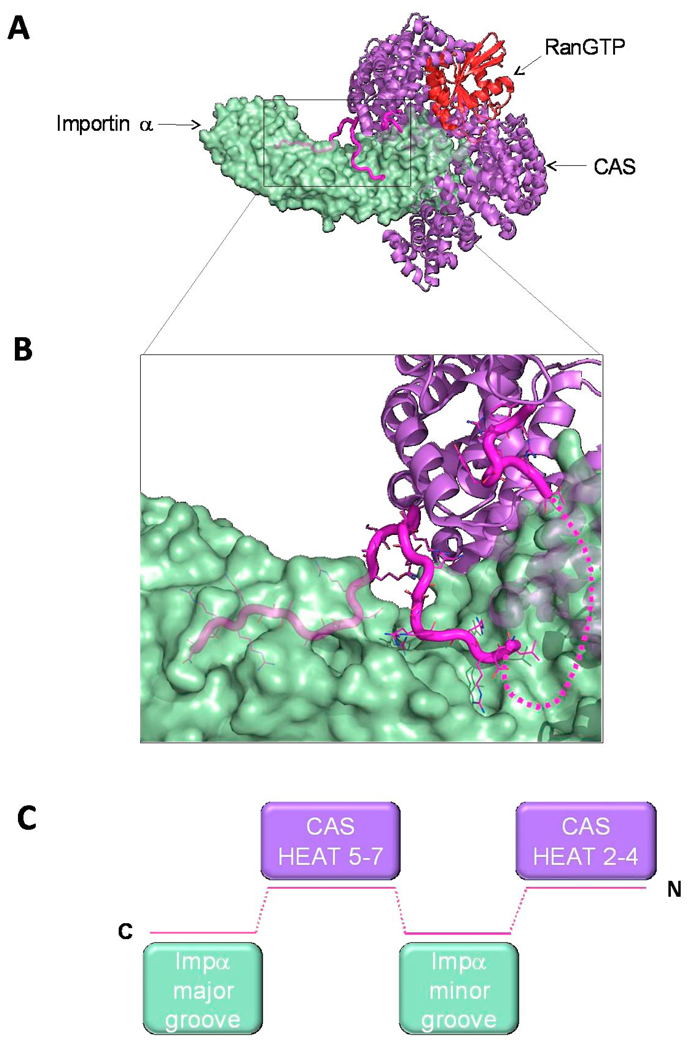
Fig. 7. Structure of the αIBB in the importin α/CAS/RanGTP export complex.
(A) Structure of the trimeric importin α export complex (pdb 1WA5) [78], which consists of Cse1p (yeast homologue of CAS), RanGTP and Kap60p (yeast homologue of importin α), in purple, red and green, respectively. The IBB-domain is colored in magenta. (B) Zoom in view of αIBB interaction with Cse1p and the NLS binding grooves of importin α. IBB residues 20–31 were not observed in the crystal structure and are modeled as a dashed line. (C) Schematic representation of the αIBB emphasizing the complex binding interactions of IBB-domain (magenta) with Cse1p (purple) and the NLS binding sites on importin α (green).
5.2. Region 1–17 of snurportin IBB-domain contains a functional Nuclear Export Sequence (NES)
Like importin α, snurportin is also constantly recycled back to the cytoplasm to replenish the pool of adaptors required for snRNP nuclear import. Recycling of snurportin is carried out by the export factor CRM1 [103] bound to RanGTP. CRM1 binds cooperatively to snurportin and RanGTP, with a Kd of complex formation of 4–5 nM [103], which is ∼5,000 times stronger than affinity for the prototypical leucine-rich NES [103]. The structural basis for high affinity binding of CRM1 to snurportin was recently unveiled by the crystal structure of the trimeric CRM1/snurportin/RanGTP complex (Fig. 8A) [104] and of an assembly intermediate consisting of CRM1 and snurportin only [56]. As shown in Fig. 8B, the N-terminal moiety of sIBB (res 1–17) contains an NES partially responsible for the interaction between snurportin and CRM1. sIBB amino acids 1–17 bind to a hydrophobic groove on CRM1 with the first N-terminal 10 amino acids folding into an α helix [56, 104] (Fig. 8B and D). The hydrophobic cleft on CRM1 is formed between the A helices from HEAT repeats 11 and 12. The width of the groove is ∼3Å wider compared to the average inter-HEAT repeat cleft, likely to accommodate the snurportin NES [56]. This binding site is essential in the overall snurportin/CRM1 complex formation as shown by multiple mutagenesis studies [56, 103, 104]. A salt bridge between CRM1 residues K534 and E575 caps the groove, which causes the remainder of the snurportin N-terminus to kink upwards [56, 104]. The NES is likely not bound to importin β during nuclear import but exposed for binding to CRM1. It is unclear whether the transfer of sIBB from importin β to CRM1 happens before, or concurrently with RanGTP binding to importin β.
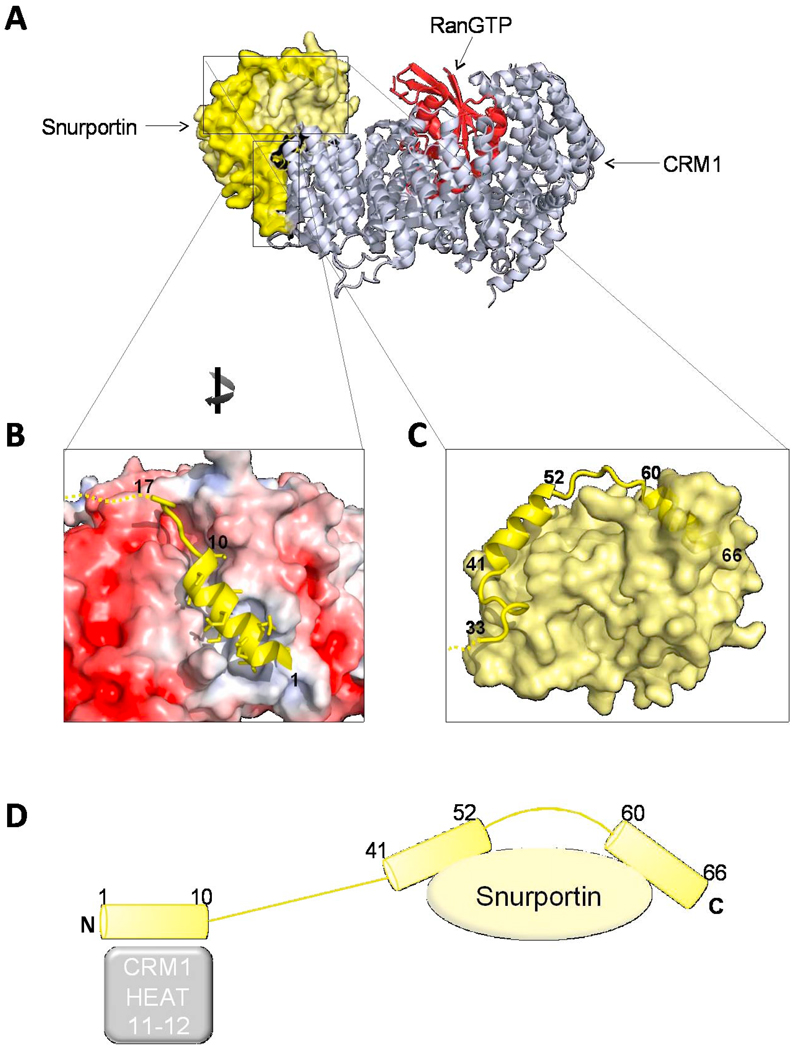
Fig. 8. Structure of the sIBB in the snurportin/CRM1/RanGTP export complex.
(A) Structure of the snurportin export complex formed by CRM1 (silver ribbon), RanGTP (red ribbon) and snurportin (shown as a light yellow surface) (pdb 3GJX) [104], with the sIBB highlighted as a yellow cartoon. (B) The N-terminus of the sIBB (shown as a yellow helix) occupies a hydrophobic cleft formed by CRM1 HEAT repeats 11–12. In the diagram, the acidic surface of CRM1 electrostatic potential map is colored in red, while the NES binding groove, which is more hydrophobic, is in white. (C) Magnification of sIBB residues 41–66 bound as an interrupted helix to the main body of snurportin. The sIBB is shown as a yellow ribbon, while snurportin main body is shown as a yellow surface. The surface exposed kink between sIBB helix 41–52 and 60–66 is a possible target for importin β. (D) Schematic representation of the sIBB-domain emphasizing its extended interactions with CRM1 (silver) and snurportin (yellow).
The remainder of the sIBB (residues 17–65) folds back to make extensive contact with the m3G-cap binding site of snurportin burying over 2800Å2 of surface area (Fig. 8C and D) [56, 104]. When comparing this export structure to that of free snurportin bound to the m3G cap [71], steric clashes occur between the m3G cap and several amino acids of the sIBB NES [104]. Three IBB residues, S20, T21, and A22 contact the m3G cap binding surface in a region that overlaps that of cargo binding. Therefore, although there has been no direct evidence that the sIBB can compete off cargo as is the case with importin α autoinhibitory state, the sIBB may still play a regulatory function in export complex formation to ensure that only cargo-free snurportin is recycled back to the cytoplasm.
5.3. Role of the IBB-domain in recycling of importin β back to the cytoplasm
RanGTP binds importin β with subnanomolar affinity [94], which helps to quickly disassemble the import complex. However, the stability of the importin β/RanGTP complex is so high that in the absence of a regulatory mechanism, GTP hydrolysis would be inhibited by importin β upon returning to the cytoplasm, thereby blocking nucleocytoplasmic transport. Therefore additional factors must aid in the dissociation of the importin β/RanGTP complex upon cytoplasmic entry. These factors are RanBP1 and RanGAP, which are cytoplasmic Ran binding proteins that activate Ran intrinsic GTPase activity, as well as the IBB-domain [29, 105]. In higher eukaryotes there are two pools of RanGAP. One is soluble and cytoplasmic, while the other is associated to the cytoplasmic nucleoporin, Nup358 [106, 107] (Fig. 6). In this way, RanGAP is positioned to interact with exiting RanGTP-containing complexes to expedite complex disassembly. In vitro, dissociation of the importin β/RanGTP complex by RanBP1 is inefficient [29, 105]. Only when αIBB is added to this trimeric complex is importin β released and GTP hydrolysis occurs. Bischoff and Gorlich determined that addition of importin α increases the dissociation of the importin β/RanGTP/RanBP1 complex by 700-fold [29]. In vivo, the dissociation of importin β/RanGTP is probably a multistep process, where GTP hydrolysis occurs after the IBB releases importin β. This is corroborated by the observation that the importin α-dependent dissociation of importin β from Ran does not require GTP hydrolysis [105].
Snurportin on the other hand probably has a different mode of export complex disassembly. As seen in the structure of snurportin bound to CRM1, a main portion of the sIBB-domain is exposed to the solvent [56, 104] (Fig. 8C and D). sIBB amino acids 52–57 form a prominent loop between helices 2 and 3, which corresponds to the C-terminal α-helix in the importin β bound sIBB structure [10]. When snurportin is recycled into the cytoplasm the loop is completely exposed to solvent, thereby allowing for binding to importin β. This binding event is thought to induce the sIBB loop to become helical, which, in the process of straightening it out, would dissociate snurportin from CRM1. Therefore snurportin itself would not change conformations, but rather the simple straightening of the sIBB kinked loop into a continuous α-helix would function as the switch between CRM1 and importin β binding. Thus the plastic structure of sIBB-domain mediates the competition between CRM1 and importin β for binding to snurportin.
6. Conclusions and perspectives
In the last 10 years there has been an explosion of new cargoes identified that bind directly to importin β. Of those, several were reported to have overlapping binding sites with the IBB-domain, while others showed new binding sites on importin β. Notably, several viral proteins were identified in the first group. For instance, Rev, a protein essential for human immunodeficiency virus type 1 (HIV-1) replication [108], was found to bind importin β via a poly-basic region spanning residues 35–50, which is also involved in RNA binding [109, 110]. In the nucleus, Rev binds to and oligomerizes in complex with viral RNA to form an export complex, which exits the nucleus in a CRM1-dependent manner [108]. The critical function played by Rev in the nucleus suggests its nuclear import is presumably highly efficient and optimized. Similarly to Rev, HIV-1 Tat [109] and human T-cell leukemia virus type 1 (HTLV-1) Rex protein [111] also possess strong nuclear import sequences. All three of these viral proteins are basic in nature (with isoelectric points all over 12), bind specifically to importin β in vitro and are efficiently imported in permeabilized cells by importin β in the absence of importin α, in a Ran-dependent manner [109, 112, 113]. It was recently shown that Rev’s non-classical NLS falls on a long α-helix [114], suggesting the actual import sequence may be part of a folded and presumably IBB-like import domain. Therefore, it is possible that in addition to cellular factors, certain viral proteins may have also developed IBB-like domains to efficiently enter the cell nucleus of infected cells. But Rev, Tat and Rex are only a small example of the many non-classical signals recently identified. Many more IBB-like sequences are likely yet to be identified both in cellular and viral proteins. Genome-wide bioinformatic prediction, together with experimental functional validation will be required to expand the list of IBB-domains. The question that remains to be answered, however, is why such a complex degree of diversification exists in the universe of nuclear localization sequences. Why have a non-classical, IBB-like import sequence in addition to classical NLSs? One possible answer to this question may come from the observation made by Timney et al. that the main limiting factor in nuclear import is the poor ability of karyopherins and NLS-cargoes to find each other in the cytoplasm [115]. Competition for binding to the right NLS in a background of overwhelming nonspecific interactions likely affects the efficiency of the import pathway more than the interaction of karyopherins with obvious partners such as the NPC and Ran. Since NLS-sequences that bind directly to importin β are usually longer and more complex than classical NLSs, it is plausible that certain viral proteins or ancient essential factors such as cyclin B1 [116] or ribosomal proteins [117] may have developed importin β-binding properties to avoid the “crowdedness” of the classical importin α-dependent import pathway. However, by doing so, they effectively contributed to increasing the redundancy of nuclear import, which in turn increased the cytoplasmic background of nonspecific interactions. Likewise, the high concentration of importin β present in the cytoplasm (∼2µM examined by Jakel et al. [76]), and the fewer number of cargoes binding directly to importin β may also represent the evolutionary advantage playing in favor of IBB-like importin β-binding NLS import sequences. In conclusion, the universe of non-classical NLSs is quickly growing. The challenge now is to understand the kinetic differences governing the nuclear import of classical, non-classical and IBB-like import sequences and how this may affect the efficiency and energetic requirement for movement through the NPC.
Acknowledgements
We thank Adem Koksal and Anshul Bhardwaj for critical reading of the manuscript. This work was supported by NIH grant GM074846-01A1 to Gino Cingolani.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kalderon D, Richardson WD, Markham AF, Smith AE. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 2.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 3.Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 4.Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 5.Sessler RJ, Noy N. A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol Cell. 2005;18:343–353. doi: 10.1016/j.molcel.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 6.Weis K, Ryder U, Lamond AI. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 7.Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Luhrmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jullien D, Gorlich D, Laemmli UK, Adachi Y. Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPalpha but not importin alpha. EMBO J. 1999;18:4348–4358. doi: 10.1093/emboj/18.15.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 10.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molecular basis for the recognition of snurportin 1 by importin beta. J Biol Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 11.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cingolani G, Bednenko J, Gillespie MT, Gerace L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol Cell. 2002;10:1345–1353. doi: 10.1016/s1097-2765(02)00727-x. [DOI] [PubMed] [Google Scholar]
- 14.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 15.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters R. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 19.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 20.Weis K. The nuclear pore complex: oily spaghetti or gummy bear? Cell. 2007;130:405–407. doi: 10.1016/j.cell.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 21.Lim RY, Ullman KS, Fahrenkrog B. Biology and biophysics of the nuclear pore complex and its components. Int Rev Cell Mol Biol. 2008;267:299–342. doi: 10.1016/S1937-6448(08)00632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahrenkrog B, Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- 23.Drummond S, Allen T. Structure, function and assembly of the nuclear pore complex. Symp Soc Exp Biol. 2004:89–114. [PubMed] [Google Scholar]
- 24.D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Martinez J, Rout MP. Nuclear pore complex biogenesis. Curr Opin Cell Biol. 2009;21:603–612. doi: 10.1016/j.ceb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci U S A. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim RY, Huang NP, Koser J, Deng J, Lau KH, Schwarz-Herion K, Fahrenkrog B, Aebi U. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc Natl Acad Sci U S A. 2006;103:9512–9517. doi: 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bischoff FR, Gorlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 30.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 34.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 35.Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards SA, Carey KL, Macara IG. Requirement of guanosine triphosphate-bound ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 38.Moroianu J, Blobel G. Protein export from the nucleus requires the GTPase Ran and GTP hydrolysis. Proc Natl Acad Sci U S A. 1995;92:4318–4322. doi: 10.1073/pnas.92.10.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci U S A. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 41.Kohler M, Speck C, Christiansen M, Bischoff FR, Prehn S, Haller H, Gorlich D, Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol Cell Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Neill RE, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 43.Meyer T, Vinkemeier U. Nucleocytoplasmic shuttling of STAT transcription factors. Eur J Biochem. 2004;271:4606–4612. doi: 10.1111/j.1432-1033.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 44.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol. 2007;14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 45.Fagerlund R, Kinnunen L, Kohler M, Julkunen I, Melen K. NF-{kappa}B is transported into the nucleus by importin {alpha}3 and importin {alpha}4. J Biol Chem. 2005;280:15942–15951. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- 46.Bednenko J, Cingolani G, Gerace L. Importin beta contains a COOH-terminal nucleoporin binding region important for nuclear transport. J Cell Biol. 2003;162:391–401. doi: 10.1083/jcb.200303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malik HS, Eickbush TH, Goldfarb DS. Evolutionary specialization of the nuclear targeting apparatus. Proc Natl Acad Sci U S A. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 49.Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 51.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 52.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 53.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 54.Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 55.Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 56.Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H, Chook YM. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458:1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 58.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 59.Fanara P, Hodel MR, Corbett AH, Hodel AE. Quantitative analysis of nuclear localization signal (NLS)-importin alpha interaction through fluorescence depolarization. Evidence for auto-inhibitory regulation of NLS binding. J Biol Chem. 2000;275:21218–21223. doi: 10.1074/jbc.M002217200. [DOI] [PubMed] [Google Scholar]
- 60.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 61.Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 62.Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 63.Fontes MR, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J Biol Chem. 2003;278:27981–27987. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- 64.Chen MH, Ben-Efraim I, Mitrousis G, Walker-Kopp N, Sims PJ, Cingolani G. Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin alpha. J Biol Chem. 2005;280:10599–10606. doi: 10.1074/jbc.M413194200. [DOI] [PubMed] [Google Scholar]
- 65.Yang SN, Takeda AA, Fontes MR, Harris JM, Jans DA, Kobe B. Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-alpha using oriented peptide library screening. J Biol Chem. doi: 10.1074/jbc.M109.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J Biol Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 67.Harreman MT, Cohen PE, Hodel MR, Truscott GJ, Corbett AH, Hodel AE. Characterization of the auto-inhibitory sequence within the N-terminal domain of importin alpha. J Biol Chem. 2003;278:21361–21369. doi: 10.1074/jbc.M301114200. [DOI] [PubMed] [Google Scholar]
- 68.Harreman MT, Hodel MR, Fanara P, Hodel AE, Corbett AH. The auto-inhibitory function of importin alpha is essential in vivo. J Biol Chem. 2003;278:5854–5863. doi: 10.1074/jbc.M210951200. [DOI] [PubMed] [Google Scholar]
- 69.Catimel B, Teh T, Fontes MR, Jennings IG, Jans DA, Howlett GJ, Nice EC, Kobe B. Biophysical characterization of interactions involving importin-alpha during nuclear import. J Biol Chem. 2001;276:34189–34198. doi: 10.1074/jbc.M103531200. [DOI] [PubMed] [Google Scholar]
- 70.Nardozzi J, Wenta N, Noriko Y, Vinkemeier W, Cingolani G. Molecular basis for the recognition of phosphorylated STAT1 by importin a5. J. Mol Biology. 2010 doi: 10.1016/j.jmb.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strasser A, Dickmanns A, Luhrmann R, Ficner R. Structural basis for m3G-cap-mediated nuclear import of spliceosomal UsnRNPs by snurportin1. EMBO J. 2005;24:2235–2243. doi: 10.1038/sj.emboj.7600701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fukuhara N, Fernandez E, Ebert J, Conti E, Svergun D. Conformational variability of nucleocytoplasmic transport factors. J Biol Chem. 2004;279:2176–2181. doi: 10.1074/jbc.M309112200. [DOI] [PubMed] [Google Scholar]
- 73.Zachariae U, Grubmuller H. Importin-beta: structural and dynamic determinants of a molecular spring. Structure. 2008;16:906–915. doi: 10.1016/j.str.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 74.Cingolani G, Lashuel HA, Gerace L, Muller CW. Nuclear import factors importin alpha and importin beta undergo mutually induced conformational changes upon association. FEBS Lett. 2000;484:291–298. doi: 10.1016/s0014-5793(00)02154-2. [DOI] [PubMed] [Google Scholar]
- 75.Bhardwaj A, Cingolani G. Conformational Selection in the Recognition of Snurportin IBB-domain by Importin beta. Biochemistry. 2010 doi: 10.1021/bi100292y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wohlwend D, Strasser A, Dickmanns A, Ficner R. Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus. J Mol Biol. 2007;374:1129–1138. doi: 10.1016/j.jmb.2007.09.065. [DOI] [PubMed] [Google Scholar]
- 78.Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 79.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nevo R, Stroh C, Kienberger F, Kaftan D, Brumfeld V, Elbaum M, Reich Z, Hinterdorfer P. A molecular switch between alternative conformational states in the complex of Ran and importin beta1. Nat Struct Biol. 2003;10:553–557. doi: 10.1038/nsb940. [DOI] [PubMed] [Google Scholar]
- 81.Zachariae U, Grubmuller H. A highly strained nuclear conformation of the exportin Cse1p revealed by molecular dynamics simulations. Structure. 2006;14:1469–1478. doi: 10.1016/j.str.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 82.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 83.Yang W, Musser SM. Nuclear import time and transport efficiency depend on importin beta concentration. J Cell Biol. 2006;174:951–961. doi: 10.1083/jcb.200605053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lyman SK, Guan T, Bednenko J, Wodrich H, Gerace L. Influence of cargo size on Ran and energy requirements for nuclear protein import. J Cell Biol. 2002;159:55–67. doi: 10.1083/jcb.200204163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lott K, Bhardwaj A, Mitrousis G, Pante N, Cingolani G. The importin beta binding domain modulates the avidity of importin beta for the nuclear pore complex. J Biol Chem. 285:13769–13780. doi: 10.1074/jbc.M109.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 87.Weis K, Dingwall C, Lamond AI. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- 88.Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cassany A, Gerace L. Reconstitution of nuclear import in permeabilized cells. Methods Mol Biol. 2009;464:181–205. doi: 10.1007/978-1-60327-461-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huber J, Dickmanns A, Luhrmann R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rollenhagen C, Muhlhausser P, Kutay U, Pante N. Importin beta-depending nuclear import pathways: role of the adapter proteins in the docking and releasing steps. Mol Biol Cell. 2003;14:2104–2115. doi: 10.1091/mbc.E02-06-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 93.Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–696. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 94.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 95.Matsuura Y, Stewart M. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 2005;24:3681–3689. doi: 10.1038/sj.emboj.7600843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol. 2000;20:5619–5630. doi: 10.1128/mcb.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gilchrist D, Mykytka B, Rexach M. Accelerating the rate of disassembly of karyopherin.cargo complexes. J Biol Chem. 2002;277:18161–18172. doi: 10.1074/jbc.M112306200. [DOI] [PubMed] [Google Scholar]
- 98.Lindsay ME, Plafker K, Smith AE, Clurman BE, Macara IG. Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell. 2002;110:349–360. doi: 10.1016/s0092-8674(02)00836-x. [DOI] [PubMed] [Google Scholar]
- 99.Sun C, Yang W, Tu LC, Musser SM. Single-molecule measurements of importin alpha/cargo complex dissociation at the nuclear pore. Proc Natl Acad Sci U S A. 2008;105:8613–8618. doi: 10.1073/pnas.0710867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 101.Hood JK, Silver PA. Cse1p is required for export of Srp1p/importin-alpha from the nucleus in Saccharomyces cerevisiae. J Biol Chem. 1998;273:35142–35146. doi: 10.1074/jbc.273.52.35142. [DOI] [PubMed] [Google Scholar]
- 102.Solsbacher J, Maurer P, Bischoff FR, Schlenstedt G. Cse1p is involved in export of yeast importin alpha from the nucleus. Mol Cell Biol. 1998;18:6805–6815. doi: 10.1128/mcb.18.11.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paraskeva E, Izaurralde E, Bischoff FR, Huber J, Kutay U, Hartmann E, Luhrmann R, Gorlich D. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J Cell Biol. 1999;145:255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324:1087–1091. doi: 10.1126/science.1173388. [DOI] [PubMed] [Google Scholar]
- 105.Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- 106.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 108.Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 109.Truant R, Cullen BR. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arnold M, Nath A, Hauber J, Kehlenbach RH. Multiple importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J Biol Chem. 2006;281:20883–20890. doi: 10.1074/jbc.M602189200. [DOI] [PubMed] [Google Scholar]
- 111.Mertz JA, Lozano MM, Dudley JP. Rev and Rex proteins of human complex retroviruses function with the MMTV Rem-responsive element. Retrovirology. 2009;6:10. doi: 10.1186/1742-4690-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Henderson BR, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 113.Palmeri D, Malim MH. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dimattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang W, Chait BT, Rout MP. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J Cell Biol. 2006;175:579–593. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moore JD, Yang J, Truant R, Kornbluth S. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jakel S, Gorlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 119.Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]