Abstract
We describe a two-dimensional capillary electrophoresis system that incorporates a replaceable enzymatic microreactor for on-line protein digestion. In this system, trypsin is immobilized on magnetic beads. At the start of each experiment, old beads are flushed to waste and replaced with a fresh plug of beads, which is captured by a pair of magnets at the distal tip of the first capillary. For analysis, proteins are separated in the first capillary. A fraction is then parked in the reactor to create peptides. Digested peptides are periodically transferred to the second capillary for separation; a fresh protein fraction is simultaneously moved to the reactor for digestion. An electrospray interface is used to introduce peptides into a mass spectrometer for analysis. This procedure is repeated for several dozen fractions under computer control. The system was demonstrated by the separation and digestion of insulin chain b oxidized and β-casein as model proteins.
Keywords: Two dimensional capillary electrophoresis, ESI-MS, Magnetic beads
1. Introduction
Two approaches are used to identify proteins: top-down and bottom-up.1 In top-down methods, intact proteins are introduced into mass spectrometer for identification.2 In bottom-up methods, proteins are digested into peptides and then analyzed by mass spectrometry.3 Because mass spectrometry of peptides is more sensitive than for whole proteins,4 bottom-up methods are more common analytical tools in proteomics.
In bottom-up analysis, complex protein samples are digested into more complex peptide mixtures, which are then subjected to two or more stages of liquid chromatographic separation before being introduced into the mass spectrometer.5 Inevitably, the peptides from a given protein are scattered throughout the multidimensional chromatographic separation. In addition, the peptides produced from high-abundance proteins are distributed throughout the chromatographic separation, obscuring peptides from low abundance proteins.
It would be beneficial to have all the peptides from a single protein introduced into the mass spectrometer in a relatively short time window. Such a procedure is commonly employed in two-dimensional-gel electrophoresis. Proteins are separated and excised from a stained gel, then digested by trypsin, and finally analyzed by mass spectrometry. The benefit of two-dimension-gel electrophoresis in proteomics study is that post-translational modification or alternative splicing results in a shift in spot position. However, two-dimensional gel electrophoresis is far too labor-intensive to be useful for high-throughput proteomics.
An alternative strategy is to incorporate on-line proteolytic digestion with some form of protein separation. In such systems, a mixture of proteins is separated, passed through a reactor containing immobilized proteolytic enzyme, and the resulting peptides analyzed by mass spectrometry. In a recent example, Yuan used size exclusion chromatography to separate proteins, which were then digested in a microreactor containing immobilized trypsin. The resulting peptides were desalted in a C8 column and separated by gradient elution reversed-phase chromatography.6
Application to multi-dimensional chromatography is complicated by incompatibility of the enzyme with some separation buffers. Instead, most applications of on-line proteolytic digestion have focused on the use of capillary electrophoresis for protein separation.7–14 In these systems, analyte undergo digestion as they pass through the microreactor, which is usually placed at the distal end of the separation capillary, before analysis by mass spectrometry.
Such systems tend to suffer from four difficulties. First, it is difficult to obtain a sufficiently long residence time in the microreactor to achieve efficient digestion; often, a relatively long microreactor and slow separation conditions are employed to provide sufficient interaction time. Second, these conditions tend to produce significant band-broadening. Third, monolithic microreactors have limited lifetime and must be periodically replaced. Fourth, there is no separation of peptides, which can complicate the detection of low abundance peptides.
To address some of these issues, we created a two-dimensional capillary electrophoresis system with a monolithic microreactor immobilized at the distal end of the first capillary.15–16 Proteins were separated in the first capillary. Protein fractions were parked in the microreactor and digested. Then, the resulting peptides were transferred into the second capillary and separated before being introduced into a mass spectrometer. A fresh plug of protein was simultaneously introduced into the microreactor and underwent digestion during the peptide separation. The system offered several advantages compared with other on-line digestion systems. First, by parking the sample in the microreactor during the second-dimension separation, quite long digestion times were produced in a short microreactor. Second, the short microreactor reduced band broadening and the system produced quite high separation efficiency in both dimensions. Third, the use of a peptide separation dimension reduced the number of peptides simultaneously introduced into the mass spectrometer, simplifying detection. However, the system had two disadvantages. First, the system employed a monolithic microreactor, which had limited lifetime and would not be suitable for application in high-throughput proteomics. Second, the separation time for our two-dimensional capillary electrophoresis system was similar to that employed in on-line microreactor employed in one-dimensional capillary electrophoresis.
Magnetic beads make an interesting alternative to monolithic reactors in capillary electrophoresis. These beads can have well-controlled surface chemistry and a large surface-to-volume ratio. They are easily manipulated with magnets, which facilitate their capture and replacement without the need for replacing the separation capillary. There have been a few reports of magnetic bead microreactors coupled to capillary electrophoresis separations.17–20
2. Materials and Methods
2.1. Materials and chemicals
TPCK-treated trypsin and insulin chain b oxidized from bovine pancreas, β-casein ≥98% (PAGE) from bovine milk, S-NHS (N-hydroxysulfosuccinimide sodium salt) ≥98.5% (HPLC), EDC (N-(3-Dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride), MES monohydrate (2-Morpholinoethanesulfonic acid Monohydrate), sodium hydroxide, ammonium acetate, and ammonium bicarbonate were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ethanolamine was purchased from Fisher Scientific (Pittsburgh, PA, USA). Carboxyl-Adembeads (diameter 100 nm) magnetic beads were purchased from Adamtech SA (Pessac, France). Triangular magnets were purchased on-line from http://www.supermagnetman.net. Methanol 100.0% was purchased from J.T. Baker (Philipsburg, NJ, USA). Fused capillaries were purchased from Polymicro Technologies (Phoenix, AZ, USA). Water was deionized by a Nano Pure Diamond system (Barnstead International, Dubuque, IA, USA).
An ammonium bicarbonate-acetate buffer was made by adding ammonium bicarbonate (10 mM, pH 8.6) into ammonium acetate (10 mM, pH 5.7) to adjust the pH value to 7.0. The electrospray sheath flow liquid contains 50% v/v methanol, 50% v/v water and 10 mM acetic acid. Buffers were passed through a 0.22 µm filter (Millipore, Bedford, MA, USA) before use.
Insulin chain b oxidized (0.6 mg/mL) and β-casein (12 mg/mL) were dissolved in ddH2O, aliquoted, and stored at −20°C. New protein samples were thawed at room temperature each day. The protein mixture was prepared by diluting the solution in ammonium bicarbonate-acetate buffer to give a final concentrations of 0.3 mg/mL (1 µM) insulin chain b oxidized and 1.2 mg/mL β-casein (5 µM).
2.2. Trypsin immobilization
The 100-nm diameter magnetic particles used in this study had a carboxylic acid functionalized surface. Trypsin was immobilized onto the beads via EDC activation.21 A 0.25 mg aliquot of the beads was washed twice with 100 µL of MES (25 mM, pH 6.0). Then the carboxyl groups were activated by adding 100 µL of EDC (50 mg/mL in MES) and 100 µL of S-NHS (50 mg/mL in MES), then incubated at 37°C for 30 minutes with slow tilt rotation. The beads were removed from the solution, washed and resuspended in 200 µL MES, then 4 mg of trypsin was added. The immobilization took 16 hours at 4°C with slow tilt rotation. The unreacted active groups were blocked by rinsing with ethanolamine (0.5 M in ddH2O) for 15 minutes under rotation. The final product was stored in 1 mL ammonium bicarbonate buffer (10 mM, pH 8.6) at 4°C. The trypsin-modified beads were synthesized every week.
A BCA protein assay kit (Thermo-Pierce, Rockford, IL, USA) was used to estimate the amount of trypsin immobilized to beads. The result indicated that about 21 µg of trypsin was immobilized onto 1 mg of beads.
2.3. Instrumentation
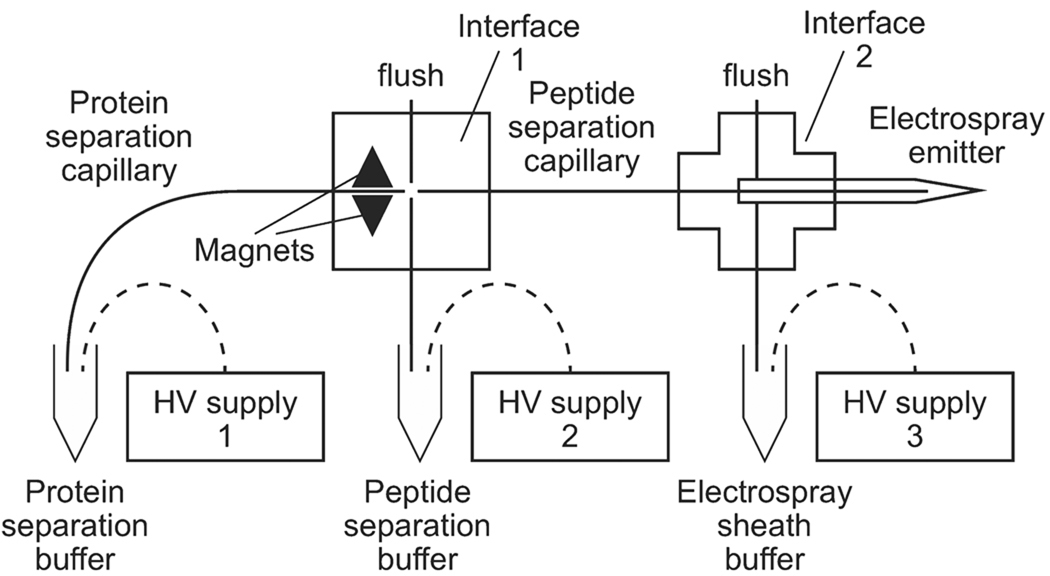
The system is shown in Fig.1. In this study, the protein separation capillary (i.d. 50 µm, o.d. 149 µm) was 36.5 cm long while the peptide separation capillary (i.d. 50 µm, o.d. 149µm) was 30.0 cm long. High voltages were provided by three CZE 1000R high-voltage power supplies (Spellman, Hauppauge, NY, USA). The electrospray emitter was borosilicate glass capillary (o.d. 1.0 mm, i.d. 0.75 mm, 10 cm, Sutter Instrument, Novato, CA, USA) pulled with a P-97 flaming/brown micropipette puller (Sutter instrument, Novato, CA, USA). The detector was an ESI Quadrupole ion trap (LCQ) mass spectrometer (Finnigan, Hercules, CA, USA). Voltage programming and data collection were controlled by LabView software. Data were processed with Matlab.
Figure 1.
Block diagram. The magnets hold the trypsin-modified magnetic beads, forming the replaceable microreactor. High voltage connections are shown as dashed curves. Details of the interface construction are provided in the supplementary material section of this paper.
2.3.1. Capillary interface
The protein and peptide separation capillaries were joined by an interface that consisted of a cross that was machined into a 0.75 inch thick Plexiglas plate. The channels were 0.037 inches deep and wide. Glass microcaps (o.d. 0.0340 inch, i.d. 0.0157 inch Drummond Scientific, Broomall, PA, USA) were glued in the channels. Fused capillaries sleeves (o.d. 363 µm, i.d. 150 µm) were glued into two of the microcaps, forming coaxial capillary guides. Features were milled into the Plexiglas plate to hold two triangular magnets (0.5625 in. length, Supermagnetman, Birmingham, AL, USA, website: www.supermagnetman.com). A piece of glass microscope slide was glued on top to seal the interface. The separation capillaries were inserted and fixed into the sleeves with epoxy glue. The gap between the two capillaries in the interface was adjusted to 30–50 µm under a microscope.
Teflon tubing was carefully twisted over the glass pipets for connections to the waste inlet and buffer reservoir. A valve was placed in-line with the waste inlet Teflon tubing to allow flushing of the system between runs. The valve was closed during the analysis.
2.3.2. CE-MS interface
The capillary electrophoresis-electrospray interface has been described in detail elsewhere.22 Briefly, a borosilicate glass capillary was pulled into an electrospray emitter with a 5–10 µm inner diameter tip. The peptide separation capillary was inserted into the electrospray emitter, which seated inside a plastic cross (IDEX, WA, USA). The end of the peptide separation capillary was about 2 mm to the emitter. One opening of the plastic cross was connected to the sheath flow liquid reservoir. The other opening was used to fill the system with sheath flow liquid before analysis, and was closed during the analysis. The sheath flow liquid level was adjusted to avoid siphoning. The inlet of the mass spectrometer was grounded.
2.4. Plug formation
Prior to bead injection, a 50-µL aliquot of a 0.5 mg/mL bead suspension was sonicated for 5 minutes to reduce aggregation. The suspension was transferred to the injection reservoir and purged into the protein separation capillary under 5-psi pressure for 2 minutes. Then, ammonium bicarbonate-acetate buffer was purged under the same conditions for another 5 minutes. The beads were trapped between the magnets and accumulated to form a dense plug 7–8 mm in length.
The beads were replaced between each run. The used ones were flushed by removing the magnets and purging 1 M NaOH, followed by water through the flush inlet by hand using a 30 mL syringe. A new plug was injected to regenerate the microreactor.
2.5. On-line digestion
The voltage program for 2D-CE and ESI has been described elsewhere.15–16, 23–24 The program has four steps. For sample introduction, the sample was placed at the inlet of the protein separation capillary. 13.3 kV was applied by HV supply 1 and 0 V was applied by HV supply 2 for five seconds.
For the preliminary separation step, the sample was replaced with running buffer. To drive the protein separation, 13.3 kV was applied by HV supply 1 and 0 V by HV supply 2 for 8 minutes.
To transfer a fraction, 13.3 kV was applied by HV supply 1 and 0 V with HV supply 2 for 5 seconds.
Finally, to separate the digested peptides, 6 kV was applied by both HV supply 1 and HV supply 2 for 60 seconds. There was no voltage drop across the protein separation capillary, which held proteins stationary during the peptide separation. Proteins present in the microreactor underwent digestion during this period.
Transfers and digestion were repeated under computer control to complete the separation, digestion, and peptide analysis of the protein sample.
At all times, 1 kV was applied at the CE-MS sheath flow reservoir to drive the electrospray. The m/z range scanned was 150 to 2000 amu.
2.6. Data acquisition and Sequest Analysis
Tandem mass spectrometry analysis was performed using the MS/MS data acquisition function in the Finnigan LCQ mass spectrometer. Spectra were taken every 1.89 s. The parameters for survey scans were set as m/z range 300–2000. For product ion scans, m/z range was set to 0–2000; mass tolerance ±0.50 Da; the m/z 1166.5 ± 0.5 was rejected, which was the triply charged peak of undigested insulin chain b oxidized; minimum MS signal required for fragmentation was 5.0×104. Two m/z range scans were collected and summed before going on to the next scan. The two most intense mass peaks were selected in every scan and CID fragmentation was set to normalized collision energy 35%. Ions that had been fragmented twice were excluded from fragmentation for 180 s.
The raw MS/MS data were converted to mzXML format file on Trans-Proteomic Pipeline (TPP, version 4.3, Jetstream revision 1) software combined with Xcalibur software and searched using Sequest version 27 against the International Protein Index bovine database (ipi.BOVINE.v3.46.fasta, European Bioinformatics Institute, http://www.ebi.ac.uk/IPI) appended to add bovine insulin chain b, allowing ±1.00 Da mass tolerance for precursor ions and ±0.0 Da mass tolerance for fragmented ions. Tryptic cleavages (K and R) with up to two missed cleavages and variable modification of cysteine oxidation (+47.9982 Da) were allowed.
The Sequest search output of peptide assignment and quantification was used as the input for pepXML module. Peptides were identified by searching the output of pepXML in Peptide Prophet and the result was used as inputs to identify proteins by the Protein Prophet.
3. Results and discussion
3.1. Analysis of a mixture of two proteins
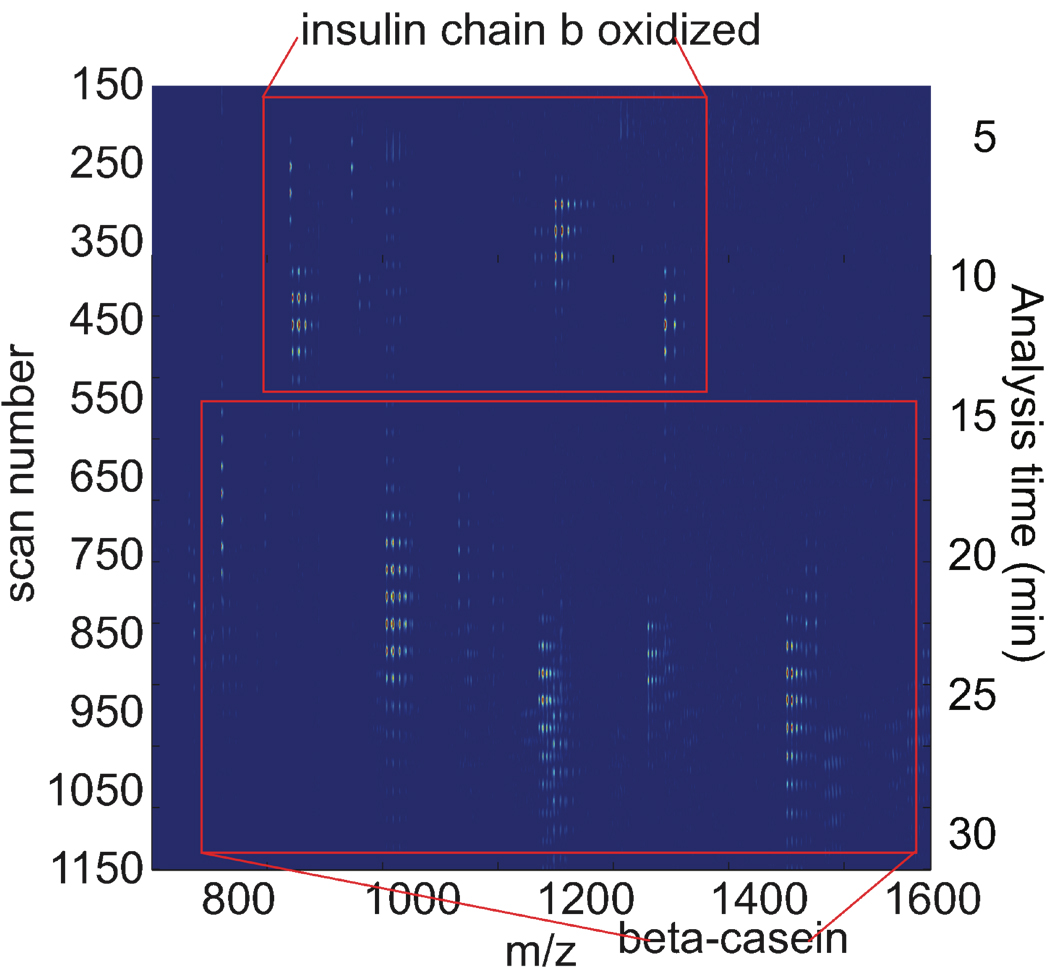
Insulin chain b oxidized (pI 7.3) and β-casein (pI 5.1) were analyzed with the system in a pH 7.0 buffer. After a preliminary separation, a series of mass spectra were recorded. Simultaneously, peptide fractions were periodically transferred to the second dimension capillary for separation. Figure 2 presents a heat-map of the raw data. In this map, successive mass spectral scans are stacked and color-coded to represent the data. The data consist of spots that repeat themselves every 34 scans, corresponding to data generated in successive transfers from capillary 1 to capillary 2. The first group of spots was generated by peptides produced by digestion of insulin chain b oxidized, from around 200 to 550 scans. The second group of spots was generated by peptides produced from β-casein, from around 550 to 1120 scans. β-casein is larger (MW 24.02 kDa), than insulin chain b oxidized (MW 3495.89 Da). β-casein was fully digested with no protein ions detected.
Figure 2.
CE-microreactor-CE-ESI-MS data of a mixture of insulin chain b oxidized (0.3 mg/mL) and β-casein (1.2 mg/mL). Injection voltage: 13.3 kV and injection time: 5 seconds.
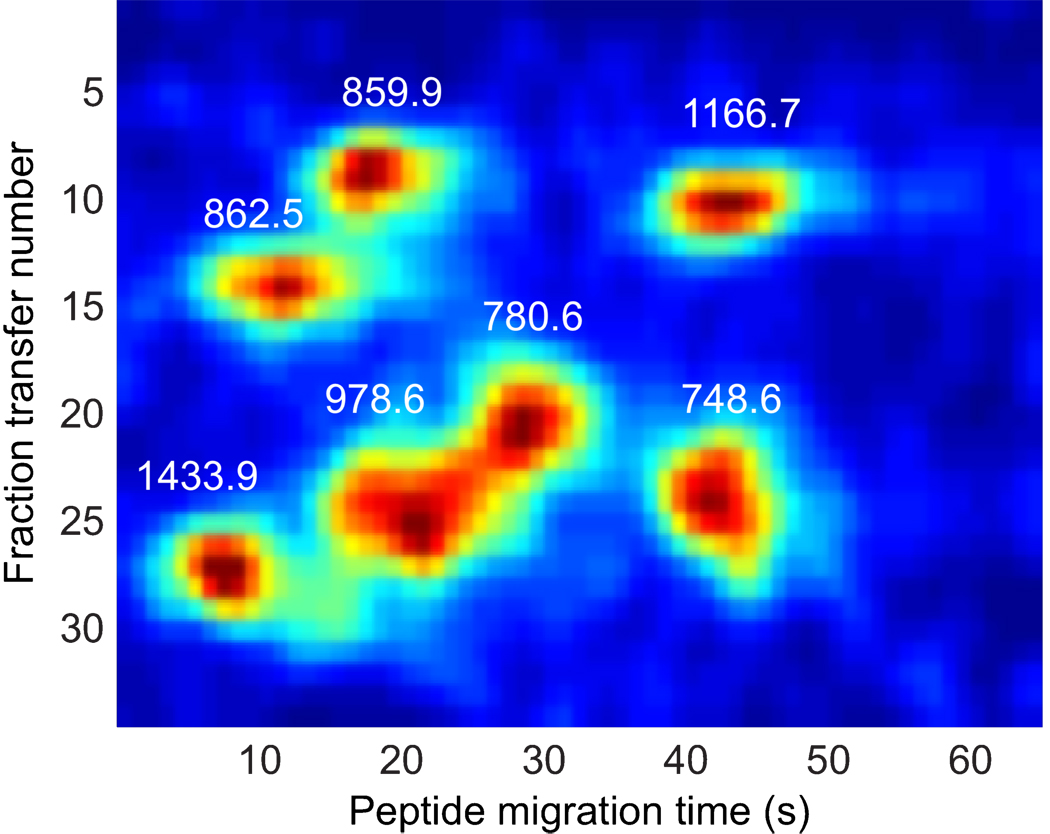
The data of figure 2 were unwrapped to present a plot of signal versus transfer number (x-axis) and peptide capillary migration time (y-axis). The data consist of a set of spots, corresponding to the migration of the tryptic digest. The data from several peptides are presented in figure 3. A two-dimensional Gaussian surface was fit to a set of selected ion reconstructed two-dimensional electropherograms. The average spot width in the peptide migration time dimension was 3.3 ± 1.0 s and in the transfer dimension was 1.3 ± 0.5 transfers. The spot capacity at any m/z value was about 125. The spot width was similar to that obtained with our monolithic reactor in the peptide separation dimension, which suggests that the magnetic bead reactor did not introduce significant band broadening. The spot width is larger in the fraction transfer dimension, which is almost certainly due to the long sample injection time.
Figure 3.
Gel image of selected ions from Figure 2. Data were smoothed by convolution with a two-dimensional Gaussian filter with width of 1-s in the peptide migration dimension and 1 transfer in the fraction transfer number.
Unlike our earlier work,16 uncoated capillaries were used in this experiment, which generated a fair amount of electro-osmotic flow. Electro-osmosis opposed the migration of both proteins and peptides, leading to longer separation times. In particular, the separation window for the peptide separation was much longer than the 65-s duration of each peptide separation, and the peptides from a given protein were observed across several peptide separations. It is clear that the system would benefit from poorer electrophoretic resolution in the peptide dimension, so that all peptides from the same protein are present in a single transfer. Most simply, a shorter capillary would speed the second dimension separation, introducing peptides from a single peptide to the mass spectrometer in a shorter time period.
The microreactor appears to be quite efficient. No undigested β-casein was detected after the 60-second incubation.
3.2. Tandem mass spectrometry analysis of protein mixture
Tandem mass spectrometry data were submitted to the TransProteomic Pipeline (TPP); the identified proteins and their probabilities are listed in table 1. TPP identified both β -casein and insulin chain b with confidences close to 100%. The sequence coverages for the two proteins are 15% and 100%.
Table 1.
Identified peptides and proteins by TPP
| Peptide sequence | Nsp adjusted probability |
Protein identified |
confidence |
|---|---|---|---|
| VLPVPQK | 0.9995 | β-casein | |
| 1.0000 | |||
| DMPIQAFLLYQEPVLGPVRGPFPIIV | 0.9656 | β-casein | |
| FVNQHLCGSHLVEALYLVCGER | 0.9995 | insulin | |
| chain b | |||
| FVNQHLCGSHLVEALYLVCGERGFFYTPKA | 0.9995 | insulin | |
| chain b | |||
| 1.0000 | |||
| GFFYTPK | 0.9692 | insulin | |
| chain b | |||
| GFFYTPKA | 0.9195 | insulin | |
| chain b |
Most of the missed peptides for β-casein were highly charged and rejected by the TPP. The tandem mass spectrometry data were manually submitted to Sequest for identification. EAMAP (645.6 Da), EMPFPK (747.6 Da) and YPVEPFTESQSLTLTDVENLHLPLPLLQSWMHQPHQPLPPTVMFPPQSVLSLSQSK (6362.3 Da) were identified as β-casein tryptic digestion products, which increased the sequence coverage to 45%.
4 Conclusion
The CE-microreactor-CE-ESI-MS offers an automated protein analysis system with replaceable magnetic bead-based microreactor. This delivers tryptic digest of a protein to the mass spectrometer within a short window. The system has operated for over two months without replacing the capillary. By clustering all peptides produced by digestion of a protein, database search scores can be used to improve protein identification scores and confidence in search results. The two-dimensional separation requires roughly 30 minutes to digest and separate 30 fractions.
Supplementary Material
Acknowledgements
We acknowledge the assistance of Dr. Martin Sadelek of the chemistry department at the University of Washington in the generation of this data. We gratefully acknowledge support from a grant from the National Institutes of Health (R33CA122900).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Han X, Aslanian A, Yates JR. Curr. Opin. Chem. Biol. 2008;12:483. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelleher NL. Anal. Chem. 2004;76:197A. [PubMed] [Google Scholar]
- 3.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat. Biotechnol. 1999;17:994. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 4.Aebersold R, Mann M. Nature. 2003;422:198. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 5.Wolters DA, Washburn MP, Yates JR. Anal. Chem. 2001;73:5683. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 6.Yuan H, Zhou Y, Zhang L, Liang Z, Zhang Y. J Chromatogr A. 2009;1216:7478. doi: 10.1016/j.chroma.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Dartiguenave C, Hamad H, Waldron KC. Anal Chim Acta. 2010;663:198. doi: 10.1016/j.aca.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 8.Amankwa LN, Kuhr WG. Anal. Chem. 1993;65:2693. [Google Scholar]
- 9.Licklider L, Kuhr WG, Lacey MP, Keough T, Purdon MP, Takigiku R. Anal. Chem. 1995;67:4170. [Google Scholar]
- 10.Bonneil E, Waldron KC. Talanta. 2000;53:687. doi: 10.1016/s0039-9140(00)00554-3. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Oleschuk R, Ouchen F, Li J, Thibault P, Harrison DJ. Rapid Commun. Mass Spectrom. 2000;14:1377. doi: 10.1002/1097-0231(20000815)14:15<1377::AID-RCM31>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Gao J, Xu J, Locascio LE, Lee CS. Anal. Chem. 2001;73:2648. doi: 10.1021/ac001126h. [DOI] [PubMed] [Google Scholar]
- 13.Sakai-Kato K, Kato M, Toyo'oka T. Anal. Chem. 2002;74:2943. doi: 10.1021/ac0200421. [DOI] [PubMed] [Google Scholar]
- 14.Kato M, Sakai-Kato K, Jin H, Kubota K, Miyano H, Toyo'oka T, Dulay TM, Zare RN. Anal. Chem. 2004;76:1896. doi: 10.1021/ac035107u. [DOI] [PubMed] [Google Scholar]
- 15.Ye M, Hu S, Schoenherr RM, Dovichi NJ. Electrophoresis. 2004;25:1319. doi: 10.1002/elps.200305841. [DOI] [PubMed] [Google Scholar]
- 16.Schoenherr RM, Ye M, Vannatta M, Dovichi NJ. Anal. Chem. 2007;79:2230. doi: 10.1021/ac061638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan TC, Clark DS, Stachowiak TB, Svec F, Fréchet JM. Anal. Chem. 2007;79:6592. doi: 10.1021/ac070705k. [DOI] [PubMed] [Google Scholar]
- 18.Rashkovetsky LG, Lyubarskaya YV, Foret F, Hughes DE, Karger BL. J. Chromatogr. A. 1997;781:197. doi: 10.1016/s0021-9673(97)00629-8. [DOI] [PubMed] [Google Scholar]
- 19.Le Nel A, Krenkova J, Kleparnik K, Smadja C, Taverna M, Viovy JL, Foret F. Electrophoresis. 2008;29:4944. doi: 10.1002/elps.200800431. [DOI] [PubMed] [Google Scholar]
- 20.Wojcik R, Vannatta M, Dovichi NJ. Anal. Chem. 2010;82:1564. doi: 10.1021/ac100029u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slovakova M, Minc N, Bilkova Z, Smadja C, Faigle W, Futterer C, Taverna M, Viovy JL. Lab Chip. 2005;5:935. doi: 10.1039/b504861c. [DOI] [PubMed] [Google Scholar]
- 22.Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Rapid Comun. Mass Spectrom. 2010;24:2554. doi: 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- 23.Kraly JR, Jones MR, Gomez DG, Dickerson JA, Harwood MM, Eggertson M, Paulson TG, Sanchez CA, Odze R, Feng Z, Reid BJ, Dovichi NJ. Anal. Chem. 2006;78:5977. doi: 10.1021/ac061029+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michels DA, Hu S, Dambrowitz KA, Eggertson MJ, Lauterbach K, Dovichi NJ. Electrophoresis. 2004;25:3098. doi: 10.1002/elps.200405939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.