Abstract
Despite recent advances and new applications of molecular and biogeochemical methodology in aquatic microbial ecology, our perception of the aquatic microbial world remains one dominated by “free-living” bacteria that account for most of the microbial activities in the pelagic zone. Recent research has, however, shown that there exist vast and hidden “microbial networks” within the water column, connected via various microhabitats such as aggregates, fecal pellets and higher organisms. Bacterial abundance within these networks may rival or exceed that of the “free-living” bacteria. Hence, what we have learned in traditional aquatic microbial ecology represents merely a fraction of the microbial world. Within these networks a bacterium can travel long distances, communicate and closely interact with other bacteria and efficiently exchange genetic information with one another. The presence of microbial networks within the water column demands better sampling strategies and a new way to understand bacterial ecology, evolution and functions within the broader context of systems biology.
Key words: bacteria, microbial networks, microhabitats, interactions, aquatic ecosystems
Although invisible to the naked eyes, bacteria are ubiquitous in all aquatic (including marine) ecosystems. The true diversity of aquatic bacteria is unknown and unknowable, but there are estimated 2 × 106 bacterial species in the ocean,1 and new strains are still being discovered frequently.2 The vast variety of bacteria performs so many critical functions in aquatic biogeochemical cycles3,4 and trophic processes5 that the biosphere will surely collapse without bacteria.
Aquatic microbial research has seen unprecedented pace of development in recent years thanks to the advances in molecular and biogeochemical techniques.6 It is therefore timely to revisit our concept of the aquatic microbial world.
The traditional view of aquatic microbial ecology presumes that “free living” bacteria contribute to the majority of bacterial production within the water body.7 A basic textbook still tells us that there are on average 105–106 ml−1 of bacteria living freely in the water,8 so a few milliliters already contain more than enough material for microbiologists to work with. Indeed, on an oceanographic expedition microbiologists often draw just a few milliliters of water from a Niskin bottle for measuring bacterial abundance and production. Meanwhile, other scientists on board may be retrieving sediment traps with abundant aggregates and fecal pellets inside them or preserving zooplankton samples or hauling in a net-full of fish. These samples are categorized separately (bacteria, aggregates, zooplankton, fish) and processed and understood within the confines of the respective disciplines that mimics the way we dissect an aquatic ecosystem.
Microhabitats in the Pelagic Environment
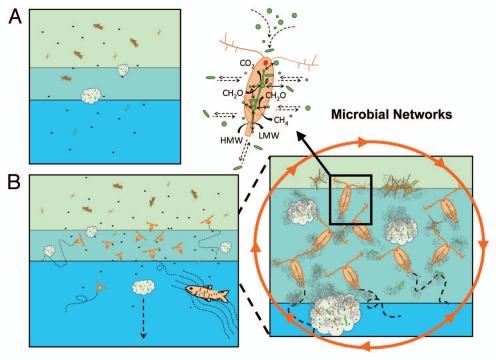
However, bacteria do not live in isolation from each other or other inhabitants. It is a common knowledge that in terrestrial systems most bacteria live on surfaces or inside a host. Research on benthic habitats and coral reefs has also focused on attached microbial consortia.9,10 Why should there be any difference in the pelagic aquatic environment? Bacteria within a water column can reach high abundances in microhabitats such as aggregates, fecal pellets, surface and gut of a zooplankter or a fish11,12 (Fig. 1). However, due to their patchy spatial and temporal distribution, they are often not sampled properly or largely ignored by aquatic microbiologists.
Figure 1.
Different conceptual views of the aquatic microbial ecosystem. In the traditional view (A), bacterial communities are mainly dominated by free-living bacteria and attached bacterial communities in microniches are rather isolated from each other. Water column stratification also limits exchanges of bacteria between water layers. In our proposed microbial networks (B), free-living and attached bacteria are tightly linked with each other via a network of microhabitats represented here by aggregates, fecal pellets, plankton and higher organisms. Mobile organisms also effectively transport bacteria over long distances and across boundaries. Within a microhabitat, such as a copepod, dense populations of diverse bacteria can closely communicate and exchange genetic information with each other, be protected from external hazards, exploit high concentration of organic matters and drive biogeochemical reactions that are otherwise not favored in the surrounding water.
Albeit limited, existing literature data show that total bacterial abundance associated with various microhabitats may rival or even exceed “free living” bacteria in the aquatic environment.11,12 What microbiologists have learned based on traditional sampling methods therefore represents only a fraction of the aquatic microbial world. These microhabitat communities of bacteria are not only important in terms of abundance and biomass, but their behavior, physiology and composition are also distinctly different from their “free living” counterpart. For examples, they have elevated enzymatic activities,13 growth and production,13 and may also experience higher grazing mortality14,15 and viral infection.16 The equivalent bacterial concentrations associated with these microhabitats are orders of magnitude higher than ambient bacterial concentrations, indicating active bacterial colonization and growth in/on these organic-rich microhabitats.11,12
Assuming an average free-living bacterial concentration of 106 ml−1, the average distance between two neighboring cells is 100 µm or approx. 100 cell lengths. This is a formidable distance for communication between two bacterial cells due to fast dissipation of chemical signals.17 In comparison, on a densely colonized aggregate surface, cells can be touching or even overlapping one another.18 Dense packing of cells on surfaces makes cell-cell interactions (both positive and negative) and communications (e.g., quorum sensing) much more feasible.19,20 The microhabitats themselves are often rich sources of organic substrates relative to the surrounding water. For example, an aggregate contains a high concentration of particulate and dissolved organic carbon.21,22 Bacteria in zooplankton's guts continuously receive organic matters ingested by the hosts.23 Some of these microhabitats also have physical-chemical characteristics different from the surrounding water. A very interesting observation is the anoxic or near anoxic condition inside an animal's gut and fecal pellet (Glud RN and Tang KW, unpublished data) and an aggregate.24 Subsequently, the high abundance of bacteria present in close proximity to each other and to the source of organic matters under specific physical-chemical conditions may allow for biogeochemical reactions that are otherwise not favored in the water column.25,26
Hitching a Ride
Only relatively recently did researchers begin to appreciate the fact that many aquatic bacteria are motile, but this motility remains quite limited.27,28 The run-and-tumble motion of individual bacteria leads to diffusion-like dispersal of a population,29 and the dispersal distance (L) can be estimated as L2 = 2Dt where D is diffusion coefficient and t is time. Even with a relatively high diffusion coefficient;30 e.g., 3.8 × 10−5 cm2s−1, the population will disperse no more than 2.6 cm per day by swimming alone. In addition to limited motility, the water column of large lakes and oceans are often vertically stratified, permanently or seasonally, such that the pycnocline further prevents mixing of bacteria between upper and lower water layers.31
Yet, bacteria seem to have an ingenious way to overcome this limitation—by hitch hiking on large particles and organisms. Aggregates and fecal pellets sink up to several hundred meters a day bringing attached bacteria with them,32 but this is primarily a one-way ride from the surface water to the deep water. Mobile organisms such as diurnally migrating zooplankton, on the other hand, move up and down across the pycnocline multiple times, working as a conveyor belt to transport bacteria in both directions.33 These long-range dispersal processes affect the distribution, resource access and genetic information exchange between different bacterial populations (see below).
Shelters for Bacteria
Death is a part of life and for bacteria there is no exception. The seemingly constant abundance of free-living bacteria in most aquatic environments, in contrast to their high growth rate, suggests that there must be a high mortality rate among them.34 Death could be due to predation,35 viral infection36 or environmental stresses.37 The many folds and turns inside an aggregate or a zooplankter could effectively shelter bacteria from external hazardous elements. Vibrio cholerae, the bacterium that causes the disease cholera, finds its ‘partner in crime’ in small crustacean zooplankton.38,39 This bacterium turns virulent when attached to chitin surfaces40 and becomes more resistant to disinfection.41 Many other bacteria attached to aggregates and zooplankton have also been found to survive harmful UV radiation, heat and chemicals.59 These findings have important implications for the spread of aquatic pathogens and water treatment strategies.
There is, however, not a strict separation between “attached” and “free” bacteria in the pelagic environment. A bacterium may attach and detach from a microhabitat, the likelihood of which depends on the bacterium's physiological conditions,42 surface grazing mortality,43,44 substrate characteristics,45 pre-existing microbial community20 and chemical cues,30 among other factors. Attached bacterial communities may also release progeny into the surrounding water.46 Consequently, there is a dynamic exchange between attached and free bacteria, and thanks to the high production rate of attached bacteria, microhabitats may be an important source of bacteria to even the free living populations.45
Because different microhabitats may emerge or disappear in a random manner, and because different bacteria may enter or exit a microhabitat at different times, there could be countless ways for bacteria to establish both short-term and long-term microbial networks. In this sense, a microhabitat in the pelagic environment is unlike the relatively more stable bacterial consortia in soil.47
Gene Exchange Market
Compared to higher organisms, the bacterial genome is very malleable and gene exchanges between bacterial cells would allow them to change and adapt to environmental conditions much more rapidly than mutation alone may allow.48 This can be achieved only when the genetic information is passed along by a mediator such as a plasmid or a virus or when two bacteria come close enough for conjugation.49 However, in the free water column the long distances between randomly moving bacteria makes gene exchanges more difficult. In contrast, when a large number of bacteria are confined within a microhabitat, this microenvironment turns into an exchange market where rapid lateral gene transfer can occur between bacteria.50 By hitchhiking on mobile organisms bacteria may even travel long distances and trade genetic information with remote populations.33
We may therefore envision that various particles and organisms within the water column constitute a hidden microbial network through which bacterial genetic information busily flows and be redistributed, leading to rapid changes in bacterial community structure and functions over ecological as well as evolutionary time scales.51
Microhabitats and the Global Carbon Cycle
Photosynthesis and respiration are the yin and yang of the carbon cycle. Bacteria account for most of the respiration in the aquatic environment returning photosynthetically fixed carbon to the atmosphere as carbon dioxide.52 Sinking detritus is a major component of the so-called biological pump for exporting fixed carbon from the euphotic zone to the deep sea, hence removing it from sea-air exchange.53 Questions, however, remain concerning bacterial turnover of particulate organic carbon within the mesopelagic zone (100–1,000 m).54 A recent modeling study suggests that free-living bacteria and attached bacteria are each responsible for turning over 38% of the detrital carbon in the mesopelagic zone and together they respire ∼85% of the exported carbon.55 Critical unknowns in the model are POC solubilization and subsequent DOC leakage due to attached bacteria and growth efficiency of particularly attached bacteria in the deep sea.56,57 That study highlights the need for proper sampling and measuring bacteria attaching to detrital particles and other microhabitats in understanding the global carbon cycle.
Conclusion
Here, we show that the traditional view of aquatic microbial ecologists is largely biased by not taking into account the true spatial and temporal dynamics of aquatic systems. Recent studies imply that bacteria associated to particles and organisms come in close contact to each other and are dispersed via multiple vectors between microhabitats—even over large distances. Hence, within the expansive and seemingly homogeneous water column there exist vast and hidden networks that bring bacteria together allowing for interactions and efficient exchange of genetic information. To better understand the adaptation, evolution and functions of distinct microbial populations, we therefore need to know their distribution and behavior in time and space and their interactions with other microbes and higher organisms within these networks. The complexity of these networks will require better sampling strategies and interpretation of bacterial data within the broader context of systems biology.58
Acknowledgements
H.P.G. received funding from the German Science Foundation (DFG-GR 1540/11-1, 12-1 and PA 1655/1-1) and the Leibniz Foundation. K.W.T. was supported by U.S. National Science Foundation grant OCE-0814558 and a visiting scientist fellowship from Leibniz Institute of Freshwater Ecology and Inland Fisheries, Berlin.
Abbreviations
- POC
particulate organic carbon
- DOC
dissolved organic carbon
Footnotes
Previously published online www.landesbioscience.com/journals/cib/article/12975
References
- 1.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Nat Acad Sci USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 3.Paerl HW, Pinckney JL. A mini-review of microbial consortia: their roles in aquatic production and biogeochemical cycling. Microb Ecol. 1996;31:225–247. doi: 10.1007/BF00171569. [DOI] [PubMed] [Google Scholar]
- 4.Grossart H-P. Ecological consequences of bacterio-plankton lifestyles: changes in concepts are needed. Env Microbiol Reports. 2010;2:706–714. doi: 10.1111/j.1758-2229.2010.00179.x. [DOI] [PubMed] [Google Scholar]
- 5.Cotner JB, Biddanda BA. Small players, large role: microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems. 2002;5:105–121. [Google Scholar]
- 6.Xu J. Microbial ecology in the age of genomics and metagenomics: concepts, tools and recent advances. Mol Ecol. 2006;15:1713–1731. doi: 10.1111/j.1365-294X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- 7.Ducklow HW. The bacteria component of the oceanic euphotic zone. FEMS Microbiol Ecol. 1999;30:1–10. [Google Scholar]
- 8.Miller CB. Biological Oceanography. Malden, MA: Blackwell Science; 2004. pp. 92–110. [Google Scholar]
- 9.Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. [Google Scholar]
- 10.Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 11.Simon M, Grossart HP, Schweitzer B, Ploug H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol. 2002;28:175–211. [Google Scholar]
- 12.Tang KW, Turk V, Grossart H-P. Linkage between crustacean zooplankton and aquatic bacteria. Aquat Microb Ecol. 2010;61:261–277. doi: 10.3354/ame01424. [DOI] [Google Scholar]
- 13.Grossart HP, Tang KW, Kiørboe T, Ploug H. Comparison of cell-specific activity between free-living and attached bacteria using isolates and natural assemblages. FEMS Microbiol Lett. 2007;266:194–200. doi: 10.1111/j.1574-6968.2006.00520.x. [DOI] [PubMed] [Google Scholar]
- 14.Caron DA. Grazing of attached bacteria by heterotrophic microflagellates. Microb Ecol. 1987;13:203–218. doi: 10.1007/BF02024998. [DOI] [PubMed] [Google Scholar]
- 15.Kiørboe T, Tang K, Grossart HP, Ploug H. Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment and grazing mortality of attached bacteria. Appl Env Microbiol. 2003;69:3036–3047. doi: 10.1128/AEM.69.6.3036-3047.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riemann L, Grossart HP. Elevated lytic phage production as a consequence of particle colonization by a marine Flavobacterium (Cellulophaga sp.) Microb Ecol. 2008;56:505–512. doi: 10.1007/s00248-008-9369-8. [DOI] [PubMed] [Google Scholar]
- 17.Atema J. Chemical signals in the marine environment: Dispersal, detection and temporal signal analysis. Proc Nat Acad Sci USA. 1995;92:62–66. doi: 10.1073/pnas.92.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiørboe T, Grossart HP, Ploug H, Tang K, Auer B. Particle-associated flagellates: swimming patterns, colonization rates and grazing on attached bacteria. Aquat Microb Ecol. 2004;35:141–152. [Google Scholar]
- 19.Gram L, Grossart HP, Schlingloff A, Kiørboe T. Production of acylated homserine lactones by Roseobacter strains isolated from marine snow. Appl Env Microb. 2002;68:4111–4116. doi: 10.1128/AEM.68.8.4111-4116.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossart HP, Kiørboe T, Tang K, Ploug H. Bacterial colonization of marine particles: growth and inter-specific interactions. Appl Env Microbiol. 2003;69:3500–3509. doi: 10.1128/AEM.69.6.3500-3509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossart HP, Simon M. Significance of limnetic organic aggregates (lake snow) for the sinking flux of particulate organic matter in a large lake. Aquat Microb Ecol. 1998;15:115–125. [Google Scholar]
- 22.Alldredge AL. Interstitial dissolved organic carbon (DOC) concentrations within sinking marine aggregates and their potential contribution to carbon flux. Limnol Oceanogr. 2000;45:1245–1253. [Google Scholar]
- 23.Tang KW. Copepods as microbial hotspots in the ocean: Effects of host feeding activities on attached bacteria. Aquat Microb Ecol. 2005;38:31–40. [Google Scholar]
- 24.Alldredge AL, Cohen Y. Can microscale chemical patches persist in the sea? Microelectrode study of marine snow and fecal pellets. Science. 1987;235:689–691. doi: 10.1126/science.235.4789.689. [DOI] [PubMed] [Google Scholar]
- 25.De Angelis MA, Lee C. Methane production during zooplankton grazing on marine phytoplankton. Limnol Oceanogr. 1994;39:1298–1308. [Google Scholar]
- 26.Tang KW, Visscher PT, Dam HG. DMSP-consuming bacteria associated with the calanoid copepod Acartia tonsa (Dana) J Exp Mar Biol Ecol. 2001;256:185–198. doi: 10.1016/s0022-0981(00)00314-2. [DOI] [PubMed] [Google Scholar]
- 27.Fenchel T. Eppur si muove: many water column bacteria are motile. Aquat Microb Ecol. 2001;24:197–201. [Google Scholar]
- 28.Grossart HP, Riemann L, Azam F. Bacterial motility in the sea and its ecological implications. Aquat Microb Ecol. 2001;25:247–258. [Google Scholar]
- 29.Berg HC. Random walks in biology. Princeton: Princeton University Press; 1993. [Google Scholar]
- 30.Kiørboe T, Grossart HP, Ploug H, Tang K. Mechanisms and rates of bacterial colonization of sinking aggregates. Appl Env Microbiol. 2002;68:3996–4006. doi: 10.1128/AEM.68.8.3996-4006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allgaier M, Grossart HP. Diversity and seasonal dynamics of Actinobacteria populations infour lakes in northeastern Germany. Appl Environ Microbiol. 2006;72:3489–3497. doi: 10.1128/AEM.72.5.3489-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turley CM, Mackie PJ. Bacteria and cyanobacterial flux to the deep NE Atlantic on sedimenting particles. Deep-Sea Res. 1995;42:1453–1474. [Google Scholar]
- 33.Grossart HP, Dziallas C, Leunert F, Tang KW. Bacteria dispersal by hitchhiking on zooplankton. Proc Nat Acad Sci USA. 2010;107:11959–11964. doi: 10.1073/pnas.1000668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ducklow HW. Production and fate of bacteria in the oceans. Bioscience. 1983;33:494–501. [Google Scholar]
- 35.Hahn MW, Höfle MG. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol Ecol. 2001;35:113–121. doi: 10.1111/j.1574-6941.2001.tb00794.x. [DOI] [PubMed] [Google Scholar]
- 36.Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea—viruses play critical roles in the structure and function of aquatic food webs. Bioscience. 1999;49:781–788. [Google Scholar]
- 37.Häder DP, Kumar HD, Smith RC, Worrest RC. Effects of solar UV radiation on aquatic ecosystems and interactions with climate change. Photochem Photobiol Sci. 2007;6:267–285. doi: 10.1039/b700020k. [DOI] [PubMed] [Google Scholar]
- 38.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Env Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cottingham KL, Chiavelli DA, Taylor RK. Environmental microbe and human pathogen: the ecology and microbiology of Vibrio cholerae. Front Ecol Env. 2003;1:80–86. [Google Scholar]
- 40.Pruzzo C, Vezzulli L, Colwell RR. Global impact of Vibrio cholerae interactions with chitin. Env Microbiol. 2008;10:1400–1410. doi: 10.1111/j.1462-2920.2007.01559.x. [DOI] [PubMed] [Google Scholar]
- 41.Chowdhury MA, Huq A, Xu B, Madeira FJ, Colwell RR. Effect of alum on free-living and copepod-associated Vibrio cholerae O1 and O139. Appl Env Microbiol. 1997;63:3323–3326. doi: 10.1128/aem.63.8.3323-3326.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yam EM, Tang KW. Effects of starvation on aggregate colonization and motility of marine bacteria. Aquat Microb Ecol. 2007;48:207–215. [Google Scholar]
- 43.Kiørboe T, Tang K, Grossart HP, Ploug H. Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment and grazing mortality of attached bacteria. Appl Env Microbiol. 2003;69:3036–3047. doi: 10.1128/AEM.69.6.3036-3047.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang KW, Grossart HP, Yam EM, Jackson GA, Ducklow HW, Kiørboe T. Mesocosm study of particle dynamics and control of particle-associated bacteria by flagellate grazing. Mar Ecol Prog Ser. 2006;325:15–27. [Google Scholar]
- 45.Grossart HP, Kiørboe T, Tang KW, Allgaier M, Yam EM, Ploug H. Interactions between marine snow and heterotrophic bacteria: aggregate formation and microbial dynamics. Aquat Microb Ecol. 2006;42:19–26. [Google Scholar]
- 46.Azam F, Smith DC. Bacterial influence on the variability in the ocean's biochemical state: a mechanistic view. In: Demers S, editor. Particle analysis in oceanography. Berlin: Springer-Verlag; 1991. pp. 213–236. [Google Scholar]
- 47.Haruta S, Kato S, Yamamoto K, Igarash Y. Intertwined interspecies relationships: approaches to untangle the microbial network. Environ Microbiol. 2009;11:2963–2969. doi: 10.1111/j.1462-2920.2009.01956.x. [DOI] [PubMed] [Google Scholar]
- 48.Lawrence JG. Gene transfer, speciation and the evolution of bacteria genomes. Curr Opin Microbiol. 1999;2:519–523. doi: 10.1016/s1369-5274(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 49.Paul JH. Microbial gene transfer: an ecological perspective. J Molec Microbiol Biotechnol. 1999;1:45–50. [PubMed] [Google Scholar]
- 50.Doolittle WF. The practice of classification and the theory of evolution and what the demise of Charles Darwin's tree of life hypothesis means for both of them. Phil Trans R Soc B. 2009;364:2221–2228. doi: 10.1098/rstb.2009.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venail PA, MacLean RC, Bouvier T, Brockhurst MA, Hochberg ME, Mouquet N. Diversity and productivity peak at intermediate dispersal rate in evolving meta-communities. Nature. 2008;452:210–215. doi: 10.1038/nature06554. [DOI] [PubMed] [Google Scholar]
- 52.Rivkin RB, Legendre L. Biogenic carbon cycling in the upper ocean: effects of microbial respiration. Science. 2001;23:2398–2400. doi: 10.1126/science.291.5512.2398. [DOI] [PubMed] [Google Scholar]
- 53.Volk T, Hoffert ML. Ocean carbon pumps: Analysis of relative strengths and efficiencies in ocean-driven atmospheric CO2 changes. In: Sundquist ET, Brocker WS, editors. The carbon cycle and atmospheric CO2: natural variations archean to present. Washington DC: American Geophysical Union; 1985. pp. 99–110. [Google Scholar]
- 54.Burd AB, Hansell DA, Steinberg DK, Anderson TR, Arístegui J, Galtar F, et al. The imbalance between geochemical and biochemical indicators of meso- and bathypelagic biological activity: What the @#! Is wrong with present calculations of carbon budgets? Deep-Sea Res II. 2010;57:1557–1571. [Google Scholar]
- 55.Anderson TR, Tang KW. Carbon cycling and POC turnover in the mesopelagic zone of the ocean: Insights from a simple model. Deep-Sea Res II. 2010;16:1581–1592. [Google Scholar]
- 56.Del Giorgio PA, Cole JJ. Bacterial growth efficiency in natural aquatic systems. Annu Rev Ecol Syst. 1998;29:503–541. [Google Scholar]
- 57.Baltar F, Arístegui J, Gasol JM, Herndl GJ. Prokaryotic carbon utilization in the dark ocean: growth efficiency, leucine-to-carbon conversion factors and their relation. Aquat Microb Ecol. 2010;60:227–232. [Google Scholar]
- 58.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 59.Tang KW, Dziallas C, Grossart H-P. Zooplankton and aggregates as refuge for aquatic bacteria: Protection from UV, heat and ozone stresses used for water treatment. Environ Microbiol. 2010 doi: 10.1111/j.1462-2920.2010.02335.x. In press. [DOI] [PubMed] [Google Scholar]