Abstract
The extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) pathway is a highly conserved signaling pathway that regulates diverse cellular processes including differentiation, proliferation, and survival. Kinase suppressor of Ras-1 (KSR1) binds each of the three ERK cascade components to facilitate pathway activation. Even though KSR1 contains a C-terminal kinase domain, evidence supporting the catalytic function of KSR1 remains controversial. In this study, we produced recombinant wild-type or kinase-inactive (D683A/D700A) KSR1 proteins in E. coli to test the hypothesis that KSR1 is a functional protein kinase. Recombinant wild-type KSR1, but not recombinant kinase-inactive KSR1, underwent autophosphorylation on serine residue(s), phosphorylated myelin basic protein (MBP) as a generic substrate, and phosphorylated recombinant kinase-inactive MAPK/ERK kinase-1 (MEK1). Furthermore, FLAG immunoprecipitates from KSR1−/− colon epithelial cells stably expressing FLAG-tagged wild-type KSR1 (+KSR1), but not vector (+vector) or FLAG-tagged kinase-inactive KSR1 (+D683A/D700A), were able to phosphorylate kinase-inactive MEK1. Since TNF activates the ERK pathway in colon epithelial cells, we tested the biological effects of KSR1 in the survival response downstream of TNF. We found that +vector and +D683A/D700A cells underwent apoptosis when treated with TNF, whereas +KSR1 cells were resistant. However, +KSR1 cells were sensitized to TNF-induced cell loss in the absence of MEK kinase activity. These data provide clear evidence that KSR1 is a functional protein kinase, MEK1 is an in vitro substrate of KSR1, and the catalytic activities of both proteins are required for eliciting cell survival responses downstream of TNF.
Keywords: KSR1, MEK, MAPK, TNF, ERK, Apoptosis
Introduction
Many cellular responses to external stimuli utilize mitogen-activated protein kinase (MAPK) pathways to carry out a diverse range of biological processes. Activation of these pathways, which are conserved in all eukaryotes, is often initiated by GTPases downstream of cell surface receptors, followed by sequential signal transduction through a three-component kinase system. One particular MAPK module consists of the protein kinases Raf, MEK, and ERK. Canonical activation of the Raf/MEK/ERK cascade occurs downstream of the small GTPase Ras to elicit a variety of cellular responses including proliferation, differentiation, and cell survival [1–4]. Since the ERK pathway is integral for many cellular events, and constitutive pathway activation is frequently concurrent with many cancers, understanding the precise mechanisms that contribute to pathway activation are essential for developing therapeutic targets that modulate this pathway [5]. Kinase suppressor of Ras-1 (KSR1), first identified through genetic screens in D. melanogaster and C. elegans, is an evolutionarily conserved protein that positively regulates the Raf/MEK/ERK cascade by functioning either upstream or in parallel with Raf-1 [6–8]. KSR1 functions as a molecular scaffold by binding several signaling components of the ERK cascade; and thus can enhance MAPK activation by regulating the efficiency of these interactions [9–11]. In addition to its scaffolding role, there is evidence that KSR1 functions as a protein kinase. The KSR1 C-terminus contains the eleven subdomains that are conserved in all protein kinases including the conserved aspartic acid and asparagine residues within subdomain VIb (HRDLKxxN motif) and the aspartic acid in subdomain VII (DFG motif) [12, 13]. However, the catalytic function of KSR1 remains controversial since mammalian KSR1 contains an arginine in place of the invariant lysine residue in subdomain II. This lysine positioned in subdomain II is involved in binding and orienting the ATP molecule to facilitate phosphotransfer of ATP γ-phosphate [14]. While lysine to arginine mutations in this position disrupt ATP binding and render many protein kinases inactive [15–18], a KSR1 splice variant is able to bind ATP when the arginine was substituted with lysine or methionine [19]. This suggests that KSR1 might utilize a different lysine, as seen with the protein kinase with no lysine-1 (WNK1) [20], or may have a structurally unique ATP-binding cleft compared to other protein kinase domains. Therefore, further investigation into KSR1 catalytic function is warranted.
Initial reports of KSR1 protein kinase activity suggest that immunoprecipitated KSR1 autophosphorylates, as well as phosphorylates and activates Raf-1, in vitro [21–23]. However, immunoprecipitated KSR1 contains additional co-precipitating protein kinases making it difficult to delineate KSR1 protein kinase activity from that of other contaminating kinases in the assay [24, 25]. Therefore, to resolve KSR1 kinase activity from other protein kinases in vitro requires isolating recombinant proteins expressed in a system with no known serine/threonine protein kinases, such as E. coli [26].
Here we report that bacterially-derived KSR1 underwent serine autophosphorylation, phosphorylated myelin basic protein (MBP) as a generic substrate, and phosphorylated recombinant kinase-inactive MEK1 (rMEK K97M). We also demonstrate that both a functional KSR1 kinase domain and MEK protein kinase activity are required for resistance to TNF-induced cell death in colon epithelial cells. Taken together, these data indicate that in addition to a scaffold, KSR1 is indeed a functional protein kinase in the ERK pathway downstream of TNF signaling.
Materials and methods
Generation of stable KSR1 cell lines
The conditionally immortalized KSR1−/− colon epithelial cell line was generated by crossing a KSR1−/− mouse with the H-2Kb–tsA58 ImmortoMouse (Charles River Laboratories International Inc., Wilmington, MA), as previously described [23, 27, 28]. N-terminally FLAG-tagged murine wild-type KSR1 or murine kinase-inactive KSR1 harboring an amino acid substitution of aspartic acid to alanine at two residues within the kinase domain that are critical for enzymatic activity (D683A/D700A) were a generous gift from Richard Kolesnick (Memorial Sloan-Kettering Cancer Center, New York, NY). Both KSR1 constructs were subcloned into the bicistronic pLZRS-IRES-GFP retroviral vector at a single EcoR1 restriction site, screened for proper orientation, and transfected into Phoenix 293 ecotropic viral packaging cells. Viral supernatants were collected and KSR1−/− colon epithelial cells were infected with virus containing empty vector (+vector), FLAG-tagged wild-type KSR1 (+KSR1), or FLAG-tagged kinase-inactive KSR1 (+D683A/D700A). Infected cells were then sorted based on GFP expression by fluorescence-activated cell sorting (FACS). Sorted cell lines were screened for KSR1 protein expression and those expressing near endogenous levels of KSR1, when compared to young adult mouse colon (YAMC) epithelial cells, were used for subsequent cell culture experiments.
Cell culture
Conditionally-immortalized colon epithelial cell lines were maintained under permissive conditions consisting of 5 units/ml of murine interferon-γ (BD Bioscience, San Jose, California) at a temperature of 33° C 5% CO2 in RPMI 1640 (Mediatech, Manassas, VA) supplemented with 5% FBS (Atlanta Biologicals, Lawrenceville, GA), 100 units/mL penicillin and streptomycin (Invitrogen, Carlsbad, California), 5 mg/mL insulin, 5 mg/mL transferrin, 5 mg/mL selenous acid (ITS) (BD Bioscience). Prior to each experiment, cells were placed overnight at non-permissive conditions at 37° C 5% CO2 in RPMI 1640 containing 0.5% FBS, 100 units/mL penicillin and streptomycin, and without interferon–γ.
TUNEL and cell loss assays
Cell lines expressing +vector, +KSR1, and +D683A/D700A were seeded in 4-well chamber slides at 5×104 cells (4-well chamberslide, Lab-Tek), and placed under non-permissive conditions overnight prior to experiments. Cells were then treated with murine TNF (Peprotech, Rocky Hill, NJ) for 8 hours. Apoptotic cells were labeled using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) (Millipore, Billerica, MA) following manufacturer’s protocol. For cell loss assays, 6-well dishes were seeded at 4×105 cells/well and maintained under permissive conditions for 24 hours. Following 16 hours under non-permissive conditions, cells were then pre-treated with vehicle (DMSO) or the MEK kinase inhibitor U0126 (Calbiochem, Darmstadt, Germany) for 45 minutes followed by TNF for 24 hours. Cells were then washed 3X in PBS, incubated in lysis buffer, and the total number of cells was determined by counting nuclei using a NucleoCounter (New Brunswick Scientific, Edison, New Jersey). Cell loss was calculated by comparing 24-hour treated cell numbers to 24-hour untreated control.
Immunoprecipitation
KSR1−/− colon epithelial cells expressing FLAG-tagged KSR1 proteins and YAMC cells expressing FLAG-tagged Raf-1 were serum starved under the non-permissive conditions for 16 hours. Cells were lysed in FLAG lysis buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and protease, phosphatase I, and phosphatase II inhibitor cocktails (Sigma). 1 mg of total protein from whole cell lysates were used for immunoprecipitation with ANTI-FLAG M2 antibody (Sigma) covalently linked to agarose beads overnight at 4°C. Immunoprecipitates were washed 3X in FLAG lysis buffer. Immunoprecipitated proteins were analyzed by Western blot analysis.
Western blot analysis
Whole cell lysates or immunoprecipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose. Membranes were then blocked for 1 hour in 5% milk at room temperature. Membranes were incubated in primary antibody at room temperature for 1 hour, washed three times in TBST, incubated with the appropriate HRP-conjugated secondary antibody for 1 hour at room temperature followed by 3 washes in TBST. Total and phospho-proteins were detected by chemiluminescence using a luminol HRP substrate. Antibodies used in these studies include: KSR1 (BD Bioscience, #611577), KSR1 C-terminus (Santa Cruz Biotechnology, #sc-1837, Santa Cruz, CA) Raf-1 (Santa Cruz Biotechnology, #sc-133), B-Raf (Santa Cruz, #sc-166), MEK1/2 (Cell Signaling, #9122, Danvers, MA), phospho-MEK1/2 (Cell Signaling, #9154), ERK1/2 (Cell Signaling, #4695), and phospho-ERK1/2 (Promega, #V803A, Madison, Wisconsin). The anti-HtpG antibody was a generous gift from Olivier Genest (NCI/NIH, Bethesda, MD)
Bacterial expression plasmids
Hexahistidine (His)-tagged and FLAG-tagged constructs of murine wild-type KSR1, murine kinase-inactive KSR1 (D683A/D700A), His-tagged wild-type Raf-1, and His-tagged kinase-inactive Raf-1 (Raf-1 K375M) were subcloned into the pET DUET (Novagen, Darmstadt, Germany) bacterial expression vector. Murine KSR1 harboring an N-terminal truncation of 521 amino acids (KSR1ΔN521), and murine KSR1 harboring a deletion of the CA1 domain (KSR1ΔCA1) were generated by PCR based mutagenesis and subcloned into the pET DUET bacterial expression vector. The bacterial expression vector containing His-tagged kinase-inactive MEK1 (MEK K97M) was a generous gift from Melanie Cobb (University of Texas Southwestern, Dallas, TX).
Bacterial protein expression
Bacterial expression constructs were transformed into E. coli BL21(DE3) (Novagen). Protein expression was induced upon the addition of 0.1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at 25° C. Bacteria were lysed using CelLytic B Plus Kit (Sigma, St Louis, MO) according to manufacturer’s protocol. rKSR1 proteins were recovered from insoluble inclusion bodies under denaturing conditions using 8 M urea and dialyzed into PBS/NaCl (100 mM). His-tagged Raf-1 and His-MEK K97M protein expression was induced as before and bacteria supernatant containing recombinant proteins were purified over Ni-MAC column (Novagen).
In vitro kinase assays
rMEK K97M phosphorylation assays using FLAG immunoprecipitates from KSR1 cells lines were washed 3X in FLAG lysis buffer containing 1M NaCl to remove co-precipitating proteins and then equilibrated in kinase assay dilution buffer (ADB) (Millipore) for 20 minutes prior to incubation with rMEK K97M in the presence Mg2+/ATP (Millipore) at 30° C for 30 minutes. The reaction was stopped upon addition of Laemmli sample buffer and heated to 95° C for 5 minutes. Western blot analysis was performed on separated samples. For in vitro kinase assays using recombinant proteins, rMEK K97M was incubated with rKSR1 or rRaf-1 proteins in the presence of ADB containing Mg2+/ATP (Millipore) at 30° C for 30 minutes. Samples were analyzed by Western blot analysis. Immunoblots are representative of 3 independent experiments. 32P incorporation assays were performed as described above in the presence of 10 μCi [γ-32P]ATP (Perkin-Elmer, Waltham, Massachusetts), separated using SDS-PAGE, transferred to PVDF membrane, and assayed by autoradiography. In vitro MAPK cascade kinase assays were conducted using recombinant or immunoprecipitated mammalian KSR1 and Raf-1 proteins, unactive recombinant wild-type MEK1 (rMEK) (Millipore), and either unactive recombinant wild-type ERK2 (rERK) (Millipore) or recombinant kinase-dead ERK2 (rERK K52R) (Millipore) all individually expressed and purified form E. coli. For some assays, recombinant constitutively active truncated Raf-1 (rCA-Raf-1) (Millipore) was used as a positive control for in vitro MAPK cascade activation. MBP phosphorylation assays were performed using the MBP Kinase Flex Assay Kit (Millipore) with rKSR1, rD683/D700A, rKSR1ΔN521, rRaf-1, or rRaf-1 K375M proteins following manufacturer’s protocol. Whole cell lysates from epidermal growth factor (EGF) treated young adult mouse colon (YAMC) cells were used as a positive control for MBP phosphorylation. Relative MBP phosphorylation was calculated by comparing recombinant proteins incubated with MBP to MBP phosphorylation when no additional recombinant proteins were present (MBP only).
Phospho-amino acid analysis (PAA)
Following autoradiography, PVDF bound 32P-KSR1 was excised and amino acids were hydrolyzed using 6 M HCl at 110° C for 1 hour. Amino acids were then lyophilized and rehydrated with pH 1.9 buffer containing PAA standards. Amino acids were spotted at a single origin on a TLC plate and were run in the first dimension at 1500 V for 25 minutes. The TLC plate was then rotated 90° and run in the second dimension in pH 3.9 buffer at 1300 V for 20 minutes. The TLC plate was dried and then sprayed with 0.25% Ninhydrin solution and incubated at 100° C until PAA standards were detectable by development of Ruhemann’s purple [29]. rKSR1 32P-phosphoaminio acids were determined via autoradiography and aligning the exposed Kodak BioMax film against the Ruhemann’s purple phosphoamino acid standards visible on the TLC plate.
2D-phosphopeptide mapping
Briefly, following an in vitro kinase assay using [γ-32P]ATP and exposure to film, the 32P-rKSR1 was excised from the PVDF membrane and wetted with MeOH. The PVDF membrane was then incubated at 37° C for 30 minutes in 50 mM NH4HCO3 was rinsed 5X with H2O, 2X with 50 mM NH4HCO3 buffer, then resuspended in 200 μl NH4HCO3 buffer containing 0.1% Tween-20. Following incubation, membrane containing 10 μg trypsin and digested for 3 hours at 37° C. Following digestion, 400 ml water was added, vortexed, and spun 5 min at 13k RPM. Supernatant was removed and lyophilized using a speed-vac. Lyophilized sample was resuspended in 400 μl of pH 1.9 buffer and lyophilized again. Lyophilized sample was resuspended in 6 μl of pH 1.9 buffer and spotted on a pre-marked TLC plate (origin) and subjected to electrophoresis at 1000 V for 30 minutes. The TLC plate was dried and placed in a chromatography buffer (75 vols. n-Butanol, 50 vols. pyridine, 15 vols. acetic acid, and 60 vols. H2O) chamber overnight and oriented such that migration in the second dimension is towards the top of the chromatography chamber. The TLC plate was dried and 32P-containing phosphopeptides visualized by autoradiography.
Statistical analysis
For the experiments in figures 2C, 5A, & 5B the analysis of variance model was used to determine significance. The p value was adjusted for multiple comparisons with the Dunnett’s post-test using a 95% confidence interval for three independent experiments. For figures 2B & 3A, statistical analyses were performed following three independent experiments using a two-tailed Student’s t-test with a 95% confidence interval. All statistical analysis was performed using Prism (GraphPad Software, Inc., La Jolla, CA).
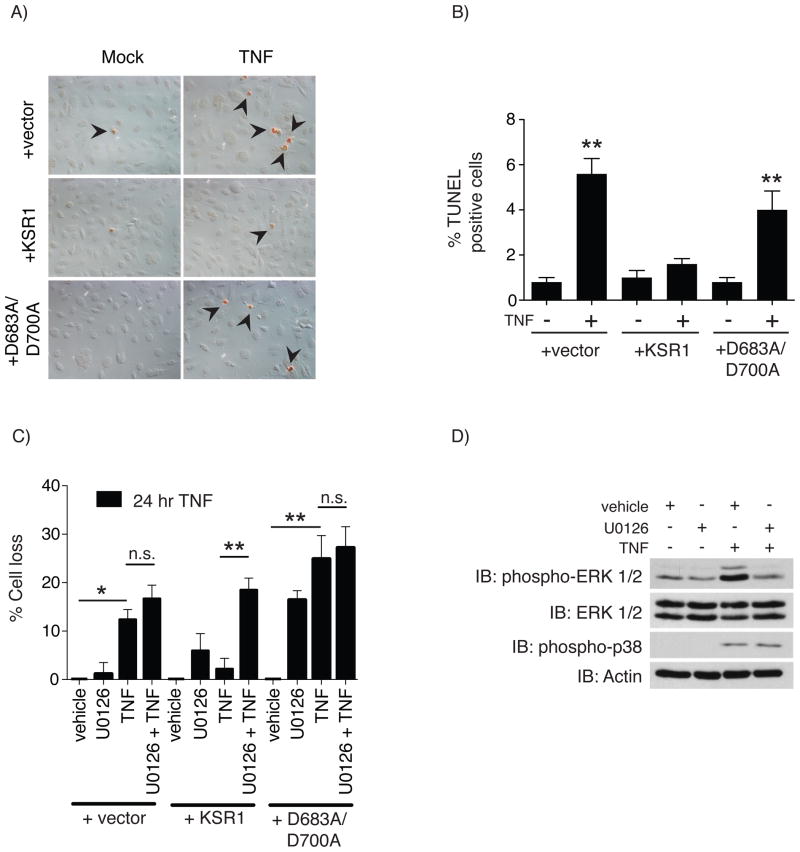
Fig. 2.
KSR1-mediated protection against TNF-induced apoptosis requires both a functional KSR1 kinase domain and MEK kinase activity. A) TNF-induced apoptosis was determined by treating serum-starved +vector, +KSR1, and +D683A/D700A cells with TNF (100 ng/ml) for 8 hours. Apoptotic nuclei were labeled using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL). B) Apoptotic cells from panel A were quantified and reported as the percentage of TUNEL positive cells out of 500 cells. Solid bars represent the mean for each condition and the error bars the SEM. C) TNF-induced cell loss was determined for cells that were pre-treated with vehicle or the MEK inhibitor U0126 (10 μM) for 45 minutes followed by TNF (100 ng/ml) for 24 hours. The total number of adherent cells was quantified and cell loss calculated as the percent of the vehicle treated control for each cell line. Assays were performed a minimum of 3 times. D) The specificity of U0126 was confirmed in +KSR1 cells pretreated with vehicle or U0126 (10 μM) for 45 minutes followed by TNF for 15 minutes and assayed for phosphorylated ERK. Protein phosphorylation and total protein were determined by Western blot analysis using the indicated antibodies. Immunoblots are representatives of at least 3 independent experiments. * = P < 0.05, ** = P < 0.01, *** = P < 0.001
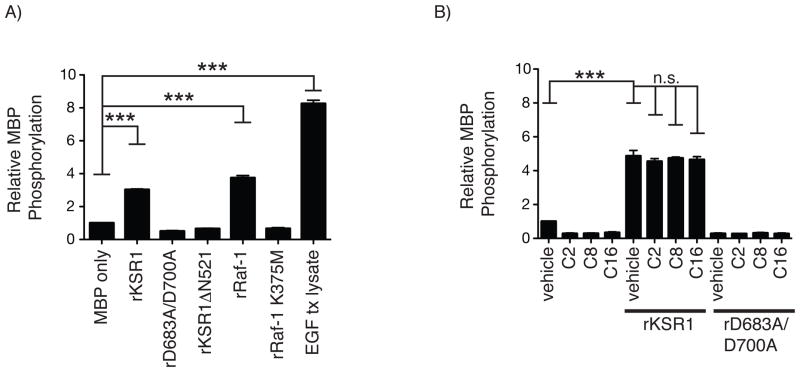
Fig. 5.
Recombinant KSR1 phosphorylates Myelin basic protein (MBP). A) Recombinant rKSR1, rD683A/D700A, rKSR1ΔN521, rRaf-1, rRaf-1 K375M, or lysate from EGF-treated cells (EGF tx lysate) were incubated with MBP in an in vitro kinase assay. Solid bars are the average relative MBP phosphorylation compared to MBP alone and error bars represent the SEM from 3 independent experiments. B) In vitro MBP kinase assay performed as before with rKSR1 or rD683A/D700A incubated with vehicle, C2-, C8-, or C16-ceramide (3 μM). MBP phosphorylation was determined as before. Solid bars are the average relative MBP phosphorylation compared to MBP alone and error bars represent the SEM from 3 independent experiments. Statistical analysis was performed using a one-way ANOVA with Dunnett’s post-test. *** = P < 0.001
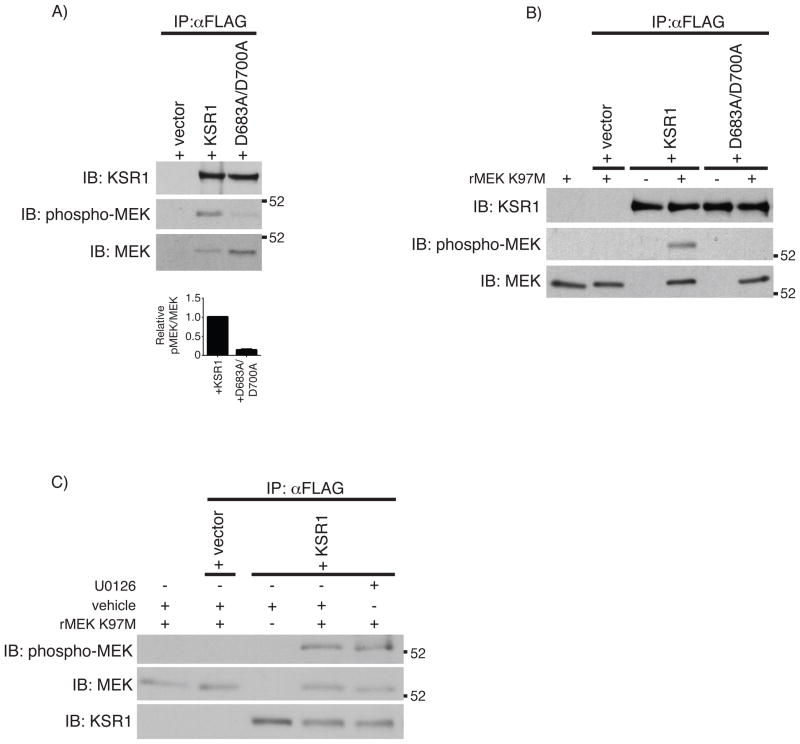
Fig. 3.
KSR1 phosphorylates MEK1 A) FLAG immunoprecipitation was performed from 1 mg protein on whole cell lysates from KSR1−/− cells expressing +vector, +KSR1, or +D683A/D700A. MEK phosphorylation was determined by Western blot analysis using the indicated antibodies. Densitometric analysis was performed on total and phosphorylated co-precipitated MEK protein and represented as the ratio of phosphorylated MEK/total MEK. Solid bars represent the mean ratio from 3 independent experiments and error bars represent the SEM. Statistical analysis was performed using a Student’s t-test comparing the pMEK/MEK ratio from +KSR1 and +D683A/D700A co-precipitations. *** P < 0.001 B) In vitro kinase assay using immunoprecipitated FLAG-tagged KSR1 proteins incubated with kinase-inactive rMEK K97M. Total protein and phosphorylated rMEK K97M were determined by Western blot analysis. C) In vitro kinase assay performed as before following a 30 minutes pre-incubation with vehicle or U0126 (10 μM). Total and phosphorylated proteins were determined by Western blot analysis. IP, immunoprecipitation; IB, immunoblot.
Results
KSR1 promotes TNF-mediated cell survival through phosphorylation of MEK1
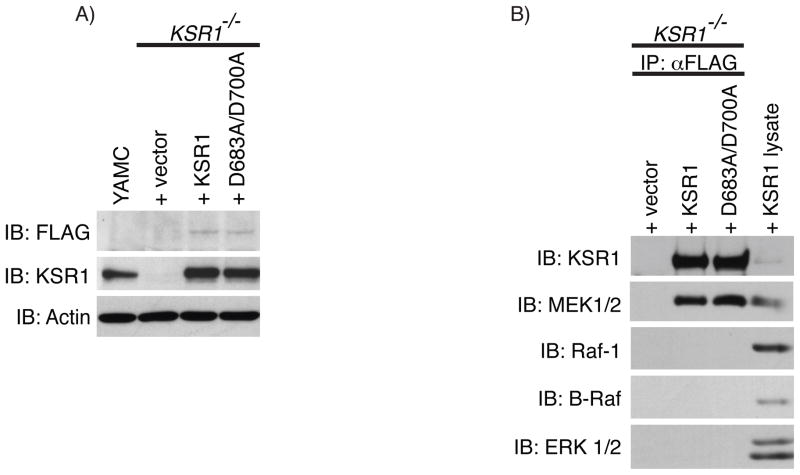
The pleiotropic pro-inflammatory cytokine, tumor necrosis factor, (TNF) elicits various cellular responses in a variety of cell types in vitro and in vivo and plays a pivotal role in chronic gastrointestinal disorders including celiac and inflammatory bowel diseases [30–32]. Previous studies from our lab show that the colon epithelium of KSR1−/− mice is sensitive to TNF-induced apoptosis compared to wild-type mice [23]. Therefore, to determine if KSR1 kinase activity is required to prevent TNF-induced cell death, we produced stable KSR1−/− colon epithelial cell lines expressing either empty vector (+vector), FLAG-tagged wild-type KSR1 (+KSR1), or FLAG-tagged kinase-inactive KSR1 containing alanine substitutions at aspartic acid residues in the HRDLKxxN and DFG motifs involved in phosphotransfer (+D683A/D700A) via retroviral infection containing bicistronic GFP. These cells were then sorted based on GFP expression to obtain cell lines expressing KSR1 at levels similar to young adult mouse colon (YAMC) epithelial cells (Fig. 1A). Since a previous report indicates that a kinase-inactivating mutation in the KSR1 kinase domain reduces KSR1 association with MEK [10], we performed FLAG immunoprecipitation on +vector, +KSR1, and +D683A/D700A cell lines and compared MEK association with KSR1 proteins. Both KSR1 proteins were equally competent to co-precipitate endogenous MEK (Fig. 1B). This indicates that the D683A/D700A kinase-inactivating mutation in KSR1 had no deleterious effects on MEK binding, consistent with a previous report [33].
Fig. 1.
Wild-type KSR1 and kinase-inactive KSR1 associate with MEK. A) KSR1 protein expression was determined by Western blot analysis on whole cell lysates from KSR1−/− mouse colon epithelial cells expressing +vector, +KSR1, or +D683A/D700A, or from YAMC cells using the indicated antibodies. B) Co-immunoprecipitation of MEK was determined by Western blot analysis on FLAG-immunoprecipitates from +vec, +KSR1, and +D683A/D700A expressing cells that were washed in high salt immunoprecipitation buffer. Whole cell lysate from +KSR1 cells was used as a control for detecting each protein probed. Immunoblots are representatives of at least 3 independent experiments. IP, immunoprecipitation; IB, immunoblot.
To determine if KSR1 was required for TNF-induced cell survival in cultured colon epithelial cells, apoptosis was assessed in TNF-treated +vector, +KSR1, and +D683A/D700A cell lines using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL). Unlike cells expressing KSR1, those expressing either +vector or +D683A/D700A were sensitive to TNF-induced apoptosis (Figs. 2A & 2B). Previous data from our lab indicate that TNF-mediated ERK activation was impaired in cells lacking KSR1 or expressing kinase-inactive KSR1 [23]. We observed a similar decrease in MEK phosphorylation in +vector or +D683A/D700A cells in response to TNF when compared to +KSR1 cells (S.1A). This does not appear to be attributed to defects in Raf protein activation (S.1B–C), nor assembly of the signaling complex containing Raf, MEK, and ERK following TNF treatment (S.1D). Therefore, we investigated whether MEK kinase activity was required for protection against TNF-induced cell death. An equal number of cells were pre-treated with vehicle or the MEK inhibitor U0126 and then treated with TNF. Cells lacking KSR1 or expressing the D683A/D700A mutant KSR1 protein were sensitive to TNF-induced cell loss, whereas +KSR1 cells were resistant (Fig. 2C). Interestingly, the +KSR1 cells were sensitized to TNF-induced cell loss following MEK inhibition, whereas U0126 did not potentiate cell loss in cells expressing +vector or +D683A/D700A. Interestingly, treatment of +D683A/D700A cells with U0126 caused increased cell loss in the absence of TNF. These data suggest that the protective effect of wild-type KSR1 in cultured colon epithelial cells is mediated through MEK activity. In a parallel experiment, U0126 effectively inhibited TNF-induced ERK phosphorylation, but not p38 MAPK phosphorylation, in +KSR1 cells, confirming its specificity for MEK (Fig. 2D). Taken together, these observations demonstrate that TNF-induced colon epithelial cell survival requires a functional KSR1 kinase domain, which in turn relies on MEK activity.
The observation that inhibiting MEK activity sensitized +KSR1 cells, but did not potentiate cell loss in +D683A/D700A or + vector cells, combined with the prior observation that kinase-inactive KSR1 expression attenuates TNF-induced ERK activation [34], suggests that KSR1 may alter MEK activation. Since MEK activation requires phosphorylation on two serine residues in the MEK activation loop [35, 36], we used a phospho-specific antibody that recognizes phosphorylated MEK in the activation loop to determine the phosphorylation state of KSR1-associated MEK. FLAG immunoprecipitations were performed on serum-starved +vector, +KSR1, and +D683A/D700A cells. MEK phosphorylation was increased when associated with wild-type KSR1 compared to kinase inactive KSR1 (Fig. 3A), demonstrating that MEK interaction with wild-type KSR1 enhances MEK phosphorylation.
Immunoprecipitated KSR1 phosphorylates recombinant kinase-dead MEK1
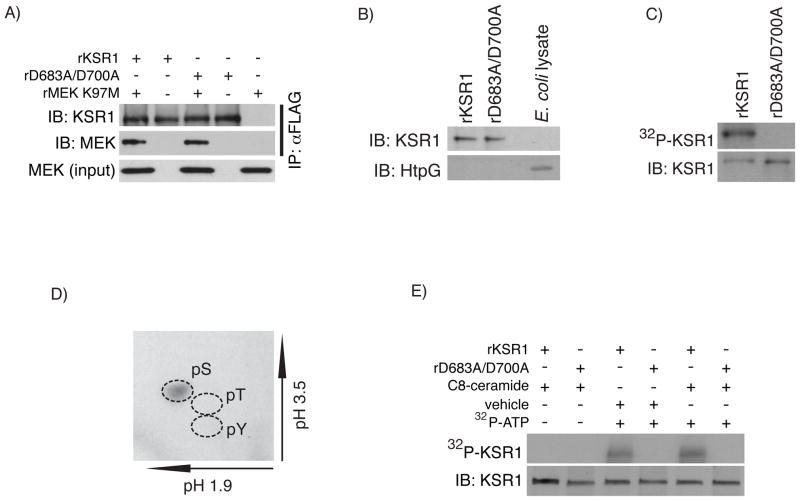
Based on our observation that MEK phosphorylation was increased when complexed with wild-type KSR1, we investigated whether MEK was a KSR1 substrate in vitro. For these assays we utilized recombinant kinase-inactive MEK1 harboring an amino acid substitution at the conserved lysine residue in subdomain II of the MEK kinase domain (rMEK K97M). FLAG immunoprecipitates from each cell line were washed in high salt buffer, to reduce co-precipitating proteins, and then incubated with rMEK K97M in the presence of ATP. Immunoprecipitates from cells expressing +KSR1, but not +vector or +D683A/D700A were able to phosphorylate rMEK K97M, as determined by Western blot analysis (Fig. 3B). Since endogenous MEK co-precipitates with KSR1 even under more stringent wash conditions, we investigated if endogenous MEK activity was responsible for phosphorylating rMEK K97M. FLAG immunoprecipitates from +KSR1 cells were pre-incubated with vehicle or U0126 then subjected to the same in vitro kinase assay using rMEK K97M. U0126 was unable to inhibit rMEK K97M phosphorylation in +KSR1 immunoprecipitates in vitro (Fig. 3C). The efficacy of U0126 in these experiments was confirmed by the ability of U0126 to block recombinant wild-type MEK (rMEK) phosphorylation of recombinant ERK (rERK) when stimulated by constitutively active recombinant Raf-1 (rCA-Raf-1) (S.2). Taken together, these data suggest that MEK is a substrate for KSR1 in vitro.
Recombinant KSR1 is a functional protein kinase
Our data utilizing FLAG immunoprecipitated KSR1 suggests that MEK is a substrate of KSR1. However, it remains possible that rMEK K97M phosphorylation is a result of co-precipitating protein kinases. Therefore, to directly assay KSR1 kinase activity, we produced recombinant wild-type KSR1 (rKSR1) or recombinant kinase-inactive KSR1 (rD683A/D700A) in a system devoid of serine/threonine protein kinases using E. coli. Since proper protein folding is a concern when expressing recombinant proteins, we first examined the ability of rKSR1 proteins to co-precipitate MEK. Consistent with our cell culture data, both rKSR1 and rD683A/D700A were equally competent in MEK binding (Fig. 4A). Given that KSR1 is known to associate with heat shock protein 90 (Hsp90) [9], and since E. coli express a homolog of Hsp90, named HtpG that possesses ATPase activity [37], we analyzed recombinant KSR1 proteins for the presence of HtpG. While HtpG was detected in E. coli lysates, we did not detect HtpG in rKSR1 or rD683A/D700A preparations (Fig. 4B). Previous data indicate that mammalian KSR1 autophosphorylates [21]. To determine if rKSR1 is capable of autophosphorylation, we performed in vitro kinase assays using [γ-32P]ATP. We found that rKSR1 but not rD683A/D700A incorporated 32P, indicating that rKSR1 undergoes autophosphorylation (Fig. 4C). To determine which amino acid residue(s) were autophosphorylated we performed phosphoamino acid analysis on rKSR1 following the [γ-32P]ATP in vitro kinase assay. We found that rKSR1 autophosphorylation occurred exclusively on serine residue(s) (Fig. 4D). Further analysis using 2D tryptic phosphopeptide mapping indicated that KSR1 autophosphorylation likely occurs on a single or limited number of serine residues since only a single migrating 32P-lableled peptide was detected (S.3A).
Fig. 4.
Recombinant KSR1 undergoes serine autophosphorylation. A) rKSR1 and rD683A/D700A proteins were incubated with rMEK K97M for 2 hours an then immunoprecipitated with αFLAG antibody. MEK co-precipitation was determined by Western blot analysis using the indicated antibodies. B) Western blot analysis of rKSR1 and rD683A/D700A proteins screened for the presence of bacterial HtpG using cell lysates from E. coli as a positive control. C) KSR1 autophosphorylation was assayed using in vitro kinase assays in which rKSR1 or rD683A/D700A were incubated in the presence of [γ-32P]ATP at 30° C for 30 minutes. rKSR1 and rD683A/D700A samples were then separated by SDS-PAGE and 32P incorporation was determined by autoradiography and total KSR1 protein detected by Western blot analysis. D) Phosphoamino acid analysis was conducted on 32P-rKSR1 following an in vitro kinase assay. Phosphorylated amino acids were determined by autoradiography. Dashed lines indicate the migration of phosphoamino acid standards. E) In vitro kinase assay with rKSR1 and rD683A/D700A in the presence of [γ-32P]ATP with vehicle or C8-ceramide (3 μM). KSR1 autophosphorylation was determined as before. Total KSR1 protein was detected by Western blot analysis. Autoradiography and immunoblots are representatives of 3 independent experiments. IP, immunoprecipitation; IB, immunoblot.
Recent data indicate that the cysteine rich domain of KSR1 mediates interactions with ceramide at the plasma membrane that is required for KSR1 kinase activity and cell proliferation. [21, 34, 38]. Therefore we determined if rKSR1 autophosphorylation was modulated by the addition of ceramide. In vitro kinase assays were performed using rKSR1 or rD683A/D700A in the presence or absence of C8-ceramide. The addition of C8-ceramide did not enhance rKSR1 autophosphorylation in this assay (Fig. 4E). Since ceramide bioactivity varies depending on the structure and length of the hydrocarbon chain [39], we tested the effect of various ceramide analogs in augmenting rKSR1 kinase activity in vitro. The addition of C2-, C8-, or C16-ceramide to the kinase reaction did not enhance rKSR1 autophosphorylation (S.3B). Collectively, these data demonstrate that recombinant KSR1 is a functional protein kinase that undergoes serine autophosphorylation, likely on a limited number of serine(s), independent of ceramide in vitro.
MBP is a commonly used substrate for protein kinase activity assays in vitro [40–42]. To determine if rKSR1 is able to phosphorylate MBP as a generic substrate, we performed in vitro kinase assays in which rKSR1 or rD683A/D700A were incubated with MBP. rKSR1, but not rD683A/D700A, was able to phosphorylate MBP similar to recombinant Raf-1 (rRaf-1) (Fig. 5A). Expression of the kinase domain alone results in a constitutively active enzyme for many protein kinases. Therefore, we examined if the KSR1 kinase domain alone (rKSR1ΔN521) was sufficient to phosphorylate MBP. Consistent with previous reports, rKSR1 kinase activity requires full length KSR1 (Fig. 5A) [22, 43]. As a positive control for maximal MBP phosphorylation, we incubated MBP with whole cell lysate from EGF treated YAMC cells (EGF tx) (Fig. 5A). These findings indicate that rKSR1 can phosphorylate MBP as a generic substrate.
We next tested if rKSR1 kinase activity towards a substrate was augmented by ceramide. rKSR1 and rD683A/D700A were incubated with MBP in the presence of C2, C8, or C16-ceramide. Phosphorylation of MBP by rKSR1 was not potentiated by any of the ceramide analogs tested suggesting that the kinase activity of recombinant KSR1 does not require ceramide in vitro (Fig. 5B). The activity of ceramide was confirmed by treating YAMC cells with each ceramide analog and ERK activation was detected by Western blot analysis (data not shown). Together, these observations indicate that only full-length wild-type rKSR1 can directly phosphorylate MBP as a substrate and that ceramide does not enhance recombinant KSR1 kinase activity.
Recombinant KSR1 phosphorylates recombinant MEK1 in vitro
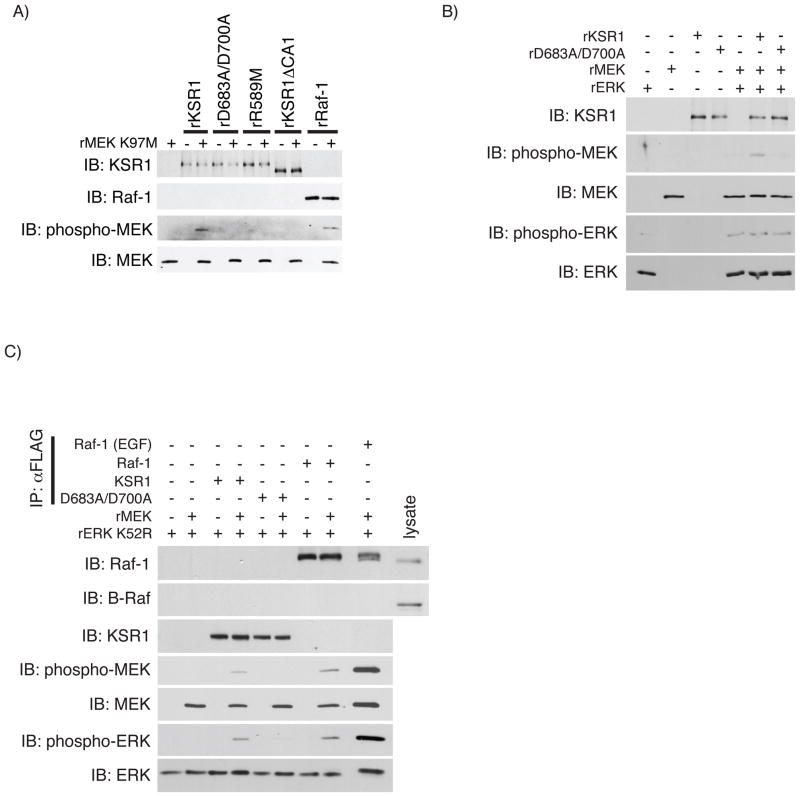
Kolesnick and co-workers recently demonstrated that mammalian KSR1 purified to homogeneity directly phosphorylates Raf-1 at Thr269 promoting Raf-1 kinase activity in vitro, consistent with their previous findings [43, 44]. Since +KSR1 cells showed enhanced endogenous MEK phosphorylation and FLAG immunoprecipitates from these cells phosphorylated rMEK K97M (Figs. 3A & 3B), we assessed whether MEK was a substrate for recombinant KSR1. In vitro kinase assays were performed incubating rMEK K97M with rKSR1, rD683A/D700A, or rRaf-1 as a positive control. Similar to rRaf-1, rKSR1 phosphorylated rMEK K97M but no kinase activity was detected for rD683A/D700A (Fig. 6A). Furthermore, no kinase activity was detected in a second kinase-inactive KSR1 mutant where the putative ATP-binding arginine was substituted with methionine (rR589M) (Fig. 6A). In the original genetic loss-of-function screen in D. melanogaster a mutation in the KSR1 CA1 domain reduced KSR1 function [6]. We generated a recombinant amino-terminal deletion of KSR1 (rKSR1ΔCA1) to test if this domain was required for KSR1 kinase activity in vitro. rKSR1ΔCA1 was unable to phosphorylate rMEK K97M, suggesting that the CA1 domain may be required structurally or facilitates KSR1 enzymatic activity through an unknown mechanism (Fig. 6A). Taken together, these data demonstrate that rKSR1 can phosphorylate rMEK K97M as a substrate consistent with the observations for KSR1 immunoprecipitated from colon epithelial cells.
Fig. 6.
Recombinant KSR1 phosphorylates rMEK K97M. A) Phosphorylation of rMEK K97M was assayed using in vitro kinase assays by incubating rMEK K97M with rKSR1, rD683A/D700A, rR589M, rKSR1ΔCA1, or rRaf-1. Phosphorylated rMEK K97M was assayed by Western blot analysis using the indicated antibodies. B) rKSR1-stimulated recombinant ERK (rERK) phosphorylation was determined using in vitro kinase assays in which rKSR1 or rD683A/D700A was incubated with rMEK in the presence rERK. Protein phosphorylation and total protein were determined by Western blot analysis using indicated antibodies. Immunoblots are representative of 3 independent experiments. C) The ability of mammalian FLAG-KSR1, FLAG-D683A/D700A, or FLAG-Raf-1 to promote rMEK kinase activity towards ERK was determined by immunoprecipitating 1 pmol of each FLAG-tagged protein and performing an in vitro kinase assay with recombinant kinase-dead ERK (rERK K52R) in the presence or absence of rMEK. Total and phospho-proteins were determined by Western blot analysis as indicated. Immunoblots are representatives of 3 independent experiments. IP, immunoprecipitation; IB, immunoblot.
While rKSR1 can phosphorylate rMEK K97M, utilizing kinase-inactive MEK did not allow for assaying rKSR1-mediated MEK activation. To determine if rKSR1 phosphorylation of rMEK is sufficient to activate rMEK, we performed an in vitro kinase assay using unactive recombinant wild-type ERK (rERK) as the wild-type rMEK substrate. While rMEK phosphorylation by rKSR1 was readily detected, it was insufficient to promote rERK phosphorylation (Fig. 6B), whereas a similar assay using recombinant Raf-1 promoted rMEK activation (S.4). We then investigated whether KSR1, D683A/D700A, or Raf-1 proteins immunoprecipitated from mammalian cells promoted rMEK kinase activity towards recombinant kinase-dead ERK (rERK K52R). We first determined the quantity of immunoprecipitated proteins to ensure equimolar amounts of KSR1 and Raf-1 proteins to be used in the kinase assay (S.5A–B). We then performed in vitro kinase assays incubating immunoprecipitated FLAG-KSR1 (1 pmol), FLAG-D683A/D700A (1 pmol), FLAG-Raf-1 (1 pmol), or FLAG-Raf-1 (1pmol) from EGF treated cells (positive control) in the presence or absence of unactive rMEK using phosphorylated rERK K52R as a readout for rMEK activity. We found that FLAG-KSR1 and FLAG-Raf-1, but not FLAG-D683A/D700A, immunoprecipitated from mammalian cells promoted rERK K52R phosphorylation by rMEK (Fig. 6C). Collectively, these data suggest that while MEK is a KSR1 substrate in vitro, activation of rMEK requires KSR1 isolated from intact mammalian cells.
Discussion
Our findings indicate that KSR1 is a functional protein kinase that, along with MEK1 protein kinase activity, is required for protection against TNF-induced cell death. The observation that MEK associated with wild-type KSR1 had increased phosphorylation, together with our in vitro kinase assays using recombinant proteins, suggest that KSR1 is capable of direct phosphorylation of MEK. Interestingly, while phosphorylation of rMEK in the MEK activation loop was detected for both recombinant and mammalian, only immunoprecipitated mammalian KSR1 was able to promote rMEK activation. With limited prior demonstration of KSR1 kinase activity, the results presented here clearly show KSR1 as a functional protein kinase that promotes cell survival in the presence of TNF. This finding has implications for chronic inflammatory diseases, such as inflammatory bowel disease (IBD), in which the cytotoxic effect of TNF contributes to epithelial cell loss and epithelial barrier dysfunction further exacerbating disease.
Activation of the ERK cascade is a key component for cell survival in a number of cell types [45–49]. While the ability of KSR1 to scaffold and sensitize the ERK pathway is known [22, 50, 51], the catalytic function of KSR1 is not well understood. We found that colon epithelial cells lacking KSR1, or stably expressing a kinase-inactive KSR1, are sensitive to TNF-induced apoptosis (Fig. 2). Interestingly, inhibition of MEK protein kinase activity sensitized +KSR1 cells to TNF-induced cell loss whereas +vector and +D683A/D700A cells had no potentiation in cell loss (Fig. 2). This suggests that the protective role of KSR1 kinase activity requires MEK kinase activity. The importance of MEK1 is appreciated from early development since MEK1−/− mice are embryonic lethal due to placental defects [52]. In addition, an inducible tissue-specific deletion of MEK1 in the epidermis causes apoptosis in keratinocytes, demonstrating a role for MEK1 in cell survival [53]. It is conceivable that wild-type KSR1 promotes cell survival by augmenting MEK kinase activity independent of KSR1 catalytic function in a manner similar to KSR1-Raf-1 interactions [24, 54]. However, this cannot fully explain our observations since +D683A/D700A associates with MEK similar to wild-type KSR1, yet +D683A/D700A-expressing cells are just as sensitive to TNF-induced apoptosis as cells lacking KSR1 (Fig. 2). We cannot rule out the possibility that expression of wild-type KSR1 induces a different protein expression profile compared to cells expressing +vector and +D683A/D700A that necessitates MEK kinase activity in TNF-mediated cell survival of +KSR1 cells.
Previous studies using KSR1 immunoprecipitates from cultured cells show that KSR1 autophosphorylates and phosphorylates Raf-1 in response to ceramide [21, 23, 38, 43]. While our results indicate that recombinant KSR1 autophosphorylates exclusively on serine, consistent with previous data [55], ceramide was not required for recombinant KSR1 protein kinase activity in vitro (Figs. 4D & 5B). Though these data seem to conflict with previous studies demonstrating a role for ceramide in mammalian KSR1 activation [21, 23, 38, 43], it may provide insight into the mechanism by which KSR1 kinase activity is regulated. For instance, in addition to associating and activating KSR1 at the plasma membrane [24, 38], KSR1 interaction with ceramide in the KSR1 CA3 domain may promote the release of a negative regulatory mechanism mediated by KSR1-associated protein(s). The 14-3-3 family of proteins bind KSR1 when phosphorylated at Ser297 and Ser392 and, like Raf-1, dissociate upon dephosphorylation by protein phosphatase 2A (PP2A) [25, 56–61]. Thus, expression of KSR1 in E. coli circumvents negative regulation by associated proteins allowing recombinant KSR1 to adopt an active conformation. In this model, a requirement for ceramide would not be essential for KSR1 activation. Alternatively, the kinase activity of KSR1 could be allosterically modulated intramolecularly. In this instance, KSR1 kinase activity could be modulated by ceramide, either directly or indirectly, to facilitate such an intramolecular interaction. In the original mutagenesis screen carried out in D. melanogaster, a weak loss of function allele mapped to a double amino acid substitution within the KSR1 CA1 domain. While this mutation was later shown to reduce KSR1 association with Raf-1 approximately two-fold, MEK phosphorylation was completely abolished following an in vitro kinase assay [62]. Interestingly, recombinant KSR1 lacking the N-terminal CA1 domain (rKSR1ΔCA1) did not function as an active kinase (Fig. 6A). These data suggest that in addition to Raf-1 association, the CA1 domain is involved in regulating the kinase activity of KSR1 and potentially contributes to Raf-1 activation directly or enhances Raf-1 activity possibly through an allosteric mechanism. In addition, the KSR1 kinase domain (rKSR1ΔN521) alone did not function as a constitutively active protein kinase (Fig. 5A), consistent with previous findings [22, 43]. This is unlike Raf-1 where expression of the kinase domain alone results in a constitutively active enzyme and indicates that KSR1 catalytic function requires a full-length protein [63, 64]. An example of such an intramolecular interaction is also observed for class II dual-specificity tyrosine phosphorylation-regulated kinases (DYRKs), which also require an N-terminal region outside the kinase domain to modulate protein kinase activity and autophosphorylation [65].
The finding that MEK1 is a KSR1 substrate raises new questions about the potential mechanisms that modulate signal transduction in the ERK pathway. The data presented here show that rKSR1 can phosphorylate rMEK1 on at least one serine residue within the MEK1 activation loop (Figs. 3B & 6A). These serine residues in the MEK activation loop are reported to be both necessary and sufficient for phosphorylation of ERK [35, 36]. Interestingly, while rKSR1 phosphorylation of rMEK was insufficient to stimulate phosphorylation of rERK2 (Fig. 6B), MEK activation was detected when using immunoprecipitated KSR1 from mammalian cells (Fig. 6C). KSR1 expressed in mammalian cells may possess post-translational modifications that promote more efficient activation of rMEK or KSR1 activity. Even so, previous observations with MEK kinase-1 (MEKK1) indicate complexity in MEK activation. While MEKK1 transfected into 293 cells readily phosphorylated MEK1 and MEK2 at these serine residues, it did not result in phosphorylation of ERK2 [66]. This data, combined with our data presented here, suggest that MEK activation is regulated, at least in part, by additional factors that likely vary with the type of stimulus or expression system. Nonetheless, the in vivo contribution of KSR1 kinase activity towards MEK may function to sensitize MEK activation by Raf-1 and lower the threshold for activation of the ERK pathway. It is also conceivable that KSR1 kinase activity alters ERK activation by maintaining a level of MEK activation loop phosphorylation once the activated Raf-KSR1-MEK complex dissociates following stimulation. The spatiotemporal control of ERK activation is known to alter the physiological responses downstream of both TNF receptor-1 and staphylococcal enterotoxin E-mediated T cell activation [2, 50].
For over a decade, KSR1 has been appreciated as a scaffold of the ERK pathway that coordinates pathway activation, yet the enzymatic contributions of KSR1 have remained controversial [10, 11, 24, 67, 68]. The data presented here demonstrate that KSR1 contains intrinsic protein kinase activity in vitro and suggests a mechanism by which KSR1 can sensitize the ERK signaling through direct phosphorylation of MEK1. KSR1 and other protein kinases have been classified as pseudokinases based on variation in conserved amino acid residue(s) within the kinase domain (reviewed in [69]). Even so, the maintenance of a highly conserved protein kinase domain throughout evolution, and the demonstration of protein kinase activity from other unique protein kinases [20, 70], imparts significance to these proteins and their potential enzymatic function. Our observation that the catalytic activity of KSR1 protects colonic epithelial cells from TNF-induced apoptosis provides a potential mechanism by which the intestinal epithelial barrier can be maintained in the presence of pathological concentrations of TNF. For patients suffering from IBD, anti-TNF therapy is beneficial and able to induce remission. Yet, the long-term side effects, and concomitant immunosuppressive therapy, highlight the need for novel strategies that reduce inflammation by promoting epithelial cell survival within the inflammatory microenvironment (reviewed in [71]). Therefore, understanding the precise mechanisms that modulate the ERK pathway downstream of TNF could lead to new therapeutic targets for patients afflicted with chronic inflammatory diseases.
Supplementary Material
MEK activation is impaired while Raf-1 activation and association with MAPK cascade components is not impaired in +D683A/D700A expressing cells. A) +vec, +KSR1, and +D683A/D700A expressing cells were treated with 100 ng/ml TNF for 10 minutes and whole cell lysates were examined for MEK phosphorylation by Western blot analysis. B) +KSR1 and +D683A/D700A expressing cells were treated with 100 ng/ml TNF for 5, 10, or 15 minutes or 10 ng/ml EGF for 5 minutes and Raf-1 phosphorylation examined by Western blot analysis. C) +KSR1 and +D683A/D700A expressing cells were treated with 100 ng/ml of TNF for 5, 10, or 15 minutes and phosphorylation of B-Raf detected by Western blot analysis. D) Association of Raf-1, MEK, and ERK with +KSR1 or +D683A/D700A was examined in untreated and cells treated with 100 ng/ml of TNF for 5 minutes. FLAG-KSR1 and FLAG-D693A/D700A were immunoprecipitated with anti-FLAG antibody and washed with immunoprecipitation buffer containing a reduced NaCl concentration. Total proteins were determined by Western blot analysis. Immunoblots are representatives of 3 independent experiments. IB, immunoblot.
U0126 blocks rCA-Raf-1 stimulated rMEK activity in vitro. In vitro kinase assay using recombinant constitutively active Raf -1 (rCA-Raf-1), recombinant MEK1 (rMEK) and recombinant ERK (rERK) in the presence of vehicle or U0126 (10 μM). Phosphorylated and total protein were determined by Western blot analysis using indicated antibodies. Immunoblot is a representative of 3 independent experiments. IB, immunoblot.
rKSR1 autophosphorylation occurs independent of ceramide on a single tryptic peptide A) Two-dimensional tryptic phosphopeptide map of rKSR1 following an in vitro kinase assay using [γ-32P]ATP. Spotted peptide origin is marked with an asterisk. B) rKSR1 autophosphorylation is not enhanced by ceramide in vitro. In vitro kinase assay using recombinant KSR1 (rKSR1) in the presence of vehicle or 3 μM C2-, C8-, or C16-ceramide and [γ-32P]ATP. Phosphorylated KSR1 was detected by autoradiography and total KSR1 protein was determined by Western blot analysis. Autoradiography and Immunoblot are representatives of at least two independent experiments. IB, immunoblot.
rRaf-1 stimulates rMEK activation in vitro. Recombinant wild-type Raf-1 subjected to an in vitro kinase assay in the presence of unactive recombinant wild-type MEK (rMEK) and unactive recombinant wild-type ERK (rERK). Phosphorylation of rMEK and rERK was determined by Western blot analysis using phospho-specific antibodies. Total protein was determined by Western blot analysis. Immunoblots are representatives of 3 independent experiments. IP, immunoprecipitation; IB, immunoblot.
Quantification of immunoprecipitated FLAG-KSR1, FLAG-D683A/D700A, and FLAG-Raf-1 proteins. A) FLAG-tagged Raf-1 was immunoprecipitated from increasing quantities of total protein (400–800 μg) from whole cell lysate, gel-loaded along with increasing amount of purified recombinant CA-Raf-1 (0.01–0.32 μg), and separated by SDS-PAGE. Densitometric measurements were obtained from Western blot analysis using an αRaf-1 antibody. Linear regression using rCA-Raf-1 as the protein standard determined the amount of immunoprecipitated FLAG-tagged Raf-1. Immunoblot is a representative of 3 different experiments. B) FLAG-tagged KSR1 was immunoprecipitated from increasing quantities of total protein (400–1000 μg) from whole cell lysate, gel-loaded along with increasing amount of purified recombinant FLAG-tagged bacterial alkaline phosphatase (BAP) (0.01–0.32 μg), and separated by SDS-PAGE. Densitometric measurements were obtained from Western blot analysis using an αFLAG antibody. Linear regression using rFLAG-BAP as the protein standard determined the amount of immunoprecipitated FLAG-tagged KSR1 protein. Immunoblots are representatives of 3 independent experiments. IP, immunoprecipitation; IB, immunoblot.
Acknowledgments
We would like to thank Philip Dubé, Stuart Hobbs, and Fang Yan for their thoughtful discussions and critique of this manuscript. This work was supported by NIH grants DK066176 and DK56008 (D.B.P)
Abbreviations
- MAPK
mitogen-activated protein kinase
- ERK
extracellular signal-regulated kinase
- KSR1
kinase suppressor of Ras-1
- MEK
MAPK/ERK kinase
- MBP
myelin basic protein
- TNF
tumor necrosis factor
- WNK1
with no lysine-1
- GFP
green fluorescent protein
- EGF
epidermal growth factor
- YAMC
young adult mouse colon
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- PP2A
protein phosphatase 2A
- MEKK1
MEK kinase-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cheng Y, Qiu F, Tashiro S-i, Onodera S, Ikejima T. ERK and JNK mediate TNFalpha-induced p53 activation in apoptotic and autophagic L929 cell death. Biochem Biophys Res Commun. 2008;376:483–488. doi: 10.1016/j.bbrc.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser GC, Yan F, Polk DB. Conversion of TNF alpha from antiproliferative to proliferative ligand in mouse intestinal epithelial cells by regulating mitogen-activated protein kinase. Exp Cell Res. 1999;249:349–358. doi: 10.1006/excr.1999.4488. [DOI] [PubMed] [Google Scholar]
- 3.Qui MS, Green SH. PC12 cell neuronal differentiation is associated with prolonged p21ras activity and consequent prolonged ERK activity. Neuron. 1992;9:705–717. doi: 10.1016/0896-6273(92)90033-a. [DOI] [PubMed] [Google Scholar]
- 4.Kanakura Y, Druker B, Wood KW, Mamon HJ, Okuda K, Roberts TM, Griffin JD. Granulocyte-macrophage colony-stimulating factor and interleukin-3 induce rapid phosphorylation and activation of the proto-oncogene Raf-1 in a human factor-dependent myeloid cell line. Blood. 1991;77:243–248. [PubMed] [Google Scholar]
- 5.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O’Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lövig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 7.Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 8.Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 9.Stewart S, Sundaram M, Zhang Y, Lee J, Han M, Guan KL. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol Cell Biol. 1999;19:5523–5534. doi: 10.1128/mcb.19.8.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu W, Fantl WJ, Harrowe G, Williams LT. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr Biol. 1998;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]
- 11.Denouel-Galy A, Douville EM, Warne PH, Papin C, Laugier D, Calothy G, Downward J, Eychène A. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr Biol. 1998;8:46–55. doi: 10.1016/s0960-9822(98)70019-3. [DOI] [PubMed] [Google Scholar]
- 12.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 13.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 14.Kamps MP, Sefton BM. Neither arginine nor histidine can carry out the function of lysine-295 in the ATP-binding site of p60src. Mol Cell Biol. 1986;6:751–757. doi: 10.1128/mcb.6.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- 16.Snyder MA, Bishop JM, McGrath JP, Levinson AD. A mutation at the ATP-binding site of pp60v-src abolishes kinase activity, transformation, and tumorigenicity. Mol Cell Biol. 1985;5:1772–1779. doi: 10.1128/mcb.5.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebina Y, Araki E, Taira M, Shimada F, Mori M, Craik CS, Siddle K, Pierce SB, Roth RA, Rutter WJ. Replacement of lysine residue 1030 in the putative ATP-binding region of the insulin receptor abolishes insulin- and antibody-stimulated glucose uptake and receptor kinase activity. Proc Natl Acad Sci USA. 1987;84:704–708. doi: 10.1073/pnas.84.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotten M, Stegmueller K, Eickhoff J, Hanke M, Herzberger K, Herget T, Choidas A, Daub H, Godl K. Exploiting features of adenovirus replication to support mammalian kinase production. Nucleic Acids Research. 2003;31:e128. doi: 10.1093/nar/gng128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller J, Cacace AM, Lyons WE, McGill CB, Morrison DK. Identification of B-KSR1, a novel brain-specific isoform of KSR1 that functions in neuronal signaling. Mol Cell Biol. 2000;20:5529–5539. doi: 10.1128/mcb.20.15.5529-5539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Yao B, Delikat S, Bayoumy S, Lin XH, Basu S, McGinley M, Chan-Hui PY, Lichenstein H, Kolesnick R. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell. 1997;89:63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 22.Xing HR, Lozano J, Kolesnick R. Epidermal growth factor treatment enhances the kinase activity of kinase suppressor of Ras. J Biol Chem. 2000;275:17276–17280. doi: 10.1074/jbc.C900989199. [DOI] [PubMed] [Google Scholar]
- 23.Yan F, John SK, Wilson G, Jones DS, Washington MK, Polk DB. Kinase suppressor of Ras-1 protects intestinal epithelium from cytokine-mediated apoptosis during inflammation. Journal of Clinical Investigation. 2004;114:1272–1280. doi: 10.1172/JCI21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaud NR, Therrien M, Cacace A, Edsall LC, Spiegel S, Rubin GM, Morrison DK. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volle DJ, Fulton JA, Chaika OV, McDermott K, Huang H, Steinke LA, Lewis RE. Phosphorylation of the kinase suppressor of ras by associated kinases. Biochemistry. 1999;38:5130–5137. doi: 10.1021/bi983050d. [DOI] [PubMed] [Google Scholar]
- 26.Rahmsdorf HJ, Pai SH, Ponta H, Herrlich P, Roskoski R, Schweiger M, Studier FW. Protein kinase induction in Escherichia coli by bacteriophage T7. Proc Natl Acad Sci USA. 1974;71:586–589. doi: 10.1073/pnas.71.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitehead RH, VanEeden PE, Noble MD, Ataliotis P, Jat PS. Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc Natl Acad Sci USA. 1993;90:587–591. doi: 10.1073/pnas.90.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corredor J, Yan F, Shen CC, Tong W, John SK, Wilson G, Whitehead R, Polk DB. Tumor necrosis factor regulates intestinal epithelial cell migration by receptor-dependent mechanisms. Am J Physiol Cell Physiol. 2003;284:C953–961. doi: 10.1152/ajpcell.00309.2002. [DOI] [PubMed] [Google Scholar]
- 29.Duclos B, Marcandier S, Cozzone AJ. Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Meth Enzymol. 1991;201:10–21. doi: 10.1016/0076-6879(91)01004-l. [DOI] [PubMed] [Google Scholar]
- 30.Breese EJ, Michie CA, Nicholls SW, Murch SH, Williams CB, Domizio P, Walker-Smith JA, MacDonald TT. Tumor necrosis factor alpha-producing cells in the intestinal mucosa of children with inflammatory bowel disease. Gastroenterology. 1994;106:1455–1466. doi: 10.1016/0016-5085(94)90398-0. [DOI] [PubMed] [Google Scholar]
- 31.Kontakou M, Sturgess RP, Przemioslo RT, Limb GA, Nelufer JM, Ciclitira PJ. Detection of interferon gamma mRNA in the mucosa of patients with coeliac disease by in situ hybridisation. Gut. 1994;35:1037–1041. doi: 10.1136/gut.35.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z, Gengaro P, Wang W, Wang X-q, Li C, Faubel S, Rivard C, Schrier RW. Role of NF-kappaB and PI 3-kinase/Akt in TNF-alpha-induced cytotoxicity in microvascular endothelial cells. Am J Physiol Renal Physiol. 2008;295:F932–941. doi: 10.1152/ajprenal.00066.2008. [DOI] [PubMed] [Google Scholar]
- 33.Xing HR, Campodonico L, Kolesnick R. The kinase activity of kinase suppressor of Ras1 (KSR1) is independent of bound MEK. J Biol Chem. 2004;279:26210–26214. doi: 10.1074/jbc.M401323200. [DOI] [PubMed] [Google Scholar]
- 34.Yan F, Polk DB. Kinase suppressor of ras is necessary for tumor necrosis factor alpha activation of extracellular signal-regulated kinase/mitogen-activated protein kinase in intestinal epithelial cells. Cancer Res. 2001;61:963–969. [PubMed] [Google Scholar]
- 35.Alessi DR, Saito Y, Campbell DG, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall CJ, Cowley S. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng CF, Guan KL. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J. 1994;13:1123–1131. doi: 10.1002/j.1460-2075.1994.tb06361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nadeau K, Das A, Walsh CT. Hsp90 chaperonins possess ATPase activity and bind heat shock transcription factors and peptidyl prolyl isomerases. J Biol Chem. 1993;268:1479–1487. [PubMed] [Google Scholar]
- 38.Yin X, Zafrullah M, Lee H, Haimovitz-Friedman A, Fuks Z, Kolesnick R. A ceramide-binding C1 domain mediates kinase suppressor of ras membrane translocation. Cell Physiol Biochem. 2009;24:219–230. doi: 10.1159/000233248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siskind LJ, Colombini M. The lipids C2- and C16-ceramide form large stable channels. Implications for apoptosis. J Biol Chem. 2000;275:38640–38644. doi: 10.1074/jbc.C000587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishimoto A, Nishiyama K, Nakanishi H, Uratsuji Y, Nomura H, Takeyama Y, Nishizuka Y. Studies on the phosphorylation of myelin basic protein by protein kinase C and adenosine 3′:5′-monophosphate-dependent protein kinase. J Biol Chem. 1985;260:12492–12499. [PubMed] [Google Scholar]
- 41.Shoji S, Ohnishi J, Funakoshi T, Fukunaga K, Miyamoto E, Ueki H, Kubota Y. Phosphorylation sites of bovine brain myelin basic protein phosphorylated with Ca2+-calmodulin-dependent protein kinase from rat brain. J Biochem. 1987;102:1113–1120. doi: 10.1093/oxfordjournals.jbchem.a122149. [DOI] [PubMed] [Google Scholar]
- 42.Erickson AK, Payne DM, Martino PA, Rossomando AJ, Shabanowitz J, Weber MJ, Hunt DF, Sturgill TW. Identification by mass spectrometry of threonine 97 in bovine myelin basic protein as a specific phosphorylation site for mitogen-activated protein kinase. J Biol Chem. 1990;265:19728–19735. [PubMed] [Google Scholar]
- 43.Zafrullah M, Yin X, Haimovitz-Friedman A, Fuks Z, Kolesnick R. Kinase suppressor of Ras transphosphorylates c-Raf-1. Biochemical and Biophysical Research Communications. 2009;390:434–440. doi: 10.1016/j.bbrc.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing HR, Kolesnick R. Kinase suppressor of Ras signals through Thr269 of c-Raf-1. J Biol Chem. 2001;276:9733–9741. doi: 10.1074/jbc.M008096200. [DOI] [PubMed] [Google Scholar]
- 45.McKay MM, Morrison DK. Caspase-dependent cleavage disrupts the ERK cascade scaffolding function of KSR1. J Biol Chem. 2007;282:26225–26234. doi: 10.1074/jbc.M702692200. [DOI] [PubMed] [Google Scholar]
- 46.Shimamura A, Ballif BA, Richards SA, Blenis J. Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr Biol. 2000;10:127–135. doi: 10.1016/s0960-9822(00)00310-9. [DOI] [PubMed] [Google Scholar]
- 47.Ripple MO, Kalmadi S, Eastman A. Inhibition of either phosphatidylinositol 3-kinase/Akt or the mitogen/extracellular-regulated kinase, MEK/ERK, signaling pathways suppress growth of breast cancer cell lines, but MEK/ERK signaling is critical for cell survival. Breast Cancer Res Treat. 2005;93:177–188. doi: 10.1007/s10549-005-4794-6. [DOI] [PubMed] [Google Scholar]
- 48.Erhardt P, Schremser EJ, Cooper GM. B-Raf inhibits programmed cell death downstream of cytochrome c release from mitochondria by activating the MEK/Erk pathway. Mol Cell Biol. 1999;19:5308–5315. doi: 10.1128/mcb.19.8.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le Gall M, Chambard JC, Breittmayer JP, Grall D, Pouysségur J, Van Obberghen-Schilling E. The p42/p44 MAP kinase pathway prevents apoptosis induced by anchorage and serum removal. Mol Biol Cell. 2000;11:1103–1112. doi: 10.1091/mbc.11.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin J, Harding A, Giurisato E, Shaw AS. KSR1 modulates the sensitivity of mitogen-activated protein kinase pathway activation in T cells without altering fundamental system outputs. Mol Cell Biol. 2009;29:2082–2091. doi: 10.1128/MCB.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fusello AM, Mandik-Nayak L, Shih F, Lewis RE, Allen PM, Shaw AS. The MAPK scaffold kinase suppressor of Ras is involved in ERK activation by stress and proinflammatory cytokines and induction of arthritis. J Immunol. 2006;177:6152–6158. doi: 10.4049/jimmunol.177.9.6152. [DOI] [PubMed] [Google Scholar]
- 52.Giroux S, Tremblay M, Bernard D, Cardin-Girard JF, Aubry S, Larouche L, Rousseau S, Huot J, Landry J, Jeannotte L, Charron J. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr Biol. 1999;9:369–372. doi: 10.1016/s0960-9822(99)80164-x. [DOI] [PubMed] [Google Scholar]
- 53.Scholl FA, Dumesic PA, Barragan DI, Harada K, Bissonauth V, Charron J, Khavari PA. Mek1/2 MAPK kinases are essential for Mammalian development, homeostasis, and Raf-induced hyperplasia. Dev Cell. 2007;12:615–629. doi: 10.1016/j.devcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Rajakulendran T, Sahmi M, Lefrançois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Mathias S, Yang Z, Kolesnick RN. Renaturation and tumor necrosis factor-alpha stimulation of a 97-kDa ceramide-activated protein kinase. J Biol Chem. 1994;269:3047–3052. [PubMed] [Google Scholar]
- 56.McPherson RA, Harding A, Roy S, Lane A, Hancock JF. Interactions of c-Raf-1 with phosphatidylserine and 14-3-3. Oncogene. 1999;18:3862–3869. doi: 10.1038/sj.onc.1202730. [DOI] [PubMed] [Google Scholar]
- 57.Xing H, Kornfeld K, Muslin AJ. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr Biol. 1997;7:294–300. doi: 10.1016/s0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- 58.Cacace AM, Michaud NR, Therrien M, Mathes K, Copeland T, Rubin GM, Morrison DK. Identification of constitutive and ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999;19:229–240. doi: 10.1128/mcb.19.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobrowsky RT, Hannun YA. Ceramide stimulates a cytosolic protein phosphatase. J Biol Chem. 1992;267:5048–5051. [PubMed] [Google Scholar]
- 60.Müller J, Ory S, Copeland T, Piwnica-Worms H, Morrison DK. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol Cell. 2001;8:983–993. doi: 10.1016/s1097-2765(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 61.Hartsough MT, Morrison DK, Salerno M, Palmieri D, Ouatas T, Mair M, Patrick J, Steeg PS. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277:32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 62.Roy F, Laberge G, Douziech M, Ferland-McCollough D, Therrien M. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 2002;16:427–438. doi: 10.1101/gad.962902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boerth NJ, Lincoln TM. Expression of the catalytic domain of cyclic GMP-dependent protein kinase in a baculovirus system. FEBS Letters. 1994;342:255–260. doi: 10.1016/0014-5793(94)80512-1. [DOI] [PubMed] [Google Scholar]
- 64.Hecht G, Pestic L, Nikcevic G, Koutsouris A, Tripuraneni J, Lorimer DD, Nowak G, Guerriero V, Elson EL, Lanerolle PD. Expression of the catalytic domain of myosin light chain kinase increases paracellular permeability. Am J Physiol. 1996;271:C1678–1684. doi: 10.1152/ajpcell.1996.271.5.C1678. [DOI] [PubMed] [Google Scholar]
- 65.Kinstrie R, Luebbering N, Miranda-Saavedra D, Sibbet G, Han J, Lochhead PA, Cleghon V. Characterization of a domain that transiently converts class 2 DYRKs into intramolecular tyrosine kinases. Sci Signal. 2010;3:ra16. doi: 10.1126/scisignal.2000579. [DOI] [PubMed] [Google Scholar]
- 66.Xu S, Robbins D, Frost J, Dang A, Lange-Carter C, Cobb MH. MEKK1 phosphorylates MEK1 and MEK2 but does not cause activation of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1995;92:6808–6812. doi: 10.1073/pnas.92.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sugimoto T, Stewart S, Han M, Guan KL. The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling Elk-1 phosphorylation from MAP kinase activation. EMBO J. 1998;17:1717–1727. doi: 10.1093/emboj/17.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Therrien M, Michaud NR, Rubin GM, Morrison DK. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 69.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends in Cell Biology. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 70.Abe Y, Matsumoto S, Wei S, Nezu K, Miyoshi A, Kito K, Ueda N, Shigemoto K, Hitsumoto Y, Nikawa J, Enomoto Y. Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J Biol Chem. 2001;276:44003–44011. doi: 10.1074/jbc.M105669200. [DOI] [PubMed] [Google Scholar]
- 71.Rutgeerts P, Van Assche G, Vermeire S. Review article: Infliximab therapy for inflammatory bowel disease--seven years on. Aliment Pharmacol Ther. 2006;23:451–463. doi: 10.1111/j.1365-2036.2006.02786.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MEK activation is impaired while Raf-1 activation and association with MAPK cascade components is not impaired in +D683A/D700A expressing cells. A) +vec, +KSR1, and +D683A/D700A expressing cells were treated with 100 ng/ml TNF for 10 minutes and whole cell lysates were examined for MEK phosphorylation by Western blot analysis. B) +KSR1 and +D683A/D700A expressing cells were treated with 100 ng/ml TNF for 5, 10, or 15 minutes or 10 ng/ml EGF for 5 minutes and Raf-1 phosphorylation examined by Western blot analysis. C) +KSR1 and +D683A/D700A expressing cells were treated with 100 ng/ml of TNF for 5, 10, or 15 minutes and phosphorylation of B-Raf detected by Western blot analysis. D) Association of Raf-1, MEK, and ERK with +KSR1 or +D683A/D700A was examined in untreated and cells treated with 100 ng/ml of TNF for 5 minutes. FLAG-KSR1 and FLAG-D693A/D700A were immunoprecipitated with anti-FLAG antibody and washed with immunoprecipitation buffer containing a reduced NaCl concentration. Total proteins were determined by Western blot analysis. Immunoblots are representatives of 3 independent experiments. IB, immunoblot.
U0126 blocks rCA-Raf-1 stimulated rMEK activity in vitro. In vitro kinase assay using recombinant constitutively active Raf -1 (rCA-Raf-1), recombinant MEK1 (rMEK) and recombinant ERK (rERK) in the presence of vehicle or U0126 (10 μM). Phosphorylated and total protein were determined by Western blot analysis using indicated antibodies. Immunoblot is a representative of 3 independent experiments. IB, immunoblot.
rKSR1 autophosphorylation occurs independent of ceramide on a single tryptic peptide A) Two-dimensional tryptic phosphopeptide map of rKSR1 following an in vitro kinase assay using [γ-32P]ATP. Spotted peptide origin is marked with an asterisk. B) rKSR1 autophosphorylation is not enhanced by ceramide in vitro. In vitro kinase assay using recombinant KSR1 (rKSR1) in the presence of vehicle or 3 μM C2-, C8-, or C16-ceramide and [γ-32P]ATP. Phosphorylated KSR1 was detected by autoradiography and total KSR1 protein was determined by Western blot analysis. Autoradiography and Immunoblot are representatives of at least two independent experiments. IB, immunoblot.
rRaf-1 stimulates rMEK activation in vitro. Recombinant wild-type Raf-1 subjected to an in vitro kinase assay in the presence of unactive recombinant wild-type MEK (rMEK) and unactive recombinant wild-type ERK (rERK). Phosphorylation of rMEK and rERK was determined by Western blot analysis using phospho-specific antibodies. Total protein was determined by Western blot analysis. Immunoblots are representatives of 3 independent experiments. IP, immunoprecipitation; IB, immunoblot.
Quantification of immunoprecipitated FLAG-KSR1, FLAG-D683A/D700A, and FLAG-Raf-1 proteins. A) FLAG-tagged Raf-1 was immunoprecipitated from increasing quantities of total protein (400–800 μg) from whole cell lysate, gel-loaded along with increasing amount of purified recombinant CA-Raf-1 (0.01–0.32 μg), and separated by SDS-PAGE. Densitometric measurements were obtained from Western blot analysis using an αRaf-1 antibody. Linear regression using rCA-Raf-1 as the protein standard determined the amount of immunoprecipitated FLAG-tagged Raf-1. Immunoblot is a representative of 3 different experiments. B) FLAG-tagged KSR1 was immunoprecipitated from increasing quantities of total protein (400–1000 μg) from whole cell lysate, gel-loaded along with increasing amount of purified recombinant FLAG-tagged bacterial alkaline phosphatase (BAP) (0.01–0.32 μg), and separated by SDS-PAGE. Densitometric measurements were obtained from Western blot analysis using an αFLAG antibody. Linear regression using rFLAG-BAP as the protein standard determined the amount of immunoprecipitated FLAG-tagged KSR1 protein. Immunoblots are representatives of 3 independent experiments. IP, immunoprecipitation; IB, immunoblot.