SUMMARY
While the small GTPase Rac1 and its effectors are well-established mediators of mitogenic and motile signaling by tyrosine-kinase receptors and have been implicated in breast tumorigenesis, little is known regarding the exchange factors (Rac-GEFs) that mediate ErbB receptor responses. Here we identify the PIP3-Gβγ-dependent Rac-GEF P-Rex1 as an essential mediator of Rac1 activation, motility, cell growth, and tumorigenesis driven by ErbB receptors in breast cancer cells. Notably, activation of P-Rex1 in breast cancer cells requires the convergence of inputs from ErbB receptors and a Gβγ- and PI3Kγ-dependent pathway. Moreover, we identified the GPCR CXCR4 as a crucial mediator of P-Rex1/Rac1 activation in response to ErbB ligands. P-Rex1 is highly overexpressed in human breast cancers and their derived cell lines, particularly those with high ErbB2 and ER expression. In addition to the prognostic and therapeutic implications, our findings reveal an ErbB effector pathway that is crucial for breast cancer progression.
Keywords: P-Rex1, Rac-GEF, Rac1, ErbB receptors, PI3Kγ, Gβγ subunits, breast cancer
INTRODUCTION
One of the hallmarks of breast cancer is the hyperactivation of ErbB receptor signaling. The human ErbB family of tyrosine-kinase (TK) receptors consists of 4 members: ErbB1 (EGFR/HER1), the orphan receptor ErbB2 (HER2), ErbB3 (HER3), and ErbB4 (HER4). The combinatorial dimerization of ErbB receptors and their distinct coupling to signaling adaptors and effectors create a complex network of signaling events that, when dysregulated, leads to uncontrolled growth and transformation [1–3]. Aberrant expression of both ErbB receptors and their growth factor activators is a common feature in the progression of many cancers. In breast cancer, overexpression of ErbB2 and ligands such as TGFα (ErbB1 ligand) or heregulins/neuregulins (ErbB3/ErbB4 ligands) occurs with high frequency [3]. Genetic abnormalities associated with breast cancer also include gain-of-function mutations of ErbB effectors, such as PI3KCA gene mutations or PTEN deletions [4, 5].
It is well established that members of the Rho family of small GTP-binding proteins mediate ErbB responses. Rac GTPases have been widely implicated in actin cytoskeleton reorganization, migration, mitogenesis, transformation, and metastasis [6]. Rac inhibition impairs breast cancer cell motility and proliferation in response to EGFR and ErbB3 ligands [7–9]. The activity of Rac is mainly regulated by Guanine nucleotide Exchange Factors (Rac-GEFs), which activate Rac by promoting the exchange of GDP by GTP, Guanine nucleotide Dissociation Inhibitors (GDIs), which limit the access of Rac to GEFs, and GTPase Activating Proteins (GAPs), which lead to Rac inactivation by accelerating its intrinsic GTPase activity [6]. TK receptors can signal through multiple mechanisms to Rac-GEFs. Most notably, many Rac-GEFs depend on the PI3K product PIP3 for their redistribution to membranes and activation [10]. Unlike Ras proteins, gain-of-function mutations in Rho GTPases are uncommon in cancer; however, there is ample evidence for hyperactivation of the Rac pathway in human cancer. For example, Rac-GAPs are down-regulated in human breast tumors [11], and aberrant overexpression of Rac-GEFs contributes to cancer progression and metastasis in various cancer types, including breast cancer [12, 13]. The Rac effector Pak1 is also hyperactive in human breast tumors and promotes anti-estrogen resistance [14–16]. Dissecting the cellular mechanisms leading to dysregulation of the Rac pathway in breast cancer is therefore highly relevant. Nonetheless, the relevance of Rac-GEFs in breast cancer progression remains elusive.
Here we report the identification of phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor-1 (P-Rex1) as an essential mediator of ErbB receptor-driven Rac responses in breast cancer models. Signals from ErbB receptors and GPCRs converge on P-Rex1 to mediate Rac1 activation. Notably, there is a remarkable up-regulation of P-Rex1 in human breast tumors, thus underscoring the potential prognostic and therapeutic implications of these findings.
RESULTS
P-Rex1 is up-regulated in breast cancer cell lines and mediates Rac activation by heregulin
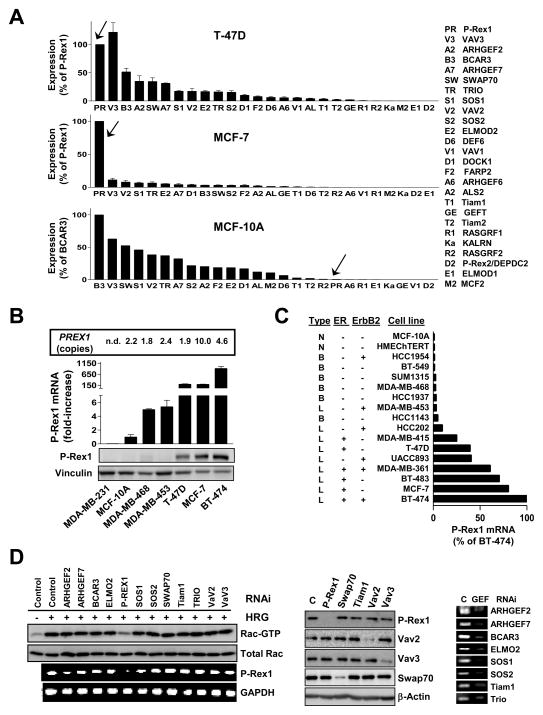
Rac1 plays essential roles in breast cancer cell motility, proliferation and tumorigenesis [6–8]. We reported that EGF and the ErbB3 ligand heregulin β1 (HRG) strongly activate Rac1 in MCF-7 and T-47D breast cancer cells [7]. EGF and HRG also activate Rac1 in other breast cancer cell lines as well as in immortalized MCF-10A mammary cells (Fig. S1A). Rac1 activation by HRG is inhibited by ectopic expression of the Rac-GAP β2-chimaerin (Fig. S1B). Activation of Rac1 by HRG in MCF-7 and T-47D cells is sustained and sensitive to the PI3K inhibitor wortmannin [7]. We aimed to identify the Rac-GEF(s) implicated in this response. To this end, we designed an array to determine the relative expression of 26 Rac-GEFs and known GEF accessory proteins in breast cancer models (“Rac-GEF array”). Surprisingly, Q-PCR analysis in MCF-7 and T-47D cells using this array revealed very high levels of P-Rex1, a PI3K- and Gβγ-regulated Rac-specific exchange factor. In striking contrast, non-transformed MCF-10A cells have negligible P-Rex1 expression (Fig. 1A). A distinct pattern of expression for Rac-GEFs was observed in MDA-MB-453, MDA-MB-468, and MDA-MB-231 breast cancer cells (Fig. S1C). P-Rex1 was originally characterized in neutrophils as a mediator of chemoattractant-induced responses via Rac2, including motility and ROS production [17–19]. To our knowledge, information on this GEF in cancer models is limited, including in breast cancer.
Figure 1. P-Rex1 is up-regulated in breast cancer cell lines and mediates the Rac1 activation by HRG.
(A) Array profiling of Rac-GEFs and accessory Rac activators in breast cell lines. Expression was determined by Q-PCR in 96-well plate arrays and normalized to GAPDH. Data were expressed as mean ± S.D. (n=3).
(B) Q-PCR analysis of P-Rex1 expression, normalized to 18S, is shown. Data, expressed as fold-change relative to MCF-10A cells, are presented as mean ± S.D. (n=3). Expression of P-Rex1 by Western blot and PREX1 gene copy number in each cell line are shown. n.d., not determined.
(C) P-Rex1 levels were obtained from Affimetrix microarray data of 17 cell lines. The tumor type from which each cell line was originated as well as the ErbB2 and ER status are listed. N, normal; B, basal type; L, luminal type.
(D) Rac-GEFs and other Rac modulators were depleted from T-47D cells using validated siRNA. After 16 h cells were serum-starved for 48 h and stimulated with HRG (10 ng/ml, 5 min). Left panel, Rac-GTP levels were determined with a PBD pull-down assay. P-Rex1 mRNA levels were determined by RT-PCR. Middle panel, depletion of Rac-GEFs as determined by Western blot. Right panel, depletion of Rac-GEFs as determined by RT-PCR. Two additional experiments gave nearly identical results. C, control RNAi.
A comparative analysis of P-Rex1 mRNA levels by Q-PCR showed that BT-474, MCF-7, and T-47D cells, which derived from luminal breast cancers, have 100–1000-fold higher P-Rex1 mRNA levels than MCF-10A cells. MDA-MB-231, a basal breast cancer derived cell line, showed essentially no P-Rex1 expression. P-Rex1 was slightly elevated in MDA-MB-453 and MDA-MB-468 cells. P-Rex1 can be readily detected in MCF-7, BT-474, and T-47D cells by Western blot (Fig. 1B). An independent microarray gene profiling analysis of 17 breast cell lines validated these findings and identified additional breast cancer cell lines with high P-Rex1 expression. Interestingly, P-Rex1 is preferentially expressed at high levels in cell lines of luminal origin, whereas none of the cell lines derived from basal breast cancers have significant P-Rex1 expression (Fig. 1C). The PREX1 locus is located in chromosome 20q13.13, a region commonly amplified in human breast tumors and cell lines [20–23]. Analysis of PREX1 copy number in genomic DNA from breast cancer cell lines revealed amplification particularly in MCF-7 cells and also in BT-474 cells, but not in T-47D cells or in other breast cancer cell lines (Fig. 1B).
To determine if P-Rex1 is implicated in Rac1 activation by HRG, Rac-GEFs with expression >10% relative to P-Rex1 were depleted from T-47D cells using validated RNAi (SMART pool ON-TARGET plus®, Dharmacon). Each RNAi duplex depleted the corresponding target by > 80% as revealed by Western blot (for those proteins that can be readily detected in T-47D cells) or RT-PCR. Remarkably, P-Rex1 RNAi depletion essentially abolished HRG-induced activation of Rac1. For all other Rac-GEFs, inhibition was <20% (Fig. 1D). These results were validated using 4 different P-Rex1 RNAi duplex sequences in T-47D cells (Fig. S1D) and MCF-7 cells (data not shown), thereby minimizing the chance of “off target” effects. The high P-Rex1 expression levels in breast cancer cell lines and the lack of redundancy with other Rac-GEFs in the HRG response were unanticipated.
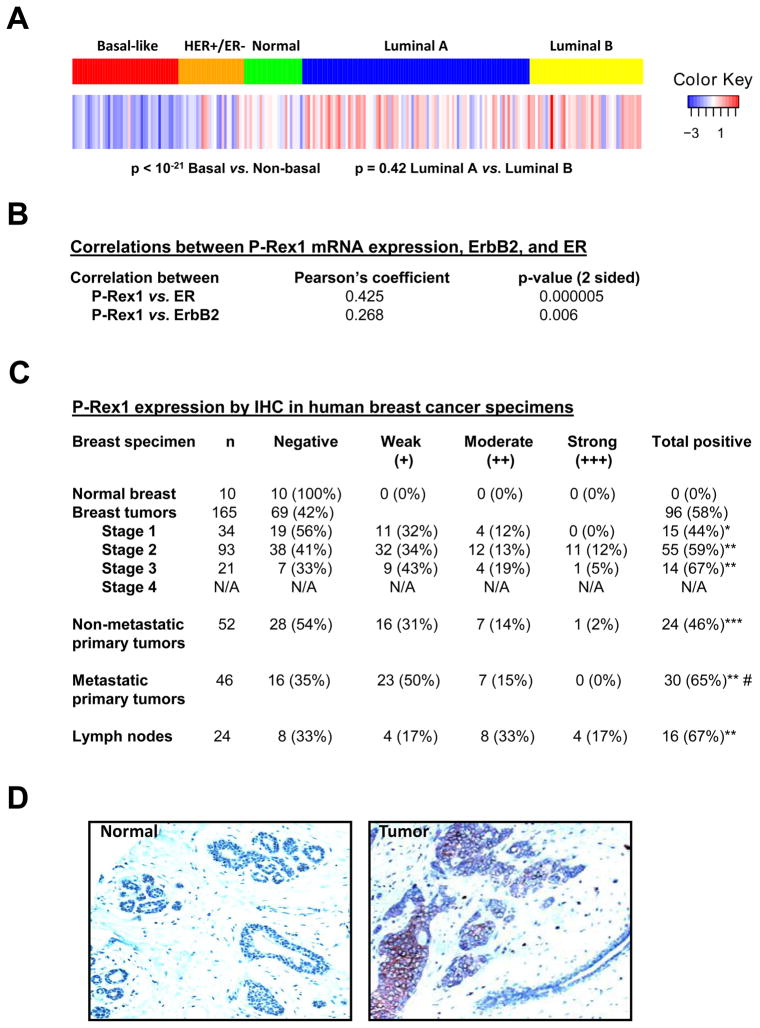
P-Rex1 is overexpressed in human breast tumors
Next, we decided to examine the expression of P-Rex1 in human breast tumors. PCR analysis of a breast cancer cDNA panel (OriGene) showed P-Rex1 up-regulation in a considerable number of tumors (Fig. S2A). As P-Rex1 is highly expressed in neutrophils [18], we needed to rule out that the P-Rex1 signal originated from neutrophils potentially present in the tumors. However, there was no correlation between the expression of P-Rex1 and the neutrophil marker myeloperoxidase. Indeed, in most cases samples with high P-Rex1 expression had very low or undetectable myeloperoxidase levels.
To further establish the clinical significance of these preliminary findings we carried out a separate analysis of P-Rex1 expression using the NKI microarray data set that established the intrinsic gene signature of 295 samples from patients [24]. Interestingly, P-Rex1 mRNA levels were particularly elevated in luminal breast cancer specimens, while the basal breast cancer group showed very low P-Rex1 levels. There were no statistical differences between luminal A and luminal B subtypes (Fig. 2A). A positive correlation between estrogen receptor (ER) and P-Rex1 expression was found (Fig. 2B), in agreement with gene expression analysis from Oncomine (Fig. S2B). P-Rex1 expression was higher in ErbB2 positive tumors (Fig. 2B). The P-Rex1-related isozyme P-Rex2a was recently reported to cooperate with PI3K signaling, and its expression levels correlate with activating mutations in the PI3KCA gene [25]. However, using the same cohort we could not find any significant association between increased P-Rex1 expression and PI3KCA mutations (data not shown).
Figure 2. P-Rex1 is overexpressed in human breast cancer.
(A) mRNA expression level of P-Rex1 in the 5 different tumor types. P-Rex1 average expression is the lowest in basal-like tumors (left) and the highest in Luminal B subtype (right). p < 10−21 basal vs. non basal.
(B) Analysis from 108 tumor samples is shown, using a Pearson correlation test.
(C) The level of P-Rex1 expression is denoted by (+).“+”, weak staining; ++, medium staining; +++, strong staining. The information on metastasis was available from 98 patients. *, p = 0.008 vs. normal; **, p < 0.005 vs. normal; ***, p = 0.004 vs. normal; #, p = 0.045 vs. non-metastatic primary tumors. Differences in the IHC staining of human breast cancer specimens were analyzed with the Fisher’s Exact (one sided) tests.
(D) Representative IHC stainings from normal mammary tissue and a P-Rex1 positive breast tumor.
To determine if P-Rex1 up-regulation also occurs in other cancer types, we used a commercial multicancer array (OriGene). Notably, P-Rex1 up-regulation could be observed in other tumor types, particularly thyroid, kidney, and prostate cancer. Although at lower frequency, some cases of high P-Rex1 levels were observed in other tumors such as esophageal, bladder, colon, endometrial, and pancreatic cancer (Fig. S2C).
Next, we screened paraffin-embedded tissue sections from 10 normal and 165 breast cancer patients by immunohistochemistry (IHC). In agreement with the cDNA array, P-Rex1 was essentially undetectable in normal mammary samples. On the other hand, P-Rex1 was detected in 58% of the tumor specimens analyzed (Fig. 2C). Importantly, P-Rex1 staining was found specifically in the tumor cells, and no appreciable P-Rex1 staining could be observed in the stroma or in normal ducts (Fig. 2D). While we observed a systematic increase in P-Rex1 levels as a function of stage (Fig. 2C), it is not statistically significant as determined by a sum of ranks analysis (p = 0.103). Nonetheless, P-Rex1 expression was statistically higher in primary tumors from patients that underwent metastasis relative to those that did not (65% vs. 46%, p = 0.045). Analysis in lymph nodes from breast cancer patients showed that 67% were P-Rex1 positive. Therefore, P-Rex1 up-regulation occurs in primary breast tumors and their metastases.
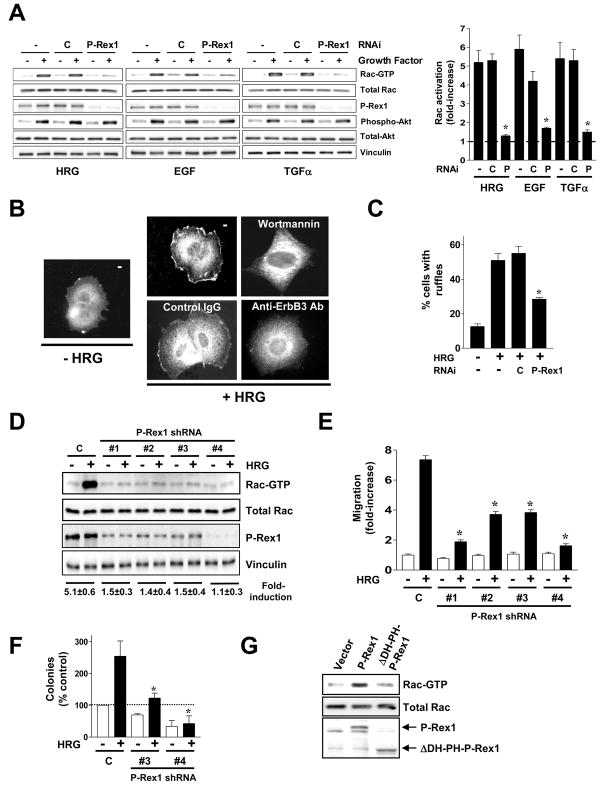
P-Rex1 mediates ErbB ligand-driven growth and migration
Then, we examined whether P-Rex1 could mediate Rac1 activation downstream of EGFR. Like HRG, the EGFR ligands EGF and TGFα caused a marked elevation in Rac-GTP levels in T-47D cells, which was reduced by silencing P-Rex1 expression using RNAi (92%, 84%, and 88% inhibition for HRG, EGF, and TGFα, respectively) (Fig. 3A). Similar inhibition (90%) was observed in MCF-7 cells (data not shown). Whereas wortmannin impairs Rac1 activation by ErbB ligands [7], activation of the PI3K effector Akt1 remained unchanged both in P-Rex1-depleted T-47D cells (Fig. 3A) and MCF-7 cells (data not shown), arguing for a PI3K-dependent but Akt-independent mechanism for P-Rex1/Rac1 activation.
Figure 3. P-Rex1 is an essential mediator of Rac responses by ErbB ligands in breast cancer cells.
(A) T-47D cells were transfected with validated P-Rex1 siRNA (P) or control duplexes (C). After 16 h cells were serum-starved for 48 h and then stimulated with HRG (10 ng/ml, 5 min), EGF (100 ng/ml, 1 min) or TGFα (10 ng/ml, 2.5 min). Densitometric values of Rac-GTP levels (normalized to total Rac) are presented as mean ± S.D. (n=5). *, p < 0.001 vs. control RNAi.
(B) Translocation of endogenous P-Rex1 by HRG in MCF-7 cells. Wortmannin (1 μM), a blocking anti-ErbB3 antibody or control IgG (10 μg/ml) were added 1 h before HRG stimulation. Similar results were observed in > 10 individual cells in at least 3 different experiments.
(C) P-Rex1 RNAi inhibits ruffle formation in T-47D cells stimulated with HRG. The percentage of cells with ruffles was determined in at least 200 cells. Results were expressed as mean ± S.E.M. of 3 independent experiments. *, p < 0.01 vs. control RNAi (C).
(D) Stable depletion of P-Rex1 in T-47D cells impairs Rac activation. P-Rex1 stably depleted T-47D cells were generated using 4 different shRNA lentiviruses (#1–4). A representative Rac-GTP pull-down assay in response to HRG is shown. Fold-induction in Rac-GTP levels normalized to total Rac, as determined by densitometry, is expressed as mean ± S.D. (n=3).
(E) Impaired motility in P-Rex1-deficient T-47D cells, as determined with a Boyden chamber. Results are presented as fold-increase relative to control cells in the absence of HRG, and expressed as mean ± S.D. of triplicate measurements. Two additional experiments gave similar results. *, p < 0.05 vs. control shRNA lentivirus (C) with HRG.
(F) Colony formation assays in P-Rex1-depleted T-47D cells. Experiments were performed in quadruplicate. Results are presented as percentage relative to control cells growing in the absence of HRG. Data are expressed as mean ± S.E.M. (n=3). *, p < 0.05 vs. control RNAi (C) with HRG.
(G) MCF-10A cells were transfected with mammalian vectors encoding either P-Rex1 (wt), ΔDH-PH-P-Rex1, or empty vector. After 16 h cells were serum starved for 48 h and Rac-GTP levels determined.
As membrane association is a pre-requisite for the activation of various Rac-GEFs, including P-Rex1 [10, 26], we asked whether HRG could relocalize P-Rex1 in breast cancer cells. Endogenous P-Rex1 distributed diffusely throughout the cytoplasm in serum-starved MCF-7 cells (Fig. 3B). HRG caused a pronounced peripheral translocation of P-Rex1, particularly to membrane ruffles. Consistent with our previous data that ErbB3, but not ErbB4, mediates Rac1 activation by HRG [7], a blocking anti-ErbB3 antibody prevented P-Rex1 translocation by HRG, whereas control IgG (Fig. 3B) or an anti-ErbB4 antibody (data not shown) did not. P-Rex1 translocation by HRG was inhibited by wortmannin, as expected from the PIP3 dependency for P-Rex1 activation [26, 27]. Using GFP-fused P-Rex1 mutants we found that deletion of the DH-PH domain tandem abolished translocation by HRG, which is consistent with the requirement of these domains for P-Rex1 activation [27, 28]. The DH-PH domain was sufficient to translocate to the cell periphery in response to HRG (Fig. S3A).
HRG causes a marked reorganization of the cytoskeleton and induces a motile response in breast cancer cells in a PI3K-Rac1 dependent manner [7, 29]. Interestingly, ruffle formation in response to HRG was markedly reduced in P-Rex1-depleted cells (Fig. 3C). To determine if P-Rex1 mediates motile responses, we generated P-Rex1 stably depleted polyclonal T-47D and MCF-7 cell lines (Fig. 3D, and Fig. S3B) using 4 different P-Rex1 shRNA lentiviruses (puromycin selected). Consistent with our transient knock-down studies, stably P-Rex1-depleted cell lines were defective in Rac1 activation (Fig. 3D, and Fig. S3B). Rac activation by HRG can be rescued by a RNAi-insensitive P-Rex1 mutant (Fig. S3C). Although no changes in the adhesive properties of the cells were detected (data not shown), we observed a significant impairment of migration in response to HRG, as determined with a Boyden chamber (Fig. 3E and Fig. S3D).
HRG has been implicated in breast cancer cell transformation [30]. T-47D cells form colonies in soft agar, and colony formation was significantly enhanced by HRG. In sharp contrast, the ability of P-Rex1-depleted cells to grow in soft agar in response to HRG was severely affected (Fig. 3F), thus arguing for a role of P-Rex1 in ErbB receptor-mediated anchorage-independent growth. Importantly, P-Rex1 shRNA depletion did not significantly affect the expression of other Rac-GEFs (Fig. S3E), therefore the effects could be attributed to specific P-Rex1 depletion.
Next, we examined the effect of overexpressing P-Rex1 in MCF-10A cells, which express very low P-Rex1 levels. MCF-10A cells were transfected with mammalian expression vectors encoding P-Rex1, a DH-PH-deleted P-Rex1 mutant, or vector alone. Overexpression of P-Rex1 elevated basal Rac-GTP levels in MCF-10A cells (2.2-fold). On the other hand, expression of ΔDH-PH-P-Rex1 did not change basal Rac-GTP levels (Fig. 3G).
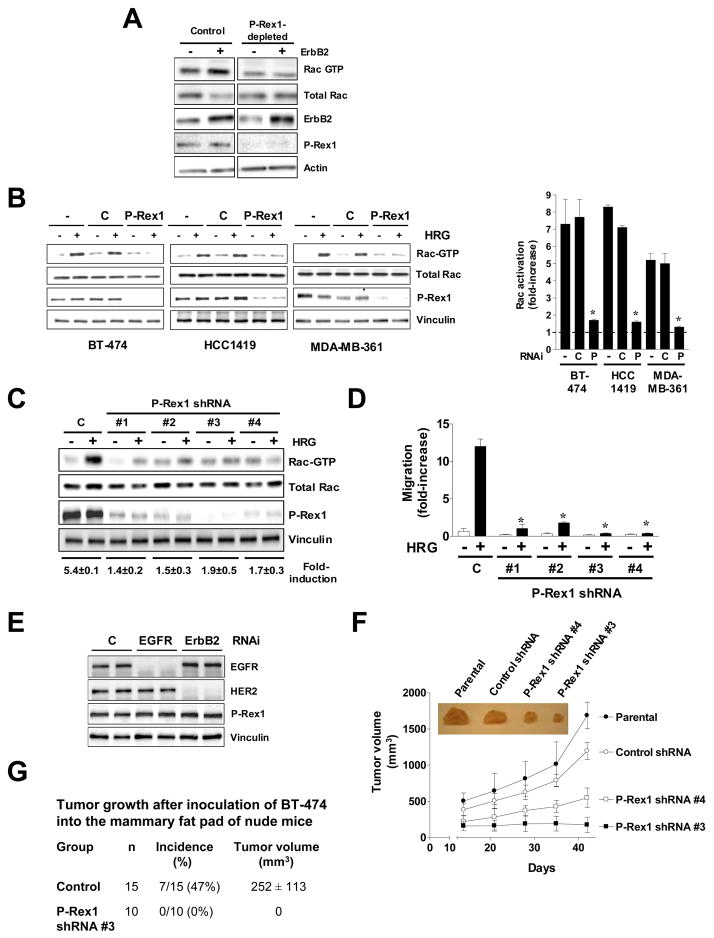
P-Rex1 is required for ErbB2-mediated migration and tumorigenesis
ErbB2 overexpression is one of the most common genetic alterations in breast cancer [1], and Rac1 is implicated in ErbB2-mediated mitogenesis and motility [7, 11, 31, 32]. As P-Rex1 is prominently overexpressed in ErbB2 positive tumors and responses by HRG are mediated by ErbB3/ErbB2 dimers [2], we speculated that P-Rex1 could be implicated in ErbB2-driven activation of Rac1. As a first approach to address this issue we overexpressed ErbB2 both in control and P-Rex1-depleted T-47D cells. As expected, ErbB2 overexpression led to elevated Rac-GTP levels in control T-47D cells. However, this effect was not observed in P-Rex1-deficient cells despite the similar levels of ErbB2 overexpression in both cell lines (Fig. 4A). Similar results were found in P-Rex-1-depleted MCF-7/ErbB2 cells (data not shown). Next, we took advantage of BT-474 cells, an ErbB2 overexpressing cell line that expresses very high P-Rex1 levels (see Fig. 1B and 1C). Transient RNAi depletion of P-Rex1 in these cells reduced Rac1 activation by HRG by 88% (Fig. 4B). Similar results were observed in two other ErbB2-positive cell lines that express very high P-Rex1 levels (HCC1419, and MDA-MB-361 cells, 90% and 93% inhibition, respectively). We then generated BT-474 cell lines in which P-Rex1 was stably silenced using shRNA lentiviruses. In agreement with the transient depletion experiments, these lines also had a defective Rac1 activation in response to HRG (80–90% inhibition) (Fig. 4C). Moreover, migration induced by HRG was impaired in P-Rex1-depleted BT-474 cells (Fig. 4D) or in P-Rex1-depleted MCF-7/ErbB2 cells (Fig. S4). Neither overexpressing ErbB2 in T-47D cells (Fig. 4A) nor silencing ErbB2 or EGFR from BT-474 cells (Fig. 4E) altered P-Rex1 expression levels, suggesting that these receptors do not modulate P-Rex1 expression.
Figure 4. P-Rex1 is required for ErbB2-mediated migration and tumorigenesis.
(A) T-47D cells (P-Rex1-depleted #3 and control from Fig. 3E) were transfected with pcDNA3-ErbB2 or empty vector. After 24 h cells were serum-starved for 48 h, and Rac-GTP levels determined.
(B) BT-474, HCC1419, or MDA-MB-361 cells were transfected with validated P-Rex1 siRNA (P) or control duplexes (C). After 16 h cells were serum-starved for 48 h and then stimulated with HRG (10 ng/ml, 5 min). Densitometric values of Rac-GTP levels (normalized to total Rac) are presented as mean ± S.D. (n=3). *, p < 0.001 vs. control RNAi.
(C) Stable depletion of P-Rex1 in BT-474 cells impairs Rac activation. P-Rex1 stably depleted BT-474 cells were generated using 4 different shRNA lentiviruses (#1–4). A representative Rac-GTP pull-down assay in response to HRG is shown. Fold-induction in Rac-GTP levels normalized to total Rac, as determined by densitometry, is expressed as mean ± S.D. (n=3).
(D) Impaired motility in P-Rex1-deficient BT-474 cells, as determined with a Boyden chamber. Results are presented as fold-increase relative to control cells in the absence of HRG, and expressed as mean ± S.D. of triplicate measurements. Two additional experiments gave similar results. *, p < 0.001 vs. control shRNA lentivirus (C) with HRG.
(E) Effect of EGFR or ErbB2 depletion on P-Rex1 levels, determined 72 h after transfection of RNAi duplexes. Duplicate samples are shown.
(F) Reduced tumorigenic potential of P-Rex1-depleted BT-474 cells in nude mice (10 mice/group). Results are expressed as mean ± S.D. *, p < 0.001 vs. control shRNA. Inset, representative tumors.
(G) Tumor growth in nude mice was monitored for 70 days after injection of control (control shRNA lentivirus) or P-Rex1-deficient BT-474 cells (stable cell line #3) into the mammary fat pad. The tumor volume shown is that at the end of the experiment. Results were expressed as mean ± S.D.
As Rac1 is implicated in ErbB2 signaling [7, 11, 31], we next examined the relevance of P-Rex1 overexpression in tumorigenesis. To this end, we assessed the effect of P-Rex1 depletion on the growth of BT-474 xenografts in nude mice. Herceptin blocks the ability of BT-474 cells to form tumors in nude mice [33], arguing that the tumorigenic capacity of these cells is dependent on ErbB2 signals. Athymic nude mice were injected s.c. with BT-474 parental cells, control lentivirus-infected BT-474 cells, or two P-Rex1-depleted BT-474 clones. While parental and control BT-474 cells readily formed tumors in nude mice, the tumorigenic ability of P-Rex1-depleted BT-474 cells was markedly impaired (Fig. 4F). We also compared the ability of control BT-474 cells and clone #3 to promote tumor formation in an orthotopic model. As shown in Fig. 4G, 47% of nude mice developed mammary tumors upon inoculation in the mammary fat pad of BT-474 cells transduced with control shRNA lentivirus, whereas none of the mice developed tumors when injected with P-Rex1-depleted BT-474 cells. Taken together, our results suggest that P-Rex1 mediates ErbB2-dependent Rac responses both in cultured breast cancer cells as well as in vivo.
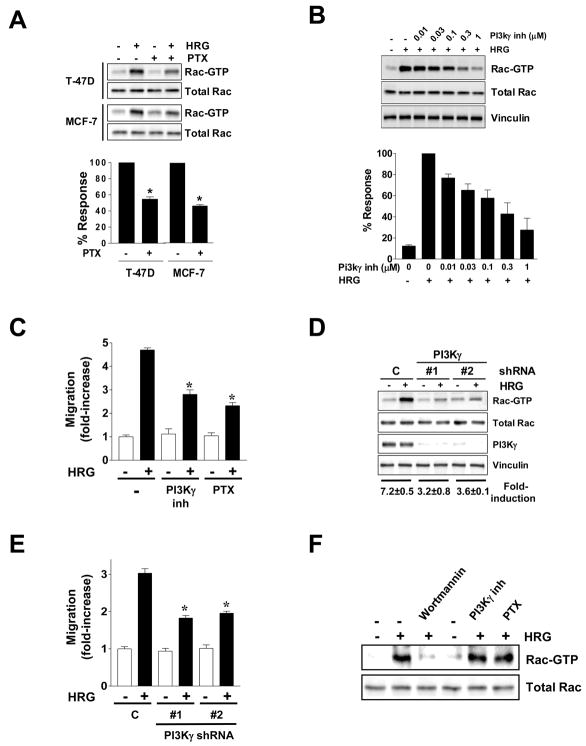
Gβγ subunits and PI3Kγ mediate the activation of the P-Rex1/Rac1 pathway by ErbB ligands
A unique feature of P-Rex1 is that it is dually regulated activated by the PI3K product PIP3 and Gβγ subunits released upon activation of Gi-coupled receptors. Gβγ alone causes partial activation of P-Rex1, and an additional PIP3 input is required for membrane targeting and full activation of this Rac-GEF [17, 27, 34]. While ErbB and other TK receptors can relay signals via transactivation of Gi-coupled receptors [35–38], a potential implication of GPCRs in the activation of P-Rex1/Rac1 signaling by ErbB receptors has not been established. First, we used pertussis toxin (PTX) to inhibit Gβγ release from heterotrimeric Gi proteins. Remarkably, Rac activation by HRG in PTX-treated T-47D and MCF-7 cells was significantly reduced (Fig. 5A). PTX also inhibited the activation of Rac1 by EGF (data not shown). Furthermore, PTX treatment reduced MCF-7 cell migration in response to HRG (see Fig. 5C). These results implicate Gβγ subunits in the activation of Rac1 by ErbB receptors in breast cancer cells expressing high P-Rex1 levels and argue for a potential transactivation via Gi-coupled receptors for signaling to P-Rex1/Rac1.
Figure 5. Involvement of Gβγ subunits and PI3Kγ in Rac activation by HRG.
(A) Serum-starved T-47D and MCF-7 cells were treated with PTX (100 ng/ml, 24 h) and then stimulated with HRG (10 ng/ml, 5 min). Top panel, representative experiments. Bottom panel, densitometric analysis. Data are expressed as % of the HRG response in the absence of PTX and presented as mean ± S.D. (n=3). *, p < 0.05 vs. control.
(B) A PI3Kγ inhibitor reduces Rac1 activation by HRG in MCF-7 cells. Densitometric analysis of the data (Rac-GTP normalized to total Rac1) is presented relative to the effect in the absence of PI3Kγ inhibitor.
(C) Inhibition of HRG-induced MCF-7 cell motility by the PI3Kγ inhibitor (1 μM) or PTX. Data from triplicate samples are presented as mean ± S.D. * p < 0.05 vs. HRG (control). Results are presented as fold-increase relative to control cells in the absence of stimuli, and expressed as mean ± S.D. of triplicate measurements. Two additional experiments gave similar results. *, p < 0.05 vs. control (C) with HRG.
(D) Stable depletion of PI3Kγ from MCF-7 cells impairs Rac1 activation and motility. MCF-7 cells were transfected with 2 different plasmids encoding PI3Kγ shRNA (#1 and #2) or a shRNA plasmid control (C) and selected with puromycin. Rac-GTP levels in response to HRG (10 ng/ml, 5 min) are shown.
(E) Impaired cell motility in PI3Kγ-depleted cells. Results are presented as fold-increase relative to control cells in the absence of stimuli, and expressed as mean ± S.D. of triplicate measurements. Data from triplicates are presented as mean ± S.D. Two additional experiments gave similar results. * p < 0.001 vs. control (C) with HRG.
(F) Serum-starved MCF-10A cells were treated with wortmannin (1 μM, 1 h), the PI3Kγ inhibitor (1 μM, 1 h) or PTX (100 ng/ml, 24 h), and Rac-GTP levels determined after stimulation with HRG.
Gβγ subunits released upon activation of Gi-coupled receptors are known to directly activate PI3Kγ, a type Ib PI3K [39]. To determine a potential implication of PI3Kγ we used the PI3Kγ inhibitor 5-quinoxalin-6-ylmethylene-thiazolidine-2,4-dione. This inhibitor dose-dependently reduced HRG-induced Rac1 activation (Fig. 5B). Moreover, MCF-7 cell migration in response to HRG was greatly reduced by the PI3Kγ inhibitor and to a similar degree as PTX (Fig. 5C). As a second approach, we stably knocked down the catalytic subunit of PI3Kγ (p110γ) from MCF-7 cells using two different shRNA constructs. These two cell lines showed deficient activation of Rac1 by HRG (~60% inhibition) (Fig. 5D) as well as a reduced migratory response (Fig. 5E). Thus, Rac1 activation by HRG in P-Rex1 expressing breast cancer cells is mediated by PI3Kγ. PTX treatment was unable to reduce further Rac-GTP levels in PI3Kγ-depleted cells (Fig. S5), arguing that Gβγ and PI3Kγ are in the same pathway.
As non-transformed MCF-10A cells express very low P-Rex1 levels (see Fig. 1B and 1C), we reasoned that Rac1 activation by HRG in these cells should be insensitive to PTX or PI3Kγ inhibition. Remarkably, and in sharp contrast to MCF-7 cells, neither PTX nor the PI3Kγ inhibitor reduced Rac1 activation by HRG in MCF-10A cells, even though wortmannin effectively impaired this response (Fig. 5F). Thus, in P-Rex1-deficient MCF-10A cells, Rac1 activation by HRG is Gβγ- and PI3Kγ-independent. These results argue for a differential utilization of Rac-GEFs in P-Rex1 positive and P-Rex1 negative cells.
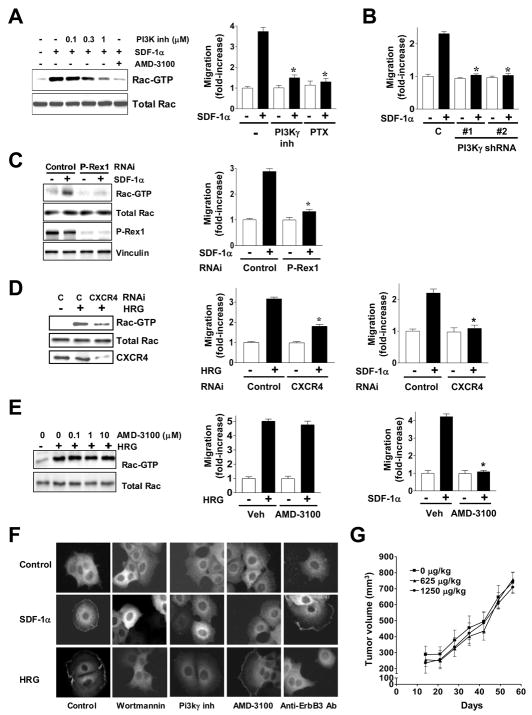
CXCR4 and EGFR are implicated in Rac1 activation by HRG in breast cancer cells
Numerous studies have implicated Gi-coupled-receptors in growth factor responses, including Rac activation and motility [35–38]. Most recently, studies in MDA-MB-435 cells (later reclassified to a melanoma cell line) showed that ErbB2-induced migration and metastasis are mediated by CXCR4, a Gi-coupled receptor for the chemokine SDF-1α/CXCL12. Both SDF-1α and its receptor are highly expressed in breast tumors and have been widely implicated in the progression of breast cancer. Moreover, there is a positive correlation between CXCR4 and ErbB2 in human breast tumors [40–42]. SDF-1α caused a strong activation of Rac1 in MCF-7 cells and consequently it induced a migratory response (Fig. 6A). Rac1 activation by SDF-1α was dose-dependently reduced by the PI3Kγ inhibitor, and as expected, the CXCR4 inhibitor AMD-3100 blocked the effect of the CXCR4 agonist (Fig. 6A, left panel). SDF-1α also stimulated MCF-7 cell migration, and this effect was blocked by the PI3Kγ inhibitor as well as by PTX (Fig. 6A, right panel). Moreover, migration induced by SDF-1α was abolished in the two PI3Kγ-depleted MCF-7 cell lines (Fig. 6B). Notably, when P-Rex-1 expression was silenced using RNAi, both SDF-1α-induced Rac1 activation (Fig. 6C, left panel) and migration (Fig. 6C, right panel) were essentially abolished. Thus, SDF-1α-induced activation of Rac1 in breast cancer cells is mediated by P-Rex1.
Figure 6. CXCR4 mediates Rac1 activation by HRG.
(A) Left panel, Rac-GTP levels in response to SDF-1α (10 nM, 5 min) in the presence of the PI3Kγ inhibitor (0.1–1 μM) or AMD-3100 (10 μg/ml, 1 h). Right panel, migration in response to SDF-1α (0–40 ng/ml) was determined using a Boyden chamber in MCF-7 cells treated with the PI3Kγ inhibitor (1 μM). Data from triplicates (fold-increase relative to control cells in the absence of stimuli) are presented as mean ± S.E.M. of 3 independent experiments. * p < 0.001 vs. controls with SDF-1α.
(B) Impaired migration of PI3Kγ-depleted MCF-7 cells (see Fig. 5) in response to SDF-1α (10 nM). Data from triplicates (fold-increase relative to control cells in the absence of stimuli) are presented as mean ± S.E.M. of 3 independent experiments. * p < 0.001 vs. controls with SDF-1α.
(C) P-Rex1 mediates SDF-1α effects. Left panel, serum-starved P-Rex1-depleted MCF-7 cell line #3 or control cells (from Fig. 3) were stimulated with SDF-1α (10 nM, 5 min) and Rac-GTP levels were then determined. Right panel, P-Rex1-depleted cells have impaired motility in response to SDF-1α (10 nM) compared to control cells. Data from triplicates (fold-increase relative to control cells in the absence of stimuli) are presented as mean ± S.E.M. of 3 independent experiments. * p < 0.001 vs. control with SDF-1α.
(D) Left panel, MCF-7 cells were transfected with either CXCR4 or control siRNA duplexes. After 16 h cells were serum-starved for 48 h, stimulated with HRG (10 ng/ml, 5 min), and Rac-GTP levels determined. Middle panel, CXCR4 depletion inhibits motility in response to HRG. Right panel, CXCR4 depletion inhibits motility in response to SDF-1α. Data from triplicates (fold-increase relative to control cells in the absence of stimuli) are presented as mean ± S.E.M. of 3 independent experiments. * p < 0.001 vs. control with HRG or SDF-1α.
(E) AMD-3100 (10 μg/ml, 1 h) does not inhibit Rac1 activation (Left panel) or migration (Middle panel) by HRG in MCF-7 cells, but it abolishes migration by SDF-1α (Right panel). Data from triplicates (fold-increase relative to vehicle in the absence of stimuli) are presented as mean ± S.E.M. of 3 independent experiments. * p < 0.01 vs. vehicle (Veh) + SDF-1α.
(F) Translocation of endogenous P-Rex1 in MCF-7 cells after stimulation with either HRG (10 ng/ml) or SDF-1α (10 nM) for 10 min, alone or in the presence of wortmannin (1 μM), the PI3Kγ inhibitor (1 μM), AMD-3100 (10 μg/ml), or an anti-ErbB3 antibody (10 μg/ml). Similar results were observed in multiple individual cells in 3 different experiments.
(G) The CXCR4 inhibitor AMD-3100 does not affect tumorigenicity of BT-474 cells in nude mice (10 mice/group). Data are expressed as mean ± S.D.
In order to determine if CXCR4 is implicated in ErbB-driven activation of Rac1, we silenced CXCR4 expression in MCF-7 cells using RNAi. Rac1 activation (Fig. 6D, left panel) and motility (Fig. 6D, middle panel) by HRG were significantly reduced (55 and 61% inhibition, respectively) in CXCR4-depleted cells. Unlike the HRG effect, migration induced by SDF-1α was essentially impaired (91% inhibition) in CXCR4-deficient cells (Fig. 6D, right panel). As Rac1 activation by HRG is rapid [7], we reasoned that most likely it does not involve the autocrine secretion of SDF-1α. In fact, blockade of CXCR4 with the specific antagonist AMD-3100 was unable to prevent HRG-induced Rac1 activation (Fig. 6E, left panel) or migration (Fig. 6E, middle panel), indicating that these effects are independent of ligand activation of CXCR4, whereas the CXCR4 antagonist abolished SDF-1α-induced migration (Fig. 6E, right panel). Moreover, AMD-3100 had no effect on HRG-induced relocalization of endogenous P-Rex1 in MCF-7 cells (Fig. 6F). In contrast, SDF-1α-induced translocation of P-Rex-1 was effectively inhibited by AMD-3100. Both HRG- and SDF-1α-induced translocation of P-Rex1 were impaired by wortmannin or the PI3Kγ inhibitor (Fig. 6F), or by PI3Kγ RNAi depletion (Fig. S6), whereas only the HRG effect was blocked by an anti-ErbB3 antibody (Fig. 6F). The lack of involvement of SDF-1α was supported by the fact that AMD-3100 was unable to inhibit BT-474 tumor growth in nude mice (Fig. 6G).
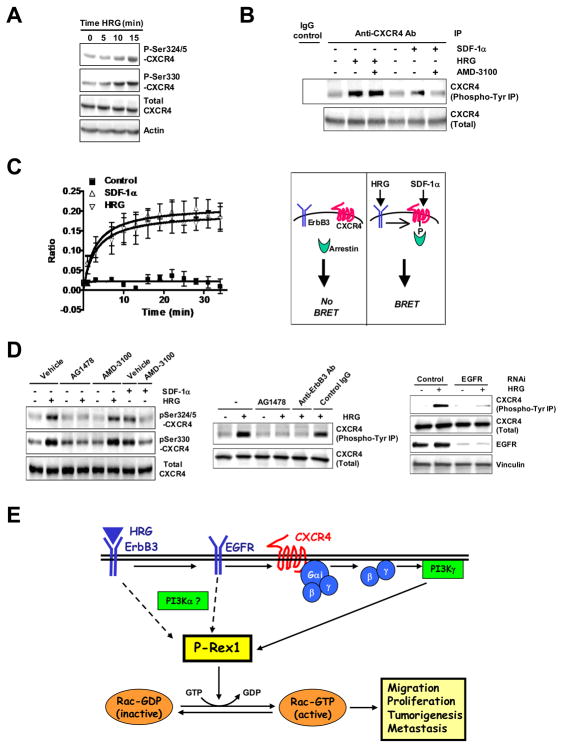
We then examined the activation status of CXCR4 in response to HRG using phospho-specific antibodies against Ser324/325 and Ser330, residues that become phosphorylated in response to SDF-1α and regulate CXCR4 signaling and trafficking [43, 44]. Notably, a significant elevation in Ser324/325- and Ser330-CXCR4 phosphorylation was detected upon HRG treatment in T-47D cells (Fig. 7A), which was similar in magnitude to that induced by SDF-1α (data not shown). Phosphorylation of these sites in CXCR4 was also observed in response to EGF (data not shown). It is known that CXCR4 becomes Tyr phosphorylated upon activation with SDF-1α [45]. We found that in MCF-7 cells CXCR4 becomes Tyr phosphorylated both in response to SDF-1α or HRG, and only the effect of SDF-1α was sensitive to AMD-3100 (Fig. 7B). Since phosphorylated (activated) CXCR4 binds arrestins, we also examined whether activation of ErbB3 receptors promotes binding of arrestin to CXCR4 in MCF-7 cells using a Bioluminescence Resonance Energy Transfer (BRET) approach [44]. Remarkably, HRG promotes the association of arrestin2 to CXCR4 in a time-dependent manner and with a magnitude similar as that observed with SDF-1α (Fig. 7C).
Figure 7. Activation of CXCR4 by HRG via EGFR.
(A) Serum-starved T-47D cells were treated with HRG (10 ng/ml) for different times and subject to Western blot with the indicated CXCR4 antibodies.
(B) After different treatments BT-474 cells were subject to immunoprecipitation (IP) with an anti-CXCR4 antibody or IgG control. Immunoprecipitates were immunobloted with either anti-phospho-Tyr or anti-CXCR4 antibodies.
(C) Left panel, MCF-7 cells were transiently transfected with RlucII-tagged CXCR4 and GFP10-arrestin2 as described in Materials and Methods. Interactions between CXCR4 and arrestin2 were measured by BRET2 following incubation with buffer (Control), SDF-1α (100 nM) or HRG (10 ng/ml). Both SDF-1α and HRG-stimulation resulted in rapid recruitment of arrestin2. Data are expressed as mean ± S.D. of triplicate samples. Two additional experiments gave similar results. Right panel, schematic representation of the BRET assay.
(D) Left panel, Effect of AG1478 (1 μM) and AMD-3100 (10 μg/ml) on SDF-1α- and HRG-induced Ser phosphorylation in CXCR4. Middle panel, Effect of AG1478 on CXCR4 Tyr phosphorylation induced by HRG. As a control, the effect of HRG is blocked by a blocking anti-ErbB3 antibody but not a control IgG (10 μg/ml, 1 h). Right panel, Effect of EGFR RNAi on HRG-induced CXCR4 Tyr phosphorylation.
(E) Model: P-Rex1 mediates inputs from ErbB receptors in breast cancer cells through the transactivation of CXCR4-Gi-PI3Kγ pathway.
In a previous study we established that EGFR was required for Rac activation by HRG in T-47D and MCF-7 cells [7]. Similar results were observed in BT-474 cells (Fig. S7). To determine whether EGFR mediates CXCR4 phosphorylation we used the EGFR inhibitor AG1478, and found that it prevented phosphorylation of CXCR4 in Ser324/325 and Ser330. AMD-3100 blocked CXCR4 Ser phosphorylation induced by SDF-1α but not by HRG (Fig. 7D, left panel). AG1478 also blocked CXCR4 Tyr phosphorylation by HRG to the same extent as an anti-ErbB3 blocking antibody (Fig. 7D, middle panel). Moreover, EGFR RNAi depletion from BT-474 cells also impaired CXCR4 Tyr phosphorylation (Fig. 7D, right panel). These results implicate EGFR in the transactivation of CXCR4 by stimulation of ErbB3 receptors. Our results not only support the concept that CXCR4 becomes activated in response to ErbB ligands independently of SDF-1α but also strongly argue for the utilization of a CXCR4-dependent pathway in the activation of P-Rex1/Rac1 by ErbB receptors.
DISCUSSION
P-Rex1 as a mediator of Rac1 activation by ErbB receptors in breast cancer cells
Our studies provide for the first time evidence that the PI3K- and Gβγ-dependent Rac-GEF P-Rex1 is an essential mediator of Rac1 activation and migration in breast cancer cells by ErbB receptors. While many GEFs for Rho GTPases were originally identified as oncogenes, they often act in cancer cells as transducers of upstream dysregulated inputs. Persistent activation of Rac may arise as a consequence of aberrant TK receptor hyperactivation or genetic alterations leading to PI3K hyperactivation (PI3KCA mutations, Pten deficiency, Ras mutations). Enhanced Rac activation due to overexpression or hyperactivation of Rac-GEFs is also frequent in cancer, as established for Vav1 in pancreatic cancer and for Vav2 in head and neck squamous carcinoma [13, 46].
The role of Rac-GEFs in human breast cancer progression remains poorly understood. The identification of P-Rex1 as a mediator of Rac1 activation in breast cancer cells was unanticipated. P-Rex1 was originally identified in neutrophils [17–19, 26], and although some evidence suggested that P-Rex1 activation could be dependent on TK activity [26, 47, 48], this GEF has been predominantly characterized as a GPCR effector. Based on the limited information on Rac-GEFs in breast cancer models, our original prediction was that Tiam1, Trio or Vav isoforms would have played a significant role in Rac1 activation by ErbB ligands. For example, Vav3 is up-regulated in human breast tumors and mediates estrogen mitogenic responses in breast cancer cells [49]. Interestingly, P-Rex1 RNAi does not affect Akt activation in breast cancer cells. Very recently, it has been shown that the P-Rex1 related isoform P-Rex2a, but not P-Rex1, inhibits Pten phosphatase activity and consequently stimulates Akt phosphorylation and cell growth in breast cancer cells [25]. Thus, different P-Rex isoforms may be implicated in breast cancer progression through remarkably distinct mechanisms. Rac-GEF utilization may be exquisitely controlled by the nature of the oncogenic input and the relative expression of exchange factors in different cancer cell lines.
While the functional relationship between ErbB2 and Rac1 activation has not been fully investigated to-date, the PI3K/Rac/Pak1 axis plays an important role in actin cytoskeleton reorganization in MCF-10A cells ectopically overexpressing ErbB2. In this case, Vav2 seems to be critical for Rac activation, which is consistent with the high expression of this GEF in MCF-10A cells (see Fig. 1A) [9, 32]. Remarkably, we found that P-Rex1 knock-down suppresses Rac1 activation and motility in cell lines overexpressing ErbB2. Moreover, silencing P-Rex1 from BT-474 cells profoundly affects their ability to form tumors in nude mice, a finding of particular relevance taking into consideration that P-Rex1 is preferentially expressed in ErbB2-positive tumors.
Overexpression of P-Rex1 in human breast cancer
Our study revealed that P-Rex1 is highly expressed in a large fraction of human breast tumors, preferentially the luminal subtype which are predominantly ER positive. Aberrant expression of P-Rex1 was detected in all stages of the disease, although there is a tendency to increase with stage. The basal subtype is characterized by a high abundance of triple negative (ER-, PR-, and ErbB2-negative) tumors and mostly do not express P-Rex1. Although a significant correlation was also found with ErbB2, it is not as strong as with ER, possibly a consequence of the relatively modest expression in the HER2+/ER− subtype. Notably, the rate of P-Rex1 positive tumors was higher in patients that developed metastasis, and ~2/3 of lymph node metastases were found to be P-Rex1 positive. A recent study showed that P-Rex1 is up-regulated in the metastatic prostate cancer cell line PC3 relative to non-metastatic LNCaP and CWR22RV1 cells, and in a small patient sample analysis higher expression was found in prostate metastasis relative to the corresponding primary prostate tumors [47]. It is conceivable that P-Rex1 overexpression confers a motile advantage required for metastatic dissemination.
The human P-Rex1 gene (PREX1) is located in chromosome 20q.13. Amplification of this region occurs in 8 to 29% of breast tumors and correlates with poor clinical prognosis [20–23]. As only a subset of breast cancer cell lines present PREX1 gene amplification, alternative transcriptional, translational or post-translational means may contribute to P-Rex1 protein up-regulation. Curiously, in silico analysis of the P-Rex1 promoter revealed multiple ER-responsive elements, and in addition estradiol-mediated proliferation and Rac activation are markedly impaired in P-Rex1-deficient T-47D cells (M.S.S. and M.G.K., unpublished observations). Estrogen promotes breast cancer proliferation via EGFR and CXCR4 transactivation [50], thus suggesting that P-Rex1 may integrate TK and GPCR inputs triggered by the estrogen response.
P-Rex1 as a mediator of CXCR4-induced Rac1 activation in breast cancer cells: integration of TK and GPCR responses
A distinctive feature of P-Rex1 is that it is synergistically regulated by PIP3 and membrane-bound Gβγ proteins via the DH-PH domain [17, 27, 34]. We found that in MCF-7 cells P-Rex1 redistributes to the plasma membrane in response to HRG via its DH-PH domain. Our PTX results established the requirement of Gβγ subunits and a transactivation mechanism involving Gi-coupled receptors in the ErbB receptor response. Curiously, multiple studies have shown that TK responses are PTX-sensitive, including those mediated by ErbB receptors [35–38]. The insensitivity to PTX in MCF-10A cells argues that Gi is not involved in mediating Rac1 activation by ErbB receptors in normal cells and possibly in P-Rex1 negative breast cancer cells.
The PIP3 component of P-Rex1 activation downstream of ErbB receptors arises largely from a Gβγ/PI3Kγ pathway. Inputs from Gβγ subunits and PI3Kγ may suffice for P-Rex1 activation in response to agonist-directed GPCR activation, as inferred from the full inhibitory effect of PI3Kγ depletion on SDF-1α-induced Rac activation and migration, and as also described in neutrophils [26]. It is conceivable that an additional input may be required for Rac activation by ErbB receptors, possibly from type Ia PI3K. Type Ia PI3Ks are indeed preferential effectors of ErbB2/ErbB3 dimers [1–3]. Thus, full activation of P-Rex1 by ErbB receptors may require the convergence of inputs from type Ia and type Ib PI3Ks as well as Gβγ subunits, as depicted in the model presented in Fig. 7E.
A remarkable finding from our studies is the association of CXCR4 with ErbB receptor-induced activation of P-Rex1/Rac signaling. CXCR4 and its ligand SDF-1α have been widely implicated in breast cancer cell proliferation, migration and invasion. CXCR4 and ErbB2 levels correlate in breast tumors [40–42]. Interestingly, in response to HRG, CXCR4 becomes phosphorylated on Ser residues that regulate signaling and trafficking of the activated receptor, and in addition it associates with arrestin, a step normally required for proper receptor signaling, internalization and degradation upon activation [44]. The inability of the CXCR4 antagonist AMD-3100 to affect P-Rex1/Rac1 activation by HRG argues against the involvement of an autocrine mechanism through SDF-1α. Notably, CXCR4 becomes Tyr phosphorylated in response to HRG. Although the implications of CXCR4 Tyr phosphorylation are not completely understood, it has been shown that CXCR4 activated by SDF-1α becomes Tyr phosphorylated, as we also show in Fig. 7B, and inhibition of Tyr phosphorylation prevents CXCR4 downstream signaling [45]. Notably, we found that EGFR is required for the activation of CXCR4 by HRG. This fits with our previous model showing that EGFR transactivation by HRG is a requirement for Rac activation [7]. One plausible scenario is that CXCR4 is a direct substrate of EGFR. CXCR4 indeed associates with other TK receptors such as IGF-RI in response to IGF, leading to CXCR4 coupling to Gi [41]. We were unable to co-IP EGFR with CXCR4 (data not shown), however CXCR4 may be phosphorylated by a TK downstream of EGFR, including soluble TKs or JAKs [51]. Disruption of this transactivation mechanism may provide an alternative means of targeting the Rac pathway.
Final remarks
In summary, the data herein suggest that P-Rex1 is essential for responses such as cell growth, migration, and tumorigenesis driven by ErbB receptors. ErbB receptors relay signals to P-Rex1/Rac1 through a Gi-PI3Kγ-dependent pathway that involves CXCR4 transactivation, strongly arguing for a role for P-Rex1 as an integrator of TK and GPCR inputs in breast cancer cells. Additionally, P-Rex1 up-regulation is a molecular signature of luminal breast tumors. In addition to the prognostic implications of these findings, our data also provide a strong basis for considering the P-Rex1/Rac pathway an attractive target for therapeutic intervention. Inhibitors of Rac-GEF/Rac interactions with anti-tumorigenic activity have been developed [52]. This may be particularly relevant for patients with ER positive tumors that develop resistance to therapies such as anti-estrogens. Rac indeed mediates anti-estrogen resistance in MCF-7 cells, and importantly, a pharmacological inhibitor of Rac abrogates anti-estrogen resistance [16], thus offering alternative therapeutic means for treatment.
MATERIALS AND METHODS
Reagents
HRG, TGFα, and SDF-1α were purchased from R&D (Minneapolis, MN). EGF was purchased from BD Biosciences (San Jose, CA). Wortmannin was from LC Laboratories (Woburn, MA). Pertussis toxin (PTX) and AMD-3100 were obtained from Sigma. Wortmannin, AG1478, 5-quinoxalin-6-ylmethylene-thiazolidine-2,4-dione, and cholera toxin were from EMD/Calbiochem (Gibbstown, NJ).
Cell culture
MCF-10A cells were obtained from the Karmanos Cancer Institute (Detroit, USA) and grown in DMEM/F12 supplemented with 10 mM HEPES, 10 μg/ml insulin, 20 ng/ml EGF, 100 ng/ml cholera toxin, 30 mM sodium bicarbonate, 0.5 μg/ml hydrocortisone, and 5% fetal horse serum. All human breast carcinoma cell lines were obtained from ATCC and grown in DMEM supplemented with 10% FBS (for MCF-7, T-47D and BT-474 cells, the medium was supplemented with 0.2 U/ml bovine insulin).
Western blot and IPs
Western blots were carried out essentially as previously described [8]. Antibodies and IP assays are described in the Supplemental data.
Pull-down assays, phalloidin staining and migration assays
These studies were carried out essentially as described [7].
Rac-GEF array
The Rac-GEF array was generated by SuperArray, and contains validated primers against Rac-GEFs and Rac-GEF modulators, and a housekeeping gene (GAPDH). Total RNA was extracted from cells using Trizol (Invitrogen). cDNA was generated using the RT2 First Strand Kit (SuperArray, Bioscience Corporation). An ABI Prism 7300 thermocycler was used for Q-PCR determinations.
Tissue arrays
P-Rex1 TaqMan primers were purchased from Applied Biosystems, mixed with TaqMan Universal Master Mix (Applied Biosystems) and added to the 96-well Human Breast Cancer TissueScan Real-Time Panel II plate (OriGene, MD). Q-PCR was performed in an ABI Prism 7300 thermocycler. Additionally, a 384-well TissueScan Cancer Survey panel (OriGene, MD) was used to determine P-Rex1 expression in 16 different tumor types, and Q-PCR was performed using an ABI Prism 7900H thermocycler. Information about the tumor samples can be found in the OriGene homepage (www.origene.com).
RNAi and generation of cell lines
siRNA delivery into breast cancer cells for transient knockdown was described previously [7]. shRNA sequences and details about the generation of cell lines and rescue experiments are described in Supplemental data.
Localization studies
MCF-7 cells in coverslides were serum-starved for 48 h, stimulated with HRG (10 ng/ml, 10 min) and fixed with 4% PFA. The anti-P-Rex1 antibody (1:250) was then added for 1 h, followed by a Cy2-conjugated goat anti-rabbit secondary antibody (1:1500; Jackson Immunoresearch Labs). Slides were mounted using Vectashield and visualized with a Nikon TE2000-U fluorescence microscope. MCF-7 cells were also transfected with pCEFL-EGFP-P-Rex1, pCEFL-EGFP-DH-PH, pCEFL-EGFP-DEP-DEP, pCEFL-EGFP-PDZ-PDZ, pCEFL-EGFP-DEP-DEP-PDZ-PDZ, or empty vector, and localization in response to HRG analyzed by real-time microscopy.
Growth in soft agar
3 × 103 cells were suspended in 0.7% granulated agar (BD Biosciences) diluted in complete medium (2X) and poured onto a 0.5% layer of agar. Fresh medium was added every 3 days, and 29 days later colonies were stained with MTT and counted.
Tumorigenesis studies
For xenograft experiments, BT-474 cells (2 × 107 cells/mouse in 300 μl Matrigel) were injected into the flanks of eight-week old female ovariectomized athymic (nude) mice (Foxnnu, Harlam Laboratories, 10 mice/group). A 17β-estradiol pellet (1.7 mg, Innovative Research of America) was implanted s.c. 7 days before injection. In some experiments, mice were injected with the CXCR4 antagonist AMD-3100 (0.625 and 1.25 mg/kg, s.c. once daily), as previously reported [53]. For orthotopic growth experiments, six-week old female SCID mice were injected into the fifth mammary gland with BT-474 cells (4×106 cells/mouse). Tumor formation was monitored by palpation and tumor volume determined with a caliper. All animal experiments were carried out in compliance with the institutions guidelines.
IHC
Detailed procedures for IHC can be found in Supplemental data.
BRET assays
Detailed procedures for BRET can be found in Supplemental data.
Statistical analysis
Differences in the IHC staining of human breast cancer specimens were analyzed with the Fisher’s Exact (one sided) tests. Microarray Pearson correlations were performed on log2 ratios from Agilent 44K two-color gene expression microarrays on 108 human breast tumors for indicated genes after whole-genome normalization. Data were analyzed using either a Student’s t-test or one-way analysis of variance (ANOVA).
HIGHLIGHTS.
The Rac-GEF P-Rex1 is overexpressed in luminal breast cancer cell lines and tumors.
P-Rex1 mediates ErbB driven activation of Rac1 and migration in breast cancer cells.
P-Rex1 is an integrator of tyrosine-kinase and GPCR inputs in breast cancer cells.
ErbB receptors signal to P-Rex1/Rac1 through a CXCR4-Gi-PI3Kγ-dependent pathway.
Supplementary Material
Acknowledgments
This work is supported by grants R01CA74197, R01CA129133, and R01CA139120 (NIH), and KG090522 (Susan Komen Foundation for the Cure) to M.G.K, and grant R01CA129626 to J.L.B. H.Y. and J.S.G. are supported by the Intramural Program, NIDCR, NIH. We thank Andy Cucchiara (UPenn) for help with statistical analyses, and Celine Lefebvre (Columbia University) for support with microarray data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5(5):341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 2.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284(1):54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 4.Bose S, et al. Reduced expression of PTEN correlates with breast cancer progression. Hum Pathol. 2002;33(4):405–9. doi: 10.1053/hupa.2002.124721. [DOI] [PubMed] [Google Scholar]
- 5.Bachman KE, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3(8):772–5. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 6.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 7.Yang C, et al. Essential role for Rac in heregulin beta1 mitogenic signaling: a mechanism that involves epidermal growth factor receptor and is independent of ErbB4. Mol Cell Biol. 2006;26(3):831–42. doi: 10.1128/MCB.26.3.831-842.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C, et al. Heregulin beta1 promotes breast cancer cell proliferation through Rac/ERK-dependent induction of cyclin D1 and p21Cip1. Biochem J. 2008;410(1):167–75. doi: 10.1042/BJ20070781. [DOI] [PubMed] [Google Scholar]
- 9.Wang SE, et al. HER2/Neu (ErbB2) signaling to Rac1-Pak1 is temporally and spatially modulated by transforming growth factor beta. Cancer Res. 2006;66(19):9591–600. doi: 10.1158/0008-5472.CAN-06-2071. [DOI] [PubMed] [Google Scholar]
- 10.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6(2):167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 11.Yang C, et al. Rac-GAP-dependent inhibition of breast cancer cell proliferation by {beta}2-chimerin. J Biol Chem. 2005;280(26):24363–70. doi: 10.1074/jbc.M411629200. [DOI] [PubMed] [Google Scholar]
- 12.Minard ME, et al. The role of the guanine nucleotide exchange factor Tiam1 in cellular migration, invasion, adhesion and tumor progression. Breast Cancer Res Treat. 2004;84(1):21–32. doi: 10.1023/B:BREA.0000018421.31632.e6. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Zapico ME, et al. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7(1):39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 14.Holm C, et al. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst. 2006;98(10):671–80. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- 15.Balasenthil S, et al. p21-activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells. J Biol Chem. 2004;279(2):1422–8. doi: 10.1074/jbc.M309937200. [DOI] [PubMed] [Google Scholar]
- 16.Felekkis KN, et al. AND-34 activates phosphatidylinositol 3-kinase and induces anti-estrogen resistance in a SH2 and GDP exchange factor-like domain-dependent manner. Mol Cancer Res. 2005;3(1):32–41. [PubMed] [Google Scholar]
- 17.Welch HC, et al. P-Rex1, a PtdIns(3,4,5)P3- and Gbetagamma-regulated guanine-nucleotide exchange factor for Rac. Cell. 2002;108(6):809–21. doi: 10.1016/s0092-8674(02)00663-3. [DOI] [PubMed] [Google Scholar]
- 18.Welch HC, et al. P-Rex1 regulates neutrophil function. Curr Biol. 2005;15(20):1867–73. doi: 10.1016/j.cub.2005.09.050. [DOI] [PubMed] [Google Scholar]
- 19.Dong X, et al. P-Rex1 is a primary Rac2 guanine nucleotide exchange factor in mouse neutrophils. Curr Biol. 2005;15(20):1874–9. doi: 10.1016/j.cub.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Hodgson JG, et al. Genome amplification of chromosome 20 in breast cancer. Breast Cancer Res Treat. 2003;78(3):337–45. doi: 10.1023/a:1023085825042. [DOI] [PubMed] [Google Scholar]
- 21.Mastracci TL, et al. Genomic alterations in lobular neoplasia: a microarray comparative genomic hybridization signature for early neoplastic proliferationin the breast. Genes Chromosomes Cancer. 2006;45(11):1007–17. doi: 10.1002/gcc.20368. [DOI] [PubMed] [Google Scholar]
- 22.Kallioniemi A, et al. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci U S A. 1994;91(6):2156–60. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonsson G, et al. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer. 2007;46(6):543–58. doi: 10.1002/gcc.20438. [DOI] [PubMed] [Google Scholar]
- 24.Fan C, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–9. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 25.Fine B, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325(5945):1261–5. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao T, et al. Signaling requirements for translocation of P-Rex1, a key Rac2 exchange factor involved in chemoattractant-stimulated human neutrophil function. J Leukoc Biol. 2007;81(4):1127–36. doi: 10.1189/jlb.0406251. [DOI] [PubMed] [Google Scholar]
- 27.Barber MA, et al. Membrane translocation of P-Rex1 is mediated by G protein betagamma subunits and phosphoinositide 3-kinase. J Biol Chem. 2007;282(41):29967–76. doi: 10.1074/jbc.M701877200. [DOI] [PubMed] [Google Scholar]
- 28.Hill K, et al. Regulation of P-Rex1 by phosphatidylinositol (3,4,5)-trisphosphate and Gbetagamma subunits. J Biol Chem. 2005;280(6):4166–73. doi: 10.1074/jbc.M411262200. [DOI] [PubMed] [Google Scholar]
- 29.Adam L, et al. Heregulin regulates cytoskeletal reorganization and cell migration through the p21-activated kinase-1 via phosphatidylinositol-3 kinase. J Biol Chem. 1998;273(43):28238–46. doi: 10.1074/jbc.273.43.28238. [DOI] [PubMed] [Google Scholar]
- 30.Atlas E, et al. Heregulin is sufficient for the promotion of tumorigenicity and metastasis of breast cancer cells in vivo. Mol Cancer Res. 2003;1(3):165–75. [PubMed] [Google Scholar]
- 31.Lee RJ, et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol. 2000;20(2):672–83. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda Y, et al. Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks transforming growth factor beta-induced cell motility. J Biol Chem. 2004;279(23):24505–13. doi: 10.1074/jbc.M400081200. [DOI] [PubMed] [Google Scholar]
- 33.Moulder SL, et al. Epidermal growth factor receptor (HER1) tyrosine kinase inhibitor ZD1839 (Iressa) inhibits HER2/neu (erbB2)-overexpressing breast cancer cells in vitro and in vivo. Cancer Res. 2001;61(24):8887–95. [PubMed] [Google Scholar]
- 34.Mayeenuddin LH, McIntire WE, Garrison JC. Differential sensitivity of P-Rex1 to isoforms of G protein betagamma dimers. J Biol Chem. 2006;281(4):1913–20. doi: 10.1074/jbc.M506034200. [DOI] [PubMed] [Google Scholar]
- 35.Luttrell LM, et al. G beta gamma subunits mediate mitogen-activated protein kinase activation by the tyrosine kinase insulin-like growth factor 1 receptor. J Biol Chem. 1995;270(28):16495–8. doi: 10.1074/jbc.270.28.16495. [DOI] [PubMed] [Google Scholar]
- 36.Johnson RM, et al. Pertussis toxin or phorbol 12-myristate 13-acetate can distinguish between epidermal growth factor- and angiotensin-stimulated signals in hepatocytes. Proc Natl Acad Sci U S A. 1986;83(7):2032–6. doi: 10.1073/pnas.83.7.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanton RC, et al. Rapid release of bound glucose-6-phosphate dehydrogenase by growth factors. Correlation with increased enzymatic activity. J Biol Chem. 1991;266(19):12442–8. [PubMed] [Google Scholar]
- 38.Hobson JP, et al. Role of the sphingosine-1-phosphate receptor EDG-1 in PDGF-induced cell motility. Science. 2001;291(5509):1800–3. doi: 10.1126/science.1057559. [DOI] [PubMed] [Google Scholar]
- 39.Andrews S, Stephens LR, Hawkins PT. PI3K class IB pathway. Sci STKE. 2007;2007(407):cm2. doi: 10.1126/stke.4072007cm2. [DOI] [PubMed] [Google Scholar]
- 40.Muller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 41.Akekawatchai C, et al. Transactivation of CXCR4 by the insulin-like growth factor-1 receptor (IGF-1R) in human MDA-MB-231 breast cancer epithelial cells. J Biol Chem. 2005;280(48):39701–8. doi: 10.1074/jbc.M509829200. [DOI] [PubMed] [Google Scholar]
- 42.Li YM, et al. Upregulation of CXCR4 is essential for HER2-mediated tumor metastasis. Cancer Cell. 2004;6(5):459–69. doi: 10.1016/j.ccr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 43.Marchese A, Benovic JL. Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J Biol Chem. 2001;276(49):45509–12. doi: 10.1074/jbc.C100527200. [DOI] [PubMed] [Google Scholar]
- 44.Busillo JM, et al. Site-specific phosphorylation of CXCR4 is dynamically regulated by multiple kinases and results in differential modulation of CXCR4 signaling. J Biol Chem. 2010;285(10):7805–17. doi: 10.1074/jbc.M109.091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vila-Coro AJ, et al. The chemokine SDF-1alpha triggers CXCR4 receptor dimerization and activates the JAK/STAT pathway. Faseb J. 1999;13(13):1699–710. [PubMed] [Google Scholar]
- 46.Patel V, et al. Persistent activation of Rac1 in squamous carcinomas of the head and neck: evidence for an EGFR/Vav2 signaling axis involved in cell invasion. Carcinogenesis. 2007;28(6):1145–52. doi: 10.1093/carcin/bgm008. [DOI] [PubMed] [Google Scholar]
- 47.Qin J, et al. Upregulation of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer metastasis. Oncogene. 2009;28(16):1853–63. doi: 10.1038/onc.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshizawa M, et al. Involvement of a Rac activator,P-Rex1, in neurotrophin-derived signaling and neuronal migration. J Neurosci. 2005;25(17):4406–19. doi: 10.1523/JNEUROSCI.4955-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee K, et al. Vav3 oncogene activates estrogen receptor and its overexpression may be involved in human breast cancer. BMC Cancer. 2008;8:158. doi: 10.1186/1471-2407-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pattarozzi A, et al. 17beta-estradiol promotes breast cancer cell proliferation-inducing stromal cell-derived factor-1-mediated epidermal growth factor receptor transactivation: reversal by gefitinib pretreatment. Mol Pharmacol. 2008;73(1):191–202. doi: 10.1124/mol.107.039974. [DOI] [PubMed] [Google Scholar]
- 51.Andl CD, et al. EGFR-induced cell migration is mediated predominantly by the JAK-STAT pathway in primary esophageal keratinocytes. Am J Physiol Gastrointest Liver Physiol. 2004;287(6):G1227–37. doi: 10.1152/ajpgi.00253.2004. [DOI] [PubMed] [Google Scholar]
- 52.Nassar N, et al. Structure-function based design of small molecule inhibitors targeting Rho family GTPases. Curr Top Med Chem. 2006;6(11):1109–16. doi: 10.2174/156802606777812095. [DOI] [PubMed] [Google Scholar]
- 53.Rubin JB, et al. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci U S A. 2003;100(23):13513–8. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.