Abstract
Entamoeba histolytica is a major health threat to people in developing countries, where it causes invasive diarrhea and liver abscesses. The study of this important human pathogen has been hindered by a lack of tools for genetic manipulation. Recently, a number of genetic approaches based on variations of the RNAi method have been successfully developed and cloning of endogenous small-interfering RNAs from E. histolytica revealed an abundant population of small RNAs with an unusual 5′-polyphosphate structure. However, little is known about the implications of these findings to amebic biology or the mechanisms of gene silencing in this organism. In this article we review the literature relevant to RNAi in E. histolytica, discuss its implications for advances in gene silencing in this organism and outline potential future directions towards understanding the repertoire of RNAi and its impact on the biology of this deep-branching eukaryotic parasite.
Keywords: Entamoeba, gene silencing, RNAi, secondary siRNA, siRNA
Background on Entamoeba histolytica
Entamoeba histolytica is a protozoan parasite that infects approximately 500 million people worldwide and has a death toll of nearly 100,000 per year [1]. It is the second leading parasitic disease in humans, occurring mainly in developing countries where access to clean water is limited. The parasite has two life stages: the cyst form is a dormant stage, which allows it to survive in challenging environments, and the trophozoite form is the active stage, which invades the host epithelial tissue causing colonic and hepatic disease [2]. Once ingested, the cyst excysts in the small intestine to the trophozoite form, proliferates and colonizes the colon. The 20 Mb genome of E. histolytica has been sequenced and is predicted to have 8201 genes [3,4]. Functional characterizations of genes encoded in the parasite have been challenging due to the polyploid nature of E. histolytica trophozoites, but recently several genetic approaches have been successfully developed, many of which are based on variations of the RNA interference (RNAi) approach [5–7].
Background on RNAi
First recognized in Caenorhabditis elegans, RNAi is a potent and specific gene down-regulation phenomenon, caused by introducing a double-stranded RNA (dsRNA) to its target gene [8]. While the exact components of the RNAi pathway can vary in different organisms, the basic mechanisms are as follows: first, the dsRNA is processed by Dicer (an RNaseIII-like endonuclease) into small-interfering RNAs (siRNAs), siRNAs are then recognized by the RNA-induced silencing complex (RISC) and a single guide strand of siRNAs is loaded. The siRNA directs the multiprotein complex to its cleavage target, resulting in mRNA down-regulation [9]. Since its initial identification, the RNAi silencing mechanism has been found to be widely conserved in most eukaryotic systems [9,10]. In the last decade the RNAi machinery and its pathways have been investigated extensively, and this pathway functions in many different cellular processes, such as development [11], differentiation [12], cancer [13] and host protection from viruses [14].
Dicer-dependent & dicer-independent siRNAs
RNAi is mediated by small RNA molecules of approximately 20–30-nt size. These small RNAs are diverse in regard to their size, specific modifications to the 5′ and 3′ ends, and functional roles in gene regulation. Classical siRNAs are generated by the protein Dicer, which processes long dsRNAs into approximately 24-nt siRNAs with 5′-P and 3′-OH features. Although classical siRNAs are Dicer-dependent, many new classes of Dicer-independent small RNAs, such as secondary siRNA, Piwi-interacting RNA (piRNA), and primal small RNA, have been recently identified from studies in C. elegans, Drosophila, rat and Schizosaccharomyces [15–18]. In C. elegans, secondary siRNAs are triggered by Dicer-produced siRNAs, but amplified by RNA-dependent RNA polymerase (RdRP), and function to augment gene silencing. These secondary siRNAs have a unique 5′-polyphosphate structure, generally start with a 5′-G (known as 22G small RNA), and cover more than 50% of genes in C. elegans, indicating their profound role in gene regulation [19]. In Drosophila and germline cells of vertebrates, piRNAs are associated with germline-specific Argonautes (AGOs) and their production is carried out by the ‘ping-pong’ mechanism, which does not involve an RdRP complex or Dicer. piRNAs play an important role in germline development and may help control transposition in germline cells [16]. In Schizosaccharomyces pombe, primal small RNAs are derived from degradation products of abundant transcripts that interact with AGO and trigger siRNA amplification and hetero-chromatin assembly within DNA repeats. A transcriptome surveillance mechanism has been proposed for the initiation of RNA silencing in this organism [18].
Gene-silencing mechanisms of RNAi
Once generated, small RNAs are loaded into the RISC or RNA-induced transcriptional silencing (RITS) complex providing target specificity. The target is silenced through either a post-transcriptional gene silencing (PTGS) or transcriptional gene silencing (TGS) mechanism. PTGS occurs through inactivation of mRNAs either by degradation (where mRNA targets are cleaved at sites of small RNA binding) or by translational repression (largely occurring in the miRNA pathway). By contrast, TGS occurs through DNA methylation or by heterochromatin formation. In plants, TGS is caused by siRNAs targeting promoter sequences and DNA methylation is found on the target gene sequences [20]. In S. pombe, TGS involves recruitment of proteins that induce DNA or histone modification and assembly of repressive chromatin, thereby causing the gene to become transcriptionally inert [21].
RNAi pathways in parasites
The rapid development of RNAi research in recent years has prompted a wave of exploration into RNAi function in non-model organisms. In fact, the finding of RNAi in Trypanosoma bruci is among the earliest identifications of the RNAi phenomenon [22]. Interestingly, two species closely related to T. brucei, Trypanosoma cruzi and Leishmania major, are RNAi negative as genome sequencing has failed to identify homologs of any essential RNAi pathway members [23]. In apicomplexan parasites, genome sequencing revealed no conventional RNAi machinery in either Plasmodium or Cryptosporidium and attempts for cloning small RNAs from Plasmodium falciparum failed to validate any siRNA/miRNA species in this parasite [24,25]. However, in Toxoplasma gondii, an AGO-like protein has been identified in the genome and recent small RNA cloning efforts confirmed that T. gondii has both siRNAs and miRNAs [26]. In the intestinal parasite Giardia lamblia, both siRNA and miRNA have been identified and have important functions in parasite biology, including regulation of antigenic variation and retrotransposon control [27–30]. Interestingly, miRNAs in this system are derived from snoRNAs [29]. In Trichomonas vaginalis, high-throughput sequencing revealed endogenous miRNA candidates [30] and genomic prediction and functional study of miRNA in this organism were also reported [31,32]. While most efforts in RNAi/miRNA study in parasitology have so far been focused on the parasite itself, some recent data began to show that parasites, such as Cryptosporidium and Toxoplasma, are able to maneuver the host cell miRNAs pathways in a way that favor parasite intracellular development [33,34].
RNAi pathway genes in the Entamoeba genome
The RNAi machinery consists of several key proteins that contribute to gene silencing, including Dicer, AGO and RdRP. Dicer is an RNaseIII endonuclease that recognizes and cleaves dsRNA into 20–30-nt duplexes [9]. Most Dicer enzymes contain two RNaseIII domains that dimerize to form a catalytic valley that accommodates and cleaves the dsRNA and a PAZ domain that binds the 3′ end of the small RNA [35]. However, there are Dicers that lack this canonical structure – most notably the two Dicer enzymes in T. brucei, which contain only two RNaseIII domains [36,37] and the Saccharomyces castellii Dicer, which contains a single RNaseIII domain and two double-stranded RNA binding domains (dsRBDs) but no PAZ domain [38]. AGO is the core component of the RISC complex and is responsible for the complex’s slicing activity [39]. AGO proteins contain four domains: N-terminal, PAZ, Mid and Piwi, with the PAZ and Piwi domains playing the greatest functional roles [39]. As in Dicer enzymes, the PAZ domain binds the small RNA while the Piwi domain contains an RNaseH-like fold through which it cleaves the target mRNA [40]. RdRP is recruited to sites of primary RNAi (initiated by Dicer produced siRNAs), and generates secondary small RNAs that amplify the initial silencing signal [15]. RdRP proteins contain catalytic domains that are similar to the β-subunit of DNA-dependent DNA polymerases [101].
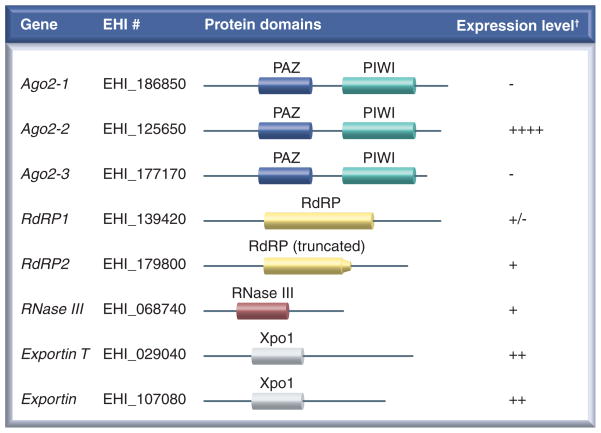
Many genes of the RNAi pathway are encoded within the E. histolytica genome including three AGO genes (EHI_125650, EHI_186850 and EHI_177170), which contain both PAZ and Piwi domains (Figure 1). Of these genes, EHI_125650 is the most highly expressed while EHI_186850 and EHI_177170 are only detectable by RT-PCR [41] [Pompey JM, Singh U, Unpublished Data]. Moreover, EHI_125650 (also referred to as EhPiwi-rp [41]), associates with 27-nt small RNAs in E. histolytica trophozoites, indicating that this protein functions in the amebic RNAi pathway [41]. There is evidence in other systems that different AGO proteins associate with specific classes of small RNAs [42,43]; therefore, one could speculate that each amebic AGO protein might associate with a different E. histolytica small RNA population. E. histolytica also contains one RdRP gene (EHI_139420) and one gene with a partial RdRP domain (EHI_179800). E. histolytica lacks a gene that fits the canonical Dicer structure. There is a single gene containing a single RNaseIII domain (EHI_068740) (Figure 1). In-depth searching of the E. histolytica genome using Hidden Markov Models did not reveal any additional RNaseIII domain proteins [Pompey JM, Singh U, Unpublished Data].
Figure 1. RNAi pathway in Entamoeba histolytica.
RNAi pathway genes with their conserved domains and expression level in trophozoites. A shorthand name for each gene is listed followed by gene IDs. All conserved domains were identified using NCBI blast tool for conserved domains and are drawn to scale. Expression data are derived from microarray experiments [56].
†There are no significant changes observed for the RNAi pathway genes when microarray gene expression data of Entamoeba histolytica under different culture conditions (heat shock, 5-azacytidine treatment, oxidative and nitrosative stresses) and different strains (200:NIH, Rahman, recent clinical isolates of E. histolytica) are compared.
Despite the key role Dicer plays in the RNAi pathway, there has been recent evidence of Dicer-independent generation of small RNAs. In S. pombe, primal RNAs were described in Dicer-deleted strains; these are thought to be derived from the degradation of single-stranded transcripts, and associate with AGO where they may play roles in heterochromatin formation [18]. miRNAs are another class of small RNAs that are processed by Dicer, yet examples of Dicer-independent biogenesis of miRNAs (or in the last case, of miRNA-like small RNAs) are found in zebrafish [44], mice [45] and fungi [46]. In zebrafish and mice, AGO2, not Dicer, is responsible for the processing and generation of the mature miR-451 species [44,45].
miRNAs modulate gene expression (in the vast majority of cases) through inhibiting translation of their mRNA targets [47,48]. Although miRNAs are structurally and functionally distinct from siRNAs, the machinery that generates miRNAs and facilitates their function is largely similar to those that produce siRNAs. miRNAs are encoded within the genome and transcribed by RNA polymerase II into long stem–loop primary-miRNA (pri-mRNA) structures. Pri-miRNAs are then recognized and cleaved by the microprocessor complex that includes Drosha, an RNaseIII endonuclease, and DGCR8, a dsRNA binding protein, and transported out of the nucleus by Exportin-5 [47,49]. In the cytoplasm, the hairpin is further processed by Dicer into the mature approximately 22-nt species and loaded into RISC. This complex locates and translationally represses the miRNA target(s) through imperfect base-paring, typically in the 3′ untranslated region [47].
Two proteins needed uniquely for the miRNA pathway (but not the siRNA pathway) are Drosha and Exportin-5. Drosha contains two tandem RNaseIII domains like Dicer but also contains a C-terminal dsRBD and an N-terminal proline-rich region [50]. No homologs of Drosha have been identified in the E. histolytica genome. Similarly, no candidates for Drosha’s partner, DGCR8, have been identified in E. histolytica. Searches of the E. histolytica genome database reveal two likely Exportin proteins, EHI_029040 and EHI_107080 (Figure 1). The former is annotated as a putative Exportin-t protein, which transports tRNAs from the nucleus into the cytoplasm [51]. EHI_107080 is annotated as a hypothetical protein but when the sequence was searched for conserved domains, an XpoI domain, commonly found in Exportin proteins, was identified. More than 30% of the E. histolytica genome contains predicted proteins of unknown function [3]; therefore, it is possible that some of these miRNA machinery proteins do exist within E. histolytica but have yet to be recognized and that sequence homology may not be sufficient to identify these proteins given the evolutionary divergence between Entamoeba and mammalian systems. miRNAs have been computationally identified in protozoan parasites, such as T. vaginalis [31], and have been sequenced in G. lamblia [52] and T. gondii [26], yet no miRNA-specific machinery has been identified within these genomes. Thus, the apparent lack of a microprocessor complex (Drosha and DGCR8) in E. histolytica does not preclude the presence of the miRNA pathway in this system.
It is well known that translationally repressed mRNA can accumulate in discrete cytoplasmic foci (P-bodies) and that the mRNA degradation proteins therein (deadenylation, decapping and degradation enzymes) are crucial in the final step of miRNA pathway [53]. Studies in mammalian cells show that AGO proteins, the key components of RISC, are not randomly distributed but are concentrated in P-bodies [54]. We have tagged the most highly expressed putative E. histolytica AGO protein, EHI_125650, and on initial analysis it does not appear to be localized in any P-body-like cytoplasmic granules [Pompey JM, Tran VG, Singh U, Unpublished Data]. A search for potential decapping enzymes indicated there are five genes in this category in the E. histolytica genome (EHI_192480, EHI_058810, EHI_169590, EHI_104780 and EHI_042970). At present, whether P-body-like structures exist in E. histolytica is an open question, as no specific antibodies exist that mark these structures in parasites. More work is needed to address these questions.
Endogenous small RNA repertoire in Entamoeba
In an effort to characterize the endogenous small RNA repertoire in E. histolytica, small RNA cloning and sequencing was performed [41]. Surprisingly, E. histolytica trophozoites were found to have multiple small RNA populations, including an abundant 27-nt small RNA population, which has an uncommon 5′-polyphosphate structure. This feature indicates that this small RNA population is generated in a Dicer-independent manner and is the first identification of 5′-polyphosphate small RNAs in a system other than C. elegans [41].
Abundant & multiple small RNA populations in Entamoeba
Several distinct bands in the small RNA range were identified (16-nt, 22-nt and 27-nt) using total RNA samples from E. histolytica trophozoites separated on a 12% denaturing polyacrylamide gel [41]. A similar profile was observed for other Entamoeba: Entamoeba dispar (an amoeba that colonizes humans but does not cause invasive disease) and Entamoeba invadens (a reptilian amoeba that causes disease similar to E. histolytica). A number of conditions (heat shock, serum starvation, oxidative and nitrosative stresses) were identified which changed the bulk populations of the small RNAs with the 22-nt population diminishing and 27-nt population increasing upon stress [Zhang H, Singh U, Unpublished Data]. Dramatic changes in the 22-nt and 27-nt populations were observed between the small RNA profiles of E. invadens trophozoites and cysts [Zhang H, Singh U, Unpublished Data] indicating that Entamoeba has conserved small RNA pathways, and that these pathways may play roles in the parasite’s response to environmental changes and regulation of stage conversion.
Small RNA cloning
Initial efforts to clone these small RNAs from a 15–30-nt size selected RNA fraction using a 5′-phosphate-dependent method failed to clone full-length small RNAs [41]. Since 5′ and 3′ modifications can inhibit ligation and prevent cloning of full-length small RNAs, the gel purified 27-nt small RNA population from E. histolytica trophozoites was characterized using several enzymatic chemical assays and found to have 5′-polyP termini and 3′-OH termini [41]. The small RNAs were then cloned using a method that is independent of the presence of a 5′-P. Once this approach was applied, the sizes of the cloned small RNAs matched the sizes revealed by Northern blot analysis and the 5′ and 3′ termini of the cloned small RNAs were confirmed [41]. Importantly, 5′-polyP siRNA have previously only been identified in secondary siRNAs from C. elegans, where they are involved in an amplified silencing mechanism [15]. The work of Zhang and colleagues thus extends the concept of amplified silencing to a single celled eukaryotic organism.
27-nt small RNAs associate with an E. histolytica AGO protein
In order to effect a silencing function, small RNAs have to associate with AGO family proteins. E. histolytica has three proteins of this family but only one (EHI_125650) is highly expressed in trophozoites. Using an N-terminal Myc-tagged EHI_125650 in E. histolytica trophozoites, immunoprecipitation (IP) with anti-Myc antibody showed that small RNAs of approximately 27-nt were specifically associated with the AGO protein [41]. The small RNAs that immunoprecipitated with EHI_125650 were subsequently shown to have 5′-polyphosphate and 3′-OH structure and a small RNA library was generated using a 5′-P independent cloning approach. Limited Sanger sequencing analysis revealed several interesting features of the endogenous small RNAs in E. histolytica. A substantial number of small RNAs mapped to coding regions (25% antisense to ORFs and 9% sense to ORFs) as well as to intergenic regions (13%). However, only a very small number mapped to retrotransposon elements (0.5%) [41]. This is in sharp contrast to small RNA sequencing profiles for other systems, including Drosophila as well as parasitic organisms, such as G. intestinalis, T. brucei and T. gondii, where the majority of small RNAs are derived from retrotransposons and repetitive elements [26,27,55]. Thus it appears that the small RNAs that associate with AGO in E. histolytica have a very different repertoire than those that associate with AGO in other systems and that this may potentially indicate a different impact on the gene regulation mechanism in E. histolytica. Pyrosequencing approaches were used to further define the small RNA repertoire associated with this AGO protein and analysis of approximately 400,000 sequences demonstrated that the mapping and genomic distribution of the small RNAs largely remains the same as what was initially noted [Zhang H, Singh U, Unpublished Data].
27-nt small RNAs that map antisense to genes are associated with silencing of the cognate gene
In order to define the potential functions of the 27-nt small RNAs, the initial focus was on small RNAs that map antisense to predicted genes. Using previously published microarray expression data [56], it was identified that genes with significant numbers of antisense small RNAs had extremely low expression values in E. histolytica HM-1:IMSS trophozoites, the strain and stage of the parasite from which small RNAs were cloned [41]. The array analysis was extended to other E. histolytica strains, including 200:NIH and Rahman, and it was observed that some genes with antisense small RNAs in HM-1:IMSS had varied expression levels in other E. histolytica strains. Northern blot analysis for a small RNA where its associated gene had variable expression among amebic strains showed that the gene and small RNA expression are inversely correlated (i.e., for a given gene, small RNAs were detectable in E. histolytica strains with low gene expression but were not detectable in E. histolytica strains with high gene expression). These data strongly suggest that small RNAs mediate target gene silencing in E. histolytica, although further work is needed to define the mechanism(s) by which this is occurring.
Potential miRNAs in E. histolytica
miRNAs are another major class of small RNAs involved in PTGS. As mentioned earlier, miRNA maturation involves multiple sequential cellular processes, beginning with initial transcription of the primary transcript, which is cleaved by Drosha into a hairpin pre-miRNA, and exported into the cytoplasm. Dicer then acts on the substrate to form the final mature miRNA. Recent studies have shown miRNAs can also be generated through non-canonical pathways that are Drosha independent, such as intron-derived miRNAs [57] and snoRNA-derived miRNAs [58], or that are Dicer independent [44,45]. Initially it was thought that miRNAs were restricted to multicellular organisms, however, recent data demonstrate that miRNAs are also present and functional in unicellular organisms, such as the green alga Chlamydomonas [59], viruses [60] and protozoan parasites (G. lamblia [29] and T. gondii [26]).
To explore the possibility whether typical miRNAs exist in E. histolytica, De et al. used a bioinformatics approach to look for miRNAs from the E. histolytica genome sequence; 17 putative candidate miRNAs were predicted [61]. However, none of these predictions were experimentally validated by either northern blot analysis or by molecular cloning and sequencing. Thus, it is still an open question whether miRNAs exist in Entamoeba and further work is needed in this area.
Gene silencing approaches in E. histolytica
Owing to the polyploid nature of the E. histolytica genome, standard molecular genetic approaches for gene knockouts are not feasible in this parasite system. Earlier attempts at gene manipulation in this organism included dominant-negative gene expression [62], antisense approaches using stable episomal transfection [63], or incubation of short antisense peptide nucleic acid oligomers with E. histolytica trophozoites [64]. In recent years, new promising approaches for gene knockdown have been explored for this organism, including the G3 strain-based silencing and variants of RNAi-based silencing (such as soaking with synthetic siRNA duplex or long dsRNA, as well as dsRNA- and shRNA-based RNAi approaches) (Table 1).
Table 1.
Summary of all gene knockdown approaches attempted in Entamoeba†
| Strategy | Vector/construct | Inducing trigger sequences | Gene targets | Knockdown efficiency | Ref. |
|---|---|---|---|---|---|
| Antisense peptide nucleic acids | N/A | 17-mers complementary to the first 17 nucleotides of NPT mRNA coding sequence (NE017ATG) or the EhErd2 mRNA (ERD17ATG) | Neomycin phosphorotransferase, NPT; EhErd2, a homolog of Erd2, (a marker for the Golgi body) | NPT activity was decreased 70%; 35% specific decrease of EhErd2 protein expression | [64] |
| N/A | The first 17 nucleotides of the coding sequence of Entamoeba histolytica Sec61α subunit gene (Eh Sec61) | EhSec61α | ~70% decrease in Sec61α levels | [85] | |
| Constitutive antisense expression‡ | pSA8 | Antisense expression of a segment of the coding gene (877 bp) for the ehcp5 | Cysteine proteinases (CPs) | Total CP activity is strongly reduced (~90% down) | [86] |
| pSA20 | Antisense expression of a segment of the coding region (842 bp) of the Igl | Lgl | 60% downregulated at protein level | [87] | |
| pAP-R2 | Antisense of gene AP-A (full gene) | AP-A | 60% downregulated at protein level | [88] | |
| pNeo-EhDead1-AS | Antisense of gene EhDead1 (full gene) | EhDead1 | RT-PCR undetectable of EhDead1 mRNA | [89] | |
| Inducible antisense expression‡ | pEhHYG-tetR-O-CAT | Sense and antisense EhCaBP (full gene) cloned into vector | EhCaBP | Decrease in EhCaBP levels (~twofold) in EhCaBP-AS cells; threefold increase in EhCaBP levels in EhCaBP-S cells; EhCaBPI protein level is ~40% that of control parasites | [90,91] |
| A 422-bp fragment from the 5′ end of the eh29 gene is cloned in antisense direction | The 29-kDa surface antigen (thiol-dependent peroxidase; Eh29) | 55% inhibition in eh29 expression | [92] | ||
| EhTMKB1-9 gene was cloned either in sense or antisense direction | EhTMKB1-9 | TMK9-AS transfectants displayed slower multiplication as the cell numbers were reduced by 40% at 72 h | [93] | ||
| EhPIG-L gene was cloned either in sense or antisense direction | EhPIG-L | A twofold reduction in EhPIG-L transcripts in EhPL-AS cells by northern blot analysis | [94] | ||
| G3§ | psAP-1, psAP-2 | 473 bp AP-A 5′ flanking region | AP-A | Complete knockdown (mRNA and protein) | [65] |
| psAP-6 | 354 bp AP-A 5′ flanking region | AP-A | Partial knockdown | [65] | |
| psAP-23 | 386 bp AP-A 5′ flanking region | AP-A | Partial knockdown | [66] | |
| pB33 | Direct ligation of Ehlgl 1 ORF to 473 bp AP-A 5′ flanking region | AP-A, Ehlgl 1, Ehlgl 2, Ehlgl 3 | Complete knockdown (mRNA and protein) | [67] | |
| pAP-CP5 | Direct ligation of EhCP-5 ORF to 473 bp AP-A 5′ flanking region | AP-A, EhCP-5 | Complete knockdown (mRNA and protein) | [67] | |
| pTL | Direct ligation of partial Ehlgl 1 ORF to 473 bp AP-A 5′ flanking region | AP-A, Ehlgl 1 | Complete knockdown (mRNA and protein) | [67] | |
| pL5 | Direct ligation of partial Ehlgl 5 ORF (311 bp) to 473 bp AP-A 5′ flanking region | AP-A, Ehlgl 5, Ehlgl 4 | Complete knockdown (mRNA and protein) | [95] | |
| psAP1Δ2600 | A psAP-1 deletion of 2.6-kb fragment containing tRNA array | AP-A | AP-A overexpression | [69] | |
| psAP1Δ2600EY1000 | Re-adding 1.0-kb EY tRNA array sequence into psAP1Δ2600 | AP-A | Complete knockdown (mRNA) | [69] | |
| Long dsRNA | pJST4 vector based | 350 bp at 5′ end of gene | Eh Diaphanous | Undetected for both RNA and protein analysis | [5] |
| 3′ end of gene (1942–2592 bp) | EhKlp5 | Protein level is significantly lowered (95% decrease) | [71] | ||
| EhKlpA1 at 5′ end (nucleotides 1–340); EhKlp2 at 3′ end (nucleotides 1278–1965); EhKlp3 at 3′ end (nucleotides 1075–1426); EhKlp4 at 3′ end (nucleotides 2060–2460) | EhKlp2–4, A1 | Approximately 40–50% knockdown for EhKlp2, 3 and 4 (mRNA level). For EhKlpA1, mRNA was 80% downregulated | [72] | ||
| 395 bp segment from the 3′ end of the EhSTIRP1 | EhSTIRP1 | mRNA for all EhSTIRP genes were drastically reduced | [73] | ||
| Short hairpin RNA | Modified pGIR310 with U6 promoter driven shRNA expression | Three shRNA constructs for PATMK at position 325, 2273, 3552 bp | Phagosome-associated TMK96 (PATMK) | Reduced PATMK protein levels for construct at position 2273, 3552, but not for construct at position 325 | [80] |
| Four shRNA constructs for Igl at positions 272, 1198,2412,2777 bp | Igl 1 | Decreased amount of Igl1 protein for construct 272, 1198, 2777 (reduced to 20–40% of control level), but construct 2412 showed no effect (95.3% to the control) | [6] | ||
| Two shRNA constructs for URE3-BP at 350, 580 bp | URE3-BP | Significant reduction of protein in both constructs (reduced to 10–15% of control level) | [6] | ||
| Two shRNA constructs for EhC2A at position 363, 502 bp | EhC2A | Construct 363 had knockdown of EhC2A protein level to 3.0% compared with control, but construct 502 had no knockdown effect on EhC2A levels | [6] | ||
| Soaking with siRNA duplex or long dsRNA | N/A | γ-tubulin siRNA (positions 170–192) | E. histolytica γ-tubulin | In the presence of 1 μg/ml of siRNA, the protein level was unaffected, with 5 μg/ml of siRNA protein level was decreased by 90% | [7,96] |
| Bacterial dsRNA expressing vector, L4440-KERP1 | For KERP1, 388 bp at 5′ end of the gene | Virulence factor KERP1 | Soaked at concentration of 5, 25 and 50 μg/ml; 34% protein reduction observed at 50 μg/ml, no obvious reduction at 5 μg/ml and 25 μg/ml | [81] | |
| Feeding with dsRNA-expressing bacteria | Bacterial dsRNA expressing vector, L4440-β-tubulin | For β-tubulin, 753 bp at 5′ end of the gene | E. histolytica β-tubulin | 58% reduction at β-tubulin mRNA level, protein level was also reduced | [81] |
| Bacterial dsRNA-expressing vector, L4440-KERP1 | For KERP1, 388 bp at 5′ end of the gene | Virulence factor KERP1 | 48% decrease of protein level, but no significant reduction of mRNA was observed | [81] | |
The silencing strategy, vector/construct, inducing trigger sequences, gene target, knockdown efficiency and reference are listed. The naming of each vector/construct and knockdown efficiency are taken from the original paper When there was no number that could be cited, whether there was partial knockdown, complete knockdown or no detectable change is indicated.
The papers on approach using dominant-negative gene overexpression are not listed here since, by definition, it does not involve a gene downregulation mechanism.
Listed are the representative papers using the same vector.
Listed are the representative constructs.
N/A: Not available.
G3 strain
A silencing phenomenon was serendipitously discovered in E. histolytica when attempts at overex-pressing a gene resulted in silencing the gene. The plasmid designed to overexpress Amoebapore A (AP-A) contained the coding region of AP-A flanked by both its 5′ and 3′ regulatory regions. However, instead of over expressing AP-A, this plasmid resulted in parasites that were permanently and stably silenced for AP-A. Silencing persisted even after the plasmid was removed and was maintained for many years [65]. Further investigation for sequences essential to this silencing process showed that silencing of AP-A can be achieved by using plasmids containing only the 5′ region of the AP-A gene promoter – this piece contains no portion of the AP-A coding region but includes an upstream region of AP-A with a truncated segment of a repetitive element and an adjacent short T-rich stretch [66]. These two components are currently believed to be essential for triggering silencing. The parasite strain in which AP-A is permanently silenced has been named G3 [65].
A number of interesting observations have been made about the silencing phenomenon in the G3 strain [67]. The silencing is inheritable and can be maintained even after removal of the plasmid: parasite clones without the initial trigger plasmid appear to retain silencing indefinitely. Silencing requires sequence homology to the initial trigger: the genes silenced in this approach have significant sequence homology to the initial ‘trigger’ used in the vector. Thus, in addition to AP-A, both Amoebapore B (AP-B) and Saposin like protein (SAPLIP1) were also downregulated in the AP-A silenced G3 trophozoites. Both AP-B and SAPLIP1 have high sequence identity in the ORF region compared with AP-A [67]. Additionally, this approach can be used to extend silencing to a second unrelated gene, although attempts in silencing a third gene have failed [67].
The silencing mechanism in G3 parasites has been extensively studied and currently a transcriptional silencing mechanism is favored. Probing for possible small RNAs revealed no detectable siRNA signal in G3 parasites and instead identified a 140-nt single-stranded RNA molecule (likely to be a short SINE1 RNA fragment) [66]. DNA methylation of EhAP-A was found to be unrelated to the silencing; sodium bisulfate treatment and DNA sequencing for both SINE1 and AP-A coding regions for cytosine methylation showed no difference between the DNA of the silenced G3 trophozoites and WT controls. Furthermore, inhibitors of DNA methylation, such as 5-azacytidine or zebularine, failed to restore the transcription of silenced genes [66]. However, histone modification is thought to be involved in the silencing as chromatin IP (ChIP) analysis using an antibody against methylated K4 of histone H3, showed a difference in methylation of histones at the AP-A locus between silenced G3 trophozoites and a non-silenced control [66]. Furthermore, a ChIP assay using custom-made antibodies against the di- and tri-methylated lysine 4 of histone H3 of E. histolytica revealed demethylation of H3K4 in the coding region of silent genes for all silenced genes [68]. Additionally, data suggested that the promoter region of silent genes is more resistant to MNase digestion, indicating that the nucleosomal organization in the promoter region of the silenced AP-A gene has been affected [68].
While the above data may point to a transcriptional silencing mechanism linked with long non-coding RNAs and histone modification, a different mechanism, trans-inactivation through tRNA gene-mediated silencing, was recently reported to be linked to the observed AP-A silencing [69]. In this study, sequence comparison between plasmid pNAP-A (that caused AP-A overexpression) and plasmid psAP1 (that caused AP-A silencing) revealed some sequence differences. The psAP1 contains an extended actin 3′ sequence, where a tRNA array comprising the glutamic acid (TTC) and tyrosine (GTA) tRNAs, resides. Testing with a series of 5′ and 3′ deletion constructs showed the 3′ end tRNA genes are sufficient to mediate AP-A trans-inactivation, while the partial SINE1 element and the T-rich region located at the 5′ flanking region of AP-A seemed to be dispensable [69].
Therefore, the molecular mechanism of AP-A silencing in E. histolytica is still open to debate. Aside from SINE element and tRNA array, a role for small RNAs in the G3 silencing method has not been definitively excluded. It is possible that the failure to detect small RNAs could be explained by technical issues and deserves to be re-explored. Given that we now know that E. histolytica has an endogenous small RNA repertoire, further experiments are needed to investigate the possibility that the small RNA pathway is linked to gene silencing in the G3 strain.
RNAi-based silencing methods
dsRNA-based RNAi approach
Long dsRNA is a potent trigger of gene silencing and is used in a number of organisms, including C. elegans [8], Drosophila [70] and Trypanosomes [22]. The first success for gene knockdown using long dsRNA strategy in E. histolytica was reported for the protein Diaphanous [5]. In this paper, the construct was designed to express 350 bp from the 5′ end of the gene and was placed as a head-to-head direction with an unrelated stuffer DNA fragment in the middle. The transcript would be expected to fold back to form a hairpin structure with a long dsRNA stem upon transcript formation. The results showed that expression of E. histolytica Diaphanous was completely inhibited (both mRNA and protein were not detectable) and the stable transformants could be maintained indefinitely without loss of silencing. This approach was further used to knockdown a number of amebic genes (KlpA1, Klp2, Klp3, Klp4 and Klp5) in subsequent studies [71,72]. In order to get gene-specific knockdown, the selected region and the length of dsRNA varied from 340 to 687 bp and the extent of mRNA downregulation achieved ranged from approximately 40% to approximately 80%. Although the dsRNA expression strategy has worked with multiple successes, no data have been presented demonstrating that downregulation is due to dsRNA being processed into siRNA.
However, a loss of dsRNA-induced gene silencing has been noted. The approach outlined above was used to downregulate expression of an E. histolytica serine, threonine, isoleucine-rich protein (EhSTIRP) [73]. Interestingly, the down-regulation of EhSTIRP reverted to wild-type levels after approximately 6 months, despite the continued presence of the correct plasmid, the drug resistance marker, and the intact dsRNA portion of the construct [74]. Several mechanistic possibilities for this reversion of dsRNA-based downregulation were investigated but no definitive explanation was identified. However, it is now clear that reversion of dsRNA-based silencing can occur in E. histolytica, similar to what has been noted in T. brucei [75].
Short hairpin RNA-based silencing
The short hairpin RNA (shRNA) approach is based on mimicking miRNAs, which naturally have a short hairpin structure. The use of shRNAs as a genetic tool to achieve gene knockdown is well documented in model systems [76–78]. The shRNA expression construct is driven by a RNA polymerase III promoter (such as the promoter for the U6 snRNA), resulting in a hairpin structure with a 19–29 bp stem and a 4–10-nt loop [79]. The nuclear-expressed shRNAs are exported into the cytoplasm and further processed by the RNAi machinery to siRNAs that mediate gene silencing.
This method has been successfully adapted in E. histolytica to knockdown protein expression of a number of amebic genes, including the E. histolytica phagocytosis-associated transmembrane kinase 96, the intermediate subunit of the galactose- and N-acetyl-D-galactosamine-inhibitable lectin, a transcription factor and a membrane binding protein [6,80]. The construct contains the E. histolytica U6 promoter followed by a gene-specific hairpin structure (29-nt complementary stem with a 9-nt loop), which resulted in downregulation of both mRNA and protein levels, with protein knockdown ranging from 72 to 97%. Several interesting observations were noted. First, not all shRNA constructs worked for gene downregulation and of the constructs that did work, not all were equally effective in mediating silencing. A 25–29-nt hairpin stem had the best silencing efficiency and was measurably better than the 22-nt length shRNA utilized in mammalian cell lines [76]. Second, the reduced mRNA level did not correlate with the level of protein knockdown. Third, small RNAs to the targeted gene were detected in the downregulated parasites, with an observed size of 27-nt. Given the fact that the most highly abundant endogenous small RNA population in Entamoeba is 27-nt, it is intriguing to note that the 27-nt small RNA had the greatest efficiency of gene downregulation.
siRNA soaking
siRNA soaking was successful for gene knockdown of E. histolytica γ-tubulin [7]. In this study, 22-nt siRNA duplexes were synthesized and added to E. histolytica culture at a final concentration of 1.5–10 μg/ml for 15 h. The level of γ-tubulin mRNA was decreased by 51% and the level of protein expression was decreased by 90% when parasites were soaked with 5 μg/ml siRNA. Additionally, these authors showed that γ-tubulin RNAi inactivation can be reversible (i.e., after washing treated parasites, γ-tubulin mRNA levels recovered 40% of wild-type levels after 72 h).
Similarly, taking advantage of intrinsic phagocytic properties of E. histolytica, a second soaking method was adapted using the feeding method from C. elegans [81]. In this study, parasites were either directly fed with dsRNA-expressing bacteria or soaked with purified dsRNA. Both treatments resulted in the downregulation of gene expression. For the feeding experiment, trophozoites were incubated with bacteria producing dsRNA. E. coli strain HT115(DE3), which lack expression of RNaseIII and thereby allow accumulation of the dsRNA, were used. Upon induction, dsRNA is produced from a dual T7 promoter construct. It was shown that mRNA level of the target gene β-tubulin was reduced by 58% and protein level was also reduced by western blot analysis. A second gene chosen for knockdown by this approach was the virulence factor, EhKERP1. No significant reduction on KERP1 mRNA was observed but protein was reduced by 48%. In addition, direct soaking of trophozoites with purified dsRNA also downregulated EhKERP1 at both the mRNA and protein levels.
There are many unknowns regarding gene silencing using the various RNAi pathway methods outlined above. In bacteria feeding, it is unclear how dsRNA are released into the cytoplasm following bacterial phagocytosis by the amoeba, as well as whether whole dsRNA or its partially degraded products are the triggers of gene silencing. For the dsRNA and duplex siRNA soaking methods, it is unknown if dsRNAs are first partially digested in the medium by nucleases secreted from parasites [82] and then the smaller RNA fragments diffuse or get imported into the amoeba, or if dsRNA can be imported either by endocytic processes or by transporter-mediated RNA entry. Furthermore, the small RNA size used for siRNA duplex soaking was 22-nt, which is much smaller than the cloned endogenous small RNAs and is also smaller than the shRNA stem used for inducing silencing. Therefore, it is possible that longer siRNA duplex sequences would increase silencing effciency since they would be more similar in size to the endogenous 27-nt small RNAs in Entamoeba; this possibility needs to be demonstrated experimentally.
Implications for gene silencing in Entamoeba
Some implications can be drawn from the current data.
The RNAi machinery is functional in Entamoeba but it appears to be divergent compared with RNAi machinery in model systems. Most components of typical RNAi pathway genes are present in the parasite genome and one AGO gene (EHI_125650) has been proven to be functional in vivo. The missing key component, Dicer, is still elusive at the moment. On the other hand, small RNA cloning efforts have revealed the existence of a complex population of small RNAs in the parasite, and these RNAs are likely to be involved in regulating gene expression. Interestingly, since the 5′-polyP small RNAs are Dicer-independent, it is possible that the parasite could have evolved some unknown mechanism to manipulate gene expression without a Dicer enzyme.
RNAi-based approaches for gene downregulation in Entamoeba are in the early stages of development. Although multiple RNAi-based approaches have been used, success with each technique has been sporadic. To date, no clear molecular mechanism has been demonstrated for gene downregulation in Entamoeba. For the dsRNA-based method, experiments are needed to verify if dsRNA is actually processed into siRNA duplexes. For the soaking method, experiments using radiolabeled duplex siRNA followed by AGO IP would answer if one strand of duplex is loaded into AGO, which AGO it is loaded into, and if polyP modifcation can modify the unwound strand of duplex. For the shRNA-based method, similar questions need to be answered: how was the shRNA processed, where is it processed, which AGO is it loaded into, are any modifcations to the 5′-end of the small RNA?
The molecular mechanism for G3-based silencing is even more elusive. Although it has turned out to be a more reliable tool for gene silencing in Entamoeba, its silencing mechanism has not been defined. Whether the silencing mechanism is transcriptional or post-transcriptional, needs to be re-addressed. Although nuclear run-on experiments have shown G3 silencing of AP-A is at the level of transcription initiation, other more sensitive approaches, such as real-time reverse transcription (RT)-PCR to detect and compare precursor and mature mRNA expression levels will strengthen this observation. However, it is worth noting that both PTGS and TGS could be contributing to silencing in G3 depending on what region of the trigger sequences are used. This would be reminiscent of the observation in plants that PTGS and TGS are mechanistically linked, where transgenes expressing dsRNA can induce PTGS when coding sequences are used but induce TGS when promoter sequences are used [20]. On the other hand, it could be similar to the antisense strand of siRNAs in human cells, which directs histone methylation and transcriptional gene silencing [83].
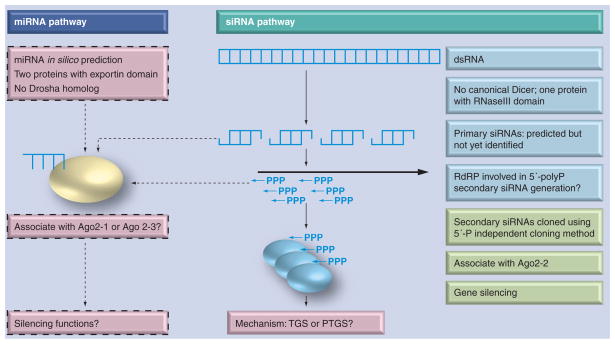
The current understanding of the small RNA pathway in E. histolytica is shown in Figure 2. Some putative miRNAs have been predicted using a bioinformatics approach; validation is needed from both cloning and functional assays. On the other hand, strong data, in particular the finding of an abundant population of 27-nt siRNAs with an unusual 5′-polyP structure, indicate that the siRNA pathway is functional in the parasite. The association of this 27-nt population with one of E. histolytica AGO proteins further validates its functional participation in RISC and in gene silencing. However, many unknowns are still left, including: the identity of the endogenous dsRNA trigger and how the trigger dsRNA is processed into siRNA; how the primary siRNAs interact with the RdRP complex and lead to the generation of secondary siRNA; whether the other two AGOs participate in any pathway; and what mechanisms do 5′-polyP siRNAs use to achieve gene silencing.
Figure 2. Potential small RNA pathways in Entamoeba.
Dotted boxes and arrows indicate the pathway is solely based on computational/speculative prediction. Color annotation: pink: processes are speculative and lack any supporting experimental data; blue: processes lack supporting experimental data but are considered highly likely based on studies in model organisms; green: processes are based on experimental data.
RdRP: RNA-dependent RNA polmerase.
Future perspectives
E. histolytica remains a significant health threat to the people living in developing countries; however, research in understanding this important human disease has been undermined by the lack of genetic tools to study this parasite. Over the recent years, new techniques for gene silencing have been developed, including the G3 silencing approach and various dsRNA and siRNA-based silencing methods, all of which have centered on small RNA molecules. Important directions for future research on the use of small RNA-based gene knockdown as well as the biological functions of small RNAs in Entamoeba include:
Understanding the amebic RNAi machinery of gene silencing
Genome sequencing has demonstrated the presence of the RNAi machinery components in this parasite, and some effort in characterizing E. histolytica AGO and RNaseIII proteins has been reported [84], but until now, it is has not been proven that a canonical Dicer exists in this parasite. Additionally, the RNAi machinery that fuels the 5′-polyP small RNA generation and function is currently unknown. The knowledge obtained in such a study will not only help us better understand how RNAi functions in the parasites, but also help to design more efficient strategies and vectors. In the future this may lead to the development of a genome-wide RNAi screening assay to systematically study the genes involved in parasite pathogenesis.
Understanding the role of small RNAs during parasite stage conversion & stress conditions, including tissue invasion
Small RNAs in developmental regulation have been evident from study of model systems and this will be an important area of future investigation in E. histolytica.
Discovering small RNAs in Entamoeba from in vivo culture
We have documented the small RNA populations in Entamoeba using in vitro cultured trophozoites. These cultures have been maintained in axenic in vitro culture conditions for many years, a condition that is known to cause the parasites to undergo changes, including the loss of the ability to encyst. It is unknown whether certain species of small RNAs have been lost compared with parasites in vivo. Therefore, clinical strains, more recently isolated and grown in xenic cultures may be used to identify small RNAs, which may be linked to host colonization and disease manifestation.
Summary
In summary, Entamoeba histolytica contains the RNAi machinery and unique small RNA populations. RNAi and its related gene-silencing techniques have facilitated parasite research and the discovery of small RNAs in this important parasite has opened a new window in the study of this human pathogen.
Acknowledgments
We thank Gretchen Ehrenkaufer and all members of Singh’s laboratory for critical reading of the manuscript.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
Justine Pompey was supported by an ASM Robert D Watkins fellowship and Hanbang Zhang and Upinder Singh were supported by NIH grants (AI-085178 and AI-53724). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.WHO. Bulletin of the World Health Organization. 1997;75:291. [PMC free article] [PubMed] [Google Scholar]
- 2.Stanley SL., Jr Amoebiasis. Lancet. 2003;361(9362):1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 3.Loftus B, Anderson I, Davies R, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433(7028):865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 4.Lorenzi HA, Puiu D, Miller JR, et al. New assembly, reannotation and analysis of the Entamoeba histolytica genome reveal new genomic features and protein content information. PLoS Negl Trop Dis. 2010;4(6):e716. doi: 10.1371/journal.pntd.0000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5▪.Kaur G, Lohia A. Inhibition of gene expression with double strand RNA interference in Entamoeba histolytica. Biochem Biophys Res Comm. 2004;320(4):1118–1122. doi: 10.1016/j.bbrc.2004.06.064. First report of using long dsRNA approach for gene silencing in Entamoeba. [DOI] [PubMed] [Google Scholar]
- 6▪.Linford AS, Moreno H, Good KR, Zhang H, Singh U, Petri WA., Jr Short hairpin RNA-mediated knockdown of protein expression in Entamoeba histolytica. BMC Microbiol. 2009;9:38. doi: 10.1186/1471-2180-9-38. A shRNA approach was established to silence a number of genes in Entamoeba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7▪.Vayssie L, Vargas M, Weber C, Guillen N. Double-stranded RNA mediates homology-dependent gene silencing of γ-tubulin in the human parasite Entamoeba histolytica. Mol Biochem Parasitol. 2004;138(1):21–28. doi: 10.1016/j.molbiopara.2004.07.005. dsRNA feeding and soaking methods are used for gene down regulation in Entamoeba. [DOI] [PubMed] [Google Scholar]
- 8.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 9.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006;50(2):81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301(5631):336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 12.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Gene Dev. 2010;24(11):1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Qu F, Morris TJ. Suppressors of RNA silencing encoded by plant viruses and their role in viral infections. FEBS Lett. 2005;579(26):5958–5964. doi: 10.1016/j.febslet.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 15▪ ▪.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in. C elegans Science. 2007;315(5809):241–244. doi: 10.1126/science.1132839. First paper on the identifcation of 5′-polyphosphate siRNA in Caenorhabditis elegans. [DOI] [PubMed] [Google Scholar]
- 16.Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 17.Lau NC, Seto AG, Kim J, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 18.Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140(4):504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu W, Shirayama M, Conte D, Jr, et al. Distinct argonaute-mediated 22g-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36(2):231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sijen T, Vijn I, Rebocho A, et al. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol. 2001;11(6):436–440. doi: 10.1016/s0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
- 21.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457(7228):413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci USA. 1998;95(25):14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6(6):509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 24.Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580(22):5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 25.Xue X, Zhang Q, Huang Y, Feng L, Pan W. No miRNA were found in plasmodium and the ones identifed in erythrocytes could not be correlated with infection. Malaria. 2008;7:47. doi: 10.1186/1475-2875-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun L, Cannella D, Ortet P, et al. A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog. 2010;6(5):e1000920. doi: 10.1371/journal.ppat.1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullu E, Lujan HD, Tschudi C. Small sense and antisense RNAs derived from a telomeric retroposon family in Giardia intestinalis. Eukaryot Cell. 2005;4(6):1155–1157. doi: 10.1128/EC.4.6.1155-1157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prucca CG, Slavin I, Quiroga R, et al. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456(7223):750–754. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- 29.Saraiya AA, Wang CC. SnoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4(11):e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen XS, Collins LJ, Biggs PJ, Penny D. High throughput genome-wide survey of small RNAs from the parasitic protists Giardia intestinalis and Trichomonas vaginalis. Genome Biol Evol. 2009;1:165–175. doi: 10.1093/gbe/evp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin WC, Li SC, Shin JW, et al. Identification of microRNA in the protist Trichomonas vaginalis. Genomics. 2009;93(5):487–493. doi: 10.1016/j.ygeno.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Lin WC, Huang KY, Chen SC, et al. Malate dehydrogenase is negatively regulated by miR-1 in Trichomonas vaginalis. Parasitol Res. 2009;105(6):1683–1689. doi: 10.1007/s00436-009-1616-5. [DOI] [PubMed] [Google Scholar]
- 33.Gong AY, Zhou R, Hu G, et al. Cryptosporidium parvum induces b7-h1 expression in cholangiocytes by downregulating microRNA-513. J Infect Dis. 2010;201(1):160–169. doi: 10.1086/648589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeiner GM, Norman KL, Thomson JM, Hammond SM, Boothroyd JC. Toxoplasma gondii infection specifcally increases the levels of key host microRNAs. PLoS ONE. 2010;5(1):e8742. doi: 10.1371/journal.pone.0008742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blaszczyk J, Tropea JE, Bubunenko M, et al. Crystallographic and modeling studies of RNAse III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9(12):1225–1236. doi: 10.1016/s0969-2126(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 36.Shi H, Tschudi C, Ullu E. An unusual dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006;12(12):2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patrick KL, Shi H, Kolev NG, Ersfeld K, Tschudi C, Ullu E. Distinct and overlapping roles for two dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc Natl Acad Sci USA. 2009;106(42):17933–17938. doi: 10.1073/pnas.0907766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drinnenberg IA, Weinberg DE, Xie KT, et al. RNAi in budding yeast. Science. 2009;326(5952):544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of argonaute and its implications for RISC slicer activity. Science. 2004;305(5689):1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 41▪▪.Zhang H, Ehrenkaufer GM, Pompey JM, Hackney JA, Singh U. Small RNAs with 5′-polyphosphate termini associate with a piwi-related protein and regulate gene expression in the single-celled eukaryote Entamoeba histolytica. PLoS Pathog. 2008;4(11):e1000219. doi: 10.1371/journal.ppat.1000219. Small RNA cloning and termini testing revealed that the amebic 27-nt siRNA have 5′-polyphosphate features. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for argonaute proteins in small RNA-directed RNA cleavage pathways. Gene Dev. 2004;18(14):1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yigit E, Batista PJ, Bei Y, et al. Analysis of the C. elegans argonaute family reveals that distinct argonautes act sequentially during RNAi. Cell. 2006;127(4):747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 44.Cifuentes D, Xue H, Taylor DW, et al. A novel miRNA processing pathway independent of dicer requires argonaute2 catalytic activity. Science. 2010;328(5986):1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent mirna biogenesis pathway that requires AGO catalysis. Nature. 2010;465(7298):584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee HC, Li L, Gu W, et al. Diverse pathways generate microRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38(6):803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579(26):5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 48.Nowotny M, Yang W. Structural and functional modules in RNA interference. Curr Opin Struct Biol. 2009;19(3):286–293. doi: 10.1016/j.sbi.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a ranGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee Y, Han J, Yeom KH, Jin H, Kim VN. Drosha in primary microRNA processing. Cold Spring Harb Symp Quant Biol. 2006;71:51–57. doi: 10.1101/sqb.2006.71.041. [DOI] [PubMed] [Google Scholar]
- 51.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60(8):1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang YQ, Chen DL, Tian HF, Zhang BH, Wen JF. Genome-wide computational identifcation of microRNAs and their targets in the deep-branching eukaryote Giardia lamblia. Comput Biol Chem. 2009;33(5):391–396. doi: 10.1016/j.compbiolchem.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 53.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 54.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8(1):9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 55.Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA. 2001;7(11):1522–1530. [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007 doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- 57.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ender C, Krek A, Friedlander MR, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32(4):519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 59.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. MiRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447(7148):1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 60.Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 61.De S, Pal D, Ghosh SK. Entamoeba histolytica: computational identification of putative microRNA candidates. Exp Parasitol. 2006;113(4):239–243. doi: 10.1016/j.exppara.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 62.Guillen N, Boquet P, Sansonetti P. The small GTP-binding protein RACG regulates uroid formation in the protozoan parasite Entamoeba histolytica. J Cell Sci. 1998;111(Pt 12):1729–1739. doi: 10.1242/jcs.111.12.1729. [DOI] [PubMed] [Google Scholar]
- 63.Petri WA, Ramakrishnan G. Applying antisense technology to the study of Entamoeba histolytica pathogenesis. Trends Microbiol. 1999;7(12):471–474. doi: 10.1016/s0966-842x(99)01626-1. [DOI] [PubMed] [Google Scholar]
- 64.Stock RP, Olvera A, Sanchez R, et al. Inhibition of gene expression in Entamoeba histolytica with antisense peptide nucleic acid oligomers. Nat Biotechnol. 2001;19(3):231–234. doi: 10.1038/85671. [DOI] [PubMed] [Google Scholar]
- 65.Bracha R, Nuchamowitz Y, Mirelman D. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot Cell. 2003;2(2):295–305. doi: 10.1128/EC.2.2.295-305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anbar M, Bracha R, Nuchamowitz Y, Li Y, Florentin A, Mirelman D. Involvement of a short interspersed element in epigenetic transcriptional silencing of the amoebapore gene in Entamoeba histolytica. Eukaryot Cell. 2005;4(11):1775–1784. doi: 10.1128/EC.4.11.1775-1784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67▪▪.Bracha R, Nuchamowitz Y, Anbar M, Mirelman D. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2006;2(5):e48. doi: 10.1371/journal.ppat.0020048. The G3 silencing approach was extended to multiple genes and proven to be a reliable gene-silencing approach in Entamoeba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huguenin M, Bracha R, Chookajorn T, Mirelman D. Epigenetic transcriptional gene silencing in Entamoeba histolytica: insight into histone and chromatin modifications. Parasitology. 2010;137(4):619–627. doi: 10.1017/S0031182009991363. [DOI] [PubMed] [Google Scholar]
- 69▪.Irmer H, Hennings I, Bruchhaus I, Tannich E. tRNA gene sequences are required for transcriptional silencing in Entamoeba histolytica. Eukaryot Cell. 2010;9(2):306–314. doi: 10.1128/EC.00248-09. Amoebapore-A (AP-A) gene silencing was linked to tRNA gene-mediated silencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95(7):1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 71.Dastidar PG, Majumder S, Lohia A. Eh klp5 is a divergent member of the kinesin 5 family that regulates genome content and microtubular assembly in Entamoeba histolytica. Cell Microbiol. 2007;9(2):316–328. doi: 10.1111/j.1462-5822.2006.00788.x. [DOI] [PubMed] [Google Scholar]
- 72.Dastidar PG, Lohia A. Bipolar spindle frequency and genome content are inversely regulated by the activity of two n-type kinesins in Entamoeba histolytica. Cell Microbiol. 2008;10(7):1559–1571. doi: 10.1111/j.1462-5822.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- 73.Macfarlane RC, Singh U. Identification of an Entamoeba histolytica serine-, threonine-, and isoleucine-rich protein with roles in adhesion and cytotoxicity. Eukaryot Cell. 2007;6(11):2139–2146. doi: 10.1128/EC.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Macfarlane RC, Singh U. Loss of dsRNA-based gene silencing in Entamoeba histolytica: implications for approaches to genetic analysis. Exp Parasitol. 2008;119(2):296–300. doi: 10.1016/j.exppara.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shi H, Chamond N, Tschudi C, Ullu E. Selection and characterization of RNA interference-defcient trypanosomes impaired in target mRNA degradation. Eukaryot Cell. 2004;3(6):1445–1453. doi: 10.1128/EC.3.6.1445-1453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specifc silencing in mammalian cells. Gene Dev. 2002;16(8):948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Siolas D, Lerner C, Burchard J, et al. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23(2):227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 78.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296(5567):550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 79.Miyagishi M, Taira K. U6 promoter-driven sirnas with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20(5):497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 80.Boettner DR, Huston CD, Linford AS, et al. Entamoeba histolytica phagocytosis of human erythrocytes involves patmk, a member of the transmembrane kinase family. PLoS Pathog. 2008;4(1):e8. doi: 10.1371/journal.ppat.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Solis CF, Santi-Rocca J, Perdomo D, Weber C, Guillen N. Use of bacterially expressed dsRNA to downregulate Entamoeba histolytica gene expression. PLoS ONE. 2009;4(12):e8424. doi: 10.1371/journal.pone.0008424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mcgugan GC, Jr, Joshi MB, Dwyer DM. Identification and biochemical characterization of unique secretory nucleases of the human enteric pathogen, Entamoeba histolytica. J Biol Chem. 2007;282(43):31789–31802. doi: 10.1074/jbc.M705975200. [DOI] [PubMed] [Google Scholar]
- 83.Weinberg MS, Villeneuve LM, Ehsani A, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12(2):256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abed M, Ankri S. Molecular characterization of Entamoeba histolytica RNAse III and ago2, two RNA interference hallmark proteins. Exp Parasitol. 2005;110(3):265–269. doi: 10.1016/j.exppara.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 85.Sanchez R, Saralegui A, Olivos-Garcia A, et al. Entamoeba histolytica: intracellular distribution of the sec61α subunit of the secretory pathway and downregulation by antisense peptide nucleic acids. Exp Parasitol. 2005;109(4):241–251. doi: 10.1016/j.exppara.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 86.Ankri S, Stolarsky T, Mirelman D. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol Microbiol. 1998;28(4):777–785. doi: 10.1046/j.1365-2958.1998.00837.x. [DOI] [PubMed] [Google Scholar]
- 87.Ankri S, Padilla-Vaca F, Stolarsky T, Koole L, Katz U, Mirelman D. Antisense inhibition of expression of the light subunit (35 kDa) of the gal/galnac lectin complex inhibits Entamoeba histolytica virulence. Mol Microbiol. 1999;33(2):327–337. doi: 10.1046/j.1365-2958.1999.01476.x. [DOI] [PubMed] [Google Scholar]
- 88.Bracha R, Nuchamowitz Y, Leippe M, Mirelman D. Antisense inhibition of amoebapore expression in Entamoeba histolytica causes a decrease in amoebic virulence. Mol Microbiol. 1999;34(3):463–472. doi: 10.1046/j.1365-2958.1999.01607.x. [DOI] [PubMed] [Google Scholar]
- 89.Lopez-Camarillo C, De La Luz Garcia-Hernandez M, Marchat LA, et al. Entamoeba histolytica ehdead1 is a conserved dead-box RNA helicase with atpase and atp-dependent RNA unwinding activities. Gene. 2008;414(1–2):19–31. doi: 10.1016/j.gene.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 90.Sahoo N, Bhattacharya S, Bhattacharya A. Blocking the expression of a calcium binding protein of the protozoan parasite Entamoeba histolytica by tetracycline regulatable antisense-RNA. Mol Biochem Parasitol. 2003;126(2):281–284. doi: 10.1016/s0166-6851(02)00284-0. [DOI] [PubMed] [Google Scholar]
- 91.Sahoo N, Labruyere E, Bhattacharya S, Sen P, Guillen N, Bhattacharya A. Calcium binding protein 1 of the protozoan parasite Entamoeba histolytica interacts with actin and is involved in cytoskeleton dynamics. J Cell Sci. 2004;117(Pt 16):3625–3634. doi: 10.1242/jcs.01198. [DOI] [PubMed] [Google Scholar]
- 92.Sen A, Chatterjee NS, Akbar MA, Nandi N, Das P. The 29-kilodalton thiol-dependent peroxidase of Entamoeba histolytica is a factor involved in pathogenesis and survival of the parasite during oxidative stress. Eukaryot Cell. 2007;6(4):664–673. doi: 10.1128/EC.00308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shrimal S, Bhattacharya S, Bhattacharya A. Serum-dependent selective expression of ehtmkb1–9, a member of Entamoeba histolytica b1 family of transmembrane kinases. PLoS Pathog. 2010;6(6):e1000929. doi: 10.1371/journal.ppat.1000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vats D, Vishwakarma RA, Bhattacharya S, Bhattacharya A. Reduction of cell surface glycosylphosphatidylinositol conjugates in Entamoeba histolytica by antisense blocking of E. histolytica glcnac-phosphatidylinositol deacetylase expression: effect on cell proliferation, endocytosis, and adhesion to target cells. Infect Immun. 2005;73(12):8381–8392. doi: 10.1128/IAI.73.12.8381-8392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bracha R, Nuchamowitz Y, Wender N, Mirelman D. Transcriptional gene silencing reveals two distinct groups of Entamoeba histolytica gal/galnac-lectin light subunits. Eukaryot Cell. 2007;6(10):1758–1765. doi: 10.1128/EC.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Solis CF, Guillen N. Silencing genes by RNA interference in the protozoan parasite Entamoeba histolytica. Methods Mol Biol. 2008;442:113–128. doi: 10.1007/978-1-59745-191-8_9. [DOI] [PubMed] [Google Scholar]
Website
- 101.RNA dependent RNA polymerase. http://pfam.sanger.ac.uk/family?acc=PF05183.