Abstract
There are well known examples in nature of microtubules dramatically changing their function by re-organizing their structure. Most interphase animal cells rely on the radial organization of the microtubule network for precise cargo delivery. Dividing cells re-organize microtubules with the help of motor proteins to form the spindle and drive the segregation of chromosomes into daughter cells. These examples present a kind of dichotomy: microtubules can be utilized as stationary tracks along which motor proteins move, or they can perform work themselves by utilizing the power of motor proteins. While both occur during mitosis, our recent findings demonstrate that both functions may occur simultaneously in interphase cells as well. We find that kinesin-1 (a motor known for its role in transporting cargo along microtubule tracks) powers microtubule sliding in non-dividing cells and this mechanism is used to form cellular protrusions.
Key words: microtubules, kinesin, microtubule motors, cytoskeleton, S2 cells
In most unpolarized cells, microtubule minus ends are tethered at the nucleation site, which is often the peri-nuclear centrosome containing gamma-tubulin, centrioles, pericentriolar material and other proteins.1–3 The growing, GTP-hydrolyzing, plus ends are found at or near the plasma membrane. This configuration allows microtubule motor proteins, most notably the ubiquitous kinesin-1 and cytoplasmic dynein, to walk long distances carrying cellular cargo either towards the nucleus by the minus-end directed dynein motor, or towards the plasma membrane by the plus-end directed kinesin-1. In polarized epithelial cells, microtubule minus ends anchor at the apical side and plus ends locate at the basal side to maintain apico-basal polarity.4 These arrangements, whether in a polarized cell or not, are essential to cellular organization.
However, microtubules are not always stationary tracks, and are often reorganized to allow them to perform work. During cell division, the microtubule network is restructured into the spindle. The process of spindle formation begins with the separation of duplicated centrosomes into opposite sides of the cell as a result of the coordination of three mitotic motors, kinesin 5, kinesin 14 and cytoplasmic dynein5–8 (Fig. 1). Spindle assembly involves the pushing apart of centrosomes by interdigitating microtubules from opposite centrosomes sliding against oneanother by the bipolar kinesin 5 motor.9–12 Upon disruption of the kinesin 5 motor, a monopolar spindle forms, resulting in a mitotic/meiotic block.13 At the same time, cytoplasmic dynein is responsible for “focusing” the spindle and contributing to the tethering of microtubule minus ends to the centrosome and the centrosomes to the cell cortex.14,15 Perturbation of cytoplasmic dynein results in splayed microtubule minus ends at the spindle poles and the dislocation of centrosomes from the spindle.7
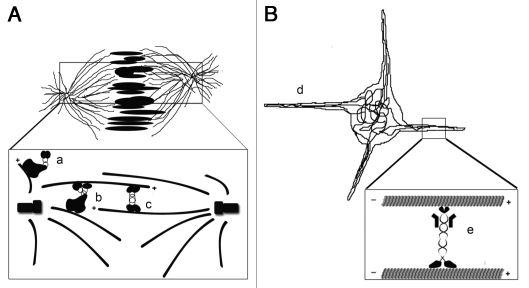
Figure 1.
Molecular motors organize microtubules to facilitate microtubule-mediated force production. (A) The mitotic spindle is organized by three motor proteins and drives the separation of chromosomes into daughter cells. The motors include the dynein/dynactin complex, which functions to anchor the centrosomes to the cell cortex (a), and anchor microtubule minus ends to the centrosome (b). The kinesin 5 motor (c) pushes the two centrosomes apart by crosslinking and sliding anti-parallel microtubules. (B) Our recent study shows that the growth of cellular processes filled with microtubule bundles (d) is powered by microtubule sliding by kinesin-1 heavy chain (e) in interphase cells.23
There are examples of motors organizing microtubules outside of the context of the spindle as well. Microtubule bundling is used by the fungus U. maydis in determining cell polarity and growth,16 and bundles are organized, at least in part, by kinesin-1. In 2006, Straube and colleagues17 showed that the C-terminus (including the putative C-terminal microtubule binding domain) of the kinesin-1 homolog is required for microtubule cross-linking in these cells. This was the first time an in vivo role for kinesin-1 in mediating microtubule-microtubule interactions was reported.17 During the initial characterization of the ubiquitous conventional kinesin (kinesin-1) in 1991, it was discovered that in vitro kinesin-1 also has the ability to slide microtubules against one another.18 Further evidence verified the existence of a C-terminal microtubule binding domain in the kinesin heavy chain.19–21
Microtubule bending and buckling, especially near the periphery, is commonly observed in all types of cultured animal cells, and has been attributed to a variety of cellular forces. Until now, studies suggesting a role for molecular motors in driving microtubule bending have implicated the minus-end directed motor dynein,17,22 based on the observation that the bending observed was mostly in the anterograde direction in each cell type. Straube and colleagues use this as evidence that only 4% of microtubule bending events in U. maydis could be attributed to the activity of a plus-end directed kinesin motor.17 Our recent study show for the first time that kinesin-1 mediated microtubule-microtubule sliding occurs ubiquitously in animal cells and, contrary to previously thought, that dynein does not drive microtubule sliding.23
We developed a new microscopic technique using photoconversion of small areas of the microtubule network to track and quantify the microtubule motion in cultured Drosophila S2 cells expressing tubulin tagged with a photoconvertible protein, Dendra2. These movements were essentially eliminated by RNAi-mediated knock-down of kinesin-1 heavy chain. Knock-down of the kinesin light chain, other kinesins or the dynein motor did not inhibit microtubule-microtubule sliding. In addition to Drosophila cells, we observed microtubule sliding in Xenopus fibroblasts and PtK2 rat kangaroo cells. Inhibition of kinesin-1 in Ptk2 cells by antibody injection blocked microtubule sliding. The advantage of our approach is that we directly tested the involvement of each motor using RNAi-mediated protein knock-down and quantified the amount of sliding using the photoconversion assay. In our study, we consistently observe a modest increase in sliding upon disruption of the dynein motor. Our results show that the dynein motor actually inhibits microtubule sliding.23 Possibly, dynein binding to cargo increases the likelihood that kinesin-1 is participating in organelle transport. Alternatively, dynein may counteract the sliding of kinesin-1 by stabilizing microtubules.
We go on to show that kinesin-1 mediated microtubule sliding is responsible for the creation of microtubule bundle-filled processes in Drosophila S2 cells when the actin cytoskeleton has been disrupted (see Fig. 1).23 Our results show that the actin network plays a critical role in the organization of the microtubule network. However, we find that in Drosphila S2 cells the actin network does not directly modulate microtubule sliding. This is not true for all cell types; our unpublished studies revealed that in certain cell types such as Xenopus melanophores, disruption of the actin network leads to a marked increase in the amount of microtubule sliding observed. A recent study of Drosphila S2 cells confirms our finding that actin-microtubule cross-talk is essential for microtubule organization.24 Applewhite and colleagues observe a global change in microtubule movement upon depletion of the actin-microtubule cross-linking spectraplakin short stop.24 Their results are not inconsistent with our finding that actin does not regulate microtubule sliding; the authors observe an overall increase in microtubule displacement, while we specifically analyzed the effect of the actin network on the sliding phenomenon.
We have identified a novel and ubiquitous mechanism used in interphase animal cells to move microtubules against oneanother. There are many ways a cell might utilize this mechanism: to drive changes in cell shape, power cytoplasmic movements, create new filament network at locations far from nucleating sites or transport multiple cargoes simultaneously on a moving short microtubule fragment. There are some exciting leads in the current literature that suggest kinesin-1 mediated microtubule sliding may drive microtubule-dependent processes. For example, microtubule sliding has been implicated in the formation of proplatelets, the precursor to blood platelets, from the parent megakaryocyte.25 Ooplasmic streaming refers to the developmental stage during Drosophila oogenesis when the cytoplasm undergoes a large-scale rotation to properly distribute mRNA and proteins to establish proper oocyte polarity, and is known to require both microtubules and the kinesin-1 heavy chain.26 Our findings set the groundwork for future studies identifying specialized cellular processes that require kinesin-1 mediated microtubule-microtubule sliding. Of particular interest will be to determine whether quiescent, undifferentiated cells sometimes use kinesin-1 to transport microtubules in addition to cargo, or if microtubule-microtubule sliding is activated by developmental or environmental cues solely in specialized cells.
Acknowledgements
The work in the Gelfand lab is supported by the grant from NIGMS (GM-52111).
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/13212
References
- 1.Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 2.Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, et al. Pericentrin and gammatubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oakley BR, Oakley CE, Yoon Y, Jung MK. Gammatubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- 4.Musch A. Microtubule organization and function in epithelial cells. Traffic. 2004;5:1–9. doi: 10.1111/j.1600-0854.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 5.Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol. 1999;1:51–54. doi: 10.1038/9025. [DOI] [PubMed] [Google Scholar]
- 6.Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, et al. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaglio T, Dionne MA, Compton DA. Mitotic spindle poles are organized by structural and motor proteins in addition to centrosomes. J Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, et al. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholey JM. Kinesin-5 in Drosophila embryo mitosis: sliding filament or spindle matrix mechanism? Cell Motil Cytoskel. 2009;66:500–508. doi: 10.1002/cm.20349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol Biol Cell. 2009;20:1749–1762. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole DG, Saxton WM, Sheehan KB, Scholey JM. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- 13.Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinberg G, Wedlich-Soldner R, Brill M, Schulz I. Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J Cell Sci. 2001;114:609–622. doi: 10.1242/jcs.114.3.609. [DOI] [PubMed] [Google Scholar]
- 17.Straube A, Hause G, Fink G, Steinberg G. Conventional kinesin mediates microtubule-microtubule interactions in vivo. Mol Biol Cell. 2006;17:907–916. doi: 10.1091/mbc.E05-06-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urrutia R, McNiven MA, Albanesi JP, Murphy DB, Kachar B. Purified kinesin promotes vesicle motility and induces active sliding between microtubules in vitro. Proc Natl Acad Sci USA. 1991;88:6701–6705. doi: 10.1073/pnas.88.15.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navone F, Niclas J, Hom-Booher N, Sparks L, Bernstein HD, McCaffrey G, et al. Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J Cell Biol. 1992;117:1263–1275. doi: 10.1083/jcb.117.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews SB, Gallant PE, Leapman RD, Schnapp BJ, Reese TS. Single kinesin molecules crossbridge microtubules in vitro. Proc Natl Acad Sci USA. 1993;90:6503–6507. doi: 10.1073/pnas.90.14.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackney DD, Stock MF. Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat Cell Biol. 2000;2:257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 22.Bicek AD, Tüzel E, Demtchouk A, Uppalapati M, Hancock WO, Kroll DM, et al. Anterograde microtubule transport drives microtubule bending in LLC-PK1 epithelial cells. Mol Biol Cell. 2009;20:2943–2953. doi: 10.1091/mbc.E08-09-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolly AL, Kim H, Srinivasan D, Lakonishok M, Larson AG, Gelfand VI. Kinesin-1 heavy chain mediates microtubule sliding to drive changes in cell shape. Proc Natl Acad Sci USA. 2010;107:12151–12156. doi: 10.1073/pnas.1004736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Applewhite DA, Grode KD, Keller D, Zadeh AD, Slep KC, Rogers SL. The spectraplakin Short stop is an actin-microtubule cross-linker that contributes to organization of the microtubule network. Mol Biol Cell. 2010;21:1714–1724. doi: 10.1091/mbc.E10-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel SR, Richardson JL, Schulze H, Kahle E, Galjart N, Drabek K, et al. Differential roles of microtubule assembly and sliding in proplatelet formation by megakaryocytes. Blood. 2005;106:4076–4085. doi: 10.1182/blood-2005-06-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palacios IM, St Johnston D. Kinesin light chain-independent function of the kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development. 2002;129:5473–5485. doi: 10.1242/dev.00119. [DOI] [PubMed] [Google Scholar]