Abstract
The prevalence of natural selection relative to genetic drift is of central interest in evolutionary biology. Depending on the distribution of fitness effects of new mutations, the importance of these evolutionary forces may differ in species with different effective population sizes. Here, we survey population genetic variation at 105 orthologous X-linked protein coding regions in Drosophila melanogaster and its sister species D. simulans, two closely related species with distinct demographic histories. We observe significantly higher levels of polymorphism and evidence for stronger selection on codon usage bias in D. simulans, consistent with a larger historical effective population size on average for this species. Despite these differences, we estimate that <10% of newly arising nonsynonymous mutations have deleterious fitness effects in the nearly neutral range (i.e., −10 < Nes < 0) in both species. The inferred distributions of fitness effects and demographic models translate into surprisingly high estimates of the fraction of “adaptive” protein divergence in both species (∼85–90%). Despite evidence for different demographic histories, differences in population size have apparently played little role in the dynamics of protein evolution in these two species, and estimates of the adaptive fraction (α) of protein divergence in both species remain high even if we account for recent 10-fold growth. Furthermore, although several recent studies have noted strong signatures of recurrent adaptive protein evolution at genes involved in immunity, reproduction, sexual conflict, and intragenomic conflict, our finding of high levels of adaptive protein divergence at randomly chosen proteins (with respect to function) suggests that many other factors likely contribute to the adaptive protein divergence signature in Drosophila.
Keywords: genome diversity, codon bias, adaptive protein evolution, selective constraint, effective population size, comparative population genetics
Introduction
A goal of population genetics is to understand the processes shaping genome evolution. Key among population genetic parameters is the effective population size, Ne, which determines the efficacy of selection relative to genetic drift acting on variation within a species (Charlesworth 2009). Theory has shown that the ratio of levels of polymorphism within species to divergence between species is sensitive to small differences in the fitness effects of mutations, s, particularly when the product of Ne and s is close to unity (Ohta 1973; Kimura 1983). In particular, species with smaller Ne are expected to accumulate a greater proportion of mildly deleterious mutations relative to species with larger Ne, leading to faster rates of evolution (Ohta 1973). Correlations between reduced Ne and faster rates of evolution have been documented in island relative to mainland species (Ohta 1993; Woolfit and Bromham 2005), in primates relative to domesticated dogs and rodents (Lindblad-Toh et al. 2005), and in broader comparisons of taxa (Popadin et al. 2007; Wright and Andolfatto 2008). Faster rates of evolution have also been documented for specific regions of the genome that are thought to have a reduced Ne, such as mitochondrial DNA (Rand and Kann 1996; Weinreich and Rand 2000), Y chromosomes (Bachtrog 2006), and other genomic regions with highly reduced recombination (Kliman and Hey 1993; Presgraves 2005; Haddrill et al. 2007; Betancourt et al. 2009; but see Bullaughey et al. 2008).
The predicted effect of Ne differences on patterns of genome evolution crucially depends on both the magnitude of these differences and the shape of the distribution of fitness effects of newly arising mutations (hereafter the “DFE”, Eyre-Walker and Keightley 2007). For example, if most mutations have selection intensity Nes = −10, these will be little affected by a 2-fold difference in Ne. On the other hand, if the distribution of Nes among mutations is exponential with mean Nes = −10, then a significant fraction of mutations will fall into the range of selection intensities (i.e., −10 < Nes < 0) that will be strongly affected by a 2-fold difference in Ne. Previous studies have suggested that a large fraction of the Drosophila genome, such as synonymous sites (Akashi 1995; Akashi and Schaeffer 1997; Maside et al. 2004; Zeng and Charlesworth 2009) and most noncoding DNA (Andolfatto 2005; Casillas et al. 2007), may be experiencing very weak selection (i.e., −5 < Nes < −1). Eyre-Walker et al. (2002) estimated that ∼20% of newly arising nonsynonymous mutations have Nes < 1 in Drosophila simulans, and Loewe et al. (2006) estimated a mean Nes ∼ −4 in D. miranda and D. pseudoobscura, implying that a substantial fraction of nonsynonymous mutations may also be weakly selected.
In this study, we have focused on comparing orthologous X-linked coding regions of Drosophila melanogaster and D. simulans, two recently diverged sister species. We chose this species pair because differences in Ne have been invoked to explain lower levels of diversity (Aquadro et al. 1988; Moriyama and Powell 1996; Andolfatto 2001; Eyre-Walker et al. 2002), higher amino acid polymorphism (Choudhary and Singh 1987; Moriyama and Powell 1996; Andolfatto 2001; Eyre-Walker et al. 2002), and weaker selection maintaining codon usage bias (Akashi 1995, 1996; Begun 1996) in D. melanogaster relative to D. simulans.
Although the focus in population genetics since the proposal of the neutral theory has been on deleterious mutations, recent evidence from Drosophila and other organisms suggests that positive selection may be playing a larger role in shaping genome evolution than previously thought (Sella et al. 2009; Pool et al. 2010). Differences in Ne are predicted to impact rates of adaptation primarily by determining the number of beneficial mutations that are introduced each generation (though fixation probabilities also weakly depend on Ne). Thus, if adaptation is mutation limited, rates of adaptation are expected to be higher in species with larger Ne, and given the higher efficacy of selection against mildly deleterious mutations, so is the proportion of divergence attributable to positive selection relative to mildly deleterious or neutral mutations.
But just how different are the effective population sizes of D. melanogaster and D. simulans and how has this impacted patterns of genome evolution in the two species? Several factors have made this question a difficult one to answer. The earliest studies suggested a 2- to 5-fold difference in Ne (Aquadro et al. 1988; Moriyama and Powell 1996; Eyre-Walker et al. 2002). However, these studies are difficult to evaluate quantitatively because they often compared different sets of genomic regions in the two species and pooled data from very different population samples and chromosomal or functional contexts. We now know that recombination rate is a strong determinant of levels of variability in Drosophila (Begun and Aquadro 1992) and that teleomeric and centromeric suppression of recombination is more pronounced in D. melanogaster than D. simulans (Sturtevant 1929; Ohnishi and Voelker 1979). We also know that variation in both species exhibits profound geographical structuring, with the most diverse populations of both species being found in East Sub-Saharan Africa (Begun and Aquadro 1993; Baudry et al. 2004, 2006). There is a large X-autosome discrepancy to this geographic structuring, with the X and autosomes having similar levels of variability in African populations, but variability on the X is reduced relative to autosomes in non-African populations (Andolfatto 2001; Kauer et al. 2002). Comparing autosomal variability in the two species is further complicated by the presence of relatively recently derived autosomal inversions in D. melanogaster that likely modify recombination rates and may increase the scale of genetic hitchhiking, at least in equatorial populations where they are found at high frequencies (Andolfatto et al. 2001; Aulard et al. 2002).
Several subsequent studies have noted similar levels of diversity on the X in African population samples of D. melanogaster and D. simulans, suggesting their effective population sizes may not be as different as initially thought (Andolfatto 2001; Haddrill et al. 2008; Nolte and Schlotterer 2008). However, each of these studies was limited by the amount and/or the nature of data. All three studies considered a relatively small number of orthologous genomic regions (10, 21, and 10 loci, respectively). In addition, selection on surveyed sites likely poses a problem for quantifying differences in population size (see Discussion). This may be particularly problematic for interpreting the study of Nolte and Schlotterer (2008) whose data set is a mixture of long intergenic, long intronic and coding loci, which are all likely to be experiencing considerable selective constraint in both species (Andolfatto 2005; Haddrill et al. 2008). However, naively comparing synonymous sites diversities (Andolfatto 2001; Haddrill et al. 2008) may also suffer from this problem if selection on codon usage is not taken into account.
Here, we extend previous studies by surveying population-level nucleotide variation at 105 orthologous coding regions in large samples (n = 20 alleles) for both species. We have surveyed the most highly recombining portion of the X chromosome to avoid complications arising from recombination rate variation or inversion polymorphism (the latter being specific to the autosomes of D. melanogaster). We use this data to compare population genetic patterns at synonymous and nonsynonymous sites in the two species and what these imply about the relative Ne of the X chromosomes, the distribution of fitness effects of newly arising nonsynonymous mutations, and patterns of protein evolution and codon usage bias in the two species. We conclude that, despite historical differences in Ne for the two species, most newly arising nonsynonymous mutations are sufficiently strongly selected that these differences have played little role in the dynamics of protein evolution.
Materials and Methods
Choice of Populations
We surveyed 20 individuals each from a Victoria Falls, Zimbabwe, Africa population of D. melanogaster, and a Madagascar population of D. simulans, both collected by Bill Ballard in 2002. These are African populations of the two species with the highest levels of variability and lowest levels of linkage disequilibrium (Begun and Aquadro 1993; Baudry et al. 2004, 2006; Haddrill, Thornton, et al. 2005; Nolte and Schlotterer 2008), suggesting that they have likely maintained large populations that have been free of bottlenecks associated with very recent range expansion.
Choice of Loci, Sequencing, Alignment, and Basic Analyses
We surveyed 105 randomly chosen (with respect to function) orthologous coding regions located between cytological positions 3D2 and 16E1 on the X chromosome of both species. This range of cytological positions was chosen because this region of the X chromosome has the highest levels of and minimal variation in rates of recombination—a major determinant of diversity levels in Drosophila (Begun and Aquadro 1992; Charlesworth 1996). Each 700- to 800-bp region was polymerase chain reaction amplified from genomic DNA extracted from single male flies, Sanger sequenced on both strands, aligned, and annotated as described in Andolfatto (2007). Sequences have been deposited in GenBank (a list of accession numbers is provided in supplementary S4, Supplementary Material online), and FASTA alignments are available at the website http://genomics.princeton.edu/AndolfattoLab/Links.html.
The estimated number of synonymous sites, nonsynonymous sites, average pairwise diversity (π), average pairwise divergence (Dxy) along a species lineage as well as counts of the number of polymorphisms (S), and fixed differences (D) were calculated using a library of Perl scripts (“Polymorphorama”) written by P.A. and D.B. For lineage-specific estimates of divergence, we reconstructed a D. melanogaster–D. simulans ancestor (ANC) sequence using the maximum-likelihood approach implemented in the ‘‘codeml’’ (for coding regions) and ‘‘baseml’’ (for short introns) programs of PAML (Yang 1997). We used the D. yakuba genome sequence as an outgroup sequence. For one locus, CG2887, we could not find a D. yakuba ortholog, and D. erecta was used instead.
We largely restricted our analyses to 4-fold degenerate synonymous sites and 0-fold degenerate nonsynonymous sites. For the 21 surveyed short (i.e. all <120 bp with median size 60 bp) introns in this data set, we masked the starting GT and ending AG bases in population genetic analyses. The number of nonsynonymous and synonymous sites was estimated using the method of Nei and Gojobori (1986). π and Dxy estimates were corrected for multiple hits using a Jukes–Cantor correction (Jukes and Cantor 1969). Multiply hit sites were included in all analyses, but insertion–deletion polymorphisms and polymorphic sites overlapping alignment gaps were excluded. For comparisons of diversity in the two species, we used all 105 coding loci, but for analyses that involved pooling data from all loci (such as comparisons of the site frequency spectrum, demographic, and selection inferences), we excluded five loci (CG1619, CG12717, CG32702, CG32790, and CG12239) that had sample sizes <20 in one or both species. Likewise, we excluded 1 of our 21 introns (CG32702) that had a sample size of <20 in D. simulans. We define preferred and unpreferred codons based on the D. melanogaster and D. simulans codon preferences identified by Vicario et al. (2007).
Estimates of Demographic and Selection Parameters
We use the approach of Keightley and Eyre-Walker (2007) to jointly estimate the distribution of fitness effects (DFEs) of newly arising deleterious nonsynonymous mutations and parameters of an instantaneous population size change model. The method relies on a suitable choice of neutral reference sites from which the demographic model can be estimated. We start by using synonymous sites for this purpose, however, given evidence for ongoing selection on synonymous sites, particularly in D. simulans, we also use the 20 short introns surveyed for comparison. To estimate the nonsynonymous divergence excess relative to the neutral model, α , we used three approaches. The first is the method proposed by Eyre-Walker and Keightley (2009), which is an extension of their 2007 method to estimate the DFE and a demographic model. To establish 95% confidence intervals (CIs), we reestimated demography and selection parameters on 200 bootstrap-replicate (by locus with replacement) data sets. For comparison, we use the maximum-likelihood method proposed by Bierne and Eyre-Walker (2004) and the estimator proposed by Fay et al. (2001), as implemented in the program DoFE (A. Eyre-Walker, unpublished data), both of which assume a constant-size population at equilibrium and that all segregating polymorphisms are neutral.
Coestimation of a demographic model is integral to the approach of Eyre-Walker and Keightley (2009). This method explicitly parameterizes a two-epoch population size change model and does not rule out the possibility that different or more complicated demographic models are more appropriate. Keightley and Eyre-Walker (2007) and Eyre-Walker and Keightley (2009) discuss issues regarding the accuracy and robustness of their method with respect to both linkage and misspecification of the chosen demographic model. Given that distinguishing among demographic models is not the focus of this analysis, we detailed estimates of demographic parameters in supplementary S2 (Supplementary Material online).
Comparing Mean Allele Frequencies with Expectations Under Neutrality
For a data set with a given number of polymorphisms, S, we simulate neutral allele frequency distributions by generating multinomial samples that distribute these polymorphisms into 19 frequency bins using the “Multinomial” function in the R statistical package (http://www.r-project.org). We compare the observed mean frequency with 10,000 simulated replicates.
Shared Derived Mutations
Private polymorphisms are defined as polymorphic mutations at a specific site that occur only in 1 of the 2 species. Private fixations are defined as derived mutations at a specific site that are shared by all sampled individuals of a single species. Shared derived polymorphisms are defined as the occurrence of the same derived polymorphism in the two species. Shared derived mutations include shared derived polymorphisms as well as derived mutations that are polymorphic in one species but fixed in the other or fixed in both lineages leading back the mel-sim ancestor. To estimate the number of shared polymorphisms and shared derived mutations expected due to multiple mutations to the same site, we generated simulated 10,000 replicates of the observed number of derived mutations (both shared and private) at each locus. The approach is similar to that employed by Clark (1997) but does not assume equal mutation rates among loci (because sites are not pooled). Our implementation does assume equal mutations rates across a given class of sites within a locus, which may underestimate the true number of multiple hits. Code to carry out these simulations is available on request to P.A.
Estimation of Species Divergence Time
As originally formulated by Hudson et al. (1987), one can estimate the splitting time of two allopatric populations T (in units of 2N generations, where N is the current population size) for n loci as
 |
(1) |
where g is the ratio of the current and ancestral population sizes and, for each locus i, Dxy,i is the average pairwise divergence between species and θW,i is Watterson’s estimator of the population mutation rate (Watterson 1975). Confidence intervals were estimated by estimating T for 1,000 replicate bootstraps of the data (by locus, with replacement).
Results
Correlated Diversity Levels in the Two Species
We examine levels of synonymous site variability in the two species at 105 orthologous X-linked coding regions. We also present data for 21 “short” (in this case, <120 bp) introns, which are thought to evolve under lower levels of selective constraint than longer introns and synonymous sites (Haddrill, Charlesworth, et al. 2005; Halligan and Keightley 2006; Parsch et al. 2010).
Locus-by-locus estimates of synonymous site and short intron diversity are strongly positively correlated in the two species (fig. 1A), and several factors may contribute to this correlation. First, there is a strong positive correlation between Dmel-Dsim divergence and diversity levels (π) in both species (Dmel: Spearman R = 0.29, P = 0.003; Dsim: Spearman R = 0.28, P = 0.004, not shown), suggesting that mutation rate variation among loci may be an important factor. However, lineage-specific divergence estimates for synonymous sites and introns are not correlated between species (fig. 1C), suggesting that mutation rate variation is a minor contributor to correlated diversity levels in the two species. A second possibility is that some proteins are more frequent targets of recurrent selective sweeps (Andolfatto 2007; Macpherson et al. 2007). In fact, lineage-specific rates of protein evolution (dn) are highly correlated in the two species (fig. 1C, Spearman, R = 0.65, P = 8.8 × 10−14), and there is a significant negative correlation between levels of synonymous site diversity and dn in both species (fig. 1D). These results implicate genetic hitchhiking as a significant contributor to correlated diversity levels in the two species. In addition, we show that a significant fraction of polymorphism within species is shared between them (see below), suggesting that shared ancestral polymorphism also contributes to correlated levels of diversity in the two species.
FIG. 1.—
Levels of diversity at orthologous loci in D. melanogaster and D. simulans. Locus by locus estimates of (A) average pairwise diversity, π, and (B) the population mutation rate, θ. Panels A and B show P-values for the hypothesis that Dmel=Dsim using Wilcoxon Matched-pairs Signed ranks tests. Filled circles indicate 4-fold synonymous sites (Syn4f, 105 loci) and grey squares indicate short introns (21 loci). In both cases, diversity estimates are significantly positively correlated in the two species (two-tailed Spearman Rank Correlation test P-values are given in panel A). C. Lineage-specific 4-fold synonymous divergence (ds_4f, open circles) is not strongly correlated between species. Lineage-specific 0-fold nonsynonymous divergence (dn_0f, filled circles) is strongly correlated in the two species. P-values are from two-tailed Spearman Rank correlation tests. D. Synonymous site diversity (π) is negatively correlated with nonsynonymous divergence per site (dn) in both species (Dmel, filled circles; Dsim, open boxes). Synonymous site diversity estimates (π) have been corrected for ds using partial regression (Andolfatto 2007). The lines (black = Dmel; grey = Dsim) indicate a lowess fit to the data and P-values are from two-tailed Spearman Rank correlation tests.
Relative Diversity Levels in the Two Species
Under the neutral theory, levels of diversity in a species are directly proportional to the species effective population size, Ne. Average levels of X-linked diversity—as measured by synonymous site π—are virtually identical in the two species (table 1, fig. 1A), and there is no significant difference between the distributions (P = 0.96 Wilcoxon matched-pairs signed-ranks test). However, nucleotide diversity can also be measured from the number of polymorphic single nucleotide polymorphisms as θW (Watterson 1975). When this is done, there are clearly higher levels of synonymous site diversity in D. simulans relative to D. melanogaster (table 1 and fig. 1B). Drosophila simulans also has higher levels of variation when comparing diversities at short introns and nonsynonymous sites (table 1). Thus, despite the two species having similar synonymous site heterozygosities (π), D. simulans clearly harbors more polymorphism, suggesting a larger Ne for the X chromosome. Naively comparing synonymous site θW suggests a ∼1.4-fold larger Ne for the X chromosome in D. simulans (table 1, but see Discussion). Our short intron data (fig. 1 and table 1) suggests a ∼1.6-fold larger Ne for the X chromosome in D. simulans.
Table 1.
Summary of Diversity Levels at Homologous Loci in Drosophila melanogaster and D. simulans
| Measure (Sites) | # Sites | D. melanogaster Lineagea | D. simulans Lineagea | Dsim/Dmel | P Valueb |
| π (long introns)c | 3,849/3,754d | 1.61 | 1.22 | 0.76 | >0.1 |
| θ (long introns)c | 3,849/3,754d | 1.69 | 2.17 | 1.29 | >0.1 |
| π (Syn4f) | 11,048 | 2.21 | 2.19 | 1.33 | 0.046 |
| θ (Syn4f) | 11,048 | 2.41 | 3.45 | 1.43 | 2.6 × 10−7 |
| π (short intron) | 1,167 | 2.21 | 2.94 | 1.33 | 0.046 |
| θ (short intron) | 1,167 | 2.39 | 3.79 | 1.59 | 3.4 × 10−3 |
| π (Nonsyn0f) | 42,629 | 0.12 | 0.17 | 1.38 | 0.024 |
| θ (Nonsyn0f) | 42,629 | 0.19 | 0.28 | 1.51 | 3.3 × 10−4 |
Weighted average × 100.
P value determined by a Wilcoxon matched-pairs signed-rank test.
Previously published data (Glinka et al. 2003; Haddrill et al. 2008).
Number of sites in D. melanogaster and D. simulans, respectively.
The difference in patterns for synonymous π and θ are explained by a general skew toward rare variants in D. simulans relative to D. melanogaster, which may reflect stronger purifying selection in D. simulans, differences in demography, or both. However, demography should affect synonymous sites and short introns similarly if both are neutral. Diversity (π) is reduced at synonymous sites and long introns relative to short introns in D. simulans but only long introns have reduced diversity relative to short introns in D. melanogaster (table 1). This suggests that although purifying selection may reduce synonymous site diversity in the D. simulans lineage, synonymous sites are less strongly constrained in the D. melanogaster lineage, consistent with a smaller Ne for the X chromosome of the latter species. Considering the population frequencies of polymorphisms also support this conclusion. Notably, short intron polymorphisms are at a significantly higher frequency than synonymous sites in D. simulans (table 3 and supplementary S1, Supplementary Material online) but not in D. melanogaster, also consistent with weaker purifying selection and a smaller Ne for the X chromosome of the latter species.
Table 3.
Mean Frequencies (Number) of Polymorphisms by Class
| Site Class | Drosophila melanogaster | D. simulans |
| Syn (4f) | 0.248 (911) | 0.138a (1326) |
| No change (4f) | 0.284 (240) | 0.131a (371) |
| PU(4f) | 0.234b (580) | 0.123ac (738) |
| UP(4f) | 0.237 (91) | 0.201d (217) |
| Nonsyn (0f) | 0.179e (243) | 0.136a (365) |
| Intron (short) | 0.254 (93) | 0.175 (157) |
NOTE.—The expected frequency under neutral equilibrium is 0.268. All classes are significantly lower than expected under neutrality (P < 0.01, by simulations), expect those that have been underlined in D. melanogaster.
Significantly lower than intron (Syn: P = 0.027; no change: P =7.8 × 10−5; Nonsyn: P = 3 × 10−4).
Significantly lower than no change (P = 0.01, two-tailed Wilcoxon test).
Significantly lower than the no change class (P = 0.04).
Significantly higher than the no change class (P = 2.4 × 10−5).
Significantly lower than the no change class (P = 0.04).
Levels of Constraint on Proteins
An obvious signature of widespread purifying selection on proteins is reduced rates of evolution at nonsynonymous sites relative to synonymous sites (Kimura 1983). Constraint on protein sequences is most often measured as the ratio dn/ds—the rate of amino acid divergence per site scaled by the local rate of synonymous (putatively neutral) divergence (Kimura 1983). Table 2 shows that the mean of dn/ds is somewhat higher in the D. simulans lineage, although the difference between lineages is not significant. This is somewhat unexpected given the evidence for a larger Ne for the X chromosome in D. simulans relative to D. melanogaster based on levels of diversity (above).
Table 2.
Constraint on Proteins on the X Chromosome of Drosophila melanogaster and D. simulans
| Measure (Sites) | D. melanogaster Lineagea | D. simulans Lineagea | Dsim/Dmel | P Valueb |
| dn(Nonsyn0f)/ds(Syn4f)c | 0.153 | 0.279 | 1.83 | 0.09 |
| dn(Nonsyn0f) % | 0.92 | 1.00 | 1.08 | >0.1 |
| ds(Syn4f) | 7.02 | 4.86 | 0.69 | 4.7 × 10−5 |
| π(Nonsyn0f)/π(Syn4f)d | 0.096 | 0.108 | 1.13 | >0.1 |
| θw(Nonsyn0f)/θw(Syn4f)d | 0.097 | 0.102 | 1.06 | >0.1 |
Note.—A Jukes–Cantor correction has been applied to dn and ds.
Weighted averages across 104 loci for which π, θ, or d > 0 in both Dmel and Dsim.
P value determined by a Wilcoxon matched-pairs signed-rank test.
Excluding one locus no synonymous divergence in Dmel.
Excluding four loci with no synonymous polymorphism (two in Dmel and two in Dsim).
Contrasting dn/ds between species, however, suffers from two problems. First, it depends on the assumption of neutrality of synonymous sites, which is almost certainly violated in Drosophila (Akashi 1995; McVean and Vieira 2001; Nielsen et al. 2007). In accordance with previous studies (Akashi 1995, 1996; Begun et al. 2007), we find ds to be significantly lower in the D. simulans lineage relative to D. melanogaster (table 2), consistent with stronger purifying selection acting on synonymous sites in D. simulans. This interpretation is strengthened by the observation that the same asymmetry in evolutionary rates is not seen for short introns (see table 1 of Parsch et al. 2010). Because we have sampled homologous coding regions in the two species, we can directly compare dn in the two lineages to look for differences in levels of constraint. Surprisingly, we see very similar mean dn in the two lineages (table 2), implying that the level of constraint on nonsynonymous sites is similar in the two species. The number of amino acid substitutions in each lineage (0f-sites: 291 Dmel: 279 Dsim and for all amino acid substitutions: 355 Dmel: 340 Dsim) is also not significantly different. Thus, the somewhat elevated dn/ds in the D. simulans lineage is caused by reduced ds rather than elevated dn.
A second problem with considering rates of amino acid divergence is that a significant fraction of this divergence may be positively selected in Drosophila (McDonald and Kreitman 1991; Fay et al. 2002; Smith and Eyre-Walker 2002; Sella et al. 2009), complicating the interpretation of “constraint.” One way to minimize this problem is to contrast levels of within-species diversity at nonsynonymous versus synonymous sites, assuming that the majority of surveyed nonsynonymous polymorphisms are either neutral or deleterious. Interestingly, we find that levels of constraint measured both as π(Nonsyn)/π(Syn) and θW(Nonsyn)/θW(Syn) are also quite similar in the two species (table 2). This implies that selection is similarly efficient in the two species at eliminating deleterious amino acid polymorphisms, which is surprising given the evidence for a larger Ne for the X chromosome in D. simulans. However, if only a small proportion of newly arising amino acid mutations are in the nearly neutral range (as suggested by π[Nonsyn]/π[Syn] in table 2), the population dynamics of the majority of nonsynonymous diversities may not be affected much by a 1.5-fold difference in Ne between species (see below).
Another approach to detecting constraint due to purifying selection is to consider the population frequencies of polymorphic mutations. Negative selection is expected to decrease the average frequency of deleterious amino acid polymorphisms relative to neutral ones. However, because demographic events, such as population growth or bottlenecks, can also affect the mean frequencies of mutations, it is useful to compare the distribution of polymorphism frequencies with those at a putatively neutral class of sites, such as synonymous sites or short introns.
In D. melanogaster, the mean frequency of synonymous polymorphisms is significantly lower than expected under neutrality (expected frequency under neutrality = 0.268, P < 0.004, by simulation, see Materials and Methods). Despite this, the mean frequency of amino acid polymorphisms is significantly lower than synonymous and intron polymorphisms (table 3 and supplementary S1, Supplementary Material online), consistent with purifying selection on nonsynonymous sites. Lower than expected polymorphism frequencies at synonymous sites may reflect demography (population expansion), selection at linked sites or purifying selection on the sites themselves. We fail to detect a significant difference in polymorphism frequencies for synonymous sites and introns (P = 0.385, Wilcoxon test), implying that demographic causes for the negative skew at synonymous sites cannot be ruled out.
In D. simulans, the situation is somewhat different (table 3 and supplementary S1, Supplementary Material online). All classes of polymorphisms in this species are at significantly lower frequencies than expected under neutrality (P < 1 × 10−4, by simulation), suggesting population expansion or purifying selection. Despite this chromosome-wide trend, intron polymorphisms are at significantly higher frequencies than both nonsynonymous and synonymous polymorphisms, suggesting purifying selection contributes to lower frequencies at the latter two classes of sites (see also Haddrill et al. 2008). The pattern at synonymous sites with respect to putative codon fitness classes will be more closely examined below.
Although evidence for purifying selection on nonsynonymous sites in both species (and synonymous sites in D. simulans) is clear from these analyses, a goal of this study is to quantify and compare the intensity of selection on deleterious mutations in the two species. This comparison is particularly difficult given the possibility of different demographic histories for the two species, as hinted by the stronger genome-wide skew toward lower allele frequencies at all classes of sites in D. simulans. Keightley and Eyre-Walker (2007) have introduced a method to estimate the distribution of fitness effects (DFEs) of newly arising mutations under a specific model of demographic history (a two-epoch population size change) that is coestimated from the data. We applied this method to our polymorphism and divergence data in D. melanogaster and D. simulans, and the results are shown in figure 2 and supplementary S2 (Supplementary Material online).
FIG. 2.—
The inferred distribution of fitness effects of newly arising nonsynonymous mutations in the D. melanogaster (Dmel) and D. simulans (Dsim) lineages. The reference sites used for demographic inference are given in parentheses. Values in each category of Ne are calculated by integrating a gamma distribution with parameters in Supplement S2.1, where Ne is the weighted average of population size along the lineage. The estimates of Keightley and Eyre-Walker (2007) using the Zimbabwe subsample of D. melanogaster data of Shapiro et al. (2007) are shown for comparison (white bars). 95% confidence limits are based one 200 replicate bootstraps of the data by locus with replacement (see Methods).
Using this approach, we estimate that the average intensity of selection on newly arising deleterious nonsynonymous mutations in the two species is higher in D. simulans (table 4). Particularly, when short introns are used as neutral reference sites, our estimates of the mean selection intensity on deleterious mutations, N × E(s), are ∼2-fold (though not significantly) higher in D. simulans, which is consistent with the inferred difference in Ne for the two species (table 1). However, estimates of the mean selection coefficient can be misleading given the shape of the inferred DFE of newly arising mutations (fig. 2). Interestingly, the estimated fraction of newly arising mutations falling in the nearly neutral range (i.e., −10 < Nes < 0) is inferred to be remarkably similar in the two species (∼7% in D. melanogaster and ∼6% in D. simulans). These estimates predict similar patterns of protein evolution in the two species, consistent with our findings of quite similar levels of constraint at nonsynonymous sites (table 2).
Table 4.
Estimates of N × E(s) in Drosophila melanogaster and D. simulans
| Species | Selected Sites | Reference Sites | N × E(s) | 95% CI |
| D. melanogaster | NonsynOf | Syn4f | 1,202 | 468–3,686 |
| NonsynOf | Short intron | 912 | 229–12,914 | |
| Syn4f | Short intron | 0.13 | 0.0–85 | |
| PU_Syn4f | Short intron | <0.1 | 0.0–0.4 | |
| D. simulans | NonsynOf | Syn4f | 8601 | 1,521–453,800 |
| NonsynOf | Short intron | 1,787 | 317–32,338 | |
| Syn4f | Short intron | 2.7 | 0.7–6.8 | |
| PU_Syn4f | Short intron | 2.9 | 1.0–4.0 |
Note.—N × E(s) is the estimated mean selection coefficient scaled by the weighted average of N under the estimated demographic model (see supplementary S2, Supplementary Material online).
Protein Divergence Excess Relative to Neutral Expectations
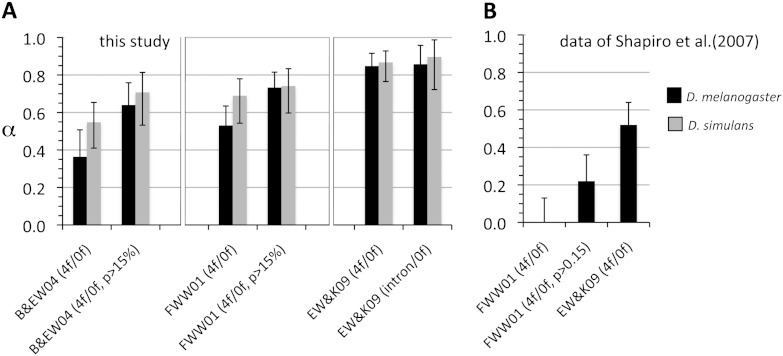
A large fraction of nonsynonymous divergence between species in the D. melanogaster species group is in excess of neutral expectations, a pattern that has been interpreted as the product of recurrent positive selection (Sella et al. 2009). Here, we compare levels of nonsynonymous divergence excess relative to neutrality, α, at 100 homologous coding regions in the D. melanogaster and D. simulans lineages. Using the maximum-likelihood approach of Bierne and Eyre-Walker (2004), 35–50% of nonsynonymous divergence is estimated to be in excess of neutral expectations in both species, with moderately higher estimates in the D. simulans lineage (fig. 3). Similarly, estimates of α using the estimator proposed by Fay et al. (2001) are 50–70% in D. melanogaster and, again, somewhat higher in D. simulans.
FIG. 3.—
Estimates of the fraction of nonsynoymus divergence excess relative to neutral expectations (a) in the D. melanogaster (black) and D. simulans (gray) lineages. B&EW: method of Bierne & Eyre-Walker (2004); FWW01: method of Fay et al. (2001); EW&K09: method of Eyre-Walker and Keightley (2009); 4f: four-fold synonymous sites; 0f: nondegenerate nonsynonymous sites; intron: short introns. The Keightley and Eyre-Walker (2007) estimates for the Zimbabwe subsample of the Shapiro et al. (2007) data set are shown in panel B. Note that the latter estimates use D. melanogaster–D. simulans divergence, rather than lineage-specific divergence.
It has been noted that including slightly deleterious mutations in the count of polymorphisms is likely to bias estimates of α downward (Charlesworth 1994; Fay et al. 2001; Charlesworth and Eyre-Walker 2008). The exclusion of low-frequency polymorphisms may remedy this to some extent by preferentially removing deleterious mutations (Fay et al. 2001). In figure 3, we see that estimates of α are much higher using the methods of Bierne and Eyre-Walker (2004) and Fay et al. (2001) if polymorphisms at <15% frequency are excluded, suggesting that segregating deleterious mutations are a factor biasing estimates downward.
However, the exclusion of low-frequency polymorphisms is an imprecise approach to correcting for the bias caused by segregating deleterious mutations and comes with a cost to statistical power (Charlesworth and Eyre-Walker 2008; Eyre-Walker and Keightley 2009). Perhaps even more of a concern, several factors are expected to bias α upward, including pooling data from regions of the genome that experience different levels of selective constraint (McDonald and Kreitman 1991; Smith and Eyre-Walker 2002; Welch 2006; Shapiro et al. 2007) and purifying selection acting on synonymous or other chosen neutral reference sites (Akashi 1995; Eyre-Walker 2002). Finally, the inference that the divergence excess is the product of positive selection depends on the assumption of a constant population size over time. Several authors have pointed out that changes in population size over time can lead to a greater accumulation of slightly deleterious mutations than expected based on current polymorphism patterns (McDonald and Kreitman 1991; Ohta 1993; Fay and Wu 2000; Eyre-Walker 2002).
Although several solutions have been proposed to address some of these problems, the most comprehensive solution to date is the approach proposed by Eyre-Walker and Keightley (2009), which explicitly accounts for deleterious mutations and demography by jointly estimating the DFE, a demographic model and α. Selection on synonymous sites will bias estimates of demography (based on reference site allele frequencies) as well as α (based on the ratio of polymorphism to divergence at reference sites). To deal with this issue as best we can, we compare estimates using synonymous sites as a reference with those using short introns as reference sites. Using this approach, we estimate α ∼85–90%, and the estimates are remarkably similar in the D. melanogaster and D. simulans lineages (fig. 3). These estimates of α are among the highest estimated in Drosophila and are considerably higher compared with a previous analysis of similar data by Eyre-Walker and Keightley (2009). This is likely due to the fact that we estimate fewer nearly neutral mutations (fig. 2) and more recent growth in D. melanogaster (supplementary S2, Supplementary Material online), implying a smaller proportion of neutral and slightly deleterious nonsynonymous substitutions to nonsynonymous divergence. This difference is discussed in greater detail below.
Patterns of Codon Usage Bias
Previous studies have suggested reduced selection to maintain codon usage bias in D. melanogaster relative to D. simulans, consistent with the former having a smaller historical Ne (Akashi 1995, 1996). McVean and Vieira (2001) and Nielsen et al. (2007) concluded that the D. melanogaster lineage shows little or no evidence for selection maintaining codon usage bias (but see Zeng and Charlesworth 2009). These previous studies have largely been based on small sets of genes, small samples of individuals, and combined genes from very different chromosomal contexts (i.e. X vs. autosome, high vs. low recombination, etc). Here, we reevaluate evidence for current and historical selection for codon usage bias in the two species in a larger data set (100 loci) with deeper sampling (n = 20 individuals) for each species.
Under one of the simplest models of selection to maintain codon bias (e.g., Bulmer 1991; Akashi 1995), synonymous codon changes can be classified into several putative fitness classes: Preferred to unpreferred (P->U) which are presumed to be deleterious; unpreferred to preferred (U->P) which are presumed to be advantageous and two “no change” classes (i.e., P->P and U->U), which are presumed to be closer to neutral than the U->P and P->U classes. Here, we examine several population genetic patterns in the context of these three putative codon change fitness classes (see Materials and Methods).
Consistent with reduced selection on codon usage bias in D. melanogaster, a larger fraction of polymorphic synonymous changes fall into the deleterious class; that is, 64% of synonymous polymorphisms are P->U, whereas 10% are U->P. In D. simulans, these proportions are 56% and 16%, respectively and highly significantly different than for D. melanogaster (P < 3.4 × 10−6, two-tailed Fisher’s Exact test). Figure 4A shows levels of diversity in D. simulans relative to D. melanogaster for the three classes of codon change. The diversity difference between D. melanogaster and D. simulans is most apparent for the U->P class and least apparent for P->U class, consistent with stronger selection on codon usage bias in the larger D. simulans lineage. In addition, the level of diversity (π) for the U->P class is 1.5-fold higher relative to the no change class in D. simulans (P = 0.009, paired Wilcoxon test), consistent with positive selection increasing diversity at this putatively advantageous class of changes relative to neutral sites (see Kimura 1983, p. 44).
FIG. 4.—
Analysis of 4-fold synonymous sites by codon change class. (A) Relative diversity in the two species. Paired Wilcoxon test P-value levels of equal diversity in Dmel and Dsim (dashed line) are: *P<2e-4; **P<2e-5; ***P<2e-7. (B) The ratio of divergence to polymorphism (D/P). Mantel-Haenzel test (with continuity correction) P-values versus the UU+PP (no change) class are P=8.5e-5 for Dmel and P=2.2e-6 for Dsim. All of the same patterns are evident when all synonymous sites are used (not shown).
Selection on U->P and P->U mutations is expected to alter the ratio of divergence to polymorphism (D/P, Akashi 1995). Figure 4B shows the D/P ratio for the three putative fitness classes of codon changes. The D/P ratio is significantly lower for the P->U class of changes compared with the no change class in both lineages (Mantel–Haenszel test, P < 8 × 10−5 and P < 2 × 10−6 for the D. melanogaster and D. simulans, respectively). This is consistent with historical purifying selection in both lineages against P->U changes. Curiously, the D/P ratio is not significantly higher for U->P changes relative to the no change class in either lineage, although there is a slight trend in that direction in both species.
A comparison of population frequencies of polymorphisms provides a window on more recent selection maintaining codon usage bias in the two species (table 3). In D. melanogaster, P->U polymorphisms are at significantly lower frequency than no-change polymorphisms. This is consistent with purifying selection acting on P->U mutations and is consistent with results based on the comparison of D/P ratios (above). In D. simulans, there is a strong skew toward rare polymorphisms for all classes compared with D. melanogaster. Despite this general pattern, P->U polymorphisms in D. simulans are also at significantly lower frequency than intronic and no change polymorphisms (table 3), consistent with ongoing purifying selection on P->U mutations.
The evidence for selection on U->P mutations based on polymorphism frequencies is less clear. In D. simulans, U->P polymorphisms are at significantly higher frequency than the no change class (table 3), which is consistent with positive selection on U->P mutations or negative selection on the no change class. However, frequencies for no change polymorphisms are significantly lower than intron polymorphisms (table 3), suggesting that some fraction of no change mutations may themselves be negatively selected in D. simulans. U->P mutations are at higher mean frequency than intron polymorphisms in D. simulans, but the difference is not significant.
The above analyses of patterns of polymorphism and divergence show that historical selection on P->U mutations is detectable in both the D. melanogaster and D. simulans lineages. To estimate the intensity of this selection, we used the method of Keightley and Eyre-Walker (2007) to estimate the DFE at synonymous sites in both species using our data for short introns as neutral reference. Using this approach, we estimate N × E(s) = −2.7 (95% CI −0.71 to −6.8) for all synonymous sites and −2.9 (95% CI −1.0 to −4.0) for P->U changes in the D. simulans lineage (table 4). In contrast, N × E(s) estimates for all synonymous sites (−0.16) and P->U changes (∼0.00) in the D. melanogaster lineage are not significantly different from 0. The results suggest weaker selection on codon usage in D. melanogaster than in D. simulans, consistent with a larger Ne for the X chromosome of the latter species.
Species Divergence Time and Shared Polymorphism
Using divergence along the D. melanogaster lineage, we estimate the species divergence time to be 7.1 (95% CI 6.1–8.3) Ne generations ago (using the estimator proposed by Hudson et al. [1987]). This divergence time implies considerable potential for a sample of 20 alleles to share a common ancestor prior to the speciation time—we estimate ∼8% of the time on the autosomes and 2% of the time on the X, by coalescent simulation. Incomplete lineage sorting should be apparent as “shared derived mutations” (defined as derived mutations that occur in both lineages). Shared derived mutations can occur either by recurrent mutation to the same site or by shared ancestry (Clark 1997). In table 5, we present an analysis of the number of shared derived mutations in our sample of alleles from both species. We detected 102 shared polymorphisms and 158 shared derived mutations (∼12% of all mutations in the D. melanogaster lineage). We estimate that ∼50% (and at least 37%) of shared derived mutations are due to incomplete lineage sorting rather than recurrent mutation. Thus, this implies that a substantial fraction of polymorphisms (∼6%) within both species were also segregating as polymorphisms in the ancestor of the two species.
Table 5.
Private and Shared Derived Mutations at 4-Fold and 0-Fold Sites
| Mutation Class | Drosophila melanogaster | D. simulans | Expected (95% CI) |
| 4-fold Syn | |||
| Private polymorphisms | 830 | 1,271 | — |
| Shared polymorphisms | 80 | 80 | 47 (35–60) |
| Private fixations | 380 | 255 | — |
| Private deriveda | 1,210 | 1,472 | — |
| Shared derivedb | 158 | 158 | 84 (66–104) |
| 0-fold Nonsyn | |||
| Private polymorphisms | 239 | 353 | — |
| Shared polymorphisms | 6 | 6 | 1.5 (0–4) |
| Private fixations | 252 | 248 | — |
| Private deriveda | 491 | 601 | — |
| Shared derivedb | 15 | 15 | 6.3 (2–12) |
Note.—Based on the analysis of 10,603 4-fold sites. Expected numbers of shared mutations due to multiple hits are based on 104 simulated replicates (see Materials and Methods).
The sum of polymorphic and fixed mutations specific to one lineage.
All mutations found in both lineage, including those that are polymorphic in one lineage but fixed in the other.
Discussion
Measuring Relative Effective Population Sizes at Sites Under Weak Selection
We have found that D. melanogaster harbors lower levels of X-linked polymorphism than D. simulans, consistent with a ∼50% larger Ne for the X chromosome of the latter species. These conclusions appear to contradict those of Nolte and Schlotterer (2008), who failed to find a significant difference in levels of variability in the two species. Andolfatto (2001) also noted similarity in levels of diversity on the X chromosome of the two species. However, both studies were based on a survey of a very small number of genomic regions, which may have afforded them little power to detect differences. In addition, the particular choice of loci of Nolte and Schlotterer (2008) (i.e., mostly long introns and intergenic regions) are likely under considerable levels of selective constraint in both species (Andolfatto 2005; Haddrill et al. 2008), and this may have further limited their ability to detect a difference in population size between the species. As an illustration, we analyzed previously published data for nine orthologous long intron regions in the two species (Glinka et al. 2003; Haddrill et al. 2008), which is similar in scale and size to the data set presented by Nolte and Schlotterer (2008). In concordance with their study, we find no significant difference in levels of diversity at long introns in the two species (table 1).
This result is not unexpected when comparing sites in two species that are under weak selection. In figure 5, we show that if 2Nes < −1 for most noncoding DNA in D. melanogaster (as suggested by the frequencies of polymorphisms in noncoding DNA, see Andolfatto 2005), there may be little power to detect a 2-fold difference in population size between the species if one existed. This provides a possible explanation for the fact that we observe a significant difference in diversity levels between the species at synonymous sites and short introns, particularly if selection is weaker for these classes of sites than at long introns (Haddrill, Charlesworth, et al. 2005; Parsch et al. 2010). This also implies that although “measured” levels of variability suggest a ∼50% difference in Ne of the X chromosome for the two species, this difference could be an underestimate given that the surveyed synonymous and short intronic sites are themselves likely to be under weak selection in one or both species. It is important to note that since we have surveyed only X-linked loci, our estimates of the relative effective population sizes may not necessarily apply to the autosomes.
FIG. 5.—
The effect of weak selection on the expected relative levels of diversity in two species with different population sizes. The x-axis corresponds to the intensity of selection in the species with the smaller population size (N1). The Y-axis plots expected relative levels of diversity in the two species. In red and purple are relative θW and π, respectively, in species with a 1.5-fold difference in population size. In blue and green are analogous expectations for a 2-fold difference in population size. Graphs are based on simulations of the Poisson Random Field model (Sawyer and Hartl 1992) using code kindly provided by C. Bustamante.
Understanding the Difference between Synonymous and Nonsynonymous Sites
A larger historical Ne for D. simulans than D. melanogaster predicts higher levels of constraint on synonymous and nonsynonymous sites in the former species. Although this prediction is largely borne out for synonymous sites, we see little evidence for differences in constraint on nonsynonymous sites in the two species, despite estimates of the mean intensity of selection on nonsynonymous sites that are ∼2× higher in D. simulans. How do we reconcile these findings? The key to understanding patterns of constraint at synonymous and nonsynonymous sites may boil down to the inferred DFE, which suggests that only a small fraction of newly arising nonsynonymous mutations are nearly neutral (−10 < Nes <0) and that the fraction of mutations in this range is similar for the two species. In contrast, synonymous sites are more weakly selected on average, and we estimate that virtually all newly arising synonymous mutations fall into the nearly neutral range.
Contrary to several previous studies (McVean and Vieira 2001; Nielsen et al. 2007), we detect significant evidence for historical selection on 4-fold synonymous P->U codon changes in the D. melanogaster lineage (i.e., the fixation index is significantly lower for P->U changes relative to the no change class). We also note that P->U polymorphisms are at significantly lower frequencies than no change synonymous polymorphisms in D. melanogaster. These findings, based on 4-fold degenerate synonymous sites, largely agree with a recent study by Zeng and Charlesworth (2009), who detect significant evidence for recent selection on 2-fold degenerate synonymous sites using polymorphism data from D. melanogaster. However, Zeng and Charlesworth (2009) also conclude that there is little evidence for population growth in D. melanogaster. In contrast to their study, which only used synonymous sites, we detect significant evidence for recent growth in D. melanogaster using data from short introns (supplementary S2, Supplementary Material online). This difference between the two studies may partly explain the fact that our estimate of the average intensity of selection on synonymous sites in D. melanogaster is smaller than that estimated by Zeng and Charlesworth (2009). Although we do detect evidence for historical selection on P->U synonymous mutations in D. melanogaster, our estimates of the mean selection intensity are lower than estimates for the D. simulans lineage.
High Estimates of “Adaptive” Protein Divergence in Both Lineages
We estimated the extent of protein divergence excess relative to neutral expectations (α) in the two species lineages using several approaches. Regardless of the method used, estimates of α appear to be consistently high in both species, although marginally higher in the D. simulans lineage. The similarity in estimates of α along these two independent lineages is surprising given the evidence for different demographic histories in the two species and suggests that the inferred protein divergence excess in these two lineages is less likely to be caused by demographic factors (see below). Several authors have noted that changes in population size may account for greater than expected divergence at nonsynonymous sites (Ohta 1993; Fay and Wu 2001; Eyre-Walker 2002). We have found that estimates of α remain high even if we account for recent 10-fold growth in both species. To substantially underestimate the fraction of protein divergence due to accumulating slightly deleterious mutations, the past population sizes of both species would have to have been considerably smaller than estimated for an appreciable amount of time. Interestingly, our finding of a substantial amount of shared polymorphism in the two species limits the extent to which the two species could have suffered drastic reductions in population size (Clark 1997), and we propose that this information could be used, in principle, to put limits on the extent of accumulation of slightly deleterious mutations. Our findings lend credence to the claim that a large fraction of protein divergence in Drosophila is indeed the product of recurrent positive selection (Sella et al. 2009) rather than a mere artifact of demography (Hughes 2007).
This said, the approaches we have used also have their limitations (Keightley and Eyre-Walker 2007). In particular, they assume that the vast majority of nonsynonymous mutations are unconditionally deleterious or advantageous, all nonsynonymous polymorphisms are deleterious and that positively selected nonsynonymous mutations are rare and strongly selected. Although this is probably a reasonable first approximation, more complicated dynamics for a significant fraction of nonsynonymous mutations, such as population structure (local adaptation), balancing selection or very weak positive selection, may have obscured a difference in the inferred efficacy of selection on nonsynonymous sites in the two species. In addition, inferences of selection on nonsynonymous sites depend on the choice of neutral reference sites. Selection on synonymous sites, in D. simulans in particular, may limit our ability to accurately quantify selection on nonsynonymous sites by obscuring evidence for purifying selection and inflating estimates of adaptive evolution. Finally, the demographic model assumed is a simple two-epoch population size model, which is likely to be quite an abstraction of the true demographic model for the species.
Keightley and Eyre-Walker (2007) introduced their approach with an analysis of population genetic data for 397 autosomal protein-coding loci in D. melanogaster produced by Shapiro et al. (2007). At first glance, the inferred DFE for our data set and the Shapiro et al. data set look similar (fig. 2). However, there are a number of key differences in both the inferred demographic model and the corresponding DFE. In particular, we estimate more recent and less extreme growth (N2/N1 = 10, t/N2 = 0.014, see supplementary A2, Supplementary Material online) than estimated for the Shapiro et al. data (N2/N1 = 20, t/N2 = 2.4, see table 4 of Keightley and Eyre-Walker 2007). In addition, we estimate the fraction of newly arising mutations for which −1< Nes < 0 to be about 2× smaller than for the Shapiro et al. data set. In fact, Keightley and Eyre-Walker’ (2007) estimate of the fraction of mutations for which −1 < Nes < 0 in the autosomal data of Shapiro et al. (0.06, see fig. 2) lies outside the 95% CI of our estimate (0.02) for the X chromosome (P < 0.005). Similarly, Keightley and Eyre-Walker’ (2007) estimate of α on the autosomes (0.52, see fig. 3) lies outside the 95% CI of our X chromosome estimate (0.85, P < 0.005).
The difference in these estimates may reflect any one, or a combination, of potentially important factors. Key among these may be an X-autosome difference in the efficacy of selection. The data of Shapiro et al. are autosomal, whereas our data are X linked. The ratio of nonsynonymous/synonymous polymorphism is elevated on the D. melanogaster autosomes relative to the X chromosome (Begun 1996; Andolfatto 2001), a pattern that might indicate less efficient purifying selection on autosomes due to recessivity of deleterious nonsynonymous mutations (Charlesworth et al. 1983; Andolfatto 2001). Common inversion polymorphisms in D. melanogaster, which are autosomal, may also increase the scale of genetic hitchhiking (by reducing the population recombination rate) thus further reducing the efficacy of selection relative to the X (Andolfatto 2001). Along similar lines, regions of highly reduced recombination in the Drosophila genome exhibit elevated levels of nonsynonymous polymorphism, consistent with reduced efficacy of selection in these regions (Presgraves 2005; Betancourt et al. 2009). Thus, an additional factor contributing to the difference in our estimates could be the local recombination rate, which varies considerably among loci in the Shapiro et al. study. In contrast, the loci surveyed in our study were chosen to maximize recombination rate and minimize rate variation. Finally, the X-autosome difference we observe may also simply reflect differences in gene ontology or expression pattern. Distinguishing between these hypotheses and a systematic X-autosome difference in the efficacy of selection may be possible using genome-wide polymorphism and divergence data.
What Drives Protein Evolution in Drosophila?
We have estimated surprisingly high levels of adaptive divergence in a collection of 100 X-linked protein coding loci (α = 0.86 and α = 0.90 for D. melanogaster and D. simulans, respectively, using short introns as reference sites, see fig. 2). Given that this set of coding regions was randomly chosen with respect to gene function, we can conclude that adaptive turnover of proteins in the Drosophila genome must be common and adaptation does not appear to be restricted to a special subset of genes. This view seems to be somewhat at odds with recent studies that have documented high rates of adaptive evolution, using similar approaches to those employed here, associated with immunity (Schlenke and Begun 2003; Lazzaro 2008; Obbard et al. 2009), reproduction and sexual conflict (Begun et al. 2000; Kern et al. 2004; Proschel et al. 2006), and intragenomic conflict (Presgraves 2007).
Intriguingly, at the 11 genes in our data set with Gene Ontology terms “defense response,” “phagocytosis,” “reproduction,” or “nuclear pore,” we estimate α = 0.95 in D. melanogaster which is significantly higher than random subsets of the data (P < 0.005, by bootstrapping with replacement). The estimate for this subset in D. simulans is also high (α = 0.93) but not significantly higher than random subsets of the data. Despite high estimates for this subset of loci, we estimate α =0.84 and α = 0.89 for the remaining 89 loci in D. melanogaster and D. simulans, respectively, suggesting adaptive divergence is nonetheless widespread among other GO categories.
We also investigated the influence of sex-biased gene expression (Gnad and Parsch 2006; Proschel et al. 2006) on our estimate from D. melanogaster. Specifically, we estimated α = 0.88 for the 11 genes with the highest male-biased expression (male/female [M/F] > 1.4) and α = 0.93 for the 11 genes with the highest female-biased expression (M/F < 0.47), and both are significantly higher than random subsets of the data (P < 0.014 and P < 0.005, respectively, by bootstrapping with replacement). In contrast, we estimate α = 0.62 at the 11 genes with the lowest bias (0.9 < M/F < 1.1 and excluding two loci with the GO term defense response), and this was not significantly lower than random subsets of the data.
Estimates of α inform us about the fraction of divergence that is adaptive but not the rate of adaptive protein evolution per se. Comparing estimates of the number of adaptive substitutions per nonsynonymous site (α × dn) among categories of genes suggests that, despite significantly higher estimates of α for the 11 immunity/reproduction/conflict proteins in D. melanogaster, the inferred rate of adaptive substitution is actually comparable with the remaining 89 proteins (supplementary S3, Supplementary Material online). In contrast, the significantly higher α estimates for the male- and female-biased sets of proteins translate to ∼2-fold higher rates of adaptation than the unbiased set of proteins (supplementary S3, Supplementary Material online). These findings support the notion that sex-biased gene expression, and potentially sexual conflict, is a major determinant of protein divergence in Drosophila (Proschel et al. 2006). This said, by our estimates, a large fraction of adaptive protein divergence (>50%) is also estimated at proteins with no sex bias in expression and no obvious role in immunity, reproduction, sexual, and/or intragenomic conflict, implying that adaptive turnover of proteins is widespread among biological functions.
Supplementary Material
Supplementary S1–S4 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
Thanks to two anonymous reviewers for helpful comments on the manuscript and Dan Davison, Molly Przeworski, Kevin Thornton, and Guy Sella for helpful discussions. Thanks to Kevin Thornton, Dan Davison, and Carlos Bustamante for help with code. This work was funded in part by National Institute of Health grant R01-GM083228 (to P.A.) and by grants R01-GM076007 and R01-GM093182 (to D.B).
References
- Akashi H. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics. 1996;144:1297–1307. doi: 10.1093/genetics/144.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Schaeffer S. Natural selection and the frequency distributions of “silent” DNA polymorphism in Drosophila. Genetics. 1997;146:295–307. doi: 10.1093/genetics/146.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol. 2001;18:279–290. doi: 10.1093/oxfordjournals.molbev.a003804. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P, et al. Inversion polymorphisms and nucleotide variability in Drosophila. Genet Res. 2001;77:1–8. doi: 10.1017/s0016672301004955. [DOI] [PubMed] [Google Scholar]
- Aquadro CF, et al. The rosy region of Drosophila melanogaster and Drosophila simulans. I. Contrasting levels of naturally occurring DNA restriction map variation and divergence. Genetics. 1988;119:875–888. doi: 10.1093/genetics/119.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulard S, et al. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genet Res. 2002;79:49–63. doi: 10.1017/s0016672301005407. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. A dynamic view of sex chromosome evolution. Curr Opin Genet Dev. 2006;16:578–585. doi: 10.1016/j.gde.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Baudry E, et al. Non-African populations of Drosophila melanogaster have a unique origin. Mol Biol Evol. 2004;21:1482–1491. doi: 10.1093/molbev/msh089. [DOI] [PubMed] [Google Scholar]
- Baudry E, et al. Contrasted polymorphism patterns in a large sample of populations from the evolutionary genetics model Drosophila simulans. Genetics. 2006;173:759–767. doi: 10.1534/genetics.105.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ. Population genetics of silent and replacement variation in Drosophila simulans and D. melanogaster: X/autosome differences? Mol Biol Evol. 1996;13:1405–1407. doi: 10.1093/oxfordjournals.molbev.a025587. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in Drosophila melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature. 1993;365:548–550. doi: 10.1038/365548a0. [DOI] [PubMed] [Google Scholar]
- Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, et al. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics. 2000;156:1879–1888. doi: 10.1093/genetics/156.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AJ, et al. Reduced effectiveness of selection caused by a lack of recombination. Curr Biol. 2009;19:655–660. doi: 10.1016/j.cub.2009.02.039. [DOI] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- Bullaughey K, et al. No effect of recombination on the efficacy of natural selection in primates. Genome Res. 2008;18:544–554. doi: 10.1101/gr.071548.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casillas S, et al. Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol Biol Evol. 2007;24:2222–2234. doi: 10.1093/molbev/msm150. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The effect of background selection against deleterious mutations on weakly selected, linked variants. Genet Res. 1994;63:213–227. doi: 10.1017/s0016672300032365. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet Res. 1996;68:131–149. doi: 10.1017/s0016672300034029. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, et al. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1983;130:113–146. [Google Scholar]
- Charlesworth J, Eyre-Walker A. The McDonald–Kreitman test and slightly deleterious mutations. Mol Biol Evol. 2008;25:1007–1015. doi: 10.1093/molbev/msn005. [DOI] [PubMed] [Google Scholar]
- Choudhary M, Singh RS. A comprehensive study of genic variation in natural populations of Drosophila melanogaster. III. Variations in genetic structure and their causes between Drosophila melanogaster and its sibling species Drosophila simulans. Genetics. 1987;117:697–710. doi: 10.1093/genetics/117.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG. Neutral behavior of shared polymorphism. Proc Natl Acad Sci U S A. 1997;94:7730–7734. doi: 10.1073/pnas.94.15.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. Changing effective population size and the McDonald–Kreitman test. Genetics. 2002;162:2017–2024. doi: 10.1093/genetics/162.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, et al. Quantifying the slightly deleterious mutation model of molecular evolution. Mol Biol Evol. 2002;19:2142–2149. doi: 10.1093/oxfordjournals.molbev.a004039. [DOI] [PubMed] [Google Scholar]
- Fay J, et al. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. The neutral theory in the genomic era. Curr Opin Genet Dev. 2001;11:642–646. doi: 10.1016/s0959-437x(00)00247-1. [DOI] [PubMed] [Google Scholar]
- Fay JC, et al. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- Glinka S, et al. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics. 2003;165:1269–1278. doi: 10.1093/genetics/165.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:2577–2579. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
- Haddrill PR, Bachtrog D, Andolfatto P. Positive and negative selection on noncoding DNA in Drosophila simulans. Mol Biol Evol. 2008;25:1825–1834. doi: 10.1093/molbev/msn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P. Patterns of intron sequence evolution in Drosophila depend upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Halligan DL, Tomaras D, Charlesworth B. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Thornton KR, Charlesworth B, Andolfatto P. Multilocus patterns on nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 2005;15:790–799. doi: 10.1101/gr.3541005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 2006;16:875–884. doi: 10.1101/gr.5022906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, et al. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor C. Evolution of protein molecules. In: Munro MN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Kauer M, et al. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics. 2002;160:247–256. doi: 10.1093/genetics/160.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177:2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern AD, et al. Molecular population genetics of male accessory gland proteins in the Drosophila simulans complex. Genetics. 2004;167:725–735. doi: 10.1534/genetics.103.020883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kliman RM, Hey J. Reduced natural selection associated with low recombination in Drosophila melanogaster. Mol Biol Evol. 1993;10:1239–1258. doi: 10.1093/oxfordjournals.molbev.a040074. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP. Natural selection on the Drosophila antimicrobial immune system. Curr Opin Microbiol. 2008;11:284–289. doi: 10.1016/j.mib.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Loewe L, et al. Estimating selection on nonsynonymous mutations. Genetics. 2006;172:1079–1092. doi: 10.1534/genetics.105.047217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson JM, et al. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics. 2007;177:2083–2099. doi: 10.1534/genetics.107.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maside X, et al. Selection on codon usage in Drosophila americana. Curr Biol. 2004;14:150–154. doi: 10.1016/j.cub.2003.12.055. [DOI] [PubMed] [Google Scholar]
- McDonald J, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McVean GA, Vieira J. Inferring parameters of mutation, selection and demography from patterns of synonymous site evolution in Drosophila. Genetics. 2001;157:245–257. doi: 10.1093/genetics/157.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama EN, Powell JR. Intraspecific nuclear DNA variation in Drosophila. Mol Biol Evol. 1996;13:261–277. doi: 10.1093/oxfordjournals.molbev.a025563. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nielsen R, et al. Maximum likelihood estimation of ancestral codon usage bias parameters in Drosophila. Mol Biol Evol. 2007;24:228–235. doi: 10.1093/molbev/msl146. [DOI] [PubMed] [Google Scholar]
- Nolte V, Schlotterer C. African Drosophila melanogaster and D. simulans populations have similar levels of sequence variability, suggesting comparable effective population sizes. Genetics. 2008;178:405–412. doi: 10.1534/genetics.107.080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, et al. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 2009;5:e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi S, Voelker RA. Comparative studies of allozyme loci in Drosophila simulans and D. melanogaster. II. Gene arrangement on the third chromosome. Jpn J Genet. 1979;54:203–209. [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Ohta T. Amino acid substitution at the Adh locus of Drosophila is facilitated by small population size. Proc Natl Acad Sci U S A. 1993;90:4548–4551. doi: 10.1073/pnas.90.10.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, et al. On the utility of short intron sequences as a reference for the detection of positive and negative selection in Drosophila. Mol Biol Evol. 2010;27:1226–1234. doi: 10.1093/molbev/msq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, et al. Population genetic inference from genomic sequence variation. Genome Res. 2010;20:291–300. doi: 10.1101/gr.079509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadin K, et al. Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc Natl Acad Sci U S A. 2007;104:13390–13395. doi: 10.1073/pnas.0701256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Recombination enhances protein adaptation in Drosophila melanogaster. Curr Biol. 2005;15:1651–1656. doi: 10.1016/j.cub.2005.07.065. [DOI] [PubMed] [Google Scholar]
- Presgraves DC. Does genetic conflict drive rapid molecular evolution of nuclear transport genes in Drosophila? Bioessays. 2007;29:386–391. doi: 10.1002/bies.20555. [DOI] [PubMed] [Google Scholar]
- Proschel M, et al. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics. 2006;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–48. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. Natural selection drives Drosophila immune system evolution. Genetics. 2003;164:1471–1480. doi: 10.1093/genetics/164.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, et al. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA, et al. Adaptive genic evolution in the Drosophila genomes. Proc Natl Acad Sci U S A. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NG, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. The genetics of Drosophila simulans. Carnegie Inst Wash. 1929;399:1–62. [Google Scholar]
- Vicario S, et al. Codon usage in twelve species of Drosophila. BMC Evol Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Rand DM. Contrasting patterns of nonneutral evolution in proteins encoded in nuclear and mitochondrial genomes. Genetics. 156:385–399. doi: 10.1093/genetics/156.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ. Estimating the genomewide rate of adaptive protein evolution in Drosophila. Genetics. 2006;173:821–837. doi: 10.1534/genetics.106.056911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfit M, Bromham L. Population size and molecular evolution on islands. Proc Biol Sci. 2005;272:2277–2282. doi: 10.1098/rspb.2005.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Andolfatto P. The impact of natural selection on the genome: emerging patterns in Drosophila and Arabidopsis. Annu Rev Ecol Evol Syst. 2008;39:193–213. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zeng K, Charlesworth B. Estimating selection intensity on synonymous codon usage in a nonequilibrium population. Genetics. 2009;183:651–662. doi: 10.1534/genetics.109.101782. 651SI–1623SI. [DOI] [PMC free article] [PubMed] [Google Scholar]