Abstract
Hedgehog (Hh) signalling is mediated through the Patched-1 (Ptch1) receptor. Hh-binding to Ptch1 blocks the inhibitory effects of Ptch1 on the activity of the transmembrane protein, Smoothened (Smo), resulting induction of target genes by the Gli-family of transcription factors. We demonstrate here that Hh-binding to Ptch1 stimulates activation of Erk1/2. This activation is insensitive to the small molecule Smo antagonists and occurs in a cell line that does not express Smo. Specifically, the C-terminus of Ptch1 harbors motifs encoding Class I and II SH3-binding sites. SH3-domain binding activity was verified using GST-c-srcSH3, -Grb2SH3 and -p85βSH3 fusion-proteins. Ectopically-expressed Grb2 or p85β could also be co-immunoprecipitated with the Ptch1 C-terminus. Addition of Shh to serum-starved human mammary epithelial cells and Shh Light II fibroblasts stimulated phosphorylation of Erk1/2. Erk1/2 activation was observed in cells where Smo activity had been inhibited using cyclopamine and in the breast epithelial cell line, MCF10A, that does not express Smo. These data reveal novel binding activities for the C-terminal region of Ptch1 and define a signalling pathway stimulated by the Hh-ligands operating independently of pathways requiring Smo.
Keywords: Hedgehog, smoothened, patched-1, Erk, SH3-domain, signal transduction
INTRODUCTION
The Hedgehog (Hh)-signalling pathway is an essential developmental pathway controlling cell fate and morphogenesis. In vertebrates, three Hh-ligands, Sonic (Shh), Indian (Ihh) and Desert Hedgehog (Dhh) (Echelard et al., 1993, Krauss et al., 1993), all closely related to Hedgehog (Hh) in Drosophila, regulate a well defined molecular-genetic signal transduction pathway. In its basic form, the 12-pass transmembrane receptor, Patched-1 (Ptch1), regulates the activity of the transcriptional effectors of Hh-signalling, Gli2 and Gli3 (Ruppert et al., 1988, Mo et al., 1997, Borycki et al., 1998, Ding et al., 1998, Sasaki et al., 1999), these latter transcription factors are closely related to Drosophila Cubitus interuptis (Ci) (Orenic et al., 1987, Orenic et al., 1990, Ruppert et al., 1990, Tiniakow and Terentieva, 1933). In the absence of Hh-ligands, the principal activity associated with Ptch1 is repression of the activity of a 7-pass transmembrane protein, Smoothened (Smo) (Alcedo et al., 1996). Inhibition of Smo by Ptch1 results in the maintenance of a cytoplasmic proteolytic complex containing Suppressor of Fused (SuFu). The proteolytic activity of this complex cleaves the C-terminal domains of the Gli proteins (Gli2 or Gli3), yielding transcriptional repressor forms (Aza-Blanc et al., 1997, Sasaki et al., 1999, Ruiz i Altaba, 1999). Upon binding of the Hh-ligands to Ptch1 (Chen and Struhl, 1996, Alcedo et al., 1996) inhibition of Smo is relieved. Smo activation, in turn, blocks the proteolytic activity of the SuFu-containing complex, thereby maintaining the Gli proteins in their transcriptional activator state and ultimately driving expression of Hh-target genes (Ohlmeyer and Kalderon, 1998, Lum et al., 2003, Pham et al., 1995, Therond et al., 1996, Hepker et al., 1997, Sisson et al., 1997, Monnier et al., 1998, Robbins et al., 1997, Forbes et al., 1993). Despite Ptch1-dependent regulation of Smo activity playing a central role in mediating Hh-signalling, the mechanisms by which this regulation occurs are not well understood.
Patched-1 is known to contain domains that suggest it may participate in the regulation of additional molecular pathways. Previous studies, for example, have defined a region in the sterol-sensing domain of Ptch1 (Loftus et al., 1997, Martin et al., 2001) that complexes cyclin B1 in a Hh-dependent manner (Barnes et al., 2001). This association suggests that the Hh-pathway may be involved in control of the cell division cycle specifically at mitosis. While genetic evidence for the significance of this Ptch1-mediated activity has not been shown, it may represent a Hh-dependent check-point control mechanism consistent with a mitogenic role for the Hh-signalling pathway (Forbes et al., 1996, Dahmane and Ruiz i Altaba, 1999, St-Jacques et al., 1999, Kenney and Rowitch, 2000, Fu et al., 2004, Mill et al., 2005). Distinct from the domain responsible for binding cyclin B1, the C-terminal cytoplasmic domain of Ptch1 binds to the molecular chaperone, Tid1 (Wakabayashi et al., 2007). Tid1 activity has been implicated in signalling pathways that involve ras (Tarunina et al., 2004, Trentin et al., 2001), Smads (Torregroza and Evans, 2006) and ErbB2 (Kim et al., 2004) as well as in the control of apoptosis (Syken et al., 1999, Edwards and Munger, 2004) and cell senescence (Tarunina et al., 2004). The significance of the interaction between Tid1 and Ptch1 has been shown using the FVB mouse strain. FVB harbors a polymorphism in the C-terminus of Ptch1 effecting its ability to complex Tid1 (Wakabayashi et al., 2007). Here, susceptibility of Kras-induced skin squamous cell carcinomas in FVB mice segregates with the mutant Ptch1 allele. How binding of the Hh-ligands to Ptch1 effects the association of Tid1 with Ptch1 and alters signalling through downstream pathways has not been characterized however.
As we published previously (Moraes et al., 2009), the role of the Hh-signalling pathway in control of the growth and morphogenesis of the mammary gland was studied using mice carrying the Mesenchymal dysplasia (mes) allele of Ptch1 (Ptch1mes) (Sweet et al., 1996). Ptch1mes lacks most of the cytoplasmic C-terminal region of Ptch1 due to a 32 bp deletion in the last exon, resulting in a frameshift and truncation of the last 220 amino acids (Makino et al., 2001). Despite this mutation, mice homozygous for the mes allele of Ptch1 are viable but sterile and exhibit polydactyly, a white belly spot, precocious hair follicle development (Nieuwenhuis et al., 2007) and, as we have recently shown, severe defects in mammary gland development during puberty (Moraes et al., 2009).
Previous studies of Ptch1mes mice showed that, in the dermis of these animals, no significant alterations were evident in the levels of Ptch1 or Gli1 (Nieuwenhuis et al., 2007), both transcriptional targets of the canonical Hh-signalling cascade (Dahmane et al., 1997, Lee et al., 1997, Alexandre et al., 1996, Marigo et al., 1996, Forbes et al., 1993). We speculated, therefore, that Hh-signalling may recruit or activate other signalling cascades through the C-terminal region of Ptch1 independent of its Smo-dependent functions. Our results show that the C-terminal region of Ptch1 binds to SH3-encoding domains of a number of proteins, including Grb2, c-src and p85β. We demonstrate further that Shh can stimulate a U0126-sensitive signalling cascade that activates Erk1/2. Furthermore, activation of Erk1/2 occurs in cell lines lacking Smo or in the presence of the small chemical inhibitors of Smo.
MATERIALS AND METHODS
Mice
Mesenchymal dysplasia (mes) mice are an inbred strain of mice harboring a 32 bp deletion of the coding region of the C-terminal cytoplasmic domain of the Ptch1 gene, resulting in truncation of the Ptch1 protein at the beginning of the cytoplastic domain (Makino et al., 2001). These mice were obtained from Jackson Labs and backcrossed onto the C57BL/6 background (Charles River) for >10 generations.
Expression Constructs
RNA from Ptch1wt/mes heterozygote animals was isolated using Trizol according to the manufacturer’s instructions. The cytoplastic domains of Ptch1wt and Ptch1mes were ampified by RT-PCR and the cDNA cloned into in frame with the pEGFP-C1 vector in order to express N-terminally GFP-tagged C-terminal domains of Ptch1wt and Ptch1mes fusion proteins. The previously described (Nieuwenhuis et al., 2007) expression constructs encoding the full length wild type or Ptch1mes Ptch1 proteins tagged at their N-terminus with GFP were a kind gift of Dr. C. C. Hui (Hospital For Sick Children Research Institute, Toronto, Canada)
Cell Culture
Human mammary gland epithelial cells (HMEC) and MCF10A cell lines were cultured in DMEM/F12 medium with 5% horse serum, 20 ng/ml EGF, 10 µg/ml insulin, 0.5 µg/ml hydrocordisone, 5 µg/ml transferin, 1 ng/ml cholora toxin, 100 µg/ml streptomycin and 100 U/ml penicillin at 37°C in a humidified atmosphere containing 5% CO2. Shh Light II fibroblasts (Sasaki et al., 1997), which harbor a Gli-responsive luciferase transgene, were obtained from ATCC. Cells were grown in DMEM/F12 medium with 10% fetal bovine serum plus 100 µg/ml streptomycin and 100 U/ml penicillin. For serum starvation, HMEC or Shh Light II cells were switched to media containing 0.1% horse serum, for 48 hours. For MCF10A cells, cells were trypisinized and replated in growth conditions for 4 hours in order for the cells to re-attach. Media was then changed to DMEM/F12 without fetal bovine serum for 24 hours.
Shh Peptide and Chemical Inhibitors
N-Shh peptide was purchased from R&D System. Before stimulation, cultured cells were serum starved using DMEM/F12 medium with 0.1% horse serum for 48 hours and thereafter stimulated with 1µg/ml of N-Shh for indicated time points. For chemical inhibitors, 5 µM of the Smo inhibitor, cyclopamine (LC Laboratories),was added to cells 24 hours before stimulation with the N-Shh peptide for HMEC and Shh Light II cells. 5 nM of the farnesyl transferase inhibitor, H-Cys-4-Abz-Met-OH (FTase II; Sigma) was added to cells 1 hour before exposure to N-Shh peptide. The MEK-specific inhibitor, U0126 (Sigma), was added to a final concentration of 10nM to cells one hour prior to addition of Shh peptide. For inhibition of N-Shh activity, 20 µl of conditioned media containing the α-Shh monoclonal antibody expressed from the hybridoma cell line, 5E1, (Developmental Studies Hybridoma Bank, University of Iowa), was mixed with the N-Shh peptide and added to serum-starved cells.
Phospho-Erk and Luciferase Assays
For phospho-Erk assays, cells were trypsinized and replated in complete medium for 4 hours. Attached cells were then switched to media lacking serum for 48 hours. Serum starved cells were then stimulated with N-Shh. For measurement of the activation of Erk1/2, cells were harvested at various times points and lysed in buffer containing 0.5% Nonidet P-40, 120 mM NaCl, 50 mM Tris, pH 8.0. Following centrifugation, 40 µg of whole cell lysate was run on 10% SDS-polyacrylamide gels and electroblotted onto nitrocellulose. Blots were then probed for using an α-phospho-Erk antibody (Cell Signalling). Blots were then stripped and reprobed with an antibody directed to total Erk (Cell Signalling).
For luciferase activity in Shh Light II cells, luciferase activity was determined as previously described (Sasaki et al., 1997). Briefly, cells were lysed in lysis buffer at room temperature for 15 minutes followed by centrifugation at 13,000rpm for 1 minute. Cell lysates were transferred to a new tube and luciferase activity was measured using Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase was used as the reporter gene and Renilla luciferase was used as control for normalization. Luciferase activity in lysates was read in plates using a Berthold Technologies microplate luminometer. Luciferase assays were performed three times with each sample assayed in triplicate. For MCF10A mammary epithelial cells, cells grown in complete medium were transfected using Lipofectamine 2000 (Invitrogen) preincubated at room temperature for 20 minutes either with 1µg of the 8xGli-Luc reporter plasmid, 1 µg of CMV-βgalactosidase (control for transfection efficiency) and either 1µg of activated Gli2 (Gli2ΔN) expression plasmid or 1µg of pcDNA3. Twenty-four hours after transfection cells were switched to serum free medium and starved for 24 hours. N-Shh peptide was added at the final concentration of 1µg /ml, grown for another 24 hours and the measured luciferase activity normalized to βgal activity.
GST Binding Assay
The GST fusion constructs encoding the SH3 domains of Grb2, p85 or c-src were kindly provided by Dr. M. Moran (University of Toronto). GST-fusion proteins were expressed in bacteria and purified on glutathione sepharose 4B (Amersham Biosciences) according to the manufacturer’s instructions. An equal amounts (2 µg) of purified GST or GST-Grb2SH3, GST-P85SH3, GST-P85SH3 were incubated with glutathione beads for 2 hours at 4°C. After washing, the beads were incubated with whole cell lysates containing GFP-Ptch1wt or GFP-Ptch1mes protein and were rocked for 2 hours at 4°C. The beads were washed five times with Nonidet P-40 lysis buffer (0.5% Nonidet P-40, 120 mM NaCl, 50 mM Tris, pH 8.0). Bound proteins were eluted by boiling in SDS buffer for 5 minutes before loading onto 10% SDS-polyacrylamide gels.
Immunoprecitations and Co-immunofluorescence
COS-1 or 293 cells were transfected with plasmids expressing triple-HA-tagged PtchC-term and either GFP-tagged full length Grb2 (Yamazaki et al., 2002)(a kind gift of Dr. L. Samelson (National Institutes of Health, Bethesda Maryland, U.S.A.), flag-tagged PIK3R2 (p85β), wild type, flag-tagged Smurf2 or the catalytically-defective Smurf2 mutant, Smurf CA ((Ogunjimi et al., 2005); the latter three expression constructs obtained from Dr. J. Wrana, Samuel Lunenfeld Res. Inst., Toronto, ON, Canada). Forty-eight hours following transfection, cell lysates were prepared as described above for the phospho-Erk analysis. Anti-GFP, anti-flag or anti-HA antibodies were added to 200 µg of lysate from transfected cells. Immune complexes were then isolated on protein G-sepharose beads and isolated proteins identified by western analysis.
HMEC cells grown on coverslips were transfected with expression vectors expressing GFP-tagged, full length Ptch1, full length Ptch1mes or GFP-fusion proteins with the C-terminal region of Ptch1 or Ptch1mes. Forty-eight hours post transfection, cells were fixed with 4% paraformaldehyde for 10 minutes at room temperature. After washing with PBS 3 times, cells were permeabilized with 0.1% Triton X-100 for 2 minutes then treated with 3% BSA. Cells were probed with α-EEA-1 (Applied Biological Materials Inc.) for 1 hour at room temperature. After three washes with PBS, cells were incubated with a TRITC-conjugated goat anti-mouse secondary antibody for 1 hour at room temperature, then washed in PBS and embedded in Vectashield mounting medium. Digital photographs were taken on a Nikon Eclipse 80i Fluorescence microscope using Q Imaging monochrome digital camera and Q Capture Pro software.
RESULTS
Canonical Hh-signalling and localization of Ptch1 appears normal in mammary cells of mice
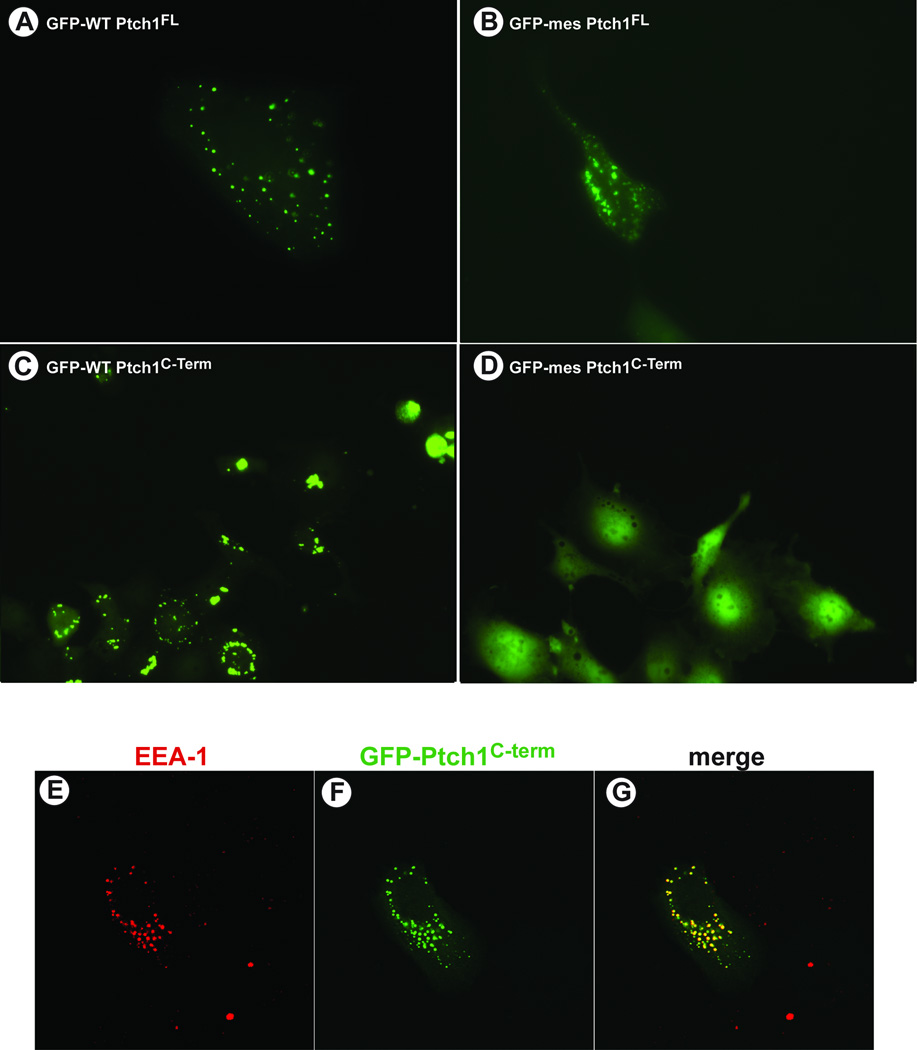
The cytoplasmic C-terminal region of Ptch1 is truncated in mes mice. While development of some tissues is altered, the overall phenotype of these animals is relatively normal suggesting that Smo-mediated Hh-signalling is generally uncompromised. We tested, therefore, if distinct activities might be attributable to the C-terminus of the mes variant of Ptch1 relative to the wild type protein. The subcellular localization of full length Ptch1 from wild type and mes mice were compared to the distribution of the isolated C-termini from these same proteins (Figure 1). Full length mutant Ptch1mes, tagged at its N-terminus with GFP, localized to vesicles similar to the localization of the full length wild type protein (Figure 1A–B). As Figure 1C showed further, the GFP-tagged C-terminal cytoplasmic domain of wild type Ptch1 also localized to vesicles in human mammary epithelial cells (HMECs). Co-immunofluorescence revealed that these vesicles harbored the endosomal marker, EEA-1 (Figure 1E–G), consistent with the previously reported cellular localization of endogenous Ptch1 (Martin et al., 2001, Incardona et al., 2002). However, in contrast to the isolated wild type C-terminus and the full length Ptch1mes protein, GFP-tagged C-terminus of Ptch1mes did not localize to any structures within the cell despite retaining the first 50 amino acids (Figure 1D). Thus, while the full length Ptch1mes protein appeared to localize normally, the C-terminus of Ptch1 from mes mice was defective for tethering to EEA1-containing vesicles.
Figure 1. The C-terminus of wild type by not mes Ptch1 localizes to vesicles.
Human mammary epithelial cells were transfected with plasmids expressing GFP-tagged (A) full length, wild type Ptch1, (B) full length mes Ptch1, (C) wild type, Ptch1C-term or (D) mes Ptch1C-term. Localization of these GFP-tagged fusion proteins was then visualized by direct immunofluorescence in live cells in culture. Both wild type and mes full length Ptch1 localize to the vesicles. Similarly, the isolated C-terminus of Ptch1 also localizes to vesicles. In contrast the C-terminus from the mes variant of Ptch1 is distributed randomly and does not localize to specific cellular structures. Human mammary epithelial cells grown on cover slips were transfected with a plasmid expressing GFP-PtchC-term. Cells were fixed and endosomal localization of EEA-1 (E) determined by indirect immunofluorescence. Localization of GFP-PtchC-term (F) was determined by direct fluorescence. The merged images (G) reveal the overlapping localization of EEA-1 and GFP-PtchC-term.
C-terminus of Patched-1 interacts with SH3-containing factors
The requirement for an intact C-terminus for localization of this isolated domain to vesicles suggested that sequences in this region might facilitate tethering of this domain to vesicles. As Figure 2 illustrates, the C-terminus of Ptch1 harbours proline-rich sequences encoding consensus binding sites for Class I and Class II SH3-domains (Feng et al., 1995, Rickles et al., 1995, Yu et al., 1994) or WW-domains (Sudol et al., 1995, Macias et al., 1996, Jager et al., 2006). These sites in Ptch1 are highly conserved among mammals and, for one of these (1288RPPPYRPRR), among vertebrates.
Figure 2. Conserved SH3- and WW-domain binding motifs in the C-terminus of Ptch1.
Comparison of the C-terminal region of Ptch1 from mouse, rat, human, chicken and frogs (Xenopus) reveal the presence of conserved Class I and Class II SH3-binding motifs.
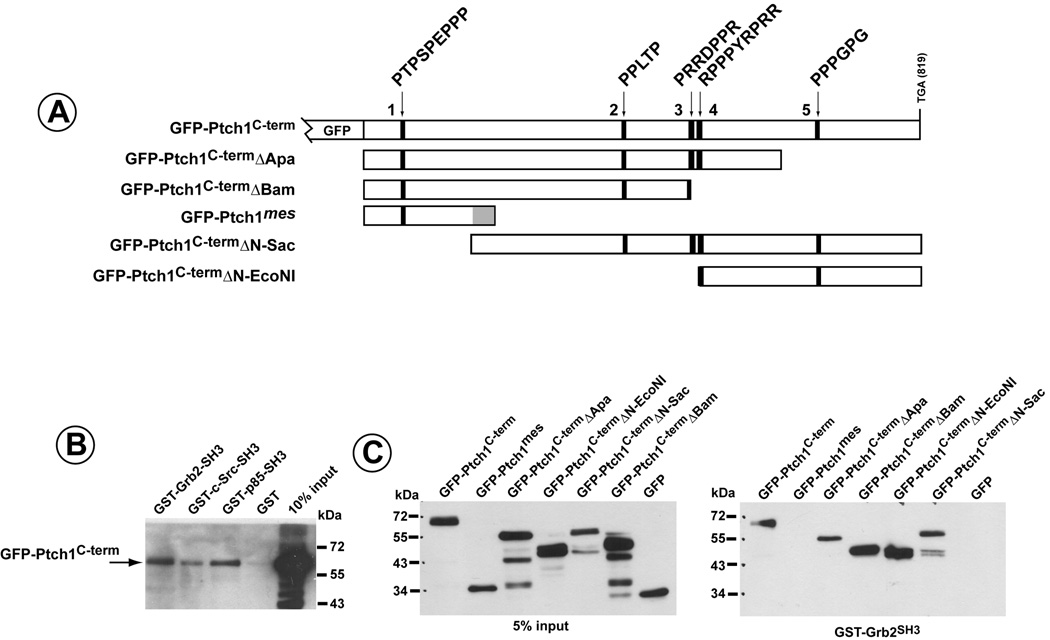
To more formally test whether Ptch1 might form complexes with SH3-containing proteins, a GFP-PtchC-term (amino acids 1163–1435) fusion protein was expressed in COS1 cells. Whole cell lysates were then added to glutathione-sepharose beads binding bacterially-expressed GST-fusion proteins encoding the SH3-domains of p85β, Grb2 and c-src. As Figure 3B illustrates, the cytoplasmic C-terminal domain of Ptch1 bound specifically to SH3-domains of Grb2, c-src and p85β but not to GST alone.
Figure 3. Association of the C-terminus of Ptch1 to the SH3-domains of Grb2, c-src and p85.
A) Schematic of GFP-fusion proteins containing the Ptch1 C-terminal region. Polyproline motifs are designated and numbered sequentially for reference. B) GFP-Ptch1C-term was expressed in HMECs and cell lysates added to GST-fusion proteins containing the SH3 domains of Grb2, c-src or p85β. GFP-Ptch1C-term could bind specifically to the SH3-domains of each of these signal transduction factors but not to GST alone. C) GFP-fusion proteins with full length Ptch1C-Term, Ptch1mes or deletions of the C-terminus were expressed in HMECs, added GST-Grb2SH3 and binding determined by western blot directed against GFP. The mes mutant of Ptch1 does not bind to the SH3-domain of Grb2. Binding of all other Ptch1 deletion mutants to Grb2SH3 reveals that Grb2SH3 binds to Ptch1 through more than one site but requires sequences deleted in the Ptch1mes mutant.
Further delineation of the regions in the C-terminus of Ptch1 required for binding to these SH3-domains is illustrated in Figure 3C. Using a series of deletion mutants of GFP-Ptch1C-term, as well as the C-terminus from Ptch1mes (see Figure 3A), binding to GST-Grb2 was determined. All of the GFP-Ptch1 C-term deletion mutants formed complexes with this SH3-domain while binding of the GFP-Ptch1 mes fusion protein could not be detected. Thus, Grb2 appeared to bind to the C-terminus of Ptch1 through at least two independent domains. However, binding required sequences downstream of the position of the altered/truncated mutant mes Ptch1 protein.
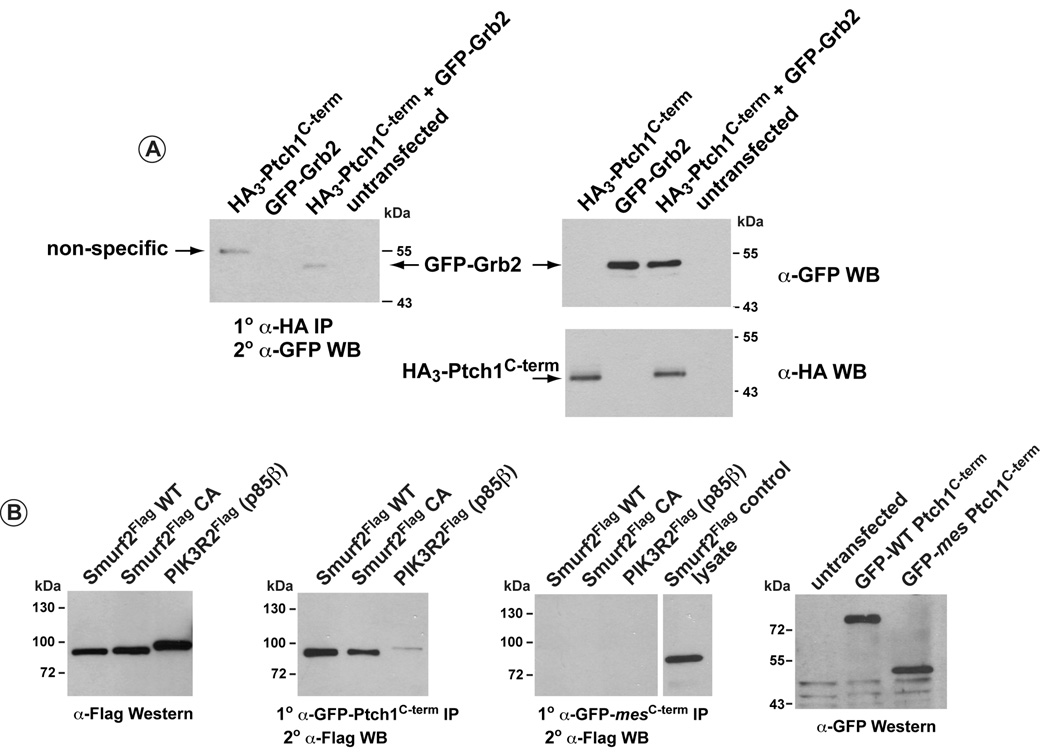
Binding of the C-terminus of Ptch1 to Grb2 was also determined in co-immunoprecipitation assays (Figure 4). GFP-tagged full length Grb2 was co-transfected with HA3-tagged Ptch1C-term. Figure 4A illustrates co-immunoprecipitation of GFP-Grb2, detected with an α-GFP antibody, following immunoprecipitation of HA3-Ptch1C-term with α-HA. Figure 4B illustrates further that flag-tagged p85β (PIK3R2) could be co-immunoprecipitated with GFP-tagged wild type Ptch1C-term but not the mes Ptch1C-term, consistent with the binding data in Figure 3B. The PPXY motif in the fourth polyproline region of the C-terminus (see Figure 2) has also been shown to mediate binding to WW-domains present in the HECT-family of E3 ubiquitin ligases (Ogunjimi et al., 2005, Wiesner et al., 2007, Qiu et al., 2000). We tested whether a candidate E3 ubiquitin ligase, Smurf2, or its catalytically-defective variant, Smurf2 CA (Ogunjimi et al., 2005), could associate with the C-terminus of Ptch1. As Figure 4B shows, Smurf2 was co-immunoprecipitated with the C-terminus of wild type but not mes Ptch1.
Figure 4. Co-immunoprecipitation of full length Grb2, p85β and Smurf2 with Ptch1C-term.
A) 293 cells were transfected with HA-tagged Ptch1C-term, GFP-tagged, full length Grb2 or both together. HA3-Ptch1C-term was then immunoprecipitated using and α-HA antibody and co-immunoprecipitated GFP-Grb2 detected by western analysis using an α-GFP antibody. Left panel shows that Grb2 is co-immunoprecipitated with the Ptch1C-term. Western blots in the right panels show the relative level of expression GFP-Grb2 (top) or HA3-Ptch1C-term (bottom).
B) 293 cells were transfected with flag-tagged Smurf2, Smurf2 CA or PIK3R2 (p85β) and GFP-Ptch1 C-term (second panel) or GFP-mes Ptch1C-term (third panel). Wild type and mes GFP-Ptch1C-term was immunoprecipitated with an α-GFP antibody. Co-immunprecipitated Smurf2, Smurf2 CA and p85β, detected by western analysis using an α-flag antibody. The second panel reveals that all three factors were co-immunprecipitated with wild type GFP-Ptch1C-term. No binding was evident for these same factors using the GFP-tagged Ptch1C-term from the mes mice (third panel). First and last panels, respectively, show similar levels of expression for flag-tagged Smurf2, Smurf2 CA and p85β or GFP-WT-Ptch1C-term and GFP-mes-Ptch1C-term.
Thus, a number of candidate proteins harbouring SH3-domains or WW-domains were able to bind to the C-terminal region of the Ptch1. However, binding of these same candidate factors to the C-terminal region of Ptch1 derived from the Ptch1mes was not apparent.
Shh stimulates activation of Erk1/2 independent Smo-mediated canonical Hh-signalling
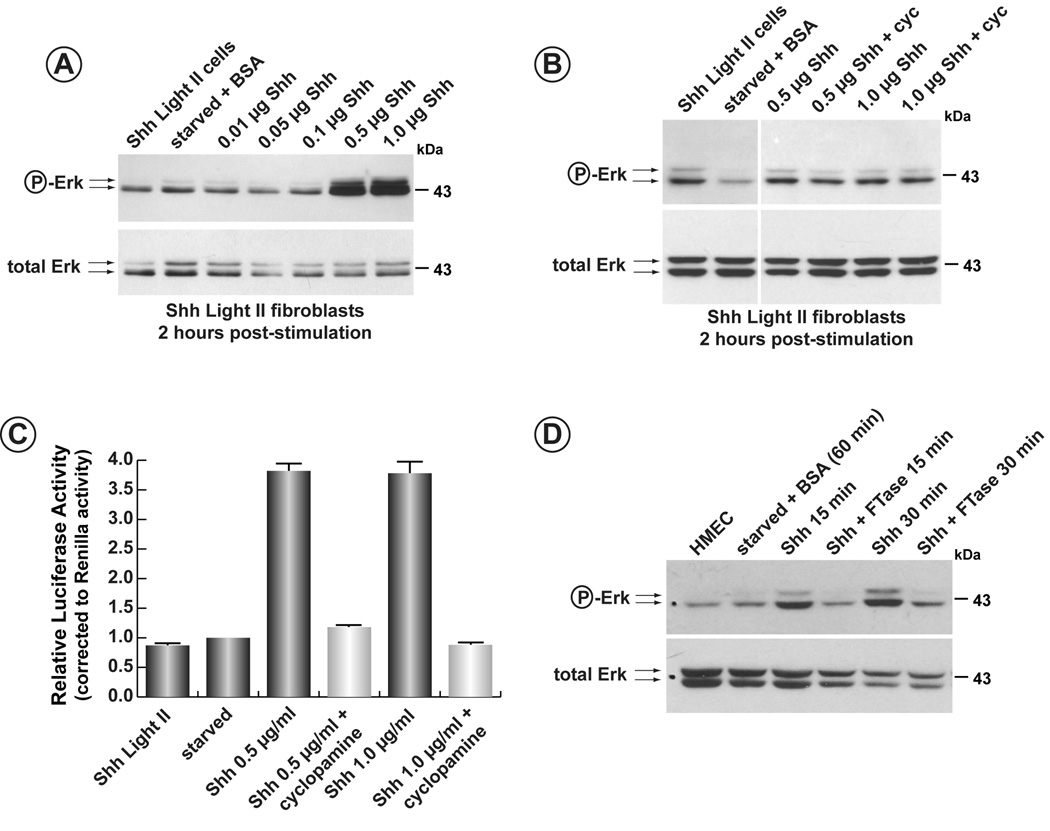
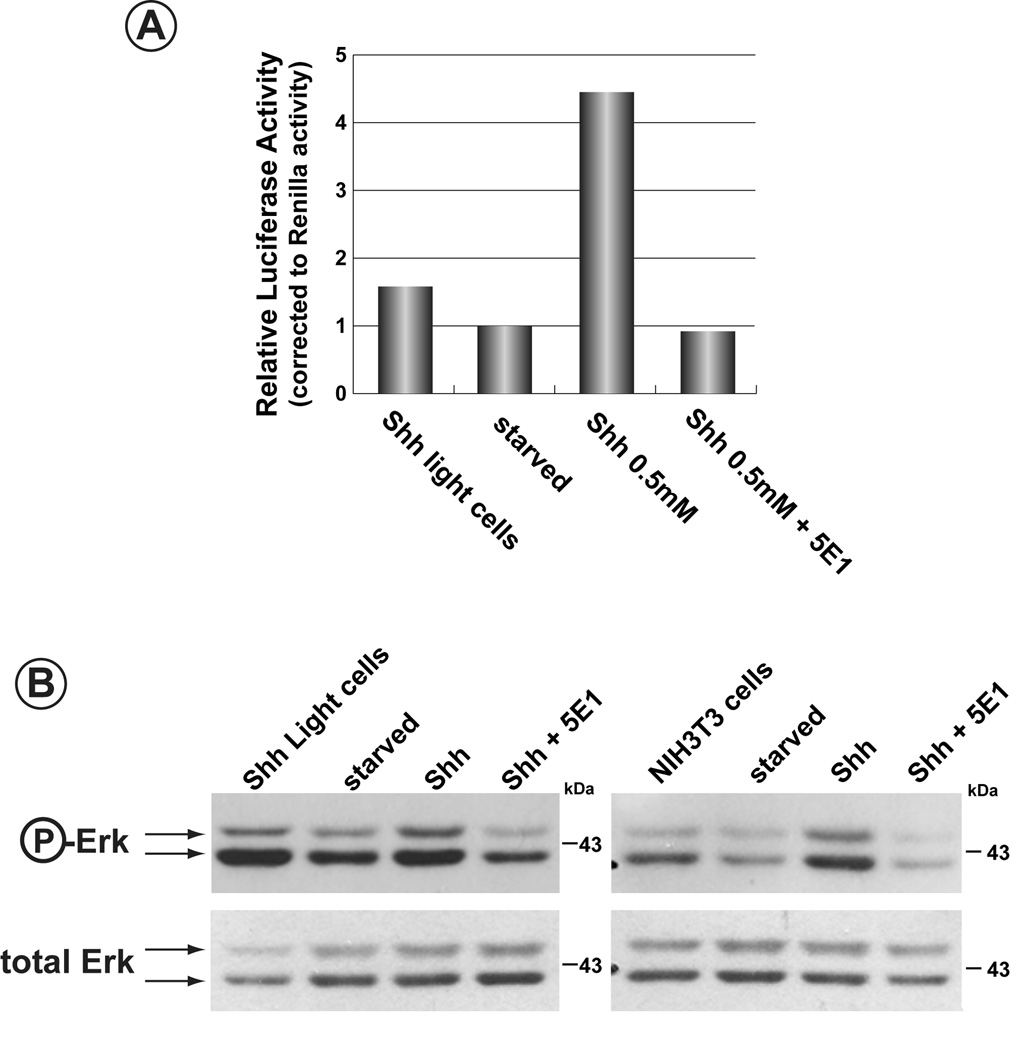
Shh Light II cells harbour a luciferase reporter transgene sensitive to stimulation by the Hh-ligands (Taipale et al., 2000). To determine if the Hh-ligands utilized signalling cascades employing SH3-containing components, serum starved Shh Light II cells were stimulated with N-Shh and assayed for phosphorylation of Erk1/2. As Figure 5A illustrates, addition of the N-Shh ligand at levels of 0.5 and 1.0 µg/ml caused a significant induction of phospho-Erk1/2 after two hours. The same concentrations of N-Shh also induced expression of luciferase from the Gli-luc reporter stably integrated in this cell line (data not shown). In order to determine that activation was independent of Smo activity, increasing concentrations of N-Shh were added to Shh Light II cells in the absence or presence of the Smo inhibitor, cyclopamine (Incardona et al., 1998, Incardona et al., 2000) (Figure 5B). Erk1/2 activation was still apparent in cyclopamine-treated cells despite the inhibition of the canonical Hh-signalling pathway, the latter verified by the inhibition of luciferase activity in these cells (Figure 5C).
Figure 5. Activation of Erk1/2 by Shh in HMECs.
A) Shh Light II cells were serum starved for 48 hours and increasing concentrations of N-Shh were added. Cells were then harvested after 2 hours and phospho-Erk1/2 detected by western blot (upper panel). The blot was then stripped and reprobed for total Erk1/2 (lower panel). B) Induction of phospho-Erk1/2 by N-Shh is repressed in the presence of FTase II. Serum-starved HMECs were induced with N-Shh in the presence or absence of the farnesyl transferase inhibitor, FTase II. Induction of phospho-Erk1/2 was blocked in the presence of this inhibitor. C) Induction of phospho-Erk1/2 is insensitive to the presence of cyclopamine. Serum-starved Shh Light II cells were stimulated in the presence or absence of the Smo- inhibitor, cyclopamine. Induction of phospho-Erk1/2 by N-Shh was unaffected by the presence of the inhibitor. D) Cyclopamine inhibits canonical Shh-signalling. The same cells used in panel C) were also assessed for induction of the Smo-mediated Hh-signalling pathway. Induction of luciferase from the Gli-luciferase reporter stably integrated in this line shows a 3.5-fold activation following addition of N-Shh. In the presence of cyclopamine, induction of luciferase is blocked. The block in the canonical Hh-pathway occurs despite induction of phospho-Erk1/2 under the same conditions.
Activation of Erk1/2 by N-Shh was also tested in human mammary epithelial cells (HMEC). As Supplementary Figure 1 illustrates, activation or Erk1/2 was detected within 2 minutes following activation by N-Shh and peaks at about 15 minutes. A second wave of stimulation occurs after 2 hours. Figure 5D also demonstrates the stimulation of phospho-Erk1/2 within 15 minutes following addition of N-Shh to serum-starved HMEC. To begin to determine the pathways leading to activation of Erk1/2, stimulation of Erk1/2 was performed in the presence of the farnesyl transferase inhibitor, FTase II. N-Shh-dependent activation of Erk1/2 was blocked in the presence of FTase II.
As Figure 6 illustrates, N-Shh-dependent activation of Erk1/2 was not due to contamination of the N-Shh preparation with factors that might signal through alternative pathways. Serum-starved Shh Light II cells or NIH3T3 fibroblasts were treated with N-Shh in the presence of the α-Shh antibody, 5E1. While Erk1/2 activation was seen for both cell types when N-Shh was added, mixing of N-Shh with the 5E1 antibody prior to addition to the cells blocked activation of Erk1/2 (Figure 6B). For Shh Light II, inhibition of Smo-mediated canonical signalling was also observed, as determined by the failure to stimulate expression of the Gli-luciferase reporter gene (Figure 6A). Thus, activation of Erk1/2 was dependent specifically on N-Shh and was not the consequence of a contaminant in the N-Shh preparation.
Figure 6. The α-Shh antibody, 5E1, blocks Shh-dependent activation of Erk1/2.
A) Specificity of N-Shh-activation of the canonical Hh-pathway was determined using an antibody, 5E1, against N-Shh. N-Shh stimulated lucferase expression from the Gli-luciferase reported gene in Shh Light II cells. This Shh-dependent induction was blocked in the presence of the 5E1 antibody. B) Induction of Erk1/2 is specific to N-Shh. In order to verify that induction of Erk1/2 by N-Shh was not due to contaminants in the N-Shh preparation, the α-Shh antibody, 5E1, was first added to N-Shh and then used to stimulate serun-starved Shh Light II cells or NIH3T3 fibroblasts. Induction of phospho-Erk1/2 by N-Shh was blocked in the presence of 5E1.
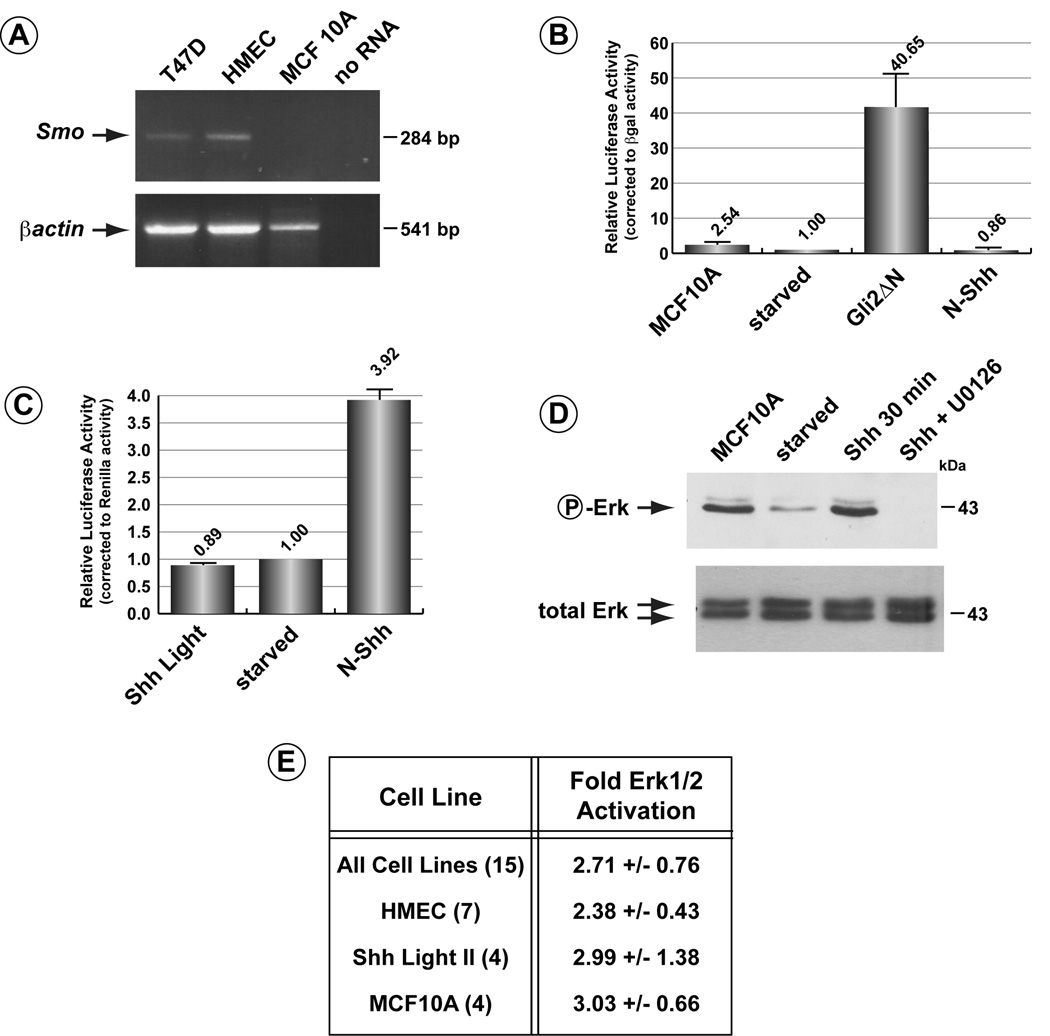
While inhibition of N-Shh-dependent activation of Erk1/2 in the presence of cyclopamine supports the notion that activation occurs through a Ptch1-mediated pathway that is independent of Smo activity, a more definitive test for this pathway was demonstrated using Smo-deficient cells. Like several other cell lines derived from mammary epithelial cells or breast cancers, the human mammary epithelial cell line, MCF10A, has been shown previously to express Ptch1, Gli2 and Gli3 but lacks Smo expression (Zhang et al., 2008). We verified by RT-PCR that MCF10A do not express detectable levels of Smo while Smo expression was detected in T47D cells, as also shown previously, as well as in HMEC (Figure 7A). The lack of Smo expression predicts that N-Shh signalling through the canonical Hh-signalling pathway would be absent. This prediction was tested in MCF10A cells transiently transfected with the Gli-luciferase reporter construct. As Figure 7B illustrates, addition of N-Shh to these cells did not cause any stimulation of the Hh-pathway reporter gene. The Gli-Luc reporter gene could be activated, however, by co-transfection with a vector expressing a constitutively active version of Gli2, Gli2ΔN. To ensure that the N-Shh used to stimulate MCF10A cells was active was determined by adding the same preparation to Shh Light II cells (Figure 7C); activation of the endogenous reporter gene was observed.
Figure 7. Shh activates Erk1/2 in the Smo-deficient cell line, MCF10A.
A) Lack of Smo expression in MCF10A cells. RT-PCR for Smo or βactin was performed on RNA isolated from the breast epithelial lines, T47D, HMEC and MCF10A. As shown previously, Smo is not expressed in MCF10A cells. B) Induction of phospho-Erk1/2 by N-Shh in MCF10A cells. Despite the lack of Smo, Erk1/2 is activated by N-Shh. The last lane shows further that induction of Erk1/2 is blocked in the presence of the MEK-inhibitor, U0126. C) Smo-deficient MCF10A cells transiently transfected with the Gli-luc reporter were stimulated with N-Shh. As expected, no activation through the canonical Hh-pathway was observed (Shh lane). Co-transfection of Gli-Luc with constitutively active Gli2ΔN did activate the reporter construct. D) That the N-Shh preparation that failed to activate the Hh-signalling pathway in MCF10A cells was active was determined by stimulating Shh Light II cells. E) Summary of levels of activation of Erk1/2 by N-Shh in all cells lines tested. Number of experiments for each cell line is denoted in brackets.
Given that MCF10A cells do not express Smo and that N-Shh does not stimulate the canonical Hh-signalling pathway, we tested next whether N-Shh in these cells could activate Erk1/2. N-Shh was added to serum starved MCF10A cells and changes in the levels of phospho-Erk1/2 determined. As Figure 7D demonstrates, stimulation of MCF10A cells with N-Shh resulted in a 3.03 +/− 0.66 fold increase in phospho-Erk1/2 levels. This level of activation was similar to that seen in HMEC and Shh Light II cells (Figure 7E). To more definitively assign the pathway induced by N-Shh leading to activation of Erk1/2, MCF10A cells were first treated with 10nM of the MEK1 inhibitor, U0126 (Duncia et al., 1998, Favata et al., 1998). As the last lane in the Figure 7D illustrates, no activation of Erk1/2 by N-Shh was evident in the presence of this MEK1 inhibitor.
Taken together, these observations reveal an apparent bifurcation of the Hh-signalling pathway at the level of Ptch1. Specifically, these data define a novel signalling cascade operating through SH3-domain-encoding factors directly stimulated by the Hh-ligands. This cascade can be stimulated by the Hh-ligands in the absence of Smo or in the presence of the small chemical inhibitors of the Smo-mediated Hh-signalling.
DISCUSSION
The requirement of the Hedgehog signalling pathway directing cell fate and morphogenesis in a large number of tissues across phyla has been well characterized. Both genetic and molecular studies have defined a relatively well conserved molecular pathway that transmits signalling upon stimulation by the Hh-ligands. Central to this signal transduction pathway is the receptor for the Hh-ligands, Ptch1, whose activity is directed toward the regulation of the activity of a 7-pass, integral membrane protein, Smo. The mechanism for this regulation has not been clearly delineated nor has a physical interaction between Ptch1 and Smo been demonstrated. Regardless, in the absence of Hh-signalling, Ptch1 acts to inhibit Smo activity, facilitating the conversion of the transcriptional effectors of the Hh pathway, Gli2/Gli3, to their repressor forms.
Additional signal transduction pathways also interact with the Hh-signalling pathway, although to date, these have typically been described in relation to changes in Smo activity . For example, evidence exists suggesting that stimulation of both heterotrimeric and small G-proteins specifically through activation of Smo activity may occur in at least some cellular contexts (Kasai et al., 2004, Riobo et al., 2006d, Oro, 2007). Likewise, potentiation of the activation of the Gli proteins may include activation of AKT via PI3k, although this may arise through pathways independent of the canonical Hh-signalling pathway (Riobo et al., 2006b). Another pathway more specifically relevant to the data presented in this paper involves MEK-1. Activation of MEK-1 was shown to synergise with the canonical Hh pathway resulting in significant enhancement of Gli-dependent transcriptional activation, the latter determined using a fibroblast cell line harboring a Gli-responsive luciferase-reporter construct (Riobo et al., 2006a). This activation was suggested to be dependent on the PKCδ activity, as determined by rottlerin-dependent repression of Hh-mediated activation of the Gli-luciferase reporter; repression of this activation was as effective as the Smo-specific inhibitor, cyclopamine. It is further interesting to note that, as was recently summarised (Riobo and Manning, 2007), these pathways may have been added to the Hh-signalling cascade evolutionarily more recently since Hh-signalling in Drosophila does not appear to invoke or depend on signalling through these mechanisms (Riobo et al., 2006c).
The observation that Ptch1 can stimulate Erk1/2 phosphorylation upon stimulation by Shh in the absence of Smo or in the presence of the Smo inhibitor, cyclopamine, suggests that the Hh-signalling pathway may have acquired additional characteristics in vertebrates where direct regulation of signalling through pathways leading to Erk1/2 activation arises. Inspection of the sequence of the C-terminus of Ptch1 from xenopus, chickens, rodents and humans reveals a highly conserved, Pro-rich sequence predicted to bind to factors harbouring SH3- and WW-domains. Additional Pro-rich motifs in this region are also conserved among mammals. While significant Pro-rich sequences exist in the Cterminal domain of Drosophila Ptch, they do not encode for the motifs observed in vertebrates, in general, and mammals in particular. The ability of these motifs in Drosophila to facilitate binding to SH3-containing factors remains to be determined.
It is clear, however, that sequences in C-terminus of murine Ptch1 facilitate association of this region to factors inhabiting EEA-1-containing vesicles. Truncations encoded by the Ptch1mes allele prevented the GFP-PtchC-term-fusion protein from localizing to these structures. Importantly, when full length Ptch1 protein, with either the wild type or Ptch1mes C-termini were expressed, both proteins localised to vesicles in HMEC's. This localization for full length Ptch1mes is consistent with the observation that in mice homozygous for the Ptch1mes allele, Hh-signalling through the canonical pathway appears to be intact in skin (Nieuwenhuis et al., 2007) and the mammary gland (H. Chang; unpublished observation). We propose that the Ptch1mes allele results in segregation at the level of Ptch1 of at least two pathways regulated by the Hh-ligands; A) Ptch1 regulates the activity of Smo in a Hh-dependent manner, although the mechanisms here remains undetermined, B) Ptch1 regulates a signalling cascade independent of Smo and that is sensitive to farnesyl transferase inhibitors and the MEK-specific inhibitor, U0126. Evidence for another Ptch1-dependent pathway regulating cell cycle progression has also been reported (Barnes et al., 2001). Based on our binding data, we suggest further that Shh-induced activation of Erk1/2 may employ a cascade of signalling factors, including MEK, that depend on SH3-containing factors binding to the C-terminus of Ptch1. It will be important to determine if differences in activation of Erk1/2 arise in cells derived from Ptch1mes/mes mice. Attempts to assay these changes were unsuccessful since serum starvation of primary mammary epithelial cells or mammary fibroblasts resulted in cell death precluding determination of changes in Erk1/2 activation upon stimulation with N-Shh (data not shown).
Our previous molecular-genetic analysis of the mes mouse revealed a mammary gland-specific role for Hh-signalling (Moraes et al., 2009). Similarly, alteration in morphogenesis of another skin appendage, the hair follicle, arises in mes mice (Nieuwenhuis et al., 2007). However, despite changes in the morphogenesis of hair follicles in the postnatal animal, expression of transcriptional targets of the canonical Hh-signalling pathway, Ptch1 and Gli1, appeared unchanged relative to wild type animals (Nieuwenhuis et al., 2007). Likewise, we also failed to detect changes in the levels of Ptch1 in epithelial cells or stromal fibroblasts isolated from the Ptch1mes mammary gland (H. Chang; unpublished observation), suggesting that canonical Hh-signalling is not significantly altered in this tissue. In contrast to these skin appendages, a reduction in the mass of white fat as well as in the size of adipose cells (non-mammary gland) was shown to be associated with a very strong increase in Ptch1 and Gli1 levels in adipocytes in Ptch1mes mice (Li et al., 2008). Furthermore, the polydactyly exhibited by mice homozygous for the mes allele of Ptch1 is consistent with altered activity of the canonical Hh-signalling pathway. Taken together, these observations suggest that altered Hh-signalling due to the truncation of the C-terminus of Ptch1 may have different effects on the growth and/or development of distinct tissues. We propose, however, that these differential effects for the Hh-pathway may arise through utilization of distinct signalling cascades working through the Ptch1 receptor upon stimulation by the Hh-ligands and that these include signalling through a ras/MEK-mediated signal transduction cascades that activates Erk1/2.
Supplementary Material
Acknowledgments
The authors which to acknowledge the thoughtful suggestions from Dr. C. Pratt and Dr. J. Hanley-Hyde. This work was supported through a grant to PAH the Canadian Institutes of Health Research (MOP-97929), and by a grant to MTL from the United States of America National Institutes of Health (P01 CA30195).
Abbreviations Used
- Shh
sonic hedgehog
- Smo
smoothened
- Ptch1
Patched-1
- Hh
Hedgehog
- HMEC
human mammary epithelial cells
- FTase II
H-Cys-4-Abz-Met-OH
- GST
glutathione S transferase
- GFP
green fluorescent protein
- MEK
mitogen-activated protein kinase kinase
- Erk
extracellular signal-regulated kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- Alexandre C, Jacinto A, Ingham PW. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Barnes EA, Kong M, Ollendorff V, Donoghue DJ. Patched1 interacts with cyclin B1 to regulate cell cycle progression. Embo J. 2001;20:2214–2223. doi: 10.1093/emboj/20.9.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki AG, Mendham L, Emerson CP., Jr Control of somite patterning by Sonic hedgehog and its downstream signal response genes. Development. 1998;125:777–790. doi: 10.1242/dev.125.4.777. [DOI] [PubMed] [Google Scholar]
- Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Lee J, Robins P, Heller P, Ruiz i Altaba A. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature. 1997;389:876–881. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui CC. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- Duncia JV, Santella JB, 3rd, Higley CA, Pitts WJ, Wityak J, Frietze WE, Rankin FW, Sun JH, Earl RA, Tabaka AC, Teleha CA, Blom KF, Favata MF, Manos EJ, Daulerio AJ, Stradley DA, Horiuchi K, Copeland RA, Scherle PA, Trzaskos JM, Magolda RL, Trainor GL, Wexler RR, Hobbs FW, Olson RE. MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg Med Chem Lett. 1998;8:2839–2844. doi: 10.1016/s0960-894x(98)00522-8. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Edwards KM, Munger K. Depletion of physiological levels of the human TID1 protein renders cancer cell lines resistant to apoptosis mediated by multiple exogenous stimuli. Oncogene. 2004;23:8419–8431. doi: 10.1038/sj.onc.1207732. [DOI] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Feng S, Kasahara C, Rickles RJ, Schreiber SL. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc Natl Acad Sci U S A. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes AJ, Lin H, Ingham PW, Spradling AC. hedgehog is required for the proliferation and specification of ovarian somatic cells prior to egg chamber formation in Drosophila. Development. 1996;122:1125–1135. doi: 10.1242/dev.122.4.1125. [DOI] [PubMed] [Google Scholar]
- Forbes AJ, Nakano Y, Taylor AM, Ingham PW. Genetic analysis of hedgehog signalling in the Drosophila embryo. Dev Suppl. 1993:115–124. [PubMed] [Google Scholar]
- Fu M, Lui VC, Sham MH, Pachnis V, Tam PK. Sonic hedgehog regulates the proliferation, differentiation, and migration of enteric neural crest cells in gut. J Cell Biol. 2004;166:673–684. doi: 10.1083/jcb.200401077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepker J, Wang QT, Motzny CK, Holmgren R, Orenic TV. Drosophila cubitus interruptus forms a negative feedback loop with patched and regulates expression of Hedgehog target genes. Development. 1997;124:549. doi: 10.1242/dev.124.2.549. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Gaffield W, Kapur RP, Roelink H. The teratogenic Veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction. Development. 1998;125:3553–3562. doi: 10.1242/dev.125.18.3553. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Gaffield W, Lange Y, Cooney A, Pentchev PG, Liu S, Watson JA, Kapur RP, Roelink H. Cyclopamine inhibition of Sonic hedgehog signal transduction is not mediated through effects on cholesterol transport. Dev Biol. 2000;224:440–452. doi: 10.1006/dbio.2000.9775. [DOI] [PubMed] [Google Scholar]
- Incardona JP, Gruenberg J, Roelink H. Sonic hedgehog induces the segregation of patched and smoothened in endosomes. Curr Biol. 2002;12:983–995. doi: 10.1016/s0960-9822(02)00895-3. [DOI] [PubMed] [Google Scholar]
- Jager M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, Kelly JW. Structure-function-folding relationship in a WW domain. Proc Natl Acad Sci U S A. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai K, Takahashi M, Osumi N, Sinnarajah S, Takeo T, Ikeda H, Kehrl JH, Itoh G, Arnheiter H. The G12 family of heterotrimeric G proteins and Rho GTPase mediate Sonic hedgehog signalling. Genes Cells. 2004;9:49–58. doi: 10.1111/j.1356-9597.2004.00701.x. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Rowitch DH. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol Cell Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Chao TH, Xiang R, Lo JF, Campbell MJ, Fearns C, Lee JD. Tid1, the human homologue of a Drosophila tumor suppressor, reduces the malignant activity of ErbB-2 in carcinoma cells. Cancer Res. 2004;64:7732–7739. doi: 10.1158/0008-5472.CAN-04-1323. [DOI] [PubMed] [Google Scholar]
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Lee J, Platt KA, Censullo P, Ruiz i Altaba A. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang H, Denhard LA, Liu LH, Zhou H, Lan ZJ. Reduced white fat mass in adult mice bearing a truncated Patched 1. Int J Biol Sci. 2008;4:29–36. doi: 10.7150/ijbs.4.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, Brown A, Ellison J, Ohno K, Rosenfeld MA, Tagle DA, Pentchev PG, Pavan WJ. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- Makino S, Masuya H, Ishijima J, Yada Y, Shiroishi T. A spontaneous mouse mutation, mesenchymal dysplasia (mes), is caused by a deletion of the most C-terminal cytoplasmic domain of patched (ptc) Dev Biol. 2001;239:95–106. doi: 10.1006/dbio.2001.0419. [DOI] [PubMed] [Google Scholar]
- Marigo V, Johnson RL, Vortkamp A, Tabin CJ. Sonic hedgehog differentially regulates expression of GLI and GLI3 during limb development. Dev Biol. 1996;180:273–283. doi: 10.1006/dbio.1996.0300. [DOI] [PubMed] [Google Scholar]
- Martin V, Carrillo G, Torroja C, Guerrero I. The sterol-sensing domain of Patched protein seems to control Smoothened activity through Patched vesicular trafficking. Curr Biol. 2001;11:601–607. doi: 10.1016/s0960-9822(01)00178-6. [DOI] [PubMed] [Google Scholar]
- Mill P, Mo R, Hu MC, Dagnino L, Rosenblum ND, Hui CC. Shh controls epithelial proliferation via independent pathways that converge on N-Myc. Dev Cell. 2005;9:293–303. doi: 10.1016/j.devcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, Joyner AL, Hui C. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- Monnier V, Dussillol F, Alves G, Lamour-Isnard C, Plessis A. Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway. Curr Biol. 1998;8:583–586. doi: 10.1016/s0960-9822(98)70227-1. [DOI] [PubMed] [Google Scholar]
- Moraes RC, Chang H, Harrington N, Landua JD, Prigge JT, Lane TF, Wainwright BJ, Hamel PA, Lewis MT. Ptch1 is required locally for mammary gland morphogenesis and systemically for ductal elongation. Development. 2009;136:1423–1432. doi: 10.1242/dev.023994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis E, Barnfield PC, Makino S, Hui CC. Epidermal hyperplasia and expansion of the interfollicular stem cell compartment in mutant mice with a C-terminal truncation of Patched1. Dev Biol. 2007;308:547–560. doi: 10.1016/j.ydbio.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, Wrana JL. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- Orenic T, Chidsey J, Holmgren R. Cell and cubitus interruptus dominant: two segment polarity genes on the fourth chromosome in Drosophila. Dev Biol. 1987;124:50–56. doi: 10.1016/0012-1606(87)90458-1. [DOI] [PubMed] [Google Scholar]
- Orenic TV, Slusarski DC, Kroll KL, Holmgren RA. Cloning and characterization of the segment polarity gene cubitus interruptus Dominant of Drosophila. Genes Dev. 1990;4:1053–1067. doi: 10.1101/gad.4.6.1053. [DOI] [PubMed] [Google Scholar]
- Oro AE. The primary cilia, a 'Rab-id' transit system for hedgehog signaling. Curr Opin Cell Biol. 2007;19:691–696. doi: 10.1016/j.ceb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A, Therond P, Alves G, Tournier FB, Busson D, Lamour-Isnard C, Bouchon BL, Preat T, Tricoire H. The Suppressor of fused gene encodes a novel PEST protein involved in Drosophila segment polarity establishment. Genetics. 1995;140:587–598. doi: 10.1093/genetics/140.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, Fang D, Hunter T, Liu YC. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- Rickles RJ, Botfield MC, Zhou XM, Henry PA, Brugge JS, Zoller MJ. Phage display selection of ligand residues important for Src homology 3 domain binding specificity. Proc Natl Acad Sci U S A. 1995;92:10909–10913. doi: 10.1073/pnas.92.24.10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riobo NA, Haines GM, Emerson CP., Jr Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 2006a;66:839–845. doi: 10.1158/0008-5472.CAN-05-2539. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2006b;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riobo NA, Lu K, Emerson CP., Jr Hedgehog signal transduction: signal integration and cross talk in development and cancer. Cell Cycle. 2006c;5:1612–1615. doi: 10.4161/cc.5.15.3130. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Manning DR. Pathways of signal transduction employed by vertebrate Hedgehogs. Biochem J. 2007;403:369–379. doi: 10.1042/BJ20061723. [DOI] [PubMed] [Google Scholar]
- Riobo NA, Saucy B, Dilizio C, Manning DR. Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci U S A. 2006d;103:12607–12612. doi: 10.1073/pnas.0600880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- Ruppert JM, Kinzler KW, Wong AJ, Bigner SH, Kao FT, Law ML, Seuanez HN, O'Brien SJ, Vogelstein B. The GLI-Kruppel family of human genes. Mol Cell Biol. 1988;8:3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert JM, Vogelstein B, Arheden K, Kinzler KW. GLI3 encodes a 190-kilodalton protein with multiple regions of GLI similarity. Mol Cell Biol. 1990;10:5408–5415. doi: 10.1128/mcb.10.10.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module--the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- Sweet HO, Bronson RT, Donahue LR, Davisson MT. Mesenchymal dysplasia: a recessive mutation on chromosome 13 of the mouse. J Hered. 1996;87:87–95. doi: 10.1093/oxfordjournals.jhered.a022981. [DOI] [PubMed] [Google Scholar]
- Syken J, De-Medina T, Munger K. TID1, a human homolog of the Drosophila tumor suppressor l(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc Natl Acad Sci U S A. 1999;96:8499–8504. doi: 10.1073/pnas.96.15.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Tarunina M, Alger L, Chu G, Munger K, Gudkov A, Jat PS. Functional genetic screen for genes involved in senescence: role of Tid1, a homologue of the Drosophila tumor suppressor l(2)tid, in senescence and cell survival. Mol Cell Biol. 2004;24:10792–10801. doi: 10.1128/MCB.24.24.10792-10801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therond PP, Knight JD, Kornberg TB, Bishop JM. Phosphorylation of the fused protein kinase in response to signaling from hedgehog. Proc Natl Acad Sci U S A. 1996;93:4224–4228. doi: 10.1073/pnas.93.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiniakow GG, Terentieva EL. Cubitus Interruptus, a New Genovariation of the Fourth Chromosome of DROSOPHILA MELANOGASTER. Genetics. 1933;18:117–120. doi: 10.1093/genetics/18.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torregroza I, Evans T. Tid1 is a Smad-binding protein that can modulate Smad7 activity in developing embryos. Biochem J. 2006;393:311–320. doi: 10.1042/BJ20050785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentin GA, Yin X, Tahir S, Lhotak S, Farhang-Fallah J, Li Y, Rozakis-Adcock M. A mouse homologue of the Drosophila tumor suppressor l(2)tid gene defines a novel Ras GTPase-activating protein (RasGAP)-binding protein. J Biol Chem. 2001;276:13087–13095. doi: 10.1074/jbc.M009267200. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Mao JH, Brown K, Girardi M, Balmain A. Promotion of Hras-induced squamous carcinomas by a polymorphic variant of the Patched gene in FVB mice. Nature. 2007;445:761–765. doi: 10.1038/nature05489. [DOI] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Zaal K, Hailey D, Presley J, Lippincott-Schwartz J, Samelson LE. Role of Grb2 in EGF-stimulated EGFR internalization. J Cell Sci. 2002;115:1791–1802. doi: 10.1242/jcs.115.9.1791. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- Zhang X, Harrington N, Moraes RC, Wu MF, Hilsenbeck SG, Lewis MT. Cyclopamine inhibition of human breast cancer cell growth independent of Smoothened (Smo) Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.