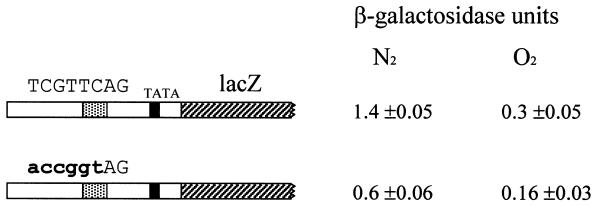Abstract
The DAN/TIR mannoprotein genes of Saccharomyces cerevisiae (DAN1, DAN2, DAN3, DAN4, TIR1, TIR2, TIR3 and TIR4) are expressed in anaerobic cells while the predominant cell wall proteins Cwp1 and Cwp2 are down-regulated. Elements involved in activation and repression of the DAN/TIR genes were defined in this study, using the DAN1 promoter as a model. Nested deletions in a DAN1/lacZ reporter pinpointed regions carrying activation and repression elements. Inspection revealed two consensus sequences subsequently shown to be independent anaerobic response elements (AR1, consensus TCGTTYAG; AR2, consensus AAAAATTGTTGA). AR1 is found in all of the DAN/TIR promoters; AR2 is found in DAN1, DAN2 and DAN3. A 120 bp segment carrying two copies of AR1 preferentially activated transcription of lacZ under anaerobic conditions. A fusion of three synthetic copies of AR1 to MEL1 was also expressed anaerobically. Mutations in either AR1 site within the 120 bp segment caused a drastic loss of expression, indicating that both are necessary for activation and implying cooperativity between adjacent transcriptional activation complexes. A single AR2 site carried on a 46 bp fragment from the DAN1 promoter activated lacZ transcription under anaerobic conditions, as did a 26 bp synthetic AR2 fragment fused to MEL1. Nucleotide substitutions within the AR2 sequence eliminated the activity of the 46 bp segment. Ablation of the AR2 sequences in the full promoter caused a partial reduction of expression. The presence of the ATTGTT core (recognized by HMG proteins) in the AR2 sequence suggests that an HMG protein may activate through AR2. One region was implicated in aerobic repression of DAN1. It contains sites for the heme-induced Mot3 and Rox1 repressors.
INTRODUCTION
Saccharomyces cerevisiae cells adapt to anaerobic growth by expressing a number of ‘anaerobic’ genes (genes which are specifically induced following oxygen depletion) (1). The regulation of some of these genes is still poorly understood, owing to a lack of knowledge about the factors and/or promoter sites which regulate their expression. However, two groups of anaerobic genes have now been identified and assigned to distinct regulons, based on the two different regulatory systems which control them. The best studied are the hypoxic genes (2–8), which are regulated by heme (9) and the Rox1 repressor (3,10,11) through one or more consensus operator sites (YYYATTGTTCTC) (4,12). Regulation occurs via control of repressor activity, with ROX1 induced by heme during aerobic growth (9,10) and repressed by the Hap1 factor during anaerobiosis (13,14). More recently a second group of heme-repressed anaerobic genes has been identified which encode cell wall mannoproteins (DAN1, DAN2, DAN3, DAN4, TIR1, TIR2, TIR3 and TIR4) (1,15–17; N.Abramova, O.Sertil, S.Mehta and C.Lowry, submitted for publication). We refer to the members of this regulon as the DAN/TIR genes. Negative regulation of some of these genes by heme in aerobic cells is quite stringent (17), resulting in undetectable levels of mRNA. The principle elements of the mechanisms which regulate them are different from those controlling the hypoxic genes (17,18).
The newly identified regulatory factors controlling expression of the DAN/TIR genes are Mox4/Upc2, a transcriptional activator (18,19), and Mox1, Mox2 and Rox7/Mot3, which are repression factors (18). Like Rox1, expression of Mot3 (20,21) is induced during aerobic growth by heme (O.Sertil, R.Kapoor, B.Cohen and C.Lowry, submitted for publication). The three Mox factors appear to be specific for the DAN/TIR regulon, while Mot3 is required for efficient repression of subsets of genes from both the hypoxic and DAN/TIR regulons (18) (O.Sertil, R.Kapoor, B.Cohen and C.Lowry, submitted for publication).
Given the number of regulatory factors controlling expression of the DAN/TIR genes, we anticipated that the promoter of DAN1 would be complex. We have identified several sequences involved in the induction of transcription under anaerobic conditions and others involved in repression under aerobic conditions. Surprisingly, we find that two different consensus sequences within the promoter serve as independent anaerobic response elements targeted by different mechanisms. One of the response elements (designated AR1) is present in all of the DAN/TIR promoters and has been implicated as the target of the Mox factors (18). The second response element (designated AR2) is targeted by a still unidentified anaerobic activation mechanism. In addition to sites mediating the two activation mechanisms, we have also identified a segment mediating aerobic repression which contains sites for Mot3 and Rox1. In addition, we have identified a second inhibitory segment which appears to be the target of a non-specific repression mechanism which may have an indirect role in maintaining aerobic repression.
MATERIALS AND METHODS
Plasmids
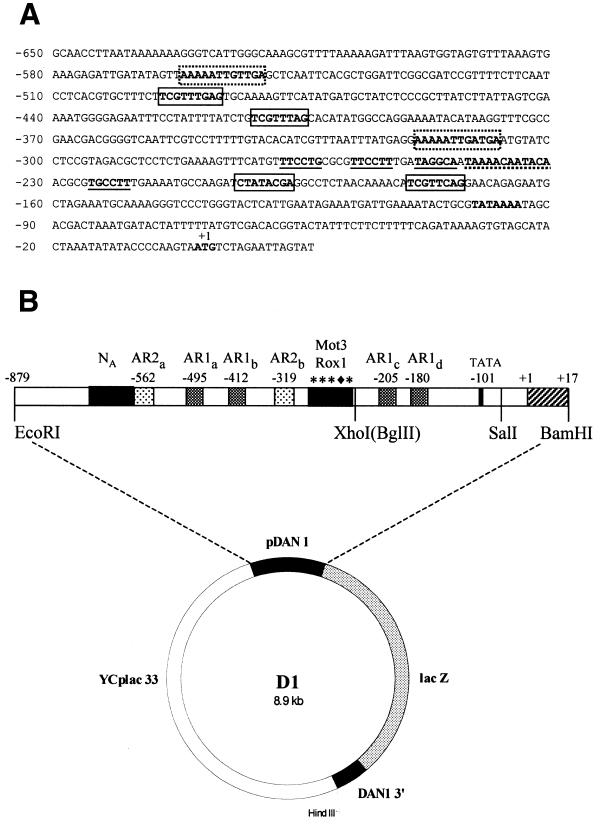
DAN1/lacZ reporter plasmids. A DAN1/lacZ reporter fusion was constructed in three steps. First the DAN1 coding region was inserted into YCplac22 (22) as a KpnI–EcoRI fragment from pBSDAN1 to create YCpDAN1-1. Then the DAN1 promoter region (–1260/+17) was positioned 5′ to the coding region on the same plasmid. For this purpose it was amplified by PCR from a library containing HindIII–NcoI inserts carried on plasmid YCplac33 (22) using the M13 reverse primer (AGCGGATAACAATTTCACACAGGA) and the non-coding strand primer A2 (5′-AGAGGGATCCATACTAATCCTAGACATTACTTGGGG), which contains a BamHI site followed by a sequence complementary to the region from –8 to +17. The PCR fragment was digested with XhoI (in the region of the polylinker adjacent to the M13 primer site) and BamHI and inserted into the SalI and BamHI sites in the polylinker region of YCpDAN1-1, creating YCpDAN1-2. To join the promoter to lacZ, a BamHI–NcoI fragment carrying the lacZ ORF was excised from the ANB1/lacZ reporter plasmid YCpAZ-H4 (13) and inserted into the BamHI (+17) and NcoI (+321) sites of YCpDAN1-2. This insertion placed lacZ in-frame with the first five amino acids of DAN1 and 5′ to the 3′-end of the DAN1 ORF, creating YCpD/Z(22). The DAN1/lacZ fusion was then placed into two other vectors. It was excised from YCpD/Z(22) with EcoRI and HindIII, generating a fragment which includes the promoter region (–879/+17) joined to lacZ flanked on the 3′-side by a small piece (+321/+408) of the DAN1 ORF, and ligated to the EcoRI and HindIII sites of YIplac128 (8) and YCplac33, creating YIpD/Z(128) and YCpD/Z(33), respectively. To facilitate mutagenesis of the promoter the BglII site at –207 of plasmid YCpD/Z(33) was converted to an XhoI site by insertion of a BglII–XhoI linker, generating plasmid D1 (Fig. 1).
Figure 1.
Plasmid D1 and the DAN1 promoter region. (A) The region of the DAN1 promoter from –879 to +17 included in the lacZ fusion is shown. AR1 sites are boxed with solid lines; AR2 sites are boxed with dotted lines; Mot3 sites are underlined with solid lines; the Rox1 site is underlined with a dotted line. (B) The D1 plasmid is shown; it contains the DAN1 promoter region joined in a coding fusion to the lacZ gene beginning with the first six amino acids of the DAN1 ORF. The DAN1 promoter region is expanded to show the position of regulatory sites. Restriction sites used in manipulation of the plasmid to generate deletions and point mutations in the promoter are indicated.
MEL1 reporter plasmids. Fusions of sequences or subfragments of the DAN1 promoter to the MEL1 gene were derived from a DAN1/MEL1 reporter fusion, which was constructed in two steps. To make use of a BamHI site at –37 in the 5′-UTR of MEL1 a BamHI site was introduced into the UTR of the DAN1 promoter segment. For this purpose the region from –879 to –1 was amplified using the coding strand primer D1 (GACGAATTCAACGATTTCAACAGTC) and non-coding strand primer D0 (GCGGCGGGATCCTACTTGGGGTATATATTTAGTATG). The PCR product was digested with EcoRI and BamHI and ligated into the same sites of D1 to create D1-X. YCpDAN1/MEL1 was then constructed by inserting the MEL1 ORF plus 37 bp of the 5′-UTR into D1-X as a BamHI–HindIII fragment from plasmid pBSKMEL1 (23) (a gift from M. Ryan and R. Morse) into the same sites of the D1-X backbone, replacing the lacZ segment. The MEL1 ORF, as configured in YCpDAN1/MEL1, was then introduced into a number of different constructs derived from D1, replacing the lacZ ORF. It was inserted as a SalI–HindIII fragment, ligated to the SalI site just 3′ to the TATA site and to the HindIII site 3′ to the lacZ segment. (AR1)3/MEL1 and AR2/MEL1 were derived from (AR1)3/lacZ and AR2/lacZ. For construction of the latter two plasmids a promoterless vector for cloning synthetic oligonucleotides was constructed by inserting an EcoRI–BstEII–XhoI linker (coding, AATTCAGCTCACCC; non-coding, TCGAGGGTGAGCTG) into plasmid C6, replacing the 46 bp segment with a BstEII site, to create D0-BST. To construct (AR1)3/lacZ a phosphorylated double-stranded oligonucleotide, containing the AR1b site and three adjacent 3′ nucleotides flanked by BstEII cohesive ends (coding, GTCACATCGTTTAGCACGTGT; non-coding, GTGACACACGTGCTAAACGAT; repeat interval 21 bp), was ligated to the BstEII site, with directional insertion (intrinsic in the BstEII site). A plasmid containing three forward-oriented concatenated copies of AR1 (repeat interval 21 bp) was isolated and converted to a MEL1 version as described above, designated (AR1)3/MEL1. AR2/MEL1, which contains a single synthetic copy of the A1 site and 14 flanking nucleotides (coding, GTCACATATAGTTAAAAATTGTTGAGCTCAAT; non-coding, GTGACATTGAGCTCAACAATTTTTAACTATAT) fused to MEL1, was constructed in analogous fashion. A control promoterless vector, D0-BST-MEL1, was constructed in analogous fashion from D0-BST.
DAN2/lacZ reporter plasmid. YCpDAN2/lacZ was constructed by inserting the promoter region of the DAN2 gene (–1105 to +3) in place of the DAN1 promoter in plasmid D1-Sal, a derivative of plasmid D1 in which a SalI–BamHI linker containing a 9 nt segment of the 5′-UTR (GATCCATTTTTGTCGACAAAAATG) had been inserted at the BamHI site. A DAN2 promoter fragment was amplified using a coding strand primer (D2A, GAGAGAATTCTACTAACGAAATGATGGCTG) containing an EcoRI site followed by homology from –1105 to –1086 and a non-coding strand primer (D2B, GAGAGTCGACTGAACTTTTGTAGATCTTTTTAGTG) containing a SalI site followed by homology from –11 to –35. The amplified promoter fragment was digested with EcoRI and SalI and ligated to the same sites of D1-Sal, placing the region from –1105 to +3 in-frame with the lacZ ORF, with an A→G substitution at –10 and an A→C substitution at –5.
Promoter deletion and subfragment plasmids. A series of plasmids derived from the DAN1/lacZ reporter plasmid was constructed containing single or double deletions of various promoter segments. To obtain the desired deletion constructs, truncated promoter fragments were generated by PCR using primers homologous to segments at the points indicated. The PCR template was either the D1 plasmid or, in the case of double deletions, a derivative deletion plasmid. For cloning purposes all of the primers contained appropriate restriction sites 5′ to the homologous sequence. Plasmid D16 carries a deletion of the segment from –207 to –107, adjacent to the TATA site at –101. For the construction a PCR fragment carrying the segment from –107 to +17 was amplified using the A2 non-coding strand primer and a coding strand primer containing an XhoI site followed by the sequence from –107 to –85. This was digested with XhoI and BamHI and ligated to the same sites in D1. Plasmid D0 carries a deletion of the entire promoter region from –897 to –107. It was constructed by excising that region from D16 with EcoRI and XhoI, end-filling with Klenow polymerase and religating.
Three sets of nested promoter deletions used in localization of regulatory sites were generated in the D1 plasmid by inserting truncated PCR fragments in place of the full-length promoter: one set of deletions extended 3′-wards from –897, one set extended 5′-wards from –107 and one set extended 5′-wards from –207. For the 3′-ward deletions, fragments of decreasing length were generated with the A2 non-coding strand primer and a set of coding strand primers, each having an EcoRI site followed by homologous sequences beginning at –762 (plasmid D2), –633 (plasmid D3), –524 (plasmid D4), –423 (plasmid D5), –354 (plasmid D6), –323 (plasmid D7), –304 (plasmid D8) or –207 (plasmid D9). The PCR fragments were digested with EcoRI and BamHI and ligated to the EcoRI and BamHI sites of D1 (located at –897 and +17). For deletions extending 5′-wards from –107 a set of fragments was generated with the A3 coding strand primer (GACGAATTCAAACGATTTCAACAGTC, homologous from –897 to –880) and a set of non-coding strand primers, each containing an XhoI site followed by homologous sequences beginning at –301 (plasmid D17), –353 (plasmid D18), –424 (plasmid D19), –547 (plasmid D20) or –547 (plasmid D24). The set of PCR fragments was digested with EcoRI and XhoI and ligated to the EcoRI and XhoI sites of D16 (located at –897 and –107). For an equivalent set of deletions extending 5′-wards from –207 the same EcoRI- and XhoI-digested PCR fragments were ligated to the EcoRI and XhoI sites of D1 (at –897 and –207).
Plasmids containing small segments of the promoter fused to lacZ were constructed by deleting the sequences flanking them from D1 or from a derivative. To construct plasmid C6, which contains the segment from –570 to –525 inserted at –107, the flanking segments, –879 to –570 and –525 to –107, were deleted, leaving the segment in the desired position. The insert for this construction was amplified from the segment between –570 and +17 in plasmid D20 (in which the segment from –525 to –107 is deleted); the primers used were a coding strand primer containing an EcoRI site followed by the sequence from –570 to –547 and the A2 non-coding strand primer. The 169 bp PCR product was digested with EcoRI and BamHI and ligated to the EcoRI and BamHI sites of D1. Plasmid C5 carries the –570 to –525 segment inserted at –207 instead of –107 and was derived in an analogous way using the same PCR primers to amplify a 269 bp fragment from plasmid D13 (in which the segment from –525 to –207 was deleted). Plasmids C1 and C2 are similar to C5 and C6, but contain the region from –633 to –525 inserted at –207 and –107, respectively. The insert for the two constructs was amplified from the D13 and D20 templates using a coding strand primer containing an EcoRI site followed by the sequence from –633 to –615 and the A2 non-coding strand primer. The two products were digested with EcoRI and BamHI and ligated to the same sites in D1. Plasmid C11 contains the segment from –524 to –403 inserted at –107. It was amplified from the –524 to –403 region using a coding strand primer containing an EcoRI site followed by a homologous sequence from –524 to –504 and a non-coding strand primer containing an XhoI site followed by homologous sequence from –403 to –425. The product was digested with EcoRI and XhoI and ligated to the same sites in D16.
Nucleotide substitutions in the DAN1 and DAN2 promoters. Putative regulatory sites were mutated in some of the plasmid constructs described above by substituting PCR-derived segments containing nucleotide substitutions. Plasmids M9 and M10 are mutant derivatives of C11 in which the AR1b sequence (–413) near the end of the –524 to –403 segment was altered. The mutation in M9 (TCGTTTAG→GTACCTAG) was created by using a substituted non-coding strand primer (GCGCTCGAGTGCTAGGTACCAGATAAAATAGGAAATTCTCCC) plus the same coding strand primer used for C11 to generate an altered fragment, which was then digested with EcoRI and XhoI and inserted into the same sites of D16. M10 contains a point mutation in AR1b (TCGTTTAG→TCATTTAG). This was introduced on a PCR fragment generated as above, but with a non-coding strand primer containing the indicated substitution GCGCTCGAGTGCTAAATGACAGATAAAATAGG.
Plasmid M11 contains a triple substitution in the AR1a site at –495 (TCTTCGTTTGAG→TCTAGATTTGAG) and was generated by insertion of two mutated fragments flanking AR1a (from –524 to –495 and from –495 to –403) in place of the wild-type –524 to –403 fragment of C11, in a triple ligation reaction. The 5′-insert was a synthetic double-stranded oligonucleotide containing the sequence from –524 to –495 flanked by EcoRI and XbaI cohesive ends (coding, AATTCCCGTTTCTTCAATCCTCACGTGCTTT; non-coding, CTAGAAAGCACGTGAGGATTGAAGAAAACGGG). The 3′-insert was a PCR product generated with the same non-coding strand primer used for the C11 construct and a coding strand primer containing an XbaI site followed by homology from –492 to –470 (GAGATCTAGATTTGAGTGCAAAAGTTCATATG). This fragment was digested with XbaI and XhoI. The two fragments were ligated together into the EcoRI- and XhoI-digested C11 backbone. In the resulting plasmid nucleotides 495–496 (TCG) have been replaced by AGA, disrupting the ARE and generating the 3′-half of the XbaI site.
Plasmids M2, M4 and M6 contain substitutions in the A1 consensus site beginning at –562. These were made using the same scheme as for C6 with substitutions in the coding strand primer. In each case a PCR fragment generated with one of the substituted primers and the A2 anchor primer was digested with EcoRI and BamHI and ligated to the D1 backbone as before. The plasmids contain the following substitutions: M2, AAAAATTGTTGA→AAAAATTATTGA, generated with coding strand primer GCGGAATTCATATAGTTAAAAATTATTGAGCTCAATTC; M4, AAAAATTGTTGA→AAAAACCGTTGA, generated with coding strand primer GCGGAATTCATATAGTTAAAAACCGTTGAGCTCAATTC; M6, AAAAATTGTTGA→GTCTCTTGTTGA, generated with coding strand primer GCGGAATTCATATAGTTGTCTCTTGTTGAGCTCAATTCACG.
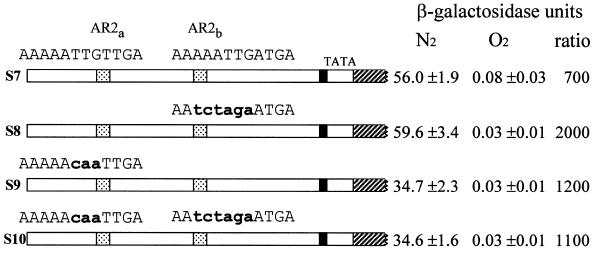
A series of plasmids containing multiple substitutions in AR2a and/or AR2b (S8, S9 and S10) and a control plasmid (S7) were constructed in the following sequence. In the first step in the series of constructs plasmids S1 and S2 were generated in parallel. S1 contains the segment –878 to –547 inserted at –207, leaving the region from –547 to –207 deleted and replaced by an XhoI site. The insertion fragment was generated by PCR using the coding strand primer A3 and the non-coding strand primer D21, digested with EcoRI and XhoI and ligated into the same sites in YCpD1. S2 is analogous to YCpS1, containing the same –878 to –547 segment but with a TTGTTG→ CAATTG substitution in the AR2a site at –557. The insertion fragment was generated using primer A3 and non-coding strand primer S2 (CAGCGTGTCTCGAGCTCAATTGTTTTTAACTATATCAATC).
In the second step of the series, plasmids S3, S4, S5 and S6 were constructed. In each case the segment from –546 to –301 was inserted into the XhoI and BamHI sites of S1 or S2. For S4 and S6 this fragment contained a mutation in the AR2b site; for S3 and S5 the unmutagenized control fragment was used. These fragments were generated using the coding strand primer S11 (GAGGTCGACACAGGCTGGATTCGGCGATC) and either the wild-type non-coding strand primer S3 (GCGGGATCCGAGCTCGAGGATACATTCATCAATTTTTC) or the AR2-substituted non-coding strand primer S4 (GCGGGATCCGAGCTCGAGGATACATTCATTCTAGATTCCTC) (this primer introduces a AAATTG→TCTAGA substitution at –317). The fragments were digested with SalI and BamHI and inserted into the XhoI and BamHI sites of S1 or S2 (the inserted fragments contained an XhoI site just 5′ to the BamHI site). In this way mutant and intact AR2a and AR2b segments were joined to generate the four possible combinations of intact and mutant sites. In the resulting plamids S3 contained intact AR2a and AR2b sites, S4 contained intact AR2a and mutant AR2b, S5 contained mutant AR2a and intact AR2b and S6 contained both mutant sites. In the final step the fragment from –301 to +17 was restored by insertion of a PCR product generated with the coding strand primer S7 (GACCTCGAGCGTAGACGCTCCTCTGAAAAG) and the non-coding strand primer A2. The product was digested with XhoI and BamHI and inserted into the same sites in S3, S4, S5 and S6 to generate S7, S8, S9 and S10, respectively.
Plasmid YCpDAN2/lacZ-AR1x contains a substitution in the AR1 site at – 847 (TCGTTCAG→ACCGGTAG) and was constructed by a triple ligation of two altered promoter subfragments (–1105 to –848 and –847 to +3) in place of the promoter region in YCpDAN2/lacZ. The fragment flanking the AR1 site on the 5′-side was generated using the D2A coding strand primer and a non-coding strand primer containing an AgeI site followed by the sequence complementary to the segment from –848 to –866 (GAGAACCGGTGGGCAATATCCAATGTTTGAC). The 3′-fragment was generated using the D2B non-coding strand primer plus a coding strand primer containing an AgeI site followed by a sequence homologous to the segment from –842 to –821 (GAGAACCGGTAGAGGGCACAGAATAAAATTTG). The two PCR fragments were digested with EcoRI and AgeI and SalI and AgeI, respectively, and ligated to the EcoRI- and SalI-digested D1-Sal backbone.
Cell growth and analysis of gene expression
For β-galactosidase assays (24) FY23 cells (25) were grown in SD medium (2% glucose) lacking uracil to maintain selection of the reporter plasmid. Overnight cultures in early stationary phase were diluted 1:25 (to ∼5 × 106/ml) and grown aerobically for 2 h and then anaerobically or aerobically for 7 h. Anaerobic conditions were imposed by bubbling cultures in sealed tubes with high purity nitrogen (99.999% according to the manufacturer, though this was not measured; very similar results were obtained with different lots). At 7 h cells are still growing in mid to late log phase under either condition. There is an ∼3 h lag in lacZ expression, but 7 h anaerobic culture is long enough to give consistent and robust expression from the DAN1/lacZ fusion. For α-galactosidase assays cells were grown in the same way and assayed as described (23). Data shown are for assays replicated three times.
RESULTS
Anaerobic response sequences in the DAN1 promoter
Expression of a DAN1/lacZ reporter (plasmid D1; Fig. 1) reporter is strictly dependent on hypoxic growth conditions, showing a >300-fold difference in expression in anaerobic and aerobic cultures (Fig. 2). Anaerobic conditions imposed by bubbling log phase cells with high purity nitrogen may not be absolute but are sufficiently stringent to observe induction either of the DAN1/lacZ reporter or of DAN1 mRNA (18). DAN1 mRNA, like the mRNAs of most of the other DAN/TIR genes, is absent from aerobic cells and is first detected after 30 min of hypoxia and fully induced by 80 min (17). DAN2 and DAN3 are expressed only after a prolonged delay, being fully induced at 3 h (N.Abramova, O.Sertil, S.Mehta and C.Lowry, submitted for publication). We earlier demonstrated that heme, which is synthesized only in aerobic cells, is the primary regulatory signal: accumulation of DAN1 mRNA is completely blocked by addition of heme (17); similarly, expression of the DAN1/lacZ fusion is >95% inhibited by heme (18), although a role for other environmental changes or for metabolites or nutrients generated or depleted during anaerobiosis has not been ruled out.
Figure 2.
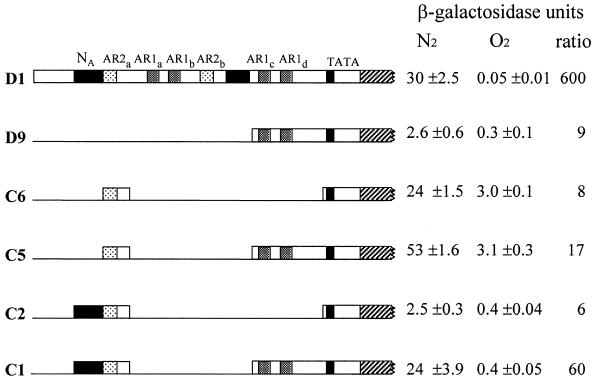
Nested deletions in the DAN1 promoter. FY23 cells carrying the D1 plasmid or derivatives carrying the indicated deletions (see Materials and Methods) were grown in SD-ura medium under aerobic and anaerobic conditions (8 h) and harvested for β-galactosidase assay, with the results shown in Miller units. The induction ratio, i.e. the ratio of activity in anaerobic cells to activity in aerobic cells, is indicated.
To locate regions containing sequences involved in anaerobic activation and/or aerobic repression of DAN1 we constructed a set of nested deletions in plasmid D1 and tested them for activity under aerobic and anaerobic growth conditions. Several segments appeared to be involved in anaerobic induction, as evidenced by reduced anaerobic expression when they were deleted (Fig. 2). These segments contained two consensus sequences, TCGTTYAG and AAAAATTGTTGA (Table 1), which were designated AR1 and AR2 (for anaerobic response element). There is at least one copy of AR1 in the promoters of each of the dan/tir genes (Table 1) and four in the DAN1 promoter. The AR2 sequence is found in DAN1, DAN2 and DAN3. Sequences within one segment (–301 to –207) also appear to be involved in aerobic repression, judging from the ∼20-fold increase in aerobic expression when it was deleted (plasmid D15; see below).
Table 1. Anaerobic induction sites in the DAN/TIR regulon.
| Gene |
ORF |
|
Sites |
|
| (A) AR1 sites | ||||
| DAN1 | YJR150C | AR1a | TCGTTAGAG | –495 to –487 |
| AR1b | TCGTTT-AG | –412 to –405 | ||
| AR1c | TCGTAT-AG (←) | –197 to –204 | ||
| AR1d | TCGTTC-AG | –180 to –173 | ||
| DAN2 | YLR037C | TCGTTC-AG | –447 to –440 | |
| DAN3 | YBR301W | TCGTTC-AG (←) | –425 to –432 | |
| TCGTTCAAG | –234 to –226 | |||
| DAN4 | YJR151C | TCGTTC-AG | –195 to –188 | |
| TIR1 | YER011W | TCGTAT-AG (←) | –520 to –527 | |
| TCGTTTAAG | –350 to –342 | |||
| TIR2 | YOR010C | TCGTAA-AG | –374 to –367 | |
| TIR3 | YIL011W | TCGTTT-AG (←) | –853 to –860 | |
| TIR4 | YOR009W | TCGTTC-AG (←) | –734 to –741 | |
| TCGTTT-AG (←) | –215 to –222 | |||
| Consensus | TCGTTYAG | |||
| (B) AR2 sites | ||||
| DAN1 | YJR150C | AR2a | AAAAATTGTTGA | –561 to –550 |
| AR2b | AAAAATTGATGA | –318 to –307 | ||
| DAN2 | YLR037C | ATAAATTGTTGA (←) | –142 to –153 | |
| DAN3 | YBR301W | CAAAAATGTTGA (←) | –571 to –581 | |
| Consensus | AAAAATTGTTGA |
Properties of the DAN/TIR anaerobic response element (AR1)
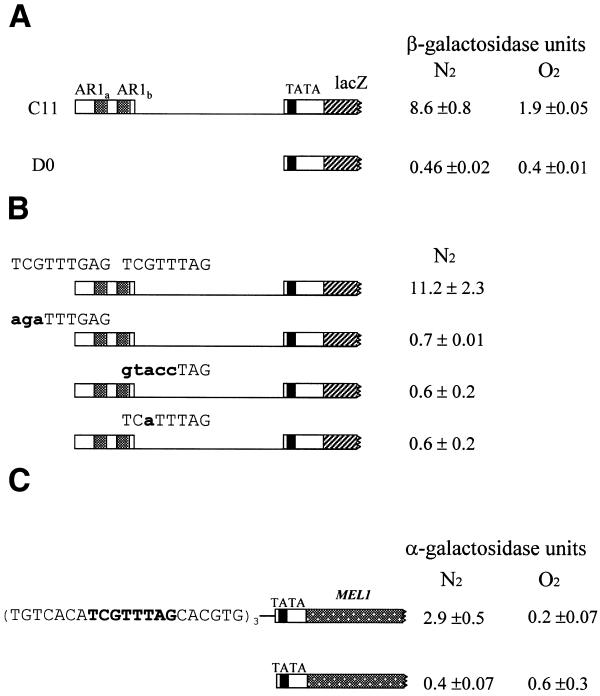
We tested for anaerobic expression from a reporter plasmid (C11) which carries AR1a and AR1b (but no AR2 sites) on a 120 bp segment (–524 to –403) inserted at the TATA site of the lacZ fusion. Reporter activity was about four times higher in anaerobic than in aerobic cells, showing that this segment contains a response element (Fig. 3A). To test the importance of the sequence in transcriptional activation we introduced nucleotide substitutions into either AR1a or AR1b in C11. Substitutions at either site caused a drastic loss of activity (Fig. 3B). We also observed a strong effect of a single substitution for the 5′-most of the two G residues in AR12 (not shown).
Figure 3.
Activation of transcription dependent on AR1 sites. FY23 cells carrying plasmids with fragments fused to the lacZ ORF or oligonucleotides fused to MEL1 (see Materials and Methods) were grown as described in Figure 2. (A) Cells were transformed with plasmid C11, which contains the promoter fragment from –524 to –403 fused at the TATA site to the lacZ reporter. Transformants were grown under aerobic and anaerobic conditions. (B) Cells transformed with plasmid C11 or mutant derivatives containing the indicated nucleotide substitutions were grown under anaerobic conditions. (C) Cells were transformed with the (AR1)3/MEL1 plasmid, which contains three copies of the AR1 sequence fused at the TATA site to the MEL1 reporter; control cells were transformed with the promoterless vector D0-BST. Transformants were grown under anaerobic and aerobic conditions as above and harvested for α-galactosidase assay as described in Materials and Methods.
The fact that mutation of either AR1 site in the –524 to –403 fragment eliminated nearly all activity showed that two copies of AR1 are required for substantial transciptional activation by the –524 to –403 segment of DAN1. This suggested that there is a cooperative interaction between neighboring activation complexes forming on this segment.
The AR1 site is sufficient for anaerobically induced promoter activity
Given that the AR1 site is necessary for expression from a DAN1 activation segment, we tested whether it is sufficient for activation in a plasmid containing three synthetic concatenated copies fused to the MEL1 gene. Cells transformed with the (AR1)3/MEL1 construct showed significant anaerobically induced activity (Fig. 3C), compared to aerobic cells. Anaerobic expression driven by AR1 implies that it is sufficient to provide a binding site for a transcriptional activator which responds to heme regulation.
The AR1 site of DAN2 is also involved in anaerobic induction
To determine whether the AR1 sequence plays an equivalent role in the promoters of other genes in the regulon we constructed two plasmids containing a fusion of the DAN2 promoter to lacZ. YCpDAN2/lacZ contains the wild-type promoter and YCpDAN2/lacZ-AR1x contains a mutated version in which the single AR1 site at –447 is disrupted by a triple nucleotide substitution. We observed weak but significant anaerobic induction from the wild-type fusion (Fig. 4). DAN2 expression is characterized by a prolonged delay (>2 h) in expression; this delay, combined with the 3–4 h lag in β-galactosidase synthesis from lacZ fusions driven by anaerobically induced promoters, may account for the relatively low output of the reporter during the 7 h incubation. Substituting 5 nt in the AR1 site caused a 60% decrease in expression (Fig. 4). This shows that the AR1 site plays a role in induction of DAN2. However, the effect of eliminating this site is much less pronounced than the effect of eliminating either of the AR1 sites in the –524 to –403 segment of DAN1 (Fig. 3); the residual activity suggests that a neighboring site(s) in the DAN2 promoter (e.g. the AR2 site at –142) plays a combinatorial role with the AR1 complex. The fact that this activity is still oxygen regulated implies that the hypothetical site is also an anaerobic response element (such as AR2; see Discussion).
Figure 4.
Importance of the AR1 site to expression of DAN2. FY23 cells transformed with YCpDAN2/lacZ or YCpDAN2/lacZ-AR1x, a derivative containing a disruption of the single AR1 site in the DAN2 promoter (–847), were grown under anaerobic and aerobic conditions and harvested for lacZ assay.
A second activation mechanism works through the AR2 response element
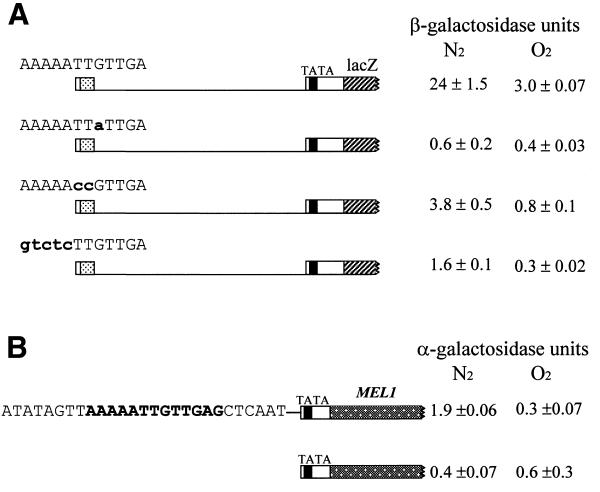
There are two copies of the AR2 consensus (AR2a and AR2b) in the DAN1 promoter and one each in those of DAN2 and DAN3 (Table 1B). AR2a contains an ATTGTT core, which is of interest because it is also the core of the binding sites of a number of HMG proteins, including Rox1. We tested a fusion of a 46 bp fragment (–570 to –525) containing AR2a to the lacZ reporter (plasmid C6) in aerobic and anaerobic cells. Expression was substantial (Fig. 5A) and was 8-fold higher in anaerobic cells than in aerobic, indicating that the segment contained an anaerobic response element. Aerobic repression of expression from C6 was not as stringent as repression of the full promoter. Nucleotide substitutions across the AR2 sequence in the C6 construct caused full or partial loss of expression (Fig. 5A), confirming its role as an activation site. These substitutions included two within the ATTGTT core, indicating the importance of this segment. Where mutations resulted in partial activity, it appeared still to be regulated, as would be expected for a mutant activation site with reduced affinity for a heme-inhibited activator, i.e. one controlled through its activation function rather than its affinity. To narrow down the segment sufficient for expression we tested a 27 bp synthetic fragment (–570 to –544) including the AR2a site and 15 flanking nucleotides in a fusion to the MEL1 gene in plasmid AR2/MEL1 (Fig. 5B). Again we observed elevated expression in response to hypoxia (Fig. 5B).
Figure 5.
Function of the AR2 site in anaerobic induction. (A) FY23 cells were transformed with the C6 plasmid, which contains the AR2 site on a 46 bp fragment, or, in descending order, by plasmid M2, M4 or M6. The latter three plasmids are derivatives of C6 generated by introduction of the indicated nucleotide substitutions within the AR2 site (M2, AAAAATTaTTGA; M4, AAAAAccGTTGA; M6, gtctcTTGTTGA). Transformed cells were grown under anaerobic and aerobic conditions and analyzed for β-galactosidase as described in Figure 2. (B) FY23 cells were transformed with the plasmid AR2/MEL1 or the MEL1 vector. Transformants were grown under anaerobic and aerobic conditions and analyzed for α-galactosidase as described in Materials and Methods.
To assess the contribution of the two AR2 sites to anaerobic induction in the full promoter we constructed derivatives of plasmid D1 containing nucleotide substitutions in either the AR2a or AR2b site, or both. Loss of AR2a caused a 40% reduction in activity, as did loss of both AR2a and AR2b; loss of AR2b alone had no effect (Fig. 6). This showed that the activation mechanism operating through the AR2a site does contribute to the activity of the full promoter. However the AR2b site appears to play no role; significantly, it differs from AR2a in the six base core (ATTGAT instead of ATTGTT). One simple explanation for anaerobic induction through the AR2 site is that it is alternately occupied by an activator during anaerobic growth and by Rox1 in aerobic cells; however, elimination of the two AR2 sites did not cause any increase in aerobic expression, arguing against a role for these sites in mediating repression.
Figure 6.
Role of AR2 sites in the full promoter. FY23 cells carrying derivatives of plasmid D1 containing nucleotide substitutions in either or both AR2 sites (AR2a and AR2b as defined in Fig. 1) and a control plasmid (S7) with normal AR2 sites were grown and analyzed as in Figure 1. All four plasmids contain identical substitutions adjacent to the AR2 sites introduced to facilitate plasmid construction.
The role of Mot3 and Rox1 sites in aerobic repression
Extensive deletions in the DAN1 promoter failed to reveal any element essential for blocking aerobic expression, i.e. none of the deletions eliminated the difference between aerobic and anaerobic expression levels. However, we would not expect any repression sites which are present to be solely responsible for blocking aerobic expression, because the Mox4 activator is itself the target of a heme-dependent repression mechanism, i.e. it is less active in aerobic cells (18). Even so, the segment between –301 and –207 does appear to play a contributory role in aerobic repression; deletion of this region from the full promoter of the D1 plasmid, generating plasmid D15, resulted in a significant increase in aerobic expression (Fig. 2) and, consequently, a decrease in the anaerobic/aerobic induction ratio from >500 to 26. This suggested that a heme-responsive repressor binds in this region. The segment contains a cluster of four moderate to high affinity sites for the heme-induced repression factor Mot3 (see Fig. 1 for sites at –268, –258, –249 and –225) (9,19,22) and a potential Rox1 site (–242), providing a likely explanation for the importance of this segment to aerobic repression. We earlier observed that deletion of MOT3 caused a substantial loss of repression of DAN1 and other DAN/TIR genes (O.Sertil, R.Kapoor, B.Cohen and C.Lowry, submitted for publication), as did deletion of ROX1, but to a lesser extent.
A non-specific inhibitory site
A comparison of anaerobic and aerobic expression from two of our lacZ constructs revealed an inhibitory segment in the region between –630 and –570: the C6 construct, which contains only the 46 bp AR2a segment (–570 to –525) fused to the TATA box, showed strong activity (Fig. 7), while the D20 construct, which contains the same segment plus the adjoining 5′-region (–878 to –571), was only weakly active. To further localize the inferred inhibitory segment we tested a plasmid (C2) which contains the AR2a segment plus the adjacent 63 bp upstream segment. Inclusion of this segment, designated NA, upstream from the AR2a element caused a strong reduction in expression of both anaerobic and aerobic activity (compare C2 and C6) (Fig. 7), indicating that a sequence within NA is targeted by a non-specific repression mechanism.
Figure 7.
Expression by plasmids containing combinations of regulatory segments. FY23 cells were transformed with various plasmids containing segments of the DAN1 promoter fused to lacZ and grown under anaerobic or aerobic conditions and analyzed as described in Materials and Methods. Cells carried D1 or plasmids containing the fragments indicated fused at –107, 5 bp upstream from the TATA site: D9, (–207 to –107); C6, (–570 to –525); C2, (–633 to –525); C5, (–570 to –525 plus –207 to –107); C1, (–633 to –525 plus –207 to –107).
Combinatorial activation and repression in minimal constructs
The DAN1 promoter contains a large number of sites which mediate the combined action of several regulators. Clearly both the AR1 and AR2 mechanisms can function independently, as demonstrated by the activity of the C6 and C11 constructs (Figs 3 and 5). To explore further how these mechanisms combine we constructed a plasmid (C5) containing the weakly active 100 bp AR1c–AR1d segment and the 46 bp AR2a segment in tandem, fused to the lacZ reporter at the TATA site (Fig. 7). The activity of C5 was twice the sum of the activities of the C6 and D9 plasmids, which contain the AR2a and AR1c–AR1d segments, respectively, indicating a modest synergy in their contributions to activation of expression. There was also a net increase in the induction ratio resulting from the combination. Another, more striking, kind of synergy between the AR2 and AR1c–AR1d segments was evident as decreased sensitivity to repression by the NA mechanism. This segment inhibited the AR1–AR2 combination far less than it did the AR2 site alone (compare C1 and C2), hence, with the inhibitory segment present the activity of the combined segments was now about eight times higher than the sum of the activities of the equivalent individual segments (compare C1 with D9 plus C2).
Comparison of the expression of these constructs under anaerobic and aerobic conditions suggested that the NA segment might contribute to heme repression in aerobic cells. As described above, inclusion of the NA segment had an equally strong effect in aerobic and anaerobic cells on activation by the AR2a segment (compare C6 and C2). However, it had a differential effect on aerobic and anaerobic expression driven by the combined AR1–AR2 segments; thus inhibition resulting from inclusion of the segment was greater in aerobic cells (∼8-fold) than in aerobic cells (∼2-fold) (compare C5 and C1). Hence, in this artificial context it appeared that the ‘non-specific’ repressor functioned to improve aerobic repression and, consequently, the anaerobic/aerobic induction ratio. Non-specific repressors have been shown to enhance induction ratios in other settings (26).
DISCUSSION
The DAN1 promoter is a mosaic of positive and negative regulatory elements, targeted in combinatorial fashion by various complexes which activate transcription during anaerobiosis and block expression during aerobic growth. Although we have not directly demonstrated the role of heme in regulating through the separate elements of the DAN1 promoter, it is clear that heme is a primary signal co-effector for the promoter as a whole, since addition of heme to anaerobic cultures causes complete loss of expression (17); the fact that heme is metabolically inert under anaerobic conditions implies that it functions directly in regulation rather than through an effect on metabolism. However, it is possible that metabolic or environmental changes associated with anaerobic growth (e.g. reduced pH, glucose depletion, alcohol production, unsaturated fatty acid, sterol depletion, etc.) are also factors in gene induction through one or more of the sites we have identified in this study.
Our observations revealed an unusual situation in which two different activation mechanisms were controlled by the same signal function on the same promoter. The first of the contributing activation mechanisms operates through the AR1 site, which appears at least once in each of the dan/tir promoters. Thus it is a common response element defining the regulon. We found that AR1 is necessary and sufficient for anaerobically induced transcriptional activation in two different experimental settings: first, AR1 site mutations in promoter segments of DAN1 and DAN2 caused loss of activity; second, there was substantial anaerobically induced expression from a fusion of concatenated AR1 sites to a MEL1 reporter [(AR1)3/MEL1] and also from lacZ reporter constructs (C11, D9, etc.) driven by promoter subsegments carrying AR1 sites.
We have reported elsewhere (18) evidence that Mox4, Mox1 and Mox2 function through the AR1 site. Mox4 is a binucleate zinc cluster protein and it might be expected to function in a dimer or a heterodimer complex. Since there was no dyad symmetry or repeated motif apparent to us in the AR1 site or its context, dimers might be expected to incorporate neighboring AR1 sites. One observation supporting this notion was that the individual complexes (presumably including Mox4) which bind to AR1 sites are not able to function independently, as shown by a nearly total loss of promoter activity when either AR1 site was eliminated from C11, indicating that the activation factor which binds to these sites functions cooperatively.
The second anaerobic induction mechanism operates independently through the AR2 site in the promoter of DAN1, and possibly in DAN2 and DAN3. This was clear first because a synthetic AR2a site stimulated anaerobic expression of reporters and second because nucleotide substitutions in the AR2a site reduced the activity of the full DAN1 promoter and eliminated the activity of a fragment carrying AR2a. Significantly, the fact that the putative AR2 activation complex activates transcription efficiently from a 46 bp fragment carrying only one AR2 site (plasmid C6) indicates that cooperative interactions are not required for it to function, in contrast to the AR1 site mechanism.
The factor(s) which activates expression through AR2 has not been identified. We speculate that it may be an HMG protein, because AR2 contains an HMG core binding sequence (ATTGTT). Mutations affecting this factor would not have shown up in our earlier search for activators of DAN1 (18), since elimination of both AR2 sites caused only a 40% reduction in expression. The AR2 site does not appear to be a target for repression by Rox1: we observed that ablation of the two AR2 sites reduced anaerobic expression as expected, but did not cause detectable loss of aerobic repression of DAN1/lacZ expression. This argues against the possibility that the AR2 activator is replaced by the repressor during aerobic growth.
There are single AR1 sites in the regulatory regions of four of the DAN/TIR genes (Table 1) and we found that the one in the DAN2 promoter is important for induction. This was surprising, since we had also found that both the AR1a and AR1b sites are strongly required for expression of a reporter construct containing the –524 to –403 segment of DAN1, implying that a single complex is ineffective and that some form of cooperativity is needed. Hence, it is not clear how single AR1 sites (in DAN2, DAN4, TIR2 and TIR3) can contribute significantly to anaerobic induction and, conversely, how loss of the AR1 site in DAN2 can have any impact (Fig. 3). One way in which a single AR1 site could participate in activation would be through an interaction with another factor bound at another site, e.g. one which binds to the AR2 site at –142. This would also explain why the residual activity is still oxygen regulated, since AR2 is also an anaerobic response site. Fortuitously, two of the DAN1 deletion constructs were analogous to the DAN2/lacZ and DAN2/lacZAR1x constructs and behaved in a similar manner: plasmid D19, which contains the AR2a site and a single AR1 site (AR1a) was moderately active, but plasmid D20, in which the AR1a site was deleted, was ∼60% less active (Fig. 2). Another factor which may join with Mox4 in activation of DAN2 is the homologous Ecm22 protein, which is needed for full expression of DAN2 and DAN3 (N.Abramova and C.Lowry, unpublished results) and is known to play a role in cell wall synthesis (27). Clearly the full set of regulators and regulatory sites controlling expression of the DAN/TIR regulon has yet to be accounted for. Some of these are likely to be involved in mediating signals other than heme, such as temperature (the TIR genes) and temporal ordering of cellular events (DAN2 and DAN3).
In several contexts the AR1 and AR2 complexes appear to act in concert, either in synthetic constructs or in the full promoter. The most striking instance of combinatorial action was provided by a construct (C1) in which a 108 bp fragment containing AR2a and the NA segment was joined to a 100 bp segment containing AR1c and AR1d; the combined segments showed strong well-regulated activity about 10 times higher than that of either segment alone (D9 and C2). The synergizing effect of the inhibitory mechanism suggests that the AR1 and AR2 mechanisms overwhelm a repressing effect when their combined strength passes a certain threshold. We observed a similar effect when we combined two AR2a sites with the NA site: in this case the activity was increased ∼20-fold by addition of the second AR2 site (data not shown). It is difficult to extrapolate from the properties of promoter subfragments to those of the full promoter, but these experiments emphasize the need to understand the balance of repression and activation in accounting for the net output of the combinatorial ensemble.
Like activation, repression of the DAN/TIR genes is also combinatorial: we earlier found that at least four heme-responsive repressors and Tup1 and Ssn6 are required to fully repress DAN1 and other members of the regulon during anaerobiosis (18). Two of these repression factors, Mox1 and Mox2, appear to function through the AR1 site, possibly through an interaction with Mox4 or by competing with it for binding to AR1. In addition, Mot3, and to a lesser extent Rox1, play a role in aerobic repression. The one region identified in our deletion study which clearly contributes to aerobic repression (–301 to –207) does contain several Mot3 sites (at least two of high affinity; 21), as well as a Rox1 site.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Steve Hanes, Randy Morse and Mike Ryan for helpful suggestions. This research was supported by grants (MCB-9723565 and MCB-9976571) from the National Science Foundation.
References
- 1.Zitomer R.S. and Lowry,C.V. (1992) Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev., 56, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowry C.V., Weiss,J.L., Walthall,D.A. and Zitomer,R.S. (1983) Modulator sequences mediate the oxygen regulation of CYC1 and a neighbouring gene in yeast. Proc. Natl Acad. Sci. USA, 80, 151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowry C.V. and Zitomer,R.S. (1984) Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc. Natl Acad. Sci. USA, 81, 6129–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowry C.V., Cerdan,M.E. and Zitomer,R.S. (1990) A hypoxic consensus operator and a constitutive activation region regulate the ANB1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 5921–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zagorec M. and Labbe-Bois,R. (1986) Negative control of yeast coproporphyrinogen oxidase synthesis by heme and oxygen. J. Biol. Chem., 261, 2506–2509. [PubMed] [Google Scholar]
- 6.Hodge M.R., Singh,K. and Cumsky,M.G. (1990) Upstream activation and repression elements control transcription of the yeast COX5b gene. Mol. Cell. Biol., 10, 5510–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turi T.G. and Loper,J.C. (1992) Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11). J. Biol. Chem., 267, 2046–2056. [PubMed] [Google Scholar]
- 8.Drgon T., Sabova,L., Nelson,N. and Kolarov,J. (1991) ADP/ATP translocator is essential only for anaerobic growth of yeast Saccharomyces cerevisiae. FEBS Lett., 289, 159–162. [DOI] [PubMed] [Google Scholar]
- 9.Lowry C.V. and Lieber,R.H. (1986) Negative regulation of the Saccharomyces cerevisiae ANB1 gene by heme, as mediated by the ROX1 gene product. Mol. Cell. Biol., 6, 4145–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowry C.V. and Zitomer,R.S. (1988) ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol. Cell. Biol., 8, 4651–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balasubramanian B., Lowry,C.V. and Zitomer,R.S. (1993) The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol. Cell. Biol., 13, 6071–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehta K.D. and Smith,M. (1989) Identification of an upstream repression site controlling the expression of an anaerobic gene (ANB1) in Saccharomyces cerevisiae. J. Biol. Chem., 264, 8670–8675. [PubMed] [Google Scholar]
- 13.Keng T. (1992) Hap1 and Rox1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 2616–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deckert J., Perini,R., Balasubramanian,B. and Zitomer,R.S. (1995) Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics, 139, 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalski L.R.Z., Kondo,K. and Inouye,M. (1995) Cold-shock induction of a family of Tip1-related proteins associated with the membrane in Saccharomyces cerevisiae. Mol. Microbiol., 15, 341–353. [DOI] [PubMed] [Google Scholar]
- 16.Donzeau M., Bourdineaud,J.P. and Lauquin,G.J. (1996) Regulation by low temperature and anaerobiosis of a yeast gene specifying a putative GPI-anchored plasma membrane protein. Mol. Microbiol., 20, 449–459. [DOI] [PubMed] [Google Scholar]
- 17.Sertil O., Cohen,B.D., Davies,K.J.D. and Lowry,C.V. (1997) The DAN1 gene of S.cerevisiae is regulated in parallel with the hypoxic genes, but by a different mechanism. Gene, 192, 199–205. [DOI] [PubMed] [Google Scholar]
- 18.Abramova N.E., Cohen,B.D., Sertil,O., Davies,K.J.A. and Lowry,C.V. (2001) Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in S. cerevisiae. Genetics, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crowley J.H., Leak,F.W., Shianna,K.V., Tove,S. and Parks,L.W. (1998) A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol ., 180, 4177–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madison J.M., Dudley,A.M. and Winston,F. (1998) Identification and analysis of Mot3, a zinc finger protein that binds to the retrotransposon Ty long terminal repeat (delta) in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 1879–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grishin A.V., Rothenberg,M., Downs,M.A. and Blumer,K.J. (1998) Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signalling in Saccharomyces cerevisiae. Genetics, 149, 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gietz R.D. and Sugino,A. (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- 23.Ryan M.P., Jones,R. and Morse,R.H. (1998) Swi-Snf complex participation in transcriptional activation at a step subsequent to activator binding. Mol. Cell. Biol., 18, 1774–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zitomer R.S., Sellers,J.W., McCarter,D.W., Hastings,G.A., Wick,P. and Lowry,C.V. (1987) Elements involved in oxygen regulation of the Saccharomyces cerevisiae CYC7 gene. Mol. Cell. Biol., 7, 2212–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winston F., Dollard,C. and Ricupero-Hovasse,S.L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast, 11, 53–55. [DOI] [PubMed] [Google Scholar]
- 26.Smart W.C., Coffman,J.A. and Cooper,T.G. (1996) Combinatorial regulation of the Saccharomyces cerevisiae CAR1 (arginase) promoter in response to multiple environmental signals. Mol. Cell. Biol., 16, 5876–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lussier M., White,A.M., Sheraton,J., Di Paolo,T., Treadwell,J., Southard,S.B., Horenstein,C.I., Chen-Weiner,J., Ram,A.F.J., Kapteyn,J.C., Roemer,T.W., Vo,D.H., Bondoc,D.C., Hall,J., Zhong,W.W., Sdicu,A.M., Davies,J., Klis,F.M., Robbins,P.W. and Bussey,H. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics, 147, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]