Abstract
A growing number of research consortia are now focused on generating antibodies and recombinant antibody fragments that target the human proteome. A particularly valuable application for these binding molecules would be their use inside a living cell, e.g., for imaging or functional intervention. Animal-derived antibodies must be brought into the cell through the membrane, whereas the availability of the antibody genes from phage display systems allows intracellular expression. Here, the various technologies to target intracellular proteins with antibodies are reviewed.
Key words: protein transfection, profection, cytosolic delivery, intracellular delivery, protein transduction domains, cell penetrating peptides, intracellular antibody, intrabodies, transbodies, live cell imaging
Introduction
Substantial progress has been made in identifying and characterizing the building blocks of cells. It has become apparent, however, that knowing the parts and their interactions is not enough to develop a full understanding of cellular processes, but localization and changes in concentrations of molecules over time need to be considered as well.1 While standard detection methods that use antibodies to localize proteins require fixation and permeabilization of cells, live cell imaging is needed to study dynamic cellular processes. Initiatives have been started that aim to generate binding molecules that target every human protein and variants, with the goal to resolve the spatiotemporal fate of proteins. Although rabbit sera raised against denatured proteins serve well on fixed tissue sections,2 their usefulness in functional approaches or analysis in a living cell is very limited. Recombinant antibodies, in particular those selected by phage display, have been proposed as an alternative3 because they can be raised to correctly folded proteins with significant throughput,4,5 and they offer additional functional approaches due to the availability of the antibody gene right from the selection. In addition, their biochemical properties can be changed at will, e.g., by fusion to other protein domains, or they can even be immediately expressed inside the living cell to induce knockdown phenotypes.6,7
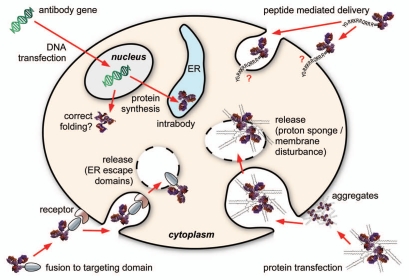
The usefulness of antibodies in visualization or the knockdown of protein functions in the living cell has been proven by microinjection,8–12 which demonstrated the stability and function of mature antibodies in the living cytoplasm. However, examples published since the first report of this method are scarce, mainly due to the very tedious process, which limits the readout assays to the microscopic observation of individual cells. It is highly desirable to have a broadly applicable method to introduce antibodies into living cells, in particular to make better use of the vast number of antibodies originating from the many ongoing “proteome binder” projects.3,13 This review evaluates the state of the art of the various approaches designed to deliver an antibody to the inside of living cells (Fig. 1).
Figure 1.
The four major approaches to deliver antibodies into a cell.
Intrabodies
Antibodies expressed in the cytosol of cells are commonly referred to as “intrabodies,” which are distinguished from “retained antibodies” that are expressed intracellularly but not in the cytosol. The introduction of antibodies into the cytosol of living cells would allow tracking at the molecular level and provide the opportunity to interfere with intracellular processes. Because of the great value of these applications, there have been many attempts to express antibodies in the cytosol of cells. In nature, however, antibodies are extracellular proteins that are part of the body's defense system and hence have evolved to be robust molecules capable of surviving in various harsh environments. Their resistance to denaturation is based on a rigid antiparallel b sheet core that is stabilized by disulfide bonds. The formation of these disulfide bonds is essential for the structural integrity and function of the majority of antibodies.14
The formation of the correct antibody conformation and disulfide bonds is assisted by endoplasmic reticulum (ER)-resident chaperones such as BiP and the ER-resident protein disulfide isomerase (PDI).15,16 Consequently, the use of intracellularly expressed antibodies was most successful when antigens in the same secretory pathway were targeted. ER retention is achieved by the addition of an ER retention sequence, e.g., the peptide KDEL, at the carboxy-terminus of the antibody. By inducing retrograde transport from the cis-golgi, the KDEL tag prevents the antibody fragment, and thus its bound antigen, from leaving the ER, which results in subsequent removal of the antigen from the cell surface by normal protein turnover. This knockdown approach is facilitated by the fact that the antibody does not need to inactivate the antigen by binding, but only needs to recognize an accessible epitope on the antigen. The method has been successfully applied to many targets, including human IL2 receptor, ErbB-2 receptor, β-amyloid precursor protein and VCAM1.6,17–21 Other subcellular compartments could be targeted by adding suitable signal sequences.19,22 Functional studies of membrane receptors or secreted proteins can thus be performed using a single standardized subcloning step after selection of a scFv antibody or Fab from a phage display library.
In contrast, the direct expression of antibody fragments in the cytoplasm of the target cell has proven to be much less reliable due to the fact that specialized chaperones needed for antibody folding are lacking and the more reducing milieu impairs the formation of disulfide bonds.23 The stability of scFvs is sufficient in less than 1% of the cases for use as a high quality intrabody.24,25 Further, every functional cytoplasmic intrabody needs to be capable of actively inhibiting the target function upon binding, a requirement that makes its selection much more tedious. Despite that, quite a number of studies of cytoplasmic proteins that were successfully targeted and function was affected,26–29 or were visualized through expression of a scFv-green fluorescent protein (GFP) fusion protein, have been reported.30
The problem of incorrect folding and reduced stability of antibodies expressed in the reducing environment of the cytosol has been tackled by different approaches. One approach focuses on determining properties that render antibodies functional in the cytosol and subsequently screening for antibodies with these particular properties. The properties of functional intrabodies for this purpose have been studied. Functional intrabodies may belong to a rare subset of antibodies that do not require intradomain disulfide bonds, as was revealed by functional assays and crystallization of cystein-deleted variants of an anti-RAS intrabody.31 By comparison of the genes of several different intrabodies, similarities that led to the definition of an intrabody consensus sequence were revealed,32,33 allowing design of specialized intrabody libraries with diversities in the range of 106–107.34
Selection of intrabodies has been performed using the intracellular antibody capture technology (IACT), which relies on a two-hybrid screen in yeast using the antigen and the scFv as a binding pair.33 Because the low efficiency with which yeast can be transformed does not allow the screening of complex libraries, a pre-selection of scFvs by panning in E. coli must be performed. The clones obtained from the panning are then subjected to a yeast two-hybrid screen and later screened in a mammalian two-hybrid system.33,35
Another strategy for selection of intrabodies makes use of the twin-arginine translocation machinery. The twin-arginine pathway (tat pathway) enables the translocation of proteins already folded in the cytoplasm through the bacterial cytoplasmic membrane.36 The intrabody selection after tat export (ISELATE) technique relies on the fusion of a tat-specific signal peptide (N-terminal) and a β-lactamase (C-terminal) to the scFv. If the scFv is correctly folded and soluble, the fusion protein is exported into the periplasm by the tat-specific signal peptide and confers ampicillin resistance to the cell. In contrast to the two-hybrid based screening, this screening strategy for intrabodies is antigen-independent.37
In contrast to the approach that focuses on selection of antibodies with specific “intrabody properties,” attempts have been made to convert arbitrary antibodies into intrabodies by fusion to another protein because the approach could be more generally applied.38 To enhance cytosolic expression, antibodies were fused to a Cκ domain,39,40 to N utilization substance A (NusA)41 or to maltose binding protein (MBP).38 Fusion of a Cκ domain to an anti-p53 scFv led to increased expression levels in the cytoplasm of mammalian cells compared to the anti-p53 scFv alone, which suggests that the Cκ fusion was less prone to degradation.39 It remains questionable whether this finding can be generalized because the fusion of another anti-p53 scFv to Cκ did not satisfyingly enhance cytoplasmic expression.42
To enhance expression of functional antibodies in the cytosol, antibodies have also been fused to solubility enhancers. NusA and MBP are among the most efficient and validated solubility enhancers for heterologous protein expression in E. coli43,44 and recombinant proteins can be prevented from aggregating if fused to NusA or MBP.44 However, the folding efficiency of a recombinant protein was found to be more dependent on the properties of the fusion partner than on the solubility enhancer, which puts the universal applicability of the method in doubt.44 Furthermore, the extent to which solubility enhancers are beneficial is not clear, as the proneness to aggregation is not necessarily an obstacle to the functionality of an intrabody. This is indicated by results obtained with a scFv prone to aggregation, which was reported to be functional by specific co-aggregation with the antigen.47
On the other hand, solubility is no guarantee of functionality. Weakened GFP fluorescence was reported to result from expression in E. coli as a MBP-fusion protein in spite of improved solubility compared to the unfused GFP.46 A study reporting the intracellular expression of a scFv-NusA fusion was carried out in a special bacterial strain with an oxidizing cytoplasm, rendering conclusions on the usefulness of this scFv as an intrabody difficult.41 Inherent properties of individual scFvs are clearly the most important factor influencing whether an antibody functions as an intrabody or not. Another possible reason for the cytosolic stability of an anti-HIV1 TAT scFv, for instance, might be the absence of a high score for “PEST” regions (proline, glutamic acid, serine and threonine rich regions) known to be responsible for rapid proteolysis.40 Some of the examples given for scFvs that exhibited improved expression upon fusion to tags were already confirmed to be well-functioning intrabodies when expressed without a fusion tag.40,48,49 Therefore, fusion to a Cκ-domain or to solubility enhancers might lead to enhanced expression or improved performance of an already confirmed functional intrabody, but this method may not be sufficient to convert all scFvs into intrabodies.
Selection of antibodies with intrabody properties, in contrast, comes with the disadvantage of a reduced diversity of the antibody repertoire. The majority of natural antibodies are expected to be non-functional if expressed in the cytosol.14 In summary, cytosolic expression of antibodies remains a method limited to individual cases, requiring some luck to identify a candidate capable of being folded correctly in the cytoplasm, binding to the target and neutralizing its function. In contrast, intracellular expression of antibodies in the ER has the potential to rapidly develop into a standard method to functionally analyze membrane proteins or the secretome.
Intracellular Antibody Delivery
Since cytosolic expression of antibodies does not reliably result in functional molecules,14 a generally applicable method that introduces antibodies into the cytosol from outside the cell would be highly desirable, and it would enable use of the growing number of antibodies that target cytoplasmic proteins. The cell membrane, however, represents a non-permissive barrier for antibodies since macromolecules depend on active uptake by the cell. Endocytosed proteins, furthermore, must still be considered “extracellular” because they do not reach the cytosol, but remain in endosomes where they are most likely destined for lysosomal degradation. In order to successfully deliver antibodies into the cytosol, cellular uptake needs to be ensured and endosomal release has to be achieved.
The focus here is on technologies that have been proposed to achieve this goal. The methods are analyzed for their potential to be generally applicable and have been divided into “protein transfection” and protein transduction domain (PTD)-based approaches. The section on protein transfection comprises approaches that employ reagents that are not genetically fused to the protein. Protein transduction approaches based on peptides, which may involve genetic fusion to the antibody fragment, were excluded in this section.
Protein transfection (profection).
Profection under scrutiny. Over the past 20 years, transfection of DNA has become a robust, standardly used technique to indirectly deliver proteins inside the cell. The direct introduction of proteins by transfection is much more challenging compared to DNA transfection. DNA has comparatively uniform physicochemical properties, but the size, structure and charge distribution of proteins vary over a wide range. Since the association of protein and transfection reagent is necessary for successful delivery to the cell, development of a standard transfection protocol suitable for every protein is difficult due to the highly diverse properties of proteins.50 Also, unlike DNA, proteins—and antibodies in particular—must retain their tertiary structure upon association with the transfection reagent. An additional difference is that transfected DNA can be amplified by replication of the plasmid, whereas there are no cellular mechanisms for amplification of transfected protein. In the case of protein transfection, the amount of protein actually delivered to the cell has to be sufficient to generate the desired effect.
In spite of the challenges, there are numerous reports of successful intracellular protein delivery. Early studies reported the co-delivery of DNA and proteins such as a transcription factor, the Cre recombinase, nuclear proteins or a polymerase into cells.51–55 Direct delivery of proteins into cells using cationic lipids and pH-responsive liposomes has been reported as well.56–61 For example, the inhibition of protein synthesis was achieved by transfection of cells with diphtheria toxin chain A (DTA) incorporated into a pH-responsive liposome. DTA alone, in contrast, did not reach the cytosol, but endosomal escape of the toxin in the absence of diphtheria toxin chain B was dependent on the transfection reagent.56 Systematic evaluation of a cationic lipid formulation composed of trifluoroacetylated lipopolyamine (TFA-DODAPL) and dioleoyl phosphatidylethanolamine (DOPE) revealed that the mixture could provide delivery of several different proteins into different cell lines.62 Many protein transfection systems that make use of lipid-based delivery reagents have since been reported, which suggests that the approach is robust (Table 1).63–71
Table 1.
Selected effects after intracellular protein delivery
| Protein | Cell line | Observed effect | Reference |
| anti-N-WASP immunoreagents | Cos-7 | Strong reduction of transferrin-endocytosis | Kessels 200263 |
| anti-EGFR phospho-Tyr845 IgG | A431 cells | Marked decrease in cell stimulation-dependent phosphorylation of Stat3 | Sato 200374 |
| anti-survivin mAb/pAb | HeLa | Microtubule defects, formation of multipolar mitotic spindles, appearance of multinucleated cells | Fortugno 2002, Fortugno 200364,75 |
| c-Fos or c-Jun antibody | quiescent PC12 | Completely blocked neurite formation | Gil 200465 |
| Anti-actin IgG | BHK | Depolymerization of cytoskeleton fibers | Dalkara 200476 |
| Anti Tubulin IgG | HeLa | ||
| Dynamitin β-galactosidase as control | Axons of Sensory neurons (primary cells) | Retrograde transport (from axon to cell body) prevented | Heerssen 200477 |
| Monoclonal anti-lamin antibody Avidin-Alexa488 | Human U87 | Localization at lamin site, formation of micronuclei (change in morphology assumed to be due to partial blockage of lamin function) if high amounts of antibody had been taken up | Didenko 200578 |
| Constitutively active recombinant MEK-1 | OK | Increased activation of endogenous ERK, translocation of PKCα to the membrane fraction, increased phosphorylation of the Na+-K+- ATPase α1 subunit, inhibition of Na+ K+-ATPase activity | Khundmiri 200566 |
| Cif | HeLa | Enlargement of the cells and formation of actin stress fibers, G2-arrest | Taieb 200667 |
| Crkl | Splenocytes (primary cells) | Significant (p < 0.00003) increase in Stat5b DNA binding ability | Laloraya 200679 |
| Rabbit anti-Cpn0585 or rabbit anti-Chlamydia EB (elementary body) antibodies | HEp-2 cells infected with C. pneumoniae | Used for visualization (cells fixed afterwards) | Cortes 200780 |
| Purified human TTase FITC-tagged antibody for determination of transfection efficiency | TTase−/− mouse LECs (lens epithelial cells) | Increased TTase activity in TTase−/− cells | Lofgren 200869 |
| β-galactosidase as a negative control | Detoxification of H2O2 in TTase loaded TTase−/− cells | ||
| Restriction endonucleases | OVCAR | Genome digestion | Geel 200981 |
| SKOV-3 | |||
| NAD+ glycohydrolase | Neurons (primary cells) | Decrease in cytosolic NAD+ | Alano 201071 |
ATPase, adenosine triphosphatase; Cif, cycle inhibiting factor; Crkl, v-crk sarcoma virus CT10 oncogene homolog (avian)-like; EGFR, epidermal growth factor receptor; ERK, extracellular signal-regulated kinase; FITC, fluorescein isothiocyanate; mAb, monoclonal antibody; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; NAD+, nicotinamide adenine dinucleotide; pAb, polyclonal antibody; PKCα, protein kinase Cα; Stat, signal transducers and activators of transcription; TTase, thioltransferase; WASP, Wiskott-Aldrich syndrome protein.
It is unclear why this approach is not more widely employed since there are numerous promising applications. One of the obstacles previously mentioned is the potential loss of protein activity upon association with the transfection reagent. Tinsley and colleagues reported the use of a polyamine reagent for the transfection of β-galactosidase and GFP into primary coronary venular endothelial cells.72,73 Contrary to expectations of a loss of protein activity, the transfection reagent had no detrimental effect on β-galactosidase, i.e., the enzyme remained fully active.72 The observation of cellular effects following protein transfection strongly supports the conclusion that delivered proteins, which include some antibody examples, are functional despite exposure to the transfection reagent.
The observation of some cellular effects also indicates the successful delivery of a sufficient amount of the proteins to the cytosol in these cases. A study describing transfection of an antibody for intracellular staining even described diffuse cytosolic distribution of the transfected antibody that depended on the amount of antibody used for transfection and suggested the presence of unbound excess antibody.82 It is therefore possible that protein transfection of antibodies may lead to “overstaining” of cells, which belies concerns of insufficient delivery.
Transfection of four different proteins was found to yield optimal results for R-phycoerythrin (R-PE), an antibody, GFP and histone at entirely different ratios of protein to transfection reagent.50 Although large differences in biochemical properties such as surface charge are seen between individual antibodies, this protein class seems to be well-suited for protein transfections due to their robustness once correctly folded. At the same time, antibodies are highly diverse in their binding specificity, which makes them useful for a vast range of applications. Indeed, many examples of antibody-profections already exist.50,62–65,74–76,78,80,82–90
Many of the proposed protein transfection systems are lipid-based, such as TFA-DODAPL:DOPE, dioctadecylglycylsperm-ine (DOGS), combinations of DOPE with CholCSper (a cationic surfactant consisting of cholesterol connected by a cysteine to carboxyspermine) or Saint-2 (N-methyl-4(dioleyl)methyl-pyridinium-chloride), as well as proprietary formulations.50,62,76,81,91,92 Lipid-based protein transfection was improved by the development of delivery systems with prolonged intracellular release and compatibility with the presence of serum.91,92 In addition to lipid-based transfection, other strategies for the intracellular delivery of proteins have been successfully employed. A polymer-based transfection that is similar to standard DNA-transfections and employs complexation of proteins with the cationic polymer polyethyleneimine (PEI) has been reported.78 Use of other carriers such as pH-sensitive carbonate apatite or various nano- and microparticles for intracellular protein delivery,93–96 as well as a method for protein transfection based on complexation with a peptide (Pep-1, amino acid sequence KET WWE TWW TEW SQP KKK RKV, pI 9.8), have also been described. Similar to cationic lipid-based transfection reagents, Pep-1 contains a hydrophobic part and a cationic part.97 This approach is a convenient transfection system compared with the substantially more sophisticated procedure required for the production of nano- or microparticles.
Intracellular protein delivery strategies can also be based on protein cationization. This technology involves chemical modification of proteins with diamines or polyamines, which results in a positive net charge of the molecule.98 The pI of the protein is artificially increased, e.g., by modification with hexamethylenediamine or by covalent linkage of PEI to the protein.99,100 Cationized proteins can then attach to the negatively charged cell surface by ionic charge interaction and are subsequently taken up by the cell.100
Mechanism and parameters affecting profection. The mechanism of protein transfection is believed to be similar to that of DNA-transfection.62,76,82 The transfection reagent and the biomolecules to be delivered form complexes with a cationic net charge that can interact electrostatically with the negatively charged proteoglycans on the cell surface.101–103 Attached complexes are then taken up by endocytosis and released from the endosomes either by membrane destabilization caused by lipids or by a “proton sponge effect.”103 The latter was proposed as a mechanism of endosomal release for transfection reagents with high buffering capacity. In the proposed process, protonable N atoms of the transfection reagent take up protons that are responsible for endosome acidification, leading to an accumulation of Cl--ions, which normally balance the increase of positive charges caused by the influx of protons for acidification, in the endosome. As a consequence, osmotic swelling of the vesicles leads to release of transfected molecules into the cytosol. If polymers are used as transfection reagents, endosome disruption is further supported by expansion of the polymeric material due to repulsive forces between the protonated N atoms.104
While the uptake mechanism and endosomal release is thought to be similar in protein- and DNA-transfections, there is a clear difference in the parameters affecting complex formation. In contrast to the importance of the charge ratio of biomolecule to transfection reagent for DNA transfections, the ratio of the surface areas is much more important for protein transfection. The association of proteins with the transfection reagent is not primarily charge-dependent because hydrophobic interactions also seem to be relevant for complex formation of proteins with cationic lipids.76
In addition to those that affect complex formation, other parameters can affect transfection efficiency. The transfection process can be subdivided into separate steps, which are (1) association of protein and transfection reagent, (2) contact with cells, (3) uptake into cells and (4) endosomal release. Parameters affecting contact-formation with cells are diffusion or sedimentation, depending on the size of the matter to be transfected. Complexes smaller than about 100 nm are subject to free Brownian motion, whereas larger particles are expected to sediment onto cells.105,106
Physical concentration of the transfection reagent at the cell surface is a crucial parameter for transfection-efficiency107 and transfection efficiency was observed to increase with complex size due to enhanced sedimentation.108,109 The transfection efficiency can be further enhanced by sedimentation of complexes onto cells employing gravitational, centrifugal or magnetic forces.107,110,111 After attachment to the cell surface, particle size and cell type influence the uptake.
Depending on their nature and size, complexes enter cells via different routes of endocytosis. Complexes with lipids (lipoplexes) may for instance be taken up by another endocytic pathway than those with polymers (polyplexes).112 The route of endocytosis is also dependent on the size of complexes. Particles smaller than 200 nm were shown to be internalized via clathrin coated pits, whereas the uptake of larger particles was mediated by caveolae.113 Endocytic uptake is also dependent on cell type. There can be redundant uptake mechanisms for the same cargo, e.g., for albumin, and the primary route of endocytic uptake can differ depending on cell type.114
Release from the transfection reagent after endosomal escape is known to be an important parameter for the efficiency of DNA-transfection.115 The release of the protein from the transfection reagent has been claimed to be of similar importance.50,91 While proteins transfected by covalently linked PEI were still functional in the cell and thus required no release from the transfection reagent,116 post-delivery kinetics appears to be a relevant parameter if proteins form a complex with the transfection reagent. As the prolonged release of proteins by CholCSper:DOPE compared to DOGS showed, different transfection reagents can result in different intracellular release profiles.91
Evaluation of profection efficiency. Although there have been attempts to use DNA-transfection reagents for the transfection of proteins, these attempts have resulted in only limited success.62 Because DNA transfection reagents and procedures must be especially tailored for proteins,62,76 there is a need for additional studies on profection parameters. Several methods are fundamental to the process of assessing and optimizing the efficiency of protein delivery. In order to determine whether proteins interact with the transfection reagent, gel shift assays similar to those used for the optimization of DNA-transfections have been employed.78 As discussed, complex formation and the nature and size of particles are crucial transfection parameters. To reproduce optimal aggregation characteristics, particle sizes and zeta potentials of the protein/transfection reagent complexes have been monitored by electrophoretic light scattering. For quality control, morphology and the supramolecular assembly characteristics have been analyzed by electron microscopy.91
Because protein transfection methods are not optimized, attention must be paid to the choice of methods in order to avoid inconclusive results. For example, there are several potential sources of error in procedures used for the detection of recombinant proteins in profected cells. It is necessary to ensure detected proteins are internalized and not only surface-attached. For internalized proteins, the possibility of fixation artifacts that impede correct microscopic localization and the uptake into dead or damaged cells must be excluded, and endosomal release must be confirmed. To reliably determine internalization, exclude fixation artifacts and ensure cell viability, the employment of reporter systems117 or live cell imaging can be employed. Endosomal escape can also be assessed by monitoring the cellular effects of the delivered proteins. A specific indicator of endosomal escape could be an antibody that binds to a distinct intracellular location that has been validated with other methods. Transfection of a fluorescent anti-nuclear pore complex antibody led to staining of a ring-like structure in cells around the nucleus.82 In contrast to DNA, some proteins such as some transcription factors and Granzyme B do not require special internalization methods;118,119 therefore, special attention should be paid to the choice of proteins that are used as model proteins in transfection studies.
Peptides as protein transduction domains—transmabs/transbodies.
Protein transduction technology emerged from the discovery that small peptide domains within the homeotic transcription protein Antennapedia and the HIV1 transcriptional activator TAT protein enabled entry into cells.120 Identification of the minimal sequence motif that confers transduction capability to these proteins led to the discovery of “cell penetrating peptides” (CPPs), which are also called “protein transduction domains” (PTDs). A large number of other PTDs of natural origin or obtained by rational design have since been described. Naturally-derived peptides include the herpes simplex virus structural protein VP22 and antimicrobial or pore-forming peptides.121 Rational design has yielded peptides such as transportan, which is a chimeric peptide composed of a neuropeptide and a wasp venom peptide toxin.122
Despite differences in amino acid sequence, most PTDs share some common physicochemical properties, such as amphipathicity and positive charge.121 The class of arginine-rich PTDs, which includes HIV1-TAT-PTD, nona-arginine and Antennapedia-PTD, is one of the best-studied and most widely used group of PTDs. Mechanistic aspects mentioned here mainly refer to this class of PTDs. As is the case with most peptidic delivery systems, the exact internalization mechanism is not known. No specific receptor responsible for the uptake of HIV1-TAT-PTD, nona-arginine or Antennapedia-PTD has been identified yet.123 However, the uptake of HIV1-TAT-PTD was blocked after incubating the target cells with trypsin, which implicates the requirement of a cell surface protein for internalization.124 Although early results that were potentially affected by methodological artifacts suggested an energy-independent mode of uptake, it is now generally accepted that PTDs are primarily taken up by endocytosis.121,125 The positive charge of the HIV1-TAT-PTD and other arginine-rich PTDs may help to concentrate the peptide on the cell surface by electrostatic interactions with negatively charged glycoproteins,124,126–128 similar to the mechanism described for transfection. Heparan sulfate proteoglycans (HSPGs) were proposed to assist in promoting attachment of HIV1-TAT protein and HIV1-TAT-PTD to the cell surface.124,127,129
Direct membrane penetration by PTDs is also considered a potential additional mechanism of uptake. Theoretical models for the interaction of HIV1-TAT-PTD with membranes have suggested the formation of transient pores of 3 nm in diameter and a concentration dependence of membrane penetration by peptides.130 Another simulation predicted the induction of membrane curvature by HIV1-TAT-PTD, potentially leading to internalization of the PTD by vesicle formation rather than pore formation.131 In experiments with model membranes, HIV1-TAT-PTD entered giant unilamellar vesicles (GUVs) when model membranes contained negative intrinsic curvature lipids such as lipids with phosphatidylethanolamine headgroups.132,133 Entry into GUVs occurred above a specific concentration threshold of peptides.133 Experiments carried out with fluorescently labeled arginine-rich PTDs applied to live cells showed a punctuate pattern at 37°C, which indicated endosomal localization of PTDs. In the absence of endocytosis, at 4°C, signals from fluorescently labeled PTDs were not absent but diffusely distributed over the cell. Internalization has consequently been suggested to occur via different routes of endocytosis, potentially including macropinocytosis, clathrin- and caveolin mediated endocytosis, as well as via a mode of entry by direct membrane penetration at high peptide concentrations.125,133–135 Although direct membrane penetration was proposed as a potential route of entry for PTDs alone, uptake of arginine-rich PTDs linked to macromolecular cargoes appears to occur solely by endocytosis.127,134,136
Since routes of entry appear to be similar for most PTDs and delivery efficiency is subject to the same limitations, the potential applications of well-characterized PTDs are worth considering, rather than searching for the “best PTD.” As Fischer suggests, mechanistic studies should focus on applications for PTDs rather than “profiling yet another CDV (cell delivery vector) in unconjugated form” and, according to him, “One of the most exciting applications of CDVs would be the delivery of antibodies against intracellular targets.”121 The appealing idea of creating “transbodies” or “transmabs” by linking a PTD to an antibody had already been promoted earlier by others.137,138 An extensive use of this exciting approach, however, has been missing so far.
Transbodies: why are they so rare? Different antibody formats have been linked to PTDs for intracellular delivery. Single chain variable fragments (scFvs), antigen-binding fragments (Fab) and whole IgGs have been linked to several PTDs, including those derived from HIV1-TAT or Antennapedia protein (Table 2).139–141
Table 2.
Transbodies
| Construct | Linkage/Production method | PTD | Cell lines | Reference |
| Fab | Conjugate | HIV1-TAT(37–62) | A431 breast carcinoma cells | Anderson 1993139 |
| F(ab′)2 | Conjugate | HIV1 TAT(37–72) | Bovine chromaffin cells | Stein 1999142 |
| whole IgG | Conjugate | KGEGAAVLLPVLLAAPG (“MTS“) | NIH 3T3 cells | Zhao 2001140 |
| whole IgG | Conjugate | R68 | HeLa cells | Chen and Erlanger 2002143 |
| MCF-7 cells | ||||
| SK-BR-3 cells | ||||
| murine lung enthothelial line | ||||
| NIH 3T3 cells | ||||
| scFv | Conjugate | HIV1 TAT (44–57) | HEK293 mice (injection) | Niesner 2002141 |
| mAbs, pAbs | Conjugate | KGEGAAVLLPVLLAAPG (“MTS“) | Human lymphoma T cells | Zhao 2003144 |
| scFv | Genetic fusion, C-terminal | HIV1 TAT(47–57) YGRKKRRQRRR | BMMC, RBL, MCF7 | Cohen-Saidon 2003145 |
| scFv, radiolabeled | Genetic fusion, 5′-terminal of scFv, produced as inclusion body | HIV1 TAT(47–57) YGRKKRRQRRR | Mice | Nakajima 2004146 |
| mAb | Conjugate | HIV1 TAT(47–57) YGRKKRRQRRR | MIN6 β cells | Ohara-Imaizumi 2004147 |
| scFv | Fusion | MTS | 293T, BT-474 and PyVmT cells | Shin 2005148 |
| Fab radiolabeled | Conjugate | HIV1 Tat protein (positions 48–60) | HeLa, rats | Kameyama 2006149 |
| Antennapedia (positions 43–58) | ||||
| HIV1 Rev protein (positions 34–50) | ||||
| Fab-Rev radiolabeled | Conjugate | HIV1 REV peptide (positions 34–50) (TRQAR RNRRR RWRER QRGC) | HeLa, rats | Kameyama 2006150 |
| IgG | Conjugate | R68 | NIH 3T3 | Chen 2006151 |
| Scfv | Genetic fusion, produced as inclusion body | HIV1 TAT(47–57) | Jurkat T cells | Theisen 2006152 |
| YGRKKRRQRRR | ||||
| Fab radiolabeled | Conjugate | TAT (GRKKRRQRRRPPQ-C-amide), REV (TRQARRNRRRRWRERQR-GC-amide) and ANP (RQIKIWFQNRRMKWKK-GC-amide) | HeLa | Kameyama 2007153 |
| mAb | Conjugate | GRKKRRQRRRPPQGYGC | MDA-MB-468 | Hu 2007154 |
| scFv | Antennapedia genetic fusion, C-terminal, immunoaffinity purification | FlRQIKIWFQNRRMKWKK | HCT-116, MRC-5 | Avignolo 2008155 |
| scFv | Genetic fusion | Penetratin | MDCK cells | Poungpair 2010156 |
Fab, fragment antigen-binding; mAbs, monoclonal antibodies; MTS, membrane transport sequence; pAbs, polyclonal antibodies; PTD, protein transduction domain; scFv, single chain fragment variable.
Antibodies or antibody fragments linked to PTDs have been used to neutralize tetanus toxin in chromaffin cells and influenza A viral activity.142,156 Further applications included interfering with the cell cycle, such as inhibition of a G1-S-phase arrest by an anti-Cdk (cyclin dependent kinase) inhibitor,154 or inhibition of cell cycle progression by an anti-cyclin D1 transbody.151 Transbodies have also been used to target apoptosis in order to promote, as well as to suppress, apoptosis.144,145 Apart from interference with intracellular processes, transbodies were used for imaging140,147 and some studies have focused on the biodistribution of radiolabeled transbodies.146,149,150 A Fab fragment conjugated to the HIV1-TAT-PTD was reported to exhibit improved retention on tumor cells.139
Despite these promising examples, a generally applicable method has yet to be defined, and a possible reason for this may be production problems. Antibodies usually need to be secreted by the producing organism. Although a stable CHO-K1 cell line that secretes TAT-EGFP has been reported,157 there are also hints at problems with secretion of TAT-fusion proteins. In a study that compared different TAT-fusion proteins (TAT-EGFP, TAT-srIκBa, TAT-RBD), the major fraction of fusion proteins was found in the cell lysate, and only a small amount of the fusion proteins was present in the supernatant.158 For several variants of HIV1-TAT-PTD with different numbers of cationic amino acids, secretion levels decreased with an increase in cationic charge.159 Like HIV1-TAT-PTD, most of the known PTDs are cationic in nature. Chemical conjugation was employed for linking antibody and PTD in most of the examples in spite of the known disadvantages associated with the method, including the inability to control stoichiometry and the precise site of linkage. Importantly, some of the few antibody-PTD fusions reported were not secreted, but produced as inclusion bodies and refolded.146,152 The production of PTD fusion proteins may further be prone to degradation by the host proteolytic system.160 Arginine-rich peptides are potential substrates of furin, an endoprotease that resides in the Golgi and cleaves the minimal recognition sequence RXXR. Degradation of the HIV1-TAT-PTD by furin has been observed.161,162 To solve this problem, a variation of the HIV1-TAT-PTD was proposed in which furin cleavage sites are mutated, but transduction capability is retained.163
An additional problem of PTDs is their generally inefficient delivery. High concentrations of PTDs are required for delivery and only small amounts of the PTDs and their cargoes are found in the cytosol. For example, only a vanishingly small fraction of a diphtheria toxin A-PTD fusion was found to reach the cytosol and delivery by the PTD appeared to be at least 10,000-fold less efficient than delivery by diphtheria toxin B.121,164 The mechanism by which PTDs and their cargoes exit the endosome is still unclear. The HIV1-TAT-PTD is believed to be mainly taken up by macropinocytosis and macropinosomes are believed to be generally “leaky”.165–167 Nevertheless, exit from the endosome remains the rate-limiting step in peptide-mediated delivery, so endosomal entrapment is still a major problem.121,168
Perspectives for the application of transbodies. If fusion to a PTD negatively affects secretion of antibodies, then this would be a serious and general disadvantage of the method. The use of universal “adaptors” that can be produced in the cytosol might circumvent the production problems. Protein A fused to HIV1-TAT-PTD (protein A-TAT) is an example of such an adaptor. While it is necessary to produce only one PTD-fusion protein, many different IgG-molecules can potentially enter cells as the cargo of protein A-TAT.169 Delivery of even larger complexes that consisted of protein A-TAT, an antibody and a third molecule captured by the antibody has been described.170 Another example of an “adaptor” is the fusion of HIV1-TAT to streptavidin (TAT-SA). Again, the production problem has to be solved for only one fusion protein that may then deliver various biotinylated cargoes into the cell.171
In order to overcome endosomal entrapment, several strategies have been proposed, including use of fusogenic lipids, membrane disruptive polymers, lysosomotropic agents, photochemical internalization and membrane disruptive peptides. A combination of PTDs with fusogenic lipids, e.g. DOPE, or membrane disruptive polymers, e.g. PEI, has been used to enhance endosomal release of nucleic acid cargoes.172 A classic approach to promote endosomal release involves the use of lysosomotropic agents such as chloroquine that prevent acidification of endosomes. At high chloroquine concentrations, endosomes can swell and rupture.173–175 Other chemicals that act via a different mechanism, including sucrose and dimethyl sulfoxide (DMSO), have also been suggested for enhancing PTD mediated delivery.176,177
Photochemical internalization enhances endosomal release by means of “photosensitizers”, which are agents that preferentially localize in cell membranes and membranes of endocytic vesicles. Upon irradiation with visible light, reactive oxygen species are formed in the vicinity of the membrane, which leads to rupture of endosomal membranes. A disadvantage of this method is the potential damage to cargo molecules that are close to the photosensitizer,178 e.g., the enzymatic activity of delivered horse radish peroxidase (HRP) was observed to decrease at higher light doses.179
A strategy derived from endosomal escape that occurs in nature involves the use of membrane disruptive peptides. Hemagglutinin (HA), one of the major surface proteins of human influenza virus, consists of two subunits that promote either cell binding (subunit HA1) or endosomal escape (subunit HA2).180,181 The HA2 subunit, which promotes endosomal release after undergoing a conformational change at acidic pH, has been employed to enhance PTD mediated delivery.167,181,182 Transduction of a TAT-Cre fusion protein was enhanced by co-incubation with a chimeric peptide consisting of TAT and the HA2 peptide.167 Increased delivery was also reported for p53 fused to HA2 at the N-terminus and nona-arginine at the C-terminus (HA2-p53-R9) compared with a construct without HA2 (p53-R11). The HA2-p53-R9 construct was employed because of solubility problems with the version containing eleven arginines (HA2-p53-R11).182 Polyhistidine was used as another membrane disrupting peptide that potentially acts via a proton sponge mechanism. By conjugation of polyhistidine to TAT (TAT-10H), the PTD-mediated delivery of DNA was observed to increase up to 7,000-fold compared with TAT alone.183
Avoiding artifacts. Technical artifacts that occurred during the collection of data on the uptake of PTDs or PTD-fusion proteins have been noted. Despite washing, peptides stayed attached to the cell surface, which led to an overestimation of internalized peptides. Surface-bound peptides cannot be distinguished from cytosolic peptides in flow cytometry and these were mistakenly reported as internalized. Misinterpretation of flow cytometry data and fixation artifacts suggested an apparent endocytosis-independent uptake of high efficiency.121,133 In 2003, Richard and colleagues showed that even mild fixation of cells caused redistribution of membrane parts, which internalized membrane-associated peptides.184 Consequently, live cell imaging became the preferred method for examination of cells.168 Reporter systems have also been used to exclude endosomally entrapped PTDs from measurement and to ensure cell viability.124 To remove cell surface bound PTDs, cells can be either treated with trypsin to digest peptides or subjected to heparin washes for removal of peptides by competitive binding.185 Fluorescence quenching of cell-surface bound PTDs by application of a membrane impermeant quencher to cells has also been used as a method to exclude external, cell-associated PTDs from measurement.185,186 Another elegant method to assay internalization based on differential fluorescent labeling of PTD and its cargo was described by Cheung and colleagues.187 Fluorescent dyes were chosen in such a way as to promote quenching of one dye by the other. Cargo and PTD were linked to each other by a disulfide bond and dequenching by separation of these two components in the reducing environment of the cytosol can be monitored as an indicator of endosomal release.
Profection vs. PTDs—strengths and weaknesses.
Profection and PTD-based delivery of antibodies have features in common, but vary in certain aspects; therefore, the two technologies have different strengths and weaknesses (Table 3). Whereas PTDs are usually covalently linked to the cargo, transfection reagents attach to their cargo only via non-covalent interactions. The latter poses a serious disadvantage if cargoes are physicochemically diverse, as proteins are. Expression of PTD fusion proteins, in turn, can be subject to production problems, which does not apply to transfection-based approaches. While PTD-linked proteins have sizes at the single molecular scale, profection delivers proteins in the form of particulate complexes. The particulate nature of cargo may be a disadvantage if therapeutic applications are envisaged since these particles are likely to be cleared by phagocytic cells of the reticuloendothelial system188 or enrich in the liver. Examples of the potential therapeutic applications of PTD-based delivery, in contrast, have already been proposed.189,190 On the other hand, larger complexes may benefit from sedimentation onto the cells and have the advantage of known increases in transfection efficiency with lower toxicity due to the requirement of decreased amounts of transfection reagent. A major problem of PTD-based delivery is its general inefficiency and endosomal entrapment. In a comparison of delivery efficiency by two profection reagents and two common PTDs, delivery mediated by a lipid based reagent was shown to be 10–20 times more efficient than HIV1-TAT-PTD.117 As the methods presented might have been studied in varying depth, the effects mentioned might not provide a balanced picture of possible side effects. Because it is likely that no method is entirely free of side effects, studies on potential side effects and reliable controls are the key to harnessing a method in an optimal way.
Table 3.
Characteristics of protein transduction domain vs. profection methods
| PTDs | Profection* | |
| Common features | Positive net charge mediates association with cell surface, endocytosis as main route of entrance | |
| Differences | PTD-fusion proteins are at a molecular scale, covalent linkage of cargo and PTD | Particulate complexes, non-covalent association of cargo and transfection reagent |
| Efficiency and endosomal release | − | + |
| Sedimentation techniques for increasing efficiency and reducing toxicity applicable? | − | + |
| Therapeutic potential | + | − |
| Cytotoxicity/known side effects | HIV1-TAT-PTD, antennapedia-PTD and nona-arginine inhibit TNF signalling by downregulation of TNF-receptors (TNF receptors are internalized without being activated),191 penetratin has inhibitory effect on NFκB,192 dose and exposure-time dependent cytotoxic effects reported for HIV1-TAT-PTD and antennapedia-PTD,193 poration and intense damage of membranes by transportan.194 | Cytotoxicity potentially increased compared with DNA-transfections as more lipids are required to complex proteins.76 |
Not including protein cationization. PTD, protein transduction domain; TNF, tumor necrosis factor.
Fusion to Targeting Proteins
A third strategy to deliver antibodies into the cytosol from the outside is based on the chemical conjugation or genetic fusion with proteins or protein domains that are naturally able to translocate through intracellular membranes, e.g., some ribonucleases or toxins. This principle has been extensively studied for immunotoxins or targeted RNases195–197 that both require cell binding and uptake into target cells by a ligand or antibody recognizing internalizing surface receptors. After endocytosis, the toxin or RNase moiety must be transferred into the cytosol in order to engage with their cellular substrates and ultimately cause cell death. Bacterial toxins like Pseudomonas exotoxin A (PE) and Corynebacterium diphtheria toxin (DT), or the plant toxin ricin from Ricinus communis naturally bind and internalize into cells, translocate through intracellular membrane barriers and inhibit protein synthesis by their catalytic domains. PE and DT contain translocation domains that undergo a pH dependent conformational change in the endosome that results in membrane insertion and cytoplasmic release of the catalytic domains.198,199 Ricin partially unfolds to translocate across the ER membrane via the Sec61p translocon, which is responsible for transfer of misfolded ER proteins back to the cytosol for degradation in proteasomes (ER-associated degradation, ERAD).200 The translocation is relatively inefficient, but only a few active toxin enzyme molecules delivered to the cytosol are needed to kill the cell.
Certain members of the RNase A superfamily are also able to translocate into the cytosol, but the underlying mechanism is not understood and no distinct translocation domains have been identified so far. It has been suggested that their strong cationic pI helps with uptake into the cell. This hypothesis is supported by studies showing that increased cationization of RNases by chemical modification enhances cytotoxicity and, therefore, cytosolic delivery.201 In addition, the involvement of distinct cellular retrograde transport mechanisms play an important role. For example, accumulation of cytotoxic RNase in the recycling receptor compartment, rather than in the late endosomes, improves cytosolic release, as do drugs that neutralize the pH of endosomes.202
Overall, the translocation of immunotoxins and targeted RNases across intracellular membranes is not very efficient and strategies to deliver other proteins by employing translocation domains of toxins are rare. The efficacy of immunotoxins can be enhanced by lysosomotropic agents like chloroquine203 or by saponins that accumulate in endosomal membranes,204,205 but interference with cellular processes may also lead to artifacts when these methods are used in cell physiological studies. Very recently, the PE translocation domain was successfully used in a tripartite construct. The NEMO-binding peptide was successfully delivered into activated endothelial cells and interfered with the nuclear factor kappaB pathway.206 Translocation adapter sequences, which are composed of cytosolic and endosomal cleavage sites flanking a viral membrane-penetrating peptide, improved the activity of immunotoxins and targeted RNases more than 100-fold207,208 and may also be suitable to deliver larger non-toxin protein moieties, including antibody fragments, into the cytosol of cells. Another new approach, receptor-mediated delivery (RMD), utilizes a variant of substance P, a neuropeptide that is rapidly internalized upon interaction with the neurokinin-1 receptor (NK1R), and was able to transfer a chemical conjugated cargo protein into the cytosol.209 In this study, an antibody was successfully transferred into the cytosol and bound specifically to its antigen, actin. Although this approach is limited to NK1R+ tissues due to overexpression of NK1R in many cancers, it can still be a valuable cell biological tool.
Outlook
The delivery of antibodies into living cells would allow study of protein function and locations in a physiological context and various cells and tissues. However, a general and facile protocol for the delivery of antibodies into a living cell has still to be developed. The most advanced method is the delivery to the ER by DNA transfection with a vector expressing a recombinant antibody fragment that typically is derived from phage display. Functional knockdowns of the antigens were shown to be broadly possible using this method, but restricted to proteins passing the ER. Direct antibody delivery to the cytoplasm is needed to complement this approach and possible solutions to various problems are evolving from a growing body of individual studies, but systematic studies must be done to identify the optimal reagents and establish a robust protocol validated for a set of different antibodies.
Efficient use of transbodies depends on solving production problems and avoiding endosomal entrapment. Among the protein delivery strategies, profection is distinct from the other approaches such as peptide-mediated transduction due to the efficient endosome escape capability generally known for transfection reagents. Several reagents have already been proposed and more than a dozen are commercially available. A systematic comparison of existing protein transfection reagents for antibodies is needed. Recent approaches employing designer translocation sequences, endosome escape domains or peptides targeting cellular receptors capable of transferring cargo proteins through intracellular membrane barriers are promising to improve the cytosolic delivery of antibodies. This would allow better use of the growing number of antibodies from proteome binder projects in the study of protein dynamics and functional knockdowns.
Acknowledgments
We gratefully acknowledge the support from the EU 7th framework programs Affinity Proteome and Affinomics.
References
- 1.Kitano H. Systems biology: A brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 2.Berglund L, Bjorling E, Jonasson K, Rockberg J, Fagerberg L, Al-Khalili Szigyarto C, et al. A whole-genome bioinformatics approach to selection of antigens for systematic antibody generation. Proteomics. 2008;8:2832–2839. doi: 10.1002/pmic.200800203. [DOI] [PubMed] [Google Scholar]
- 3.Taussig MJ, Stoevesandt O, Borrebaeck CA, Bradbury AR, Cahill D, Cambillau C, et al. ProteomeBinders: Planning a European resource of affinity reagents for analysis of the human proteome. Nat Methods. 2007;4:13–17. doi: 10.1038/nmeth0107-13. [DOI] [PubMed] [Google Scholar]
- 4.Mersmann M, Meier D, Mersmann J, Helmsing S, Nilsson P, Graslund S, et al. Towards proteome scale antibody selections using phage display. N Biotechnol. 2010;27:118–128. doi: 10.1016/j.nbt.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Pershad K, Pavlovic JD, Graslund S, Nilsson P, Colwill K, Karatt-Vellatt A, et al. Generating a panel of highly specific antibodies to 20 human SH2 domains by phage display. Protein Eng Des Sel. 2010;23:279–288. doi: 10.1093/protein/gzq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böldicke T. Blocking translocation of cell surface molecules from the ER to the cell surface by intracellular antibodies targeted to the ER. J Cell Mol Med. 2007;11:54–70. doi: 10.1111/j.1582-4934.2007.00002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dübel S. Antibody production, human, recombinant. In: Flickinger MC, editor. Encyclopedia of industrial biotechnology, bioprocess, bioseparation and cell technology. Vol. 1. Wiley VCH; 2010. pp. 351–365. [Google Scholar]
- 8.Gawlitta W, Osborn M, Weber K. Coiling of intermediate filaments induced by microinjection of a vimentin-specific antibody does not interfere with locomotion and mitosis. Eur J Cell Biol. 1981;26:83–90. [PubMed] [Google Scholar]
- 9.Lin JJ, Feramisco JR. Disruption of the in vivo distribution of the intermediate filaments in fibroblasts through the microinjection of a specific monoclonal antibody. Cell. 1981;24:185–193. doi: 10.1016/0092-8674(81)90514-6. [DOI] [PubMed] [Google Scholar]
- 10.Blose SH, Meltzer DI, Feramisco JR. 10 nm filaments are induced to collapse in living cells microinjected with monoclonal and polyclonal antibodies against tubulin. J Cell Biol. 1984;98:847–858. doi: 10.1083/jcb.98.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warn RM, Flegg L, Warn A. An investigation of microtubule organization and functions in living Drosophila embryos by injection of a fluorescently labeled antibody against tyrosinated alpha-tubulin. J Cell Biol. 1987;105:1721–1730. doi: 10.1083/jcb.105.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kallajoki M, Harborth J, Weber K, Osborn M. Microinjection of a monoclonal antibody against SPN antigen, now identified by peptide sequences as the NuMA protein, induces micronuclei in PtK2 cells. J Cell Sci. 1993;104:139–150. doi: 10.1242/jcs.104.1.139. [DOI] [PubMed] [Google Scholar]
- 13.Dubel S, Stoevesandt O, Taussig MJ, Hust M. Generating recombinant antibodies to the complete human proteome. Trends Biotechnol. 2010;28:333–339. doi: 10.1016/j.tibtech.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Stocks M. Intrabodies as drug discovery tools and therapeutics. Curr Opin Chem Biol. 2005;9:359–365. doi: 10.1016/j.cbpa.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Kirkpatrick RB, Ganguly S, Angelichio M, Griego S, Shatzman A, Silverman C, et al. Heavy-chain dimers as well as complete antibodies are efficiently formed and secreted from Drosophila via a bip-mediated pathway. J Biol Chem. 1995;270:19800–19805. doi: 10.1074/jbc.270.34.19800. [DOI] [PubMed] [Google Scholar]
- 16.Mayer M, Kies U, Kammermeier R, Buchner J. BiP and PDI cooperate in the oxidative folding of antibodies in vitro. J Biol Chem. 2000;275:29421–29425. doi: 10.1074/jbc.M002655200. [DOI] [PubMed] [Google Scholar]
- 17.Beerli RR, Wels W, Hynes NE. Intracellular expression of single chain antibodies reverts ErbB-2 transformation. J Biol Chem. 1994;269:23931–23936. [PubMed] [Google Scholar]
- 18.Richardson JH, Sodroski JG, Waldmann TA, Marasco WA. Phenotypic knockout of the high-affinity human interleukin 2 receptor by intracellular single-chain antibodies against the alpha subunit of the receptor. Proc Natl Acad Sci USA. 1995;92:3137–3141. doi: 10.1073/pnas.92.8.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kontermann R. Intrabodies as therapeutic agents. Methods. 2004;34:163–170. doi: 10.1016/j.ymeth.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Paganetti P, Calanca V, Galli C, Stefani M, Molinari M. beta-site specific intrabodies to decrease and prevent generation of Alzheimer's Abeta peptide. J Cell Biol. 2005;168:863–868. doi: 10.1083/jcb.200410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strebe N, Guse A, Schungel M, Schirrmann T, Hafner M, Jostock T, et al. Functional knockdown of VCAM-1 at the posttranslational level with ER retained antibodies. J Immunol Methods. 2008;341:30–40. doi: 10.1016/j.jim.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Persic L, Roberts A, Wilton J, Cattaneo A, Bradbury A, Hoogenboom HR. An integrated vector system for the eukaryotic expression of antibodies or their fragments after selection from phage display libraries. Gene. 1997;187:9–18. doi: 10.1016/s0378-1119(96)00628-2. [DOI] [PubMed] [Google Scholar]
- 23.Biocca S, Ruberti F, Tafani M, Pierandrei-Amaldi P, Cattaneo A. Redox state of single chain Fv fragments targeted to the endoplasmic reticulum, cytosol and mitochondria. Biotechnology. 1995;13:1110–1115. doi: 10.1038/nbt1095-1110. [DOI] [PubMed] [Google Scholar]
- 24.Auf der Maur A, Tissot K, Barberis A. Antigen-independent selection of intracellular stable antibody frameworks. Methods. 2004;34:215–224. doi: 10.1016/j.ymeth.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Visintin M, Quondam M, Cattaneo A. The intracellular antibody capture technology: Towards the high-throughput selection of functional intracellular antibodies for target validation. Methods. 2004;34:200–214. doi: 10.1016/j.ymeth.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Biocca S, Pierandrei-Amaldi P, Campioni N, Cattaneo A. Intracellular immunization with cytosolic recombinant antibodies. Biotechnology. 1994;12:396–399. doi: 10.1038/nbt0494-396. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka T, Rabbitts TH. Intrabodies based on intracellular capture frameworks that bind the RAS protein with high affinity and impair oncogenic transformation. EMBO J. 2003;22:1025–1035. doi: 10.1093/emboj/cdg106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrad U, Manteuffel R. Immunomodulation of phytohormones and functional proteins in plant cells. Trends Plant Sci. 2001;6:399–402. doi: 10.1016/s1360-1385(01)02043-x. [DOI] [PubMed] [Google Scholar]
- 29.Lo AS, Zhu Q, Marasco WA. Intracellular antibodies (intrabodies) and their therapeutic potential. Handb Exp Pharmacol. 2008:343–373. doi: 10.1007/978-3-540-73259-4_15. [DOI] [PubMed] [Google Scholar]
- 30.Alting-Mees MA, Risseeuw EP, Liu E, Desautels M, Crosby WA, Hemmingsen SM. Intracellular expression of recombinant antibody fluorescent protein fusions for localization of target antigens in Schizosaccharomyces pombe. Methods Mol Biol. 2006;313:97–105. doi: 10.1385/1-59259-958-3:097. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka T, Rabbitts TH. Functional intracellular antibody fragments do not require invariant intra-domain disulfide bonds. J Mol Biol. 2008:794–757. doi: 10.1016/j.jmb.2007.11.085. [DOI] [PubMed] [Google Scholar]
- 32.Tse E, Lobato MN, Forster A, Tanaka T, Chung GTY, Rabbitts TH. Intracellular antibody capture technology: Application to selection of intracellular antibodies recognising the BCR-ABL oncogenic protein. J Mol Biol. 2002:85–94. doi: 10.1006/jmbi.2002.5403. [DOI] [PubMed] [Google Scholar]
- 33.Visintin M, Settani G, Maritan A, Graziosi S, Marks JD, Cattaneo A. The intracellular antibody capture technology (IACT): Towards a consensus sequence for intracellular antibodies. J Mol Biol. 2002;317:73–83. doi: 10.1006/jmbi.2002.5392. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka T, Chung GTY, Forster A, Lobato MN, Rabbitts TH. De novo production of diverse intracellular antibody libraries. Nuc Acids Res. 2003;31:23. doi: 10.1093/nar/gng023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stocks M. Intrabodies: Production and promise. Drug Discovery Today. 2004;9:960–966. doi: 10.1016/S1359-6446(04)03269-6. [DOI] [PubMed] [Google Scholar]
- 36.Lee PA, Tullman-Ercek D, Georgiou G. The bacterial twin-arginine translocation pathway. Annu Rev Microbiol. 2006:373–395. doi: 10.1146/annurev.micro.60.080805.142212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher A, DeLisa MP. Efficient isolation of soluble intracellular single-chain antibodies using the twin-arginine translocation machinery. J Mol Biol. 2009;385:299–311. doi: 10.1016/j.jmb.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaki-Loewenstein S, Zfania R, Hyland S, Wels WS, Benhar I. A universal strategy for stable intracellular antibodies. J Immunol Meth. 2005;303:19–39. doi: 10.1016/j.jim.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Cohen PA, Mani JC, Lane DP. Characterization of a new intrabody directed against the N-terminal region of human p53. Oncogene. 1998;17:2445–2456. doi: 10.1038/sj.onc.1202190. [DOI] [PubMed] [Google Scholar]
- 40.Mhashilkar AM, Bagley J, Chen SY, Szilvay AM, Helland DG, Marasco WA. Inhibition of HIV-1 Tat-mediated LTR transactivation and HIV-1 infection vy anti-Tat single chain intrabodies. EMBO J. 1995;14:1542–1551. doi: 10.1002/j.1460-2075.1995.tb07140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng L, Baumann U, Reymond JL. Production of a functional catalytic antibody ScFv-NusA fusion protein in bacterial cytoplasm. J Biochem. 2003;133:577–581. doi: 10.1093/jb/mvg074. [DOI] [PubMed] [Google Scholar]
- 42.Jannot CB, Hynes NE. Characterization of scFv-421, a single-chain antibody targeted to p53. Biochem Biophys Res Commun. 1997;230:242–246. doi: 10.1006/bbrc.1996.5930. [DOI] [PubMed] [Google Scholar]
- 43.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nallamsetty S, Waugh DS. Solubility-enhancing proteins MBP and NusA play a passive role in the folding of their fusion partners. Protein Expr Purif. 2006;45:175–182. doi: 10.1016/j.pep.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 45.Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol. 2006;17:353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Gräslund S. Protein production and purification. Nat Meth. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cardinale A, Filesi I, Biocca S. Aggresome formation by anti-Ras intracellular scFv fragments—the fate of the antigen antibody complex. Eur J Biochem. 2001:268–277. doi: 10.1046/j.1432-1033.2001.01876.x. [DOI] [PubMed] [Google Scholar]
- 48.Martineau P, Jones P, Winter G. Expression of an antibody fragment at high levels in the bacterial cytoplasm. J Mol Biol. 1998;280:117–127. doi: 10.1006/jmbi.1998.1840. [DOI] [PubMed] [Google Scholar]
- 49.Bach H, Mazor Y, Shaky S, Shoham-Lev A, Berdichevsky Y, Gutnick DL, et al. Escherichia coli maltose-binding protein as a molecular chaperone for recombinant intracellular cytoplasmic single-chain antibodies. J Mol Biol. 2001;312:79–93. doi: 10.1006/jmbi.2001.4914. [DOI] [PubMed] [Google Scholar]
- 50.Weill CO, Biri S, Adib A, Erbacher P. A practical approach for intracellular protein delivery. Cytotechnology. 2008:41–48. doi: 10.1007/s10616-007-9102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaneda Y, Iwai K, Uchida T. Increased expression of DNA cointroduced with nuclear protein in adult rat liver. Science. 1989;243:375–378. doi: 10.1126/science.2911748. [DOI] [PubMed] [Google Scholar]
- 52.Debs RJ, Freedman LP, Edmunds S, Gaensler KL, Duzgunes N, Yamamoto KR. Regulation of gene-expression invivo by liposome-mediated delivery of a purified transcription factor. J Biol Chem. 1990;265:10189–10192. [PubMed] [Google Scholar]
- 53.Kato K, Nakanishi M, Kaneda Y, Uchida T, Okada Y. Expression of hepatitis B virus surface antigen in adult rat liver. Co-introduction of DNA and nuclear protein by a simplified liposome method. J Biol Chem. 1991;266:3361–3364. [PubMed] [Google Scholar]
- 54.Baubonis W, Sauer B. Genomic targeting with purified Cre recombinase. Nucleic Acids Res. 1993;21:2025–2029. doi: 10.1093/nar/21.9.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao X, Huang L. Cytoplasmic expression of a reporter gene by co-delivery of T7 RNA polymerase and T7 promoter sequence with cationic liposomes. Nucleic Acids Res. 1993;21:2867–2872. doi: 10.1093/nar/21.12.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu CJ, Dijkstra J, Lai MZ, Hong K, Szoka FC. Efficiency of cytoplasmic delivery by pH-sensitive liposomes to cells in culture. Pharm Res. 1990;7:824–843. doi: 10.1023/a:1015908831507. [DOI] [PubMed] [Google Scholar]
- 57.Nair S, Zhou X, Huang L, Rouse BT. Class I restricted CTL recognition of a soluble protein delivered by liposomes containing lipophilic polylysines. J Immunol Methods. 1992:237–243. doi: 10.1016/0022-1759(92)90145-j. [DOI] [PubMed] [Google Scholar]
- 58.Lin MF, DaVolio J, Garcia R. Cationic liposome-mediated incorporation of prostatic acid phosphatase protein into human prostate carcinoma cells. Biochem Biophys Res Commun. 1993;192:413–419. doi: 10.1006/bbrc.1993.1431. [DOI] [PubMed] [Google Scholar]
- 59.Sells MA, Li J, Chernoff J. Delivery of protein into cells using polycationic liposomes. Biotechniques. 1995;19:72–76. [PubMed] [Google Scholar]
- 60.Lee KD, Oh YK, Portnoy DA, Swanson JA. Delivery of macromolecules into cytosol using liposomes containing hemolysin from Listeria monocytogenes. J Biol Chem. 1996;271:7249–7252. [PubMed] [Google Scholar]
- 61.Lamartina S, Roscilli G, Rinaudo D, Delmastro P, Toniatti C. Lipofection of purified adeno-associated virus Rep68 protein: toward a chromosome-targeting nonviral particle. J Virol. 1998;72:7653–7658. doi: 10.1128/jvi.72.9.7653-7658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zelphati O, Wang Y, Kitada S, Reed JC, Felgner PL, Corbeil J. Intracellular delivery of proteins with a new lipid-mediated delivery system. J Biol Chem. 2001;276:35103–35110. doi: 10.1074/jbc.M104920200. [DOI] [PubMed] [Google Scholar]
- 63.Kessels MM, Qualmann B. Syndapins integrate N-WASP in receptor-mediated endocytosis. EMBO J. 2002;21:6083–6094. doi: 10.1093/emboj/cdf604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fortugno P, Beltrami E, Plescia J, Fontana J, Pradhan D, Marchisio PC, et al. Regulation of survivin function by Hsp90. Proc Natl Acad Sci USA. 2003;100:13791–13796. doi: 10.1073/pnas.2434345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gil GA, Bussolino DF, Portal MM, Pecchio AA, Renner ML, Borioli GA, et al. c-Fos activated phospholipid synthesis is required for neurite elongation in differentiating PC12 cells. Mol Biol Cell. 2004;15:1881–1894. doi: 10.1091/mbc.E03-09-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khundmiri SJ, Dean WL, McLeish KR, Lederer ED. Parathyroid hormone-mediated regulation of Na+-K+-ATPase requires ERK-dependent translocation of protein kinase Calpha. J Biol Chem. 2005;280:8705–8713. doi: 10.1074/jbc.M408606200. [DOI] [PubMed] [Google Scholar]
- 67.Taieb F, Nougayrede JP, Watrin C, Samba-Louaka A, Oswald E. Escherichia coli cyclomodulin Cif induces G2 arrest of the host cell cycle without activation of the DNA-damage checkpoint-signalling pathway. Cell Microbiol. 2006;8:1910–1921. doi: 10.1111/j.1462-5822.2006.00757.x. [DOI] [PubMed] [Google Scholar]
- 68.Hostetter DR, Loeb CR, Chu F, Craik CS. Hip is a pro-survival substrate of granzyme B. J Biol Chem. 2007;282:27865–27874. doi: 10.1074/jbc.M704312200. [DOI] [PubMed] [Google Scholar]
- 69.Lofgren S, Fernando MR, Xing KY, Wang Y, Kuszynski CA, Ho YS, et al. Effect of thioltransferase (glutaredoxin) deletion on cellular sensitivity to oxidative stress and cell proliferation in lens epithelial cells of thioltransferase knockout mouse. Invest Ophthalmol Vis Sci. 2008;49:4497–4505. doi: 10.1167/iovs.07-1404. [DOI] [PubMed] [Google Scholar]
- 70.Luk KC, Song C, O'Brien P, Stieber A, Branch JR, Brunden KR, et al. Exogenous alpha-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc Natl Acad Sci USA. 2009;106:20051–20056. doi: 10.1073/pnas.0908005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alano CC, Garnier P, Ying W, Higashi Y, Kauppinen TM, Swanson RA. NAD+ depletion is necessary and sufficient for poly(ADP-ribose) polymerase-1-mediated neuronal death. J Neurosci. 2010;30:2967–2978. doi: 10.1523/JNEUROSCI.5552-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tinsley JH, Hawker J, Yuan Y. Efficient protein transfection of cultured coronary venular endothelial cells. Am J Physiol Heart Circ Physiol. 1998:1873–1878. doi: 10.1152/ajpheart.1998.275.5.H1873. [DOI] [PubMed] [Google Scholar]
- 73.Tinsley JH, Zawieja DC, Wu MH, Ustinova EE, Xu W, Yuan SY. Protein transfection of intact microvessels specifically modulates vasoreactivity and permeability. J Vasc Res. 2001:444–452. doi: 10.1159/000051077. [DOI] [PubMed] [Google Scholar]
- 74.Sato K, Nagao T, Iwasaki T, Nishihira Y, Fukami Y. Src-dependent phosphorylation of the EGF receptor Tyr-845 mediates Stat-p21waf1 pathway in A431 cells. Genes Cells. 2003;8:995–1003. doi: 10.1046/j.1356-9597.2003.00691.x. [DOI] [PubMed] [Google Scholar]
- 75.Fortugno P, Wall NR, Giodini A, O'Connor DS, Plescia J, Padgett KM, et al. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci. 2002;115:575–585. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- 76.Dalkara D, Zuber G, Behr JP. Intracytoplasmic delivery of anionic proteins. Mol Ther. 2004;9:964–969. doi: 10.1016/j.ymthe.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 77.Heerssen HM, Pazyra MF, Segal RA. Dynein motors transport activated Trks to promote survival of target-dependent neurons. Nat Neurosci. 2004;7:596–604. doi: 10.1038/nn1242. [DOI] [PubMed] [Google Scholar]
- 78.Didenko VV, Ngo H, Baskin DS. Polyethyleneimine as a transmembrane carrier of fluorescently labeled proteins and antibodies. Anal Biochem. 2005:168–173. doi: 10.1016/j.ab.2005.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laloraya M, Davoodi-Semiromi A, Kumar GP, McDuffie M, She JX. Impaired Crkl expression contributes to the defective DNA binding of Stat5b in nonobese diabetic mice. Diabetes. 2006;55:734–741. doi: 10.2337/diabetes.55.03.06.db05-1059. [DOI] [PubMed] [Google Scholar]
- 80.Cortes C, Rzomp KA, Tvinnereim A, Scidmore MA, Wizel B. Chlamydia pneumoniae inclusion membrane protein Cpn0585 interacts with multiple Rab GTPases. Infect Immun. 2007;75:5586–5596. doi: 10.1128/IAI.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Geel TM, Meiss G, van der Gun BT, Kroesen BJ, de Leij LF, Zaremba M, et al. Endonucleases induced TRAIL-insensitive apoptosis in ovarian carcinoma cells. Exp Cell Res. 2009;315:2487–2495. doi: 10.1016/j.yexcr.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 82.Weill CO, Biri S, Erbacher P. Cationic lipid-mediated intracellular delivery of antibodies into live cells. Biotechniques. 2008;44:7–9. doi: 10.2144/000112832. [DOI] [PubMed] [Google Scholar]
- 83.Shimura H, Miura-Shimura Y, Kosik KS. Binding of tau to heat shock protein 27 leads to decreased concentration of hyperphosphorylated tau and enhanced cell survival. J Biol Chem. 2004;279:17957–17962. doi: 10.1074/jbc.M400351200. [DOI] [PubMed] [Google Scholar]
- 84.Nagai J, Christensen EI, Morris SM, Willnow TE, Cooper JA, Nielsen R. Mutually dependent localization of megalin and Dab2 in the renal proximal tubule. Am J Physiol Renal Physiol. 2005;289:569–576. doi: 10.1152/ajprenal.00292.2004. [DOI] [PubMed] [Google Scholar]
- 85.Takami T, Terai S, Yokoyama Y, Tanimoto H, Tajima K, Uchida K, et al. Human homologue of maid is a useful marker protein in hepatocarcinogenesis. Gastroenterology. 2005;128:1369–1380. doi: 10.1053/j.gastro.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 86.Cassinelli G, Lanzi C, Petrangolini G, Tortoreto M, Pratesi G, Cuccuru G, et al. Inhibition of c-Met and prevention of spontaneous metastatic spreading by the 2-indolinone RPI-1. Mol Cancer Ther. 2006;5:2388–2397. doi: 10.1158/1535-7163.MCT-06-0245. [DOI] [PubMed] [Google Scholar]
- 87.Ying M, Chen B, Tian Y, Hou Y, Li Q, Shang X, et al. Nuclear import of human sexual regulator DMRT1 is mediated by importin-beta. Biochim Biophys Acta. 2007;1773:804–813. doi: 10.1016/j.bbamcr.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 88.Cassinelli G, Favini E, Degl'Innocenti D, Salvi A, De Petro G, Pierotti MA, et al. RET/PTC1-driven neoplastic transformation and proinvasive phenotype of human thyrocytes involve Met induction and beta-catenin nuclear translocation. Neoplasia. 2009;11:10–21. doi: 10.1593/neo.08916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duerr CU, Zenk SF, Chassin C, Pott J, Gutle D, Hensel M, et al. O-antigen delays lipopolysaccharide recognition and impairs antibacterial host defense in murine intestinal epithelial cells. PLoS Pathog. 2009;5:1000567. doi: 10.1371/journal.ppat.1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee Y, Ishii T, Kim HJ, Nishiyama N, Hayakawa Y, Itaka K, et al. Efficient delivery of bioactive antibodies into the cytoplasm of living cells by charge-conversional polyion complex micelles. Angew Chem Int Ed Engl. 2010;49:2552–2555. doi: 10.1002/anie.200905264. [DOI] [PubMed] [Google Scholar]
- 91.Dalkara D, Chandrashekhar C, Zuber G. Intracellular protein delivery with a dimerizable amphiphile for improved complex stability and prolonged protein release in the cytoplasm of adherent cell lines. J Control Release. 2006;116:353–359. doi: 10.1016/j.jconrel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 92.van der Gun BTF, Monami A, Laarmann S, Raskó T, Slaska-Kiss K, Weinhold E, et al. Serum insensitive, intranuclear protein delivery by the multipurpose cationic lipid SAINT-2. J Control Release. 2007:228–238. doi: 10.1016/j.jconrel.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 93.Lee AL, Wang Y, Ye WH, Yoon HS, Chan SY, Yang YY. Efficient intracellular delivery of functional proteins using cationic polymer core/shell nanoparticles. Biomaterials. 2008;29:1224–1232. doi: 10.1016/j.biomaterials.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 94.Jung S, Huh S, Cheon YP, Park S. Intracellular protein delivery by glucose-coated polymeric beads. Chem Commun (Camb) 2009;7:5003–5005. doi: 10.1039/b906268h. [DOI] [PubMed] [Google Scholar]
- 95.Sanchez-Martin RM, Alexander L, Muzerelle M, Cardenas-Maestre JM, Tsakiridis A, Brickman JM, et al. Microsphere-mediated protein delivery into cells. Chembiochem. 2009;10:1453–1456. doi: 10.1002/cbic.200900136. [DOI] [PubMed] [Google Scholar]
- 96.Tada S, Chowdhury EH, Cho CS, Akaike T. pH-sensitive carbonate apatite as an intracellular protein transporter. Biomaterials. 2010;31:1453–1459. doi: 10.1016/j.biomaterials.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 97.Morris MC, Depollier J, Mery J, Heitz F, Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 98.Herve F, Ghinea N, Scherrmann JM. CNS Delivery Via Adsorptive Transcytosis. AAPS J. 2008;10:455–472. doi: 10.1208/s12248-008-9055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Triguero D, Buciak JB, Yang J, Pardridge WM. Blood-brain barrier transport of cationized immunoglobulin G: Enhanced delivery compared to native protein. Proc Natl Acad Sci USA. 1989:4761–4765. doi: 10.1073/pnas.86.12.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Futami J, Kitazoe M, Maeda T, Nukui E, Sakaguchi M, Kosaka J, et al. Intracellular delivery of proteins into mammalian living cells by polyethyleneimine-cationization. J Biosci Bioeng. 2005;99:95–103. doi: 10.1263/jbb.99.95. [DOI] [PubMed] [Google Scholar]
- 101.Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci USA. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kopatz I, Remy JS, Behr JP. A model for non-viral gene delivery: Through syndecan adhesion molecules and powered by actin. J Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 103.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 104.Behr JP. The proton sponge: A trick to enter cells the viruses did not exploit. Chimia. 1997;51:34–36. [Google Scholar]
- 105.Chuck AS, Clarke MF, Palsson BO. Retroviral infection is limited by Brownian motion. Hum Gene Ther. 1996;7:1527–1534. doi: 10.1089/hum.1996.7.13-1527. [DOI] [PubMed] [Google Scholar]
- 106.Erbacher P, Zou S, Bettinger T, Steffan AM, Remy JS. Chitosan-based vector/DNA complexes for gene delivery: Biophysical characteristics and transfection ability. Pharm Res. 1998;15:1332–1339. doi: 10.1023/a:1011981000671. [DOI] [PubMed] [Google Scholar]
- 107.Luo D, Saltzman WM. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotechnol. 2000;18:893–895. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- 108.Ross PC, Hui SW. Lipoplex size is a major determinant of in vitro lipofection efficiency. Gene Ther. 1999;6:651–659. doi: 10.1038/sj.gt.3300863. [DOI] [PubMed] [Google Scholar]
- 109.Duan Y, Zhang S, Wang B, Yang B, Zhi D. The biological routes of gene delivery mediated by lipid-based non-viral vectors. Expert Opin Drug Deliv. 2009;6:1351–1361. doi: 10.1517/17425240903287153. [DOI] [PubMed] [Google Scholar]
- 110.Bunnell BA, Muul LM, Donahue RE, Blaese RM, Morgan RA. High-efficiency retroviral-mediated gene transfer into human and nonhuman primate peripheral blood lymphocytes. Proc Natl Acad Sci USA. 1995;92:7739–7743. doi: 10.1073/pnas.92.17.7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Plank C, Schillinger U, Scherer F, Bergemann C, Remy JS, Krötz F, et al. The magnetofection method: Using magnetic force to enhance gene delivery. Biol Chem. 2003:737–747. doi: 10.1515/BC.2003.082. [DOI] [PubMed] [Google Scholar]
- 112.Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12:468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 113.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol. 2007:603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bally MB, Harvie P, Wong FMP, Kong S, Wasan EK, Reimer DL. Biological barriers to cellular delivery of lipid-based DNA carriers. Adv Drug Deliv Rev. 1999;38:291–315. doi: 10.1016/s0169-409x(99)00034-4. [DOI] [PubMed] [Google Scholar]
- 116.Futami J, Yamada H. Design of cytotoxic ribonucleases by cationization to enhance intracellular protein delivery. Curr Pharm Biotechnol. 2008;9:180–184. doi: 10.2174/138920108784567326. [DOI] [PubMed] [Google Scholar]
- 117.Ye D, Xu D, Singer AU, Juliano RL. Evaluation of strategies for the intracellular delivery of proteins. Pharm Res. 2002;19:1302–1309. doi: 10.1023/a:1020346607764. [DOI] [PubMed] [Google Scholar]
- 118.Prochiantz A, Joliot A. Can transcription factors function as cell-cell signalling molecules? Nat Rev Mol Cell Biol. 2003;4:814–819. doi: 10.1038/nrm1227. [DOI] [PubMed] [Google Scholar]
- 119.Bird CH, Sun J, Ung K, Karambalis D, Whisstock JC, Trapani JA, et al. Cationic sites on granzyme B contribute to cytotoxicity by promoting its uptake into target cells. Mol Cell Biol. 2005;25:7854–7867. doi: 10.1128/MCB.25.17.7854-7867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Prochiantz A. Messenger proteins: Homeoproteins, TAT and others. Curr Opin Cell Biol. 2000;12:400–406. doi: 10.1016/s0955-0674(00)00108-3. [DOI] [PubMed] [Google Scholar]
- 121.Fischer PM. Cellular uptake mechanisms and potential therapeutic utility of peptidic cell delivery vectors: Progress 2001–2006. Med Res Rev. 2007;27:755–795. doi: 10.1002/med.20093. [DOI] [PubMed] [Google Scholar]
- 122.Pooga M, Hallbrink M, Zorko M, Langel U. Cell penetration by transportan. Faseb J. 1998;12:67–77. doi: 10.1096/fasebj.12.1.67. [DOI] [PubMed] [Google Scholar]
- 123.Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 124.Gump JM, June RK, Dowdy SF. Revised role of glycosaminoglycans in TAT protein transduction domain-mediated cellular transduction. J Biol Chem. 2010;285:1500–1507. doi: 10.1074/jbc.M109.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nakase I, Takeuchi T, Tanaka G, Futaki S. Methodological and cellular aspects that govern the internalization mechanisms of arginine-rich cell-penetrating peptides. Adv Drug Deliv Rev. 2008;60:598–607. doi: 10.1016/j.addr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 126.Belting M. Heparan sulfate proteoglycan as a plasma membrane carrier. Trends Biochem Sci. 2003;28:145–151. doi: 10.1016/S0968-0004(03)00031-8. [DOI] [PubMed] [Google Scholar]
- 127.Console S, Marty C, Garcia-Echeverria C, Schwendener R, Ballmer-Hofer K. Antennapedia and HIV transactivator of transcription (TAT) “protein transduction domains” promote endocytosis of high molecular weight cargo upon binding to cell surface glycosaminoglycans. J Biol Chem. 2003;278:35109–35114. doi: 10.1074/jbc.M301726200. [DOI] [PubMed] [Google Scholar]
- 128.Kosuge M, Takeuchi T, Nakase I, Jones AT, Futaki S. Cellular internalization and distribution of arginine-rich peptides as a function of extracellular peptide concentration, serum and plasma membrane associated proteoglycans. Bioconjug Chem. 2008;19:656–664. doi: 10.1021/bc700289w. [DOI] [PubMed] [Google Scholar]
- 129.Tyagi M, Rusnati M, Presta M, Giacca M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J Biol Chem. 2001;276:3254–3261. doi: 10.1074/jbc.M006701200. [DOI] [PubMed] [Google Scholar]
- 130.Herce HD, Garcia AE. Molecular dynamics simulations suggest a mechanism for translocation of the HIV-1 TAT peptide across lipid membranes. Proc Natl Acad Sci USA. 2007;104:20805–20810. doi: 10.1073/pnas.0706574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yesylevskyy S, Marrink SJ, Mark AE. Alternative mechanisms for the interaction of the cell-penetrating peptides penetratin and the TAT peptide with lipid bilayers. Biophys J. 2009;97:40–49. doi: 10.1016/j.bpj.2009.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ciobanasu C, Siebrasse JP, Kubitscheck U. Cell-penetrating HIV1 TAT peptides can generate pores in model membranes. Biophys J. 2010;99:153–162. doi: 10.1016/j.bpj.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schmidt N, Mishra A, Lai GH, Wong GC. Arginine-rich cell-penetrating peptides. FEBS Lett. 2010;584:1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 134.Maiolo JR, Ferrer M, Ottinger EA. Effects of cargo molecules on the cellular uptake of arginine-rich cell-penetrating peptides. Biochim Biophys Acta. 2005;1712:161–172. doi: 10.1016/j.bbamem.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 135.Fretz MM, Penning NA, Al-Taei S, Futaki S, Takeuchi T, Nakase I, et al. Temperature-, concentration- and cholesterol-dependent translocation of L- and D-octa-arginine across the plasma and nuclear membrane of CD34+ leukaemia cells. Biochem J. 2007;403:335–342. doi: 10.1042/BJ20061808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tunnemann G, Martin RM, Haupt S, Patsch C, Edenhofer F, Cardoso MC. Cargo-dependent mode of uptake and bioavailability of TAT-containing proteins and peptides in living cells. FASEB J. 2006;20:1775–1784. doi: 10.1096/fj.05-5523com. [DOI] [PubMed] [Google Scholar]
- 137.Heng BC, Cao T. Making cell-permeable antibodies (Transbody) through fusion of protein transduction domains (PTD) with single chain variable fragment (scFv) antibodies: Potential advantages over antibodies expressed within the intracellular environment (Intrabody) Medical Hypotheses. 2005;6:1105–1108. doi: 10.1016/j.mehy.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 138.Muller S, Zhao Y, Brown TL, Morgan AC, Kohler H. TransMabs: cell penetrating antibodies, the next generation. Expert Opin Biol Ther. 2005;5:237–241. doi: 10.1517/14712598.5.2.237. [DOI] [PubMed] [Google Scholar]
- 139.Anderson DC, Nichols E, Manger R, Woodle D, Barry M, Fritzberg AR. Tumor cell retention of antibody Fab fragments is enhanced by an attached HIV TAT protein-derived peptide. Biochem Biophys Res Commun. 1993;194:876–884. doi: 10.1006/bbrc.1993.1903. [DOI] [PubMed] [Google Scholar]
- 140.Zhao Y, Lou D, Burkett J, Kohler H. Chemical engineering of cell penetrating antibodies. J Immunol Methods. 2001;254:137–145. doi: 10.1016/s0022-1759(01)00410-0. [DOI] [PubMed] [Google Scholar]
- 141.Niesner U, Halin C, Lozzi L, Gunthert M, Neri P, Wunderli-Allenspach H, et al. Quantitation of the tumor-targeting properties of antibody fragments conjugated to cell-permeating HIV-1 TAT peptides. Bioconjug Chem. 2002;13:729–736. doi: 10.1021/bc025517+. [DOI] [PubMed] [Google Scholar]
- 142.Stein S, Weissb A, Adermannc K, Lazarovicid P, Hochmane J, Wellhöner H. A disulfide conjugate between anti-tetanus antibodies and HIV (37–72) Tat neutralizes tetanus toxin inside chromaffin cells. FEBS Lett 1999;458:383–386. doi: 10.1016/s0014-5793(99)01186-2. [DOI] [PubMed] [Google Scholar]
- 143.Chen BX, Erlanger BF. Intracellular delivery of monoclonal antibodies. Immunol Lett. 2002;84:63–67. doi: 10.1016/s0165-2478(02)00146-3. [DOI] [PubMed] [Google Scholar]
- 144.Zhao Y, Brown TL, Kohler H, Muller S. MTS-conjugated-antiactive caspase 3 antibodies inhibit actinomycin D-induced apoptosis. Apoptosis. 2003;8:631–637. doi: 10.1023/A:1026139627930. [DOI] [PubMed] [Google Scholar]
- 145.Cohen-Saidon C, Nechushtan H, Kahlon S, Livni N, Nissim A, Razin E. A novel strategy using single-chain antibody to show the importance of Bcl-2 in mast cell survival. Blood. 2003;102:2506–2512. doi: 10.1182/blood-2002-12-3921. [DOI] [PubMed] [Google Scholar]
- 146.Nakajima O, Hachisuka A, Okunuki H, Takagi K, Teshima R, Sawada JI. Method for delivering radiolabeled single-chain Fv antibody to the brain. J Health Sci. 2004;50:159–163. [Google Scholar]
- 147.Ohara-Imaizumi M, Nishiwaki C, Kikuta T, Kumakura K, Nakamichi Y, Nagamatsu S. Site of docking and fusion of insulin secretory granules in live MIN6 beta cells analyzed by TAT-conjugated anti-syntaxin 1 antibody and total internal reflection fluorescence microscopy. J Biol Chem. 2004;279:8403–8418. doi: 10.1074/jbc.M308954200. [DOI] [PubMed] [Google Scholar]
- 148.Shin I, Edl J, Biswas S, Lin PC, Mernaugh R, Arteaga CL. Proapoptotic activity of cell-permeable anti-Akt single-chain antibodies. Cancer Res. 2005;65:2815–2824. doi: 10.1158/0008-5472.CAN-04-2898. [DOI] [PubMed] [Google Scholar]
- 149.Kameyama S, Horie M, Kikuchi T, Omura T, Takeuchi T, Nakase I, et al. Effects of cell-permeating peptide binding on the distribution of 125I-labeled Fab fragment in rats. Bioconjug Chem. 2006;17:597–602. doi: 10.1021/bc050258k. [DOI] [PubMed] [Google Scholar]
- 150.Kameyama S, Okada R, Kikuchi T, Omura T, Nakase I, Takeuchi T, et al. Distribution of immunoglobulin Fab fragment conjugated with HIV-1 REV peptide following intravenous administration in rats. Mol Pharm. 2006;3:174–180. doi: 10.1021/mp050064m. [DOI] [PubMed] [Google Scholar]
- 151.Chen BC, Erlanger BF. Cell cycle inhibition by an anti-cyclin D1 antibody chemically modified for intracellular delivery. Cancer Lett. 2006;244:71–75. doi: 10.1016/j.canlet.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 152.Theisen DM, Pongratz C, Wiegmann K, Rivero F, Krut O, Kronke M. Targeting of HIV-1 Tat traffic and function by transduction-competent single chain antibodies. Vaccine. 2006;24:3127–3136. doi: 10.1016/j.vaccine.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 153.Kameyama S, Horie M, Kikuchi T, Omura T, Tadokoro A, Takeuchi T, et al. Acid wash in determining cellular uptake of Fab/cell-permeating peptide conjugates. Biopolymers. 2007;88:98–107. doi: 10.1002/bip.20689. [DOI] [PubMed] [Google Scholar]
- 154.Hu M, Chen P, Wang J, Scollard DA, Vallis KA, Reilly RM. 123I-labeled HIV-1 tat peptide radioimmunoconjugates are imported into the nucleus of human breast cancer cells and functionally interact in vitro and in vivo with the cyclin-dependent kinase inhibitor, p21(WAF-1/Cip-1) Eur J Nucl Med Mol Imaging. 2007;34:368–377. doi: 10.1007/s00259-006-0189-0. [DOI] [PubMed] [Google Scholar]
- 155.Avignolo C, Bagnasco L, Biasotti B, Melchiori A, Tomati V, Bauer I, et al. Internalization via Antennapedia protein transduction domain of an scFv antibody toward c-Myc protein. FASEB J. 2008;22:1237–1245. doi: 10.1096/fj.07-8865com. [DOI] [PubMed] [Google Scholar]
- 156.Poungpair O, Pootong A, Maneewatch S, Srimanote P, Tongtawe P, Songserm T, et al. A human single chain transbody specific to matrix protein (M1) interferes with the replication of influenza A virus. Bioconjug Chem. 2010;21:1134–1141. doi: 10.1021/bc900251u. [DOI] [PubMed] [Google Scholar]
- 157.Barka T, Gresik ES, Henderson SC. Production of cell lines secreting TAT fusion proteins. J Histochem Cytochem. 2004;52:469–477. doi: 10.1177/002215540405200405. [DOI] [PubMed] [Google Scholar]
- 158.Koutsokeras A, Kabouridis PS. Secretion and uptake of TAT-fusion proteins produced by engineered mammalian cells. Biochim Biophys Acta. 2009;1790:147–153. doi: 10.1016/j.bbagen.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shaw PA, Catchpole IR, Goddard CA, Colledge WH. Comparison of protein transduction domains in mediating cell delivery of a secreted CRE protein. Biochemistry. 2008;47:1157–1166. doi: 10.1021/bi701542p. [DOI] [PubMed] [Google Scholar]
- 160.Chauhan A, Tikoo A, Kapur AK, Singh M. The taming of the cell penetrating domain of the HIV Tat: myths and realities. J Control Release. 2007;117:148–162. doi: 10.1016/j.jconrel.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nakayama K. Furin: A mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tikhonov I, Ruckwardt TJ, Berg S, Hatfield GS, David Pauza C. Furin cleavage of the HIV-1 Tat protein. FEBS Lett. 2004;565:89–92. doi: 10.1016/j.febslet.2004.03.079. [DOI] [PubMed] [Google Scholar]
- 163.Flinterman M, Farzaneh F, Habib N, Malik F, Gaken J, Tavassoli M. Delivery of therapeutic proteins as secretable TAT fusion products. Mol Ther. 2009;17:334–342. doi: 10.1038/mt.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Falnes PO, Wesche J, Olsnes S. Ability of the Tat basic domain and VP22 to mediate cell binding, but not membrane translocation of the diphtheria toxin A-fragment. Biochemistry. 2001;40:4349–4358. doi: 10.1021/bi002443l. [DOI] [PubMed] [Google Scholar]
- 165.Norbury CC, Hewlett LJ, Prescott AR, Shastri N, Watts C. Class I MHC presentation of exogenous soluble antigen via macropinocytosis in bone marrow macrophages. Immunity. 1995;3:783–791. doi: 10.1016/1074-7613(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 166.Meier O, Boucke K, Hammer SV, Keller S, Stidwill RP, Hemmi S, et al. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J Cell Biol. 2002;158:1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 168.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: From molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mie M, Takahashi F, Funabashi H, Yanagida Y, Aizawa M, Kobatake E. Intracellular delivery of antibodies using TAT fusion protein A. Biochem Biophys Res Commun. 2003;310:730–734. doi: 10.1016/j.bbrc.2003.09.071. [DOI] [PubMed] [Google Scholar]
- 170.Mie M, Mori K, Funabashi H, Kobatake E. Delivery of antibody-captured proteins into living cells using PTD-fused protein A. Biotechnol Lett. 2006;28:1209–1214. doi: 10.1007/s10529-006-9076-9. [DOI] [PubMed] [Google Scholar]
- 171.Albarran B, To R, Stayton PS. A TAT-streptavidin fusion protein directs uptake of biotinylated cargo into mammalian cells. Protein Eng Des Sel. 2005;18:147–152. doi: 10.1093/protein/gzi014. [DOI] [PubMed] [Google Scholar]
- 172.El-Sayed A, Futaki S, Harashima H. Delivery of Macromolecules Using arginine-rich cell-penetrating peptides: Ways to overcome endosomal entrapment. AAPS J. 2009;11:13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Maxfield FR. Weak bases and ionophores rapidly and reversibly raise the pH of endocytic vesicles in cultured mouse fibroblasts. J Cell Biol. 1982;95:676–681. doi: 10.1083/jcb.95.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Erbacher P, Roche AC, Monsigny M, Midoux P. Putative role of chloroquine in gene transfer into a human hepatoma cell line by DNA lactosylated polylysine complexes. Exp Cell Res. 1996;225:186–194. doi: 10.1006/excr.1996.0169. [DOI] [PubMed] [Google Scholar]
- 175.Ciftci K, Levy RJ. Enhanced plasmid DNA transfection with lysosomotropic agents in cultured fibroblasts. Int J Pharm. 2001:81–92. doi: 10.1016/s0378-5173(01)00623-8. [DOI] [PubMed] [Google Scholar]
- 176.Caron NJ, Quenneville SP, Tremblay JP. Endosome disruption enhances the functional nuclear delivery of Tat-fusion proteins. Biochem Biophys Res Commun. 2004;319:12–20. doi: 10.1016/j.bbrc.2004.04.180. [DOI] [PubMed] [Google Scholar]
- 177.Wang H, Zhong CY, Wu JF, Huang YB, Liu CB. Enhancement of TAT cell membrane penetration efficiency by dimethyl sulphoxide. J Control Release. 2010;143:64–70. doi: 10.1016/j.jconrel.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 178.Hogset A, Prasmickaite L, Selbo PK, Hellum M, Engesaeter BO, Bonsted A, et al. Photochemical internalisation in drug and gene delivery. Adv Drug Deliv Rev. 2004;56:95–115. doi: 10.1016/j.addr.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 179.Berg K, Selbo PK, Prasmickaite L, Tjelle TE, Sandvig K, Moan J, et al. Photochemical internalization: a novel technology for delivery of macromolecules into cytosol. Cancer Res. 1999;59:1180–1183. [PubMed] [Google Scholar]
- 180.Maeda T, Kawasaki K, Ohnishi S. Interaction of influenza virus hemagglutinin with target membrane lipids is a key step in virus-induced hemolysis and fusion at pH 5.2. Proc Natl Acad Sci USA. 1981;78:4133–4137. doi: 10.1073/pnas.78.7.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza hemagglutinin at the Ph of membrane-fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 182.Michiue H, Tomizawa K, Wei FY, Matsushita M, Lu YF, Ichikawa T, et al. The NH2 terminus of influenza virus hemagglutinin-2 subunit peptides enhances the antitumor potency of polyarginine-mediated p53 protein transduction. J Biol Chem. 2005;280:8285–8289. doi: 10.1074/jbc.M412430200. [DOI] [PubMed] [Google Scholar]
- 183.Lo SL, Wang S. An endosomolytic Tat peptide produced by incorporation of histidine and cysteine residues as a nonviral vector for DNA transfection. Biomaterials. 2008;29:2408–2414. doi: 10.1016/j.biomaterials.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 184.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, et al. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 185.Patel LN, Zaro JL, Shen WC. Cell penetrating peptides: Intracellular pathways and pharmaceutical perspectives. Pharm Res. 2007;24:1977–1992. doi: 10.1007/s11095-007-9303-7. [DOI] [PubMed] [Google Scholar]
- 186.Drin G, Cottin S, Blanc E, Rees AR, Temsamani J. Studies on the internalization mechanism of cationic cell-penetrating peptides. J Biol Chem. 2003;278:31192–31201. doi: 10.1074/jbc.M303938200. [DOI] [PubMed] [Google Scholar]
- 187.Cheung JC, Kim Chiaw P, Deber CM, Bear CE. A novel method for monitoring the cytosolic delivery of peptide cargo. J Control Release. 2009;137:2–7. doi: 10.1016/j.jconrel.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 188.Poznansky MJ, Juliano RL. Biological approaches to the controlled delivery of drugs: A critical review. Pharmacol Rev. 1984;36:277–336. [PubMed] [Google Scholar]
- 189.Fonseca SB, Pereira MP, Kelley SO. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv Drug Deliv Rev. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 190.Rapoport M, Lorberboum-Galski H. TAT-based drug delivery system—new directions in protein delivery for new hopes? Expert Opin Drug Deliv. 2009;6:453–463. doi: 10.1517/17425240902887029. [DOI] [PubMed] [Google Scholar]
- 191.Fotin-Mleczek M, Welte S, Mader O, Duchardt F, Fischer R, Hufnagel H, et al. Cationic cell-penetrating peptides interfere with TNF signalling by induction of TNF receptor internalization. J Cell Sci. 2005;118:3339–3351. doi: 10.1242/jcs.02460. [DOI] [PubMed] [Google Scholar]
- 192.Letoha T, Kusz E, Papai G, Szabolcs A, Kaszaki J, Varga I, et al. In vitro and in vivo nuclear factorkappaB inhibitory effects of the cell-penetrating penetratin peptide. Mol Pharmacol. 2006;69:2027–2036. doi: 10.1124/mol.105.019653. [DOI] [PubMed] [Google Scholar]
- 193.Cardozo AK, Buchillier V, Mathieu M, Chen J, Ortis F, Ladriere L, et al. Cell-permeable peptides induce dose- and length-dependent cytotoxic effects. Biochim Biophys Acta. 2007;1768:2222–2234. doi: 10.1016/j.bbamem.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 194.Mueller J, Kretzschmar I, Volkmer R, Boisguerin P. Comparison of cellular uptake using 22 CPPs in 4 different cell lines. Bioconjug Chem. 2008;19:2363–2374. doi: 10.1021/bc800194e. [DOI] [PubMed] [Google Scholar]
- 195.Hetzel C, Bachran C, Tur MK, Fuchs H, Stocker M. Improved immunotoxins with novel functional elements. Curr Pharm Des. 2009;15:2700–2711. doi: 10.2174/138161209788923930. [DOI] [PubMed] [Google Scholar]
- 196.Rybak SM, Arndt MA, Schirrmann T, Dubel S, Krauss J. Ribonucleases and immunoRNases as anticancer drugs. Curr Pharm Des. 2009;15:2665–2675. doi: 10.2174/138161209788923921. [DOI] [PubMed] [Google Scholar]
- 197.Schirrmann T, Krauss J, Arndt MA, Rybak SM, Dubel S. Targeted therapeutic RNases (ImmunoRNases) Expert Opin Biol Ther. 2009;9:79–95. doi: 10.1517/14712590802631862. [DOI] [PubMed] [Google Scholar]
- 198.Wedekind JE, Trame CB, Dorywalska M, Koehl P, Raschke TM, McKee M, et al. Refined crystallographic structure of Pseudomonas aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J Mol Biol. 2001;314:823–837. doi: 10.1006/jmbi.2001.5195. [DOI] [PubMed] [Google Scholar]
- 199.Perier A, Chassaing A, Raffestin S, Pichard S, Masella M, Menez A, et al. Concerted protonation of key histidines triggers membrane interaction of the diphtheria toxin T domain. J Biol Chem. 2007;282:24239–24245. doi: 10.1074/jbc.M703392200. [DOI] [PubMed] [Google Scholar]
- 200.Hartley MR, Lord JM. Cytotoxic ribosome-inactivating lectins from plants. Biochim Biophys Acta. 2004;1701:1–14. doi: 10.1016/j.bbapap.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 201.Futami J, Maeda T, Kitazoe M, Nukui E, Tada H, Seno M, et al. Preparation of potent cytotoxic ribonucleases by cationization: Enhanced cellular uptake and decreased interaction with ribonuclease inhibitor by chemical modification of carboxyl groups. Biochemistry. 2001;40:7518–7524. doi: 10.1021/bi010248g. [DOI] [PubMed] [Google Scholar]
- 202.Rodriguez M, Torrent G, Bosch M, Rayne F, Dubremetz JF, Ribo M, et al. Intracellular pathway of Onconase that enables its delivery to the cytosol. J Cell Sci. 2007;120:1405–1411. doi: 10.1242/jcs.03427. [DOI] [PubMed] [Google Scholar]
- 203.Casellas P, Bourrie BJ, Gros P, Jansen FK. Kinetics of cytotoxicity induced by immunotoxins. Enhancement by lysosomotropic amines and carboxylic ionophores. J Biol Chem. 1984;259:9359–9364. [PubMed] [Google Scholar]
- 204.Heisler I, Sutherland M, Bachran C, Hebestreit P, Schnitger A, Melzig MF, et al. Combined application of saponin and chimeric toxins drastically enhances the targeted cytotoxicity on tumor cells. J Control Release. 2005;106:123–137. doi: 10.1016/j.jconrel.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 205.Bachran C, Sutherland M, Heisler I, Hebestreit P, Melzig MF, Fuchs H. The saponin-mediated enhanced uptake of targeted saporin-based drugs is strongly dependent on the saponin structure. Exp Biol Med. 2006;231:412–420. doi: 10.1177/153537020623100407. [DOI] [PubMed] [Google Scholar]
- 206.Sehnert B, Burkhardt H, Nimmerjahn F, Pohle S, Wessels J, Vestweber D, et al. A Tissue-specific NFkappaB inhibitor ameliorates inflammatory joint diseases. Ann Rheum Dis. 2010;69:45. [Google Scholar]
- 207.Fuchs H, Bachran C, Li T, Heisler I, Dürkop H, Sutherland M. A cleavable molecular adapter reduces side effects and concomitantly enhances efficacy in tumor treatment by targeted toxins in mice. J Control Release. 2007;117:342–350. doi: 10.1016/j.jconrel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 208.Hetzel C, Bachran C, Fischer R, Fuchs H, Barth S, Stöcker M. Small cleavable adapters enhance the specific cytotoxicity of a humanized immunotoxin directed against CD64-positive cells. J Immunother. 2008;3:370–376. doi: 10.1097/CJI.0b013e31816a2d23. [DOI] [PubMed] [Google Scholar]
- 209.Rizk SS, Luchniak A, Uysal S, Brawley CM, Rock RS, Kossiakoff AA. An engineered substance P variant for receptor-mediated delivery of synthetic antibodies into tumor cells. Proc Natl Acad Sci USA. 2009;106:11011–11015. doi: 10.1073/pnas.0904907106. [DOI] [PMC free article] [PubMed] [Google Scholar]