Abstract
Objectives and Methods
Alternative splicing provides proteomic diversity that can have profound effects. The extent, pattern, and roles of alternative splicing in pancreatic cancer have not been systematically investigated. We have utilized a spliceoform-specific microarray and polymerase chain reaction to evaluate all known splice variants in human pancreatic cancer cell lines representing a spectrum of differentiation, from near-normal HPDE6 to Capan-1 and poorly differentiated MiaPaCa2 cells. Validation of altered spliceoforms was verified in primary cancer specimens and normal pancreatic ductal cells. Additionally, expression of 92 spliceosomal genes were examined to better understand the mechanism for observed differences in mRNA splicing.
Results
A statistically significant reduction in alternative splicing was found in the pancreatic cancer cell lines compared to HPDE6 cells. Many splice variants identified in Capan-1 and MiaPaCa2 cells were observed in Grade 3 and Grade 4 tumors. Analysis of genes encoding spliceosomal proteins revealed that 28 of 92 genes had significantly decreased expression in cancer compared to normal pancreas.
Conclusion
Pancreatic cancer has reduced alternative splicing diversity compared to normal pancreas. This is demonstrated in both cell lines and primary tumors, with the loss in splicing diversity correlated with relative reduction in expression of spliceosomal genes.
Keywords: Alternative splicing, microarray, pancreatic cancer
Introduction
Although there have been major advances in recent years in our knowledge of molecular events occurring in pancreatic cancer, the prognosis for patients diagnosed with this devastating disease has not been substantially improved. This relates to the typical late stage of presentation and diagnosis of pancreatic cancer, after local invasion and/or distant metastasis have occurred. Splicing of pre-mRNA to generate multiple transcripts from a common precursor may provide a new, potentially rewarding area of investigation, one with implications for improved diagnostics, prognostics, and even the development of new therapeutic agents. Approximately 95% of the multi-exon genes in the human genome exhibit alternative splicing and in most cases the resulting transcripts are expressed in different cell and tissues types1,2, thus the presence of splice variants may serve as realistic candidates for disease biomarkers that could be monitored by simple clinical assays.
Alternative splicing represents an important mechanism for the expression of structurally-and functionally-distinct proteins3. The effects of alternative splicing may result in a spectrum of results from complete loss of function, to subtle or difficult-to-detect effects, or possibly to altering the location, stability or translation of a transcript4, 5. Many alternative splicing events modify the coding sequence, while others can result in premature termination or nonsense-mediated decay6. It is now understood that alternative splicing is a highly coordinated process that can be abnormally regulated in human diseases7. The expression of alternative or even tumor-specific splice variants has been shown to significantly affect cancer biology such as cell proliferation, cell motility, apoptosis, cell cycle control, and even drug responsiveness8. Each of the “hallmarks of cancer”9 can be affected or sustained by at least a few recognized cancer-associated, functional splice variants.
Several therapeutic drug candidates currently undergoing preclinical evaluation for pancreatic cancer are known to target genes that have multiple isoforms. Among these are mitogen-activated protein kinase kinase (MEK1/2)10, aurora kinases11, TGFβ, VEGF12 and EGFR13. Without taking into consideration the presence of these variant spliceoforms and the regions of the genes targeted by the drugs, the biological effects could easily vary from those predicted, and even become deleterious14,15. Understanding the presence of specific splice variants may, therefore, provide an important complement to the effective clinical management of patients with cancer.
In this report, we have characterized patterns of expression of alternative spliceoforms and components of the splicing machinery in pancreatic cancer cells in culture, and then validated the presence of these unique splice variants in primary tumors.
Experimental Procedures
Pancreatic cancer cell lines and tissue
The human pancreatic cancer cell lines, Capan-1 and MiaPaCa2, were obtained from the American Type Culture Collection (ATCC) and were grown according to recommendations. The morphology and growth characteristics of these cell lines were stable and consistent. The non-transformed human pancreatic ductal cell line, HPDE6, was generously provided by Dr. M.S. Tsao (Ontario Cancer Institute, Canada) and was cultured as previously described16, 17. All cell lines were tested and found to be negative for pathogen contamination.
Specimens representing normal pancreas and pancreatic cancer were acquired after appropriate patient consent and approval of the Mayo Clinic Institutional Review Board. Specimens were coded and anonymized.
RNA Isolation
Cells were harvested with cell dissociation solution and 107 cells/ml were lysed in Trizol (Invitrogen). RNA was purified from each Trizol extract using the Qiagen's RNEasy Kit following manufacturer's protocol including the optional on-column DNA digestion step. (Qiagen, Valencia, CA.). Integrity and concentration of RNA samples were established by the Agilent 2100 Bioanalyzer and the Nanodrop ND-1000 spectrophotometer measurements (Nanodrop, Wilmington, DE).
Genome-wide splice arrays
A human genome-wide splicing microarray was designed from a reference transcriptome database, SEHS1, created using Build 35 of the NCBI genome assembly and by aligning multiple databases (RefSeq, mRNAs, ESTs and Alternative Splice Database (ASD))10 The 148,693 oligonucleotides on the microarray included probes for alternatively spliced exon-exon and exon-intron junctions as well as probes for single-exon genes. With this diverse junctional probe set, all splice variants annotated in Build 35 of the NCBI genome could be effectively evaluated. This includes splice variants that have exon skips, exon trims or extensions, alternative first and last exons, and intron retentions or intron splice events. The probes had an average length of 38 nucleotides. Variability in melting temperatures was optimized by varying probe lengths between 34 and 42 nucleotides and isothermally balancing junctional probes across the splice sites. Sense strand oligonucleotides with a 10-base polyT linker at the 32 end were synthesized by NimbleGen Systems using photodeposition chemistry and digital light projection. We included multiple probes per gene in order to target exon junctions unique to alternative splice products and to differentiate these from what is believed to represent the normal gene product from splice variants.
RNA labeling and Hybridization for Microarray Analysis
Aliquots of total RNA were used to produce cDNA using the Invitrogen Superscript™ Double Stranded cDNA Synthesis Kit (Invitrogen, Carlsbad, CA.). Briefly, direct one color labeling was performed by combining 40 μl of Cy3 labeled random primers with 1 ug of cDNA in 80 μl of water, heated to 98 °C for 5 min, and chilled in an ice water bath. An aliquot of 10 ul of 50× dNTP mix was added to each tube followed by 8 μl of water and 2 μl of high concentration Klenow fragment (40 U/μl). Reactions were incubated at 37 °C for 2 h, precipitated with isopropanol, and evaporated to dryness using a SpeedVap on low heat for 5 min. Samples were resuspended in 20 μl water to determine dye incorporation and concentration by spectrophotometry. For hybridization, 13 μg of Cy3-labeled sample was added to 40 μl of 2× Nimblegen hybridization buffer, and heated at 95 °C for 5 min. Samples were loaded onto the microarrays through the MAUI Mixer sample ports and hybridized at 42 °C for 16–20 hr in a MAUI Hybridization System (Biomicro, Salt Lake City, Utah). The arrays were washed three times, dried in an Array-Go Round, and scanned with an Axon scanner (Molecular Devices, Sunnyvale, CA).
Microarray Data Analysis
Scanned tab-delimited data files were analyzed in SpliceFold™, a microarray analysis package that implements several algorithms for measuring alternative splicing and that has been adapted to address exon skips and inclusions, alternative first and last exons, introns spliced and retained, and alternative acceptor and donor sites10. These arrays were single-color; thus median normalization was done on every array in order to balance any bias in dye incorporation or scanner efficiency. The methods include ASPIRE18, Splicing Index19 and Splice Ratio10. We obtained 24 measurements per splice event, including replicate experiments and replicate spots to obtain statistical significance in the filtering criteria for minimal signal, direction and magnitude of change. With a large number of measurements, and with replicates, the microarray data exhibited high statistical significance. For each sample, the data included two types of technical replicates: duplicate spots on the microarrays as well as experimental replicates hybridized on different dates, leading to four-fold redundancy in measurement. To count a change in transcription or splicing as sample-specific, we required that the same change occur in all four technical replicates as compared to the other samples. As a control, we compared the pair of experimental replicates for each sample to determine the variability in transcription and splicing that might have occurred by chance. Lastly, all raw data files have been uploaded and made publically available in GEO, accession number GPL4742.
PCR (Conventional and Quantitative)
High quality purified RNA was used to generate cDNA by the iScript cDNA synthesis kit (Biorad, Hercules, CA.) which combines random hexamers and oligo-dT primers. Approximately 25 ng of cDNA template was used for either standard RT-PCR or 10 ng for quantitative PCR reactions. Standard RT-PCR conditions generally ranging from 28–30 cycles dependent upon the transcript examined. A custom Taqman low density array (TLDA, Applied Biosystems) containing Taqman primers and probes for the commercially available 92 genes that have been identified within the spliceosome (Jurica et. al. and Zhou et al.) was designed and run following standard TLDA specifications. Normalization used three different housekeeping genes in order to account for gene-to-gene differences within the tissue samples: GAPDH, Beta Actin, and HNRPA2B1 ribosomal protein. Differences in the Ct cycles for these housekeeping genes between normal tissue and grades 3 and 4 pancreatic cancer specimens were within the precision of the TaqMan reader. The relative quantification of the gene expression changes were analyzed according to ΔΔCt method20 using the ABI's SDS RQ Manager 1.2 software with additional manual curations. These data were then subject to a one-way Model 1 ANOVA with FWER false-discovery correction to obtain a list of spliceoform differences that were significant.
Results
Genome-wide screen of alternative splicing in pancreatic cancer
Using alternative splicing microarrays, levels of expression of transcripts were measured for human pancreatic cancer cell lines reflecting different stages of differentiation. The pancreatic cancer cell lines consisted of the non-transformed and near-normal human pancreatic ductular epithelial cell line, HPDE6, the well-differentiated pancreatic cancer cell line, Capan-1, and the poorly differentiated pancreatic cancer cell line, MiaPaCa2. Comparison of data from HPDE6 versus Capan-1 and MiaPaCa2 cells, provided a near-normal/tumor comparison; while comparison of data from MiaPaCa2 versus Capan-1 cells suggested changes that may be unique to high-grade tumors. Raw microarray data were analyzed in SpliceFold™, a software package specifically designed for the analysis of alternative splicing, and showed a clear trend in both gene expression and regulation of RNA splicing. Of note, human pancreatic cell lines were used in all microarray experiments due to the large amount and high quality of cDNA required to conduct multiple replicate experiments.
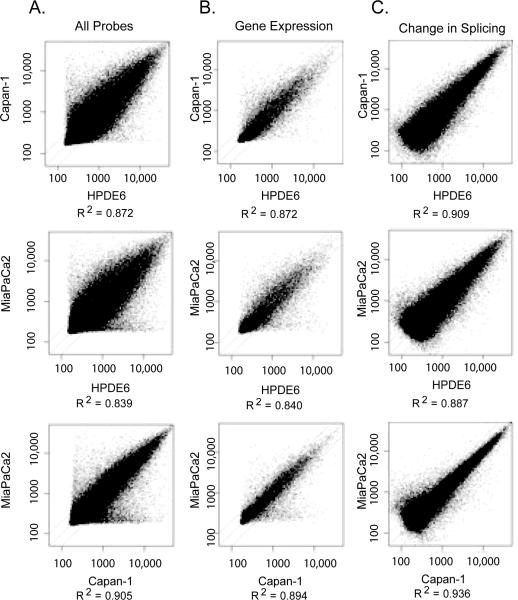
Scatter plots comparing data from near-normal HPDE6 cells versus poorly differentiated MiaPaCa2 cells, and moderately differentiated Capan1 cells vs. poorly differentiated MiaPaCa2 cells showed `wing' effects along the x and y axes. These data points reflected genes and splice events with apparent binary behavior, either being turned on or off in one sample compared to the other (Figure 1a). The intensity of each probe on the microarray reflects both gene expression and splicing regulation. In the analysis, levels of gene expression were normalized in order to evaluate the splicing variation. Using this analysis, regulated splicing accounted for up to 58.7% of the variance in probe expression measurements in HPDE6 cells compared to Capan-1 cells, up to 59.1% of the variance compared to MiaPaCa2 cells (Figure 1A), and up to 60.7% in tumor progression from Capan-1 cells to MiaPaCa2 cells. The remaining differences resulted from changes in gene transcription (Figure 1B). The data from the two pancreatic cancer cell lines correlated more closely with each other than with the near-normal cell line (bottom row Figure 1 compared to the upper rows of Figure 1). (Figure 1C).
Figure 1. Variation in RNA transcripts found in pancreatic cancer cell lines reflecting varied degrees of differentiation.
A) Shown are expression levels for all of the probes found on the splicing array, with signal intensities plotted along X- and Y-axes. The `wings' along the axes correspond to probes specific for genes and spliceoforms that were turned on or off in nearly a binary fashion. B) Shown are relative levels of gene expression observed in pancreatic cancer cell lines. C) Shown are relative levels of expression of spliceoforms observed in the pancreatic cancer cell lines after normalizing the data for levels of gene expression.
The greatest changes in gene expression patterns were seen in the HPDE6 cell line relative to the Capan-1 and MiaPaCa2 cells, while the latter 2 cell lines had minor differences. Focusing on individual cell lines and splicing events, HPDE6 cells had the most overall variety in spliceoforms, with MiaPaCa2 cells displaying the second highest variety in splicing events. Surprisingly, Capan-1 cells had the fewest unique splicing events compared to the other two cell lines. Categorizing this into specific types of splicing events, the HPDE6 cells, followed by the poorly differentiated MiaPaCa2 cells, had the most dramatic change in overall regulation of splicing21, The HPDE6 cell line had the greatest number of alternative last exons, 52 exon trims and retained introns compared to the two tumor cell lines, Capan-1 and MiaPaCa2 cells. Comparatively fewer genes and splice events were unique to Capan-1 cells, reflecting moderate differentiation. Using replicate data as a control, the empirical probability of splicing events occurring by chance with the selected cutoff criteria was zero for all but intron retentions, which had p ≤ 9.39E-5. Trends were common across multiple cutoff values for change, 2-fold, 1.4-fold, and 1.2-fold, and the weak criterion that all technical replicates agree in direction without regard to magnitude of change. Even at the weakest filtering criterion tested, the empirical p-value was ≤ 0.00342 for all splice events.
Validation of alternative spliceoforms in graded patient samples
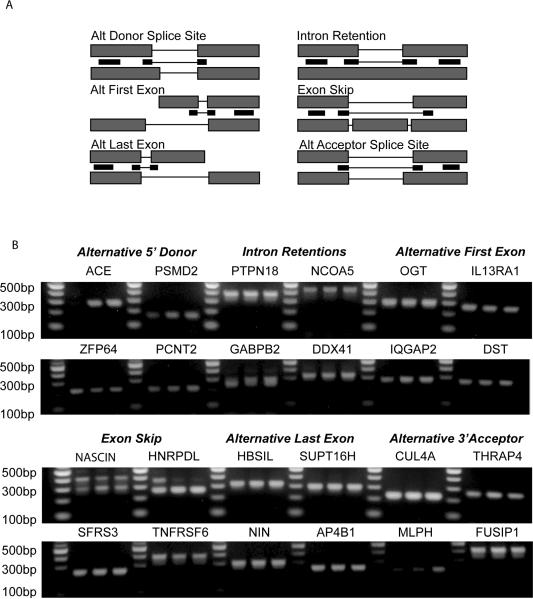
To validate the presence of alternatively spliced transcripts based on the microarray expression data, the raw expression data was reanalyzed in SpliceFold™. Specifically, we applied the ASPIRE algorithm which identifies splicing changes between two samples where two alternative splice forms are differentially expressed, with one splice form up-regulated and the other splice form down-regulated. This algorithm focuses on absolute expression levels of the two splice forms and is a very stringent test that can be useful for identifying the most pronounced splicing changes. Based on this analysis, 100 splicing events were identified that had a minimum signal intensity on the microarray of 300 and a minimum fold-change of 1.4. RT-PCR primers were designed to validate all splicing types, however primer pairs that amplified only the short form were utilized so that the RT-PCR analysis could be performed in a high throughput manner. From the 100 primer pairs designed, 77 primers amplified the expected length transcripts. Representative examples of twenty-four splicing events that encompass the six different modes of alternative splicing in the three pancreatic cell lines are shown in Figure 2. These examples are representative of the abundant short form spliceoforms that are present in pancreatic cancer.
Figure 2. RT-PCR validation of alternative splicing events predicted by the splicing array data.
A) Shown is a general schematic for design of PCR primers utilized to detect short spliceoforms representing each type of alternative splicing event. B) For each noted gene and type of alternative splicing event, primers designed similar to the schematic above were applied to pancreatic cancer cell lines HPDE6, Capan-1, and MiaPaCa2 (left to right, respectively). This analysis supported the presence of each type of splicing event that was predicted to exist.
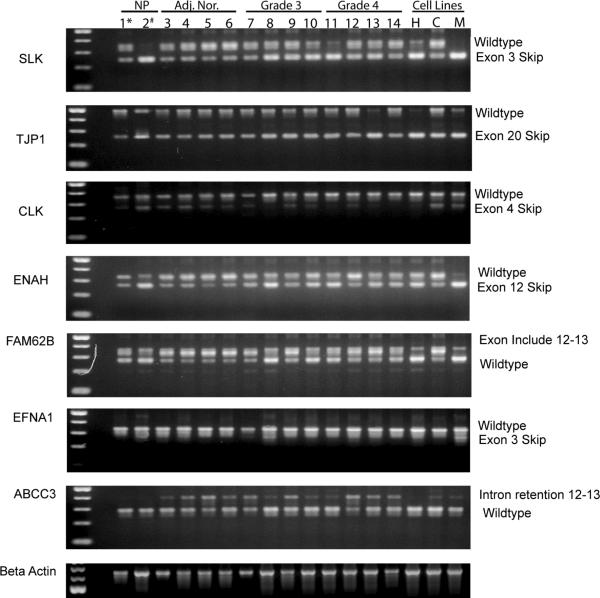
Since the purpose of this study was to identify novel signatures of spliceoforms correlating with the different stages of pancreatic cancer, we chose to characterize additional spliceoforms that were selected by using the values determined by the Splicing Index. The Splicing Index looks at a change in the relative abundance of two splice forms in two samples. It requires that only one of the splice isoforms is expressed in either sample, and may lead to large scores in cases where a splice isoform is highly expressed in one sample but not detectable in the other sample. The expression patterns as predicted by the Splicing Index values in Table 2 correlated with the patterns seen in the RT-PCR experiments. For example, the splice variant of the SLK gene skipping exon 3 was downregulated in Capan-1 cells with respect to HPDE cells, whereas the wild type variant was upregulated in Capan-1 cells compared to HPDE cells. As shown in Figure 3, we screened: 1) normal whole pancreatic tissue, 2) isolated ductal cells from normal pancreas, 3) histologically normal whole pancreatic tissue adjacent to tumor, 4) grade 3 and grade 4 ductalar pancreatic adenocarcinomas and 5) the three pancreatic cancer cell lines that we initially used in the spliceoform microarray experiments. The predicted spliceoform expression patterns that were observed in the HPDE6 cell line were present in the normal ductal cells and in the normal whole pancreatic tissue samples. Interestingly, the normal tissue adjacent to pancreatic cancer had spliceoform expression patterns that correlated more closely with the Capan-1 and MiaPaCa2 cell lines than with the HPDE6 cell line. The spliceoforms that were confirmed in Capan-1 and MiaPaCa2 cells were consistent with the graded patient tumor samples, however, the differences in expression pattern varied when compared to the human primary clinical samples. These differences could be due in part to the small sample size or, more likely, the differences between the homogenous cell lines versus the cellular heterogeneity of the tumor samples.
Table 2.
Predicted differences in spliceoform expression patterns based on the results of the Splicing Index Algorithm
| HPDE6 (H) | Capan-1 (C) | MiaPaCa2 (M) | C vs H | M vs H | M vs C | Type | Accession | |
|---|---|---|---|---|---|---|---|---|
| SLK | 4132 | 7590 | 473 | 1.84 | −8.74 | −16.06 | Wildtype | NM_014720 |
| 5034 | 3968 | 16766 | −1.27 | 3.33 | 4.23 | Exon skip: ex 3, 49 nt | D86959 | |
|
| ||||||||
| TJP1 | 3282 | 15129 | 11622 | 4.61 | 3.54 | −1.30 | Wildtype | NM_003257 |
| 22726 | 21149 | 11599 | −1.07 | −1.96 | −1.82 | Exon skip: ex 20, 240 nt | NM_175610 | |
|
| ||||||||
| CLK2 | 4721 | 2281 | 2962 | −2.07 | −1.59 | 1.30 | Wildtype | NM_003993 |
| 1153 | 1541 | 2169 | 1.34 | 1.88 | 1.41 | Exon skip: ex 4, 85 nt | NM_001291 | |
|
| ||||||||
| ENAH | 3431 | 8246 | 1471 | 2.40 | −2.33 | −5.60 | Wildtype | NM_001008493 |
| 16311 | 6372 | 18392 | −2.56 | 1.13 | 2.89 | Exon Skip: ex 12, 63 nt | NM_008212 | |
|
| ||||||||
| FAM62B | 3705 | 8799 | 3415 | 2.37 | −1.08 | −2.58 | Exon Include: btwn ex13–14,63 nt | BC013957 |
| 2768 | 1765 | 9796 | −1.57 | 3.54 | 5.55 | Wildtype | NM_020728 | |
|
| ||||||||
| ABCC3 | 346 | 220 | 219 | −1.57 | −1.58 | −1.00 | Intron Retention | BC046126 |
| 542 | 904 | 315 | 1.67 | −1.72 | −2.87 | Wildtype | NM_003786 | |
|
| ||||||||
| EFNA1 | 5168 | 1669 | 833 | −3.10 | −6.20 | −2.00 | Wildtype | NM_004428 |
| 1266 | 344 | 576 | −3.68 | −2.20 | 1.67 | Exon skip: ex 3, 66 nt | NM_182685 | |
Figure 3. Expression of alternative spliceoforms in patient samples.
Spliceoforms that were predicted to exist by the Splicing Index algorithm applied to the pancreatic cancer cell lines were analyzed by RT-PCR in normal pancreas (NP*), isolated ductal cells from normal pancreas (NP#), adjacent normal pancreatic tissue (Adj. Nor.), and grade 3 and 4 primary pancreatic carcinomas, as well as the pancreatic cancer cell lines HPDE6 (H), Capan1 (C), and MiaPaCa2 (M). This supported the presence of the predicted spliceoforms in patient samples.
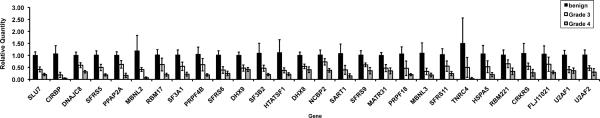
Since the microarray results identified unique patterns of spliceoforms for the different cell lines, with the presence of these spliceoforms and their patterns of expression validated in primary tumor tissue, we hypothesized that the splicing machinery itself could be abnormal in cancer cells. Using the proteomic data generated by Zhou et al.22, 23, a Taqman Low Density array that contained primers and probes for 92 out of the 300 characterized proteins found in the spliceosome was designed. Quantitative PCR was performed on the pancreatic cell lines24 and on graded patient samples. As shown in Figure 4, proteins involved in the splicing process have dramatically different gene expression levels in primary pancreatic cancer samples compared to adjacent normal tissue. These trends were consistent in the grade 3 and 4 clinical cancer samples and in the cancer cell lines24. Out of the 92 spliceosomal genes assayed, 28 genes were significantly different. Two trends were observed: for five splice factors, after the initial decrease in expression, the levels between the grade 3 and 4 tumors remained constant. For the remaining twenty-three splice factors, the levels of expression continued to decrease from grade 3 to grade 4 (ex. CIRBP, MBNL2, and TNRC4). This suggests that there are specific cancer-associated changes of certain spliceosomal proteins, rather than a general increase or decrease in the expression of all components of the splicing machinery.
Figure 4. Quantitative PCR expression profiles for selected spliceosomal transcripts.
cDNA from six normal pancreatic specimens and from pancreatic cancer specimens from five patients with grade 3 tumors and from five patients with grade 4 tumors were analyzed by quantitative PCR, with the relative quantities of product normalized relative to three different housekeeping genes (GAPDH, beta actin, and HNRPA2B1 ribosomal protein). After performing a one way-ANOVA and FWER false-discovery correction, 28 of 92 genes that were tested were identified to be significantly different in normal tissue relative to the tumors. Many of these genes also varied with tumor grade. Shown are values representing means ± 95% confidence interval.
Discussion
Splicing of primary transcripts from the genetic code is essential to the normal function of a cell. The spliceosome, which essentially regulates the shuffling of genetic material, is one of the most structurally and functionally complex components of the cell. Accurate pre-mRNA splicing is necessary for the production of diverse products of a single gene that are often critical for normal cellular development and function. The myriad of protein isoforms can also change with neoplastic progression; in such a setting, one would anticipate cancer-associated splicing changes. In this report, we have characterized genome-wide splicing events in pancreatic cancer cell lines, MiaPaCa and Capan-1, compared to the near-normal pancreatic ductal cell line, HPDE6, using a custom splice variant microarray that measures 98,000 spliceoforms. Splicing variation changes were then validated by RT-PCR of 100 splice variants that were differentially expressed between these cell lines. Validation was performed in the original pancreatic cell lines, as well as in whole tissue and isolated ducts from normal pancreas, histologically normal pancreatic tissue adjacent to tumors, and primary pancreatic adenocarcinomas.
Our data, from both cell lines and human pancreatic tissue, revealed the largest diversity in alternative spliceoforms in the normal cells and tissue, while pancreatic cancer and cancer cell lines had a sharp reduction in the number of spliceoforms present. This observation was first recognized on the genome-wide custom splicing array and then verified by RT-PCR. Of note the histologically normal pancreatic tissue adjacent to cancer had an intermediate pattern between cancer (least splicing variation) and true normal pancreas (most splicing variation). While the number of spliceoforms was limited in the pancreatic cancer cell lines, some of these splice variants appeared to be unique to the tumors cells.
The encouraging results from the microarray validation experiments and the changes in splice factor expression patterns in patient samples motivated us to further test the predicted variants in patient samples. The presence of the variants was consistent with the predicted results, however the relative levels of expression in the clinical samples were often not as predicted by the cell line data: there was splicing variation between individuals with the same diagnosis, as well as variation between diagnostic groups. For example, the tumor cell lines and the graded patient samples contained the predicted splice variants but the absolute levels of expression were not as expected. Also, in addition to the predicted variants, the tumor samples contained variants that had not been identified in the microarray analysis of the cell lines. Since homogeneous cell lines were used in the initial microarray screen, expression patterns for spliceoforms that are present in clinical (and more complex) samples might not have been detected. However, in the tissue validation studies, the presence of the additional spliceoforms are most likely due to the splicing diversity in the complex mixture of cells found within the tumor tissue. Though these results were not broadly generalizable, some patients' samples did display the expression trends predicted by the cell line expression data, thus supporting the use of the alternative splicing microarray for detecting differences in the presence and expression of spliceoforms that might be unique to the various stages of pancreatic cancer in patient samples.
Similar findings have been described in splicing studies of colon, prostate, and bladder cancers25, where the variance in splicing ranged from lowest to highest in the following order: technical replicates, inter-cell line variation, intra-patient tumor variation, and inter-patient tumor variation. The study also found larger and more significant differences in splice indices between normal samples from colon, bladder, and prostate than between normal and cancer samples.
In order to discover why there was a widespread reduction in splicing diversity in pancreatic cancer, we analyzed the expression differences for 92 genes associated with the spliceosome. Thirty percent (28 of 92 genes) of the spliceosome genes tested had decreased expression in pancreatic cancer samples compared to normal pancreas. Several of the key splice factors that are responsible for initiating the splice complex and for the formation of the lariat structure had decreased expression. Interestingly, none of the splice factors were significantly over-expressed in the tumors. A cancer-associated decrease of expression of splicing initiation genes and genes that regulate splicing function would certainly fit with the dramatic loss of splicing variation that we detect in pancreatic cancer cell lines and primary tumors.
Regulation of the splicing machinery may vary according to the type of cancer, as studies of ovarian tumors found that regulatory splicing factors, (SFRS1 and SFRS2) show a marked increase in expression compared to normal tissue, while the expression other regulatory splicing factors (SFRS5 and SFRS6) remained constant26. In our study of pancreatic cancer, the expression level of these splicing factors (SFRS 1, 2, 5 and 6) were decreased in tumors. Another factor in considering the different levels of splice variants in different cancers is the fact that regulation of the spliceosome can very according to the underlying tissue type, with the normal human brain having the greatest splicing diversity of organs thus far tested27. Thus, splice variation and the changes in regulation of the splicing machinery may not be comparable across diseases and across organs.
We have described the loss of splicing diversity associated with pancreatic cancer cell lines in the initial genome-wide scan and then validated the loss of diversity in clinical pancreatic cancer samples. Lastly, we described the loss of expression of the many spliceoform components and regulatory factors. The finding of intermediate loss of splicing diversity in the normal appearing tissue adjacent to cancer suggests that dysfunction of the splicing machinery may occur relatively early in the process of neoplastic progression, before morphologic changes are apparent microscopically.
Several recent studies have characterized the mammalian pancreatic cancer transcriptome using RNA and DNA sequencing techniques28–30. Jones et al, sequenced 20,735 transcripts from 24 advanced pancreatic adenocarcinomas and concluded that 160 missense mutations that can be created by intron retentions or splice site changes were predicted to contribute to tumorigenesis31. Based on these results, it is quite apparent how prevalent alternative splicing and variants are, but one should have caution when interpreting alternative splicing results. The differences that are seen in sequencing transcriptomes from heterogenous tumor samples may be an under-representation of the most informative splice variants since there is high variability between patient samples. In order to fully understand the importance of alternative splicing and pancreatic cancer, a combined approach using the information from the alternative splicing microarray, followed by transcriptome sequencing, may be the most comprehensive way to identify informative splice variants
Table 1.
Alternative splicing events specific for each cell line and for pairwise sample comparison
| SPLICING EVENT | HPDE6 | Capan-1 | MiaPaCa2 | Capan-1 vs. HPDE6 | MiaPaCa2 vs. HPDE6 | MiaPaCa2 vs. Capan-1 |
|---|---|---|---|---|---|---|
| Exon Skips | 130 | 20 | 78 | 269 | 344 | 301 |
| Exon Inclusions | 130 | 50 | 53 | 326 | 350 | 186 |
|
| ||||||
| Alternative First Exon | 192 | 61 | 121 | 649 | 714 | 460 |
| Alternative Last Exon | 302 | 57 | 143 | 353 | 463 | 316 |
|
| ||||||
| 5' Exon Trim | 126 | 27 | 76 | 248 | 331 | 196 |
| 5' Exon Extensions | 141 | 32 | 59 | 242 | 302 | 173 |
| 3' Exon Trim | 128 | 25 | 61 | 256 | 322 | 198 |
| 3' Exon Extension | 146 | 26 | 62 | 241 | 318 | 201 |
|
| ||||||
| Intron Splices | 126 | 14 | 45 | 261 | 317 | 184 |
| Intron Retentions | 144 | 25 | 101 | 271 | 391 | 253 |
After median normalization of all probes, splicing events were determined from samples that displayed a minimum signal intensity of 225 and a minimum splice fold change of 1.4.
Acknowledgements
This work was supported by grants from the National Institutes of Health, DK46577 and DK32878 (L.J.M.), a Canary Foundation/ American Cancer Society Postdoctoral Fellowship (P.E.C.), the Mayo Clinic/Arizona State University Seed Grant (P.E.C., L.J.M.), and additional funds from the Mayo Clinic and the Fiterman Foundation (L.J.M.). The authors would like to thank A. Beck, L. Bruins, and D. Johnson for technical support, and Dr. G. Hayes for helpful discussions.
Funding Sources: National Institutes of Health, DK46577 and DK32878 (L.J.M.), a Canary Foundation/American Cancer Society Postdoctoral Fellowship (P.E.C.),
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson JM, Castle J, Garrett-Engele P, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Barash Y, Calarco JA, Gao W, et al. Deciphering the splicing code. Nature. 465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 3.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Modrek B, Lee C. A Genomic View of Alternative Splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 5.Ladd A, Cooper T. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biology. 2002;3:reviews0008. doi: 10.1186/gb-2002-3-11-reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green RE, Lewis BP, Hillman RT, et al. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19:i118–121. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- 7.Yeakley JM, Fan JB, Doucet D, et al. Profiling alternative splicing on fiber-optic arrays. Nat Biotechnol. 2002;20:353–8. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- 8.Bracco L, Kearsey J. The relevance of alternative RNA splicing to pharmacogenomics. Trends in Biotechnology. 2003;21:346–353. doi: 10.1016/S0167-7799(03)00146-X. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 10.Bingham JL, Carrigan PE, Miller LJ, et al. Extent and Diversity of Human Alternative Splicing Established by Complementary Database Annotation and Microarray Analysis. OMICS: A Journal of Integrative Biology. 2008;12:83–92. doi: 10.1089/omi.2007.0041. [DOI] [PubMed] [Google Scholar]
- 11.Warner S, Gray PJ, Von Hoff DD. Tubulin-associated drug targets: Aurora kinases, Polo-like kinases, and others. Semin Oncol. 2006;33:426–48. doi: 10.1053/j.seminoncol.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Frelin C, Ladoux A, D'angelo G, et al. Vascular endothelial growth factors and angiogenesis. Ann Endocrinol (Paris) 2000;61:70–4. [PubMed] [Google Scholar]
- 13.Laurent-Puig P, Lievre A, Blons H. Mutations and Response to Epidermal Growth Factor Receptor Inhibitors. Clin Cancer Res. 2009;15:1133–1139. doi: 10.1158/1078-0432.CCR-08-0905. [DOI] [PubMed] [Google Scholar]
- 14.Akgul C, Moulding DA, Edwards SW. Alternative splicing of Bcl-2-related genes: functional consequences and potential therapeutic applications. Cellular and Molecular Life Sciences. 2004;61:2189–2199. doi: 10.1007/s00018-004-4001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuperlovic-Culf M, Belacel N, et al. Data analysis of alternative splicing microarrays. Drug Discovery Today. 2006;11 doi: 10.1016/j.drudis.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa T, Duguid WPP, Rosenberg L, et al. Long Term culture and immortalization of epithelial cells from normal adult human pancreatic ducts by transfection with E6E7 gene of human papilloma virus 16. Am. J. Pathol. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- 17.Ouyang H, Mou L-j, Luk C, et al. Immortal Human Pancreatic Duct Epithelial Cell Lines with Near Normal Genotype and Phenotype. Am J Pathol. 2000;157:1623–1631. doi: 10.1016/S0002-9440(10)64800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ule J, Ule A, Spencer J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 19.Srinivasan K, Shiue L, Hayes JD, et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–359. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Livak K, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;4:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Choudhury A, Moniaux N, Winpenny JP, et al. Human MUC4 mucin cDNA and its variants in pancreatic carcinoma. J Biochem (Tokyo) 2000;128:233–43. doi: 10.1093/oxfordjournals.jbchem.a022746. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Licklider LJ, Gygi SP, et al. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 23.Rappsilber J, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Research. 2002;12:1231–45. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abetamann V, Kern HF, Elsasser HP. Differential expression of the hyaluronan receptors CD44 and RHAMM in human pancreatic cancer cells. Clin Cancer Res. 1996;2:1607–18. [PubMed] [Google Scholar]
- 25.Thorsen K, Brems-Eskildsen AS, Modin C, et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol Cell Proteomics. 2008;7:1214–24. doi: 10.1074/mcp.M700590-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Fischer DC, Noack K, Runnebaum IB, et al. Expression of splicing factors in human ovarian cancer. Oncol Rep. 2004;11:1085–90. [PubMed] [Google Scholar]
- 27.Dehay C. Transcriptional regulation and alternative splicing make for better brains. Neuron. 2009;62:455–7. doi: 10.1016/j.neuron.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 30.Aguirre A, Brennan C, Bailey G, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proceedings of the National Academy of Sciences. 2004;101:9067–72. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones S, Zhang X, Parsons DW, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]