Abstract
Biochemical systems biology augments more traditional disciplines, such as genomics, biochemistry and molecular biology, by championing (i) mathematical and computational modeling; (ii) the application of traditional engineering practices in the analysis of biochemical systems; and in the past decade increasingly (iii) the use of near-comprehensive data sets derived from ‘omics platform technologies, in particular “downstream” technologies relative to genome sequencing, including transcriptomics, proteomics and metabolomics. The future progress in understanding biological principles will increasingly depend on the development of temporal and spatial analytical techniques that will provide high-resolution data for systems analyses. To date, particularly successful were strategies involving (a) quantitative measurements of cellular components at the mRNA, protein and metabolite levels, as well as in vivo metabolic reaction rates, (b) development of mathematical models that integrate biochemical knowledge with the information generated by high-throughput experiments, and (c) applications to microbial organisms. The inevitable role bioinformatics plays in modern systems biology puts mathematical and computational sciences as an equal partner to analytical and experimental biology. Furthermore, mathematical and computational models are expected to become increasingly prevalent representations of our knowledge about specific biochemical systems.
1. Introduction
The term “systems biology” has emerged recently to describe the frontier of cross-disciplinary research in biology [1–5]. This term was propelled into the mainstream merely ten years ago [1–3], coinciding with the completion of the Human Genome Project (HGP) [6, 7] and the concomitant emergence of ‘omics technologies, namely transcriptomics [8, 9], proteomics [10], and metabolomics [11, 12]. However, the origins of modern systems biology can be traced back to the middle of last century [13–15], with history that is both conceptually complex and institutionally convoluted. For example, a general systems theory was developed and applied to biology in late 1960's [14, 15]. Independently, the theory of metabolic control was developed, and metabolic flux was recognized as a “systemic property” [16–18]. Here, we focus on the reemergence of “systems thinking” linked to the post-genomic era and the development of global molecular profiling methods collectively known as ‘omics technologies. The discussion of systems biology in the broader historical context can be found elsewhere (see, e.g., [4, 19] and references therein).
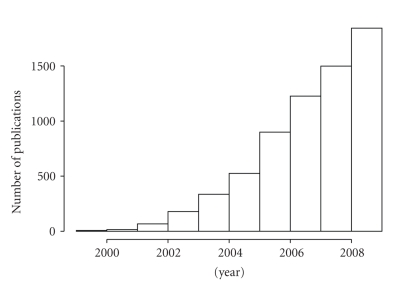
Interest in systems biology has increased rapidly in the past decade, as evidenced by the number of referencing publications (Figure 1). Systems biology has fuzzy boundaries and overlaps with several emerging, post-genomic fields, such as synthetic biology [20–24], systems microbiology [25], systems biotechnology [26, 27], integrative biology [26, 28], systems biomedicine [29], and metagenomics [25, 30]. Numerous definitions of systems biology have been proposed [1, 2, 4, 5, 31], but to date, there is no universally accepted definition—reflecting the difficulty in defining a heterogeneous school of thought by a comprehensive yet concise definition. Each of the proposed definitions, however, revolves around a fundamental understanding of biological systems based on the underlying component interactions (molecular interactions, in the case of biochemical systems biology). In a broad sense, the same is the goal of more traditional disciplines, such as molecular biology, genomics, and biochemistry. Hence, the question “what is new in Systems Biology?” has been extensively discussed (see, e.g., [1–5, 31, 32]). Furthermore, it has been argued that systems biology is an approach, rather than a scientific discipline in the traditional sense [31–33]. While the room for future debate on these questions remains, it is clear that systems biology fundamentally depends on the applications of mathematical and computational modeling. As the computational applications in biology are most often associated with the province of bioinformatics, another relevant question is: “what is the relationship between systems biology and bioinformatics?”. Here, we address this question by focusing on the recent, post-HGP history, and the reemergence of biochemical systems biology.
Figure 1.
The number of publications referencing “systems biology” in the PubMed database by year (2000–2009). In 2009, over 1,500 such publications appeared in PubMed.
2. From Genomics to Systems Biology
The term “genomics” was coined by Thomas Roderick in 1986, and soon after was adopted as the name of the new journal aimed to support the new discipline of genome mapping and sequencing [34]. This was a time of great excitement and profound transformation in biology brought about by the development of increasingly efficient methods for DNA sequencing [35–37]. At the time, the call for the sequencing of the human genome was gaining momentum [38, 39], and in 1988, the National Research Council of the US Academy of Sciences recommended the initiation of the Human Genome Project [39]. The HGP, completed a decade later, was an enormous success, thus validating the new discipline of genomics. It rallied the scientific community in unprecedented ways, from being a global collaboration of 20 sequencing centers from six countries to opening new horizons in large-scale biology [39]. The momentum of the HGP has spurred a plethora of genome-sequencing projects of other organisms, including plants, animals, and microorganisms. In the early phases, the sequencing projects focused mainly on mapping, sequencing, and identifying genes [40]. As the various genome-sequencing projects gathered momentum, it has become clear that collected genome sequences were only revealing more of hidden complexity, and are opening new and deeper biological questions [40, 41]. As a result, an increasing emphasis was placed upon the relationship between the sequence and function, and the field of genomics started to differentiate into “sequence genomics” and “functional genomics” [40, 42, 43].
The early view underpinning genomics was that the genome, the ultimate sequence map of the organism's DNA, is “a rosetta stone from which the complexities of gene expression in development can be translated and the genetic mechanisms of disease interpreted” [34]. This simplistic view rested on the deterministic concept of a gene and its role in determining biological function and organism's phenotype, the notion of which was tacitly extended to the entire genome. The degree of elusiveness of the gene concept has become fully apparent only in the last decade [44–48], based on the analysis of sequenced genomes, and extensive studies of the transcriptome with new techniques (such is cap-analysis gene expression (CAGE) and tiling arrays [49, 50]). Several facts highlight the complexity of the relationship between the organism's phenotype and its genome: (i) less than 2% of human DNA directly encodes proteins [51], (ii) the genomes of eukaryotic organisms are nearly entirely transcribed [50, 52], (iii) a massive amount of noncoding RNA transcripts identified in higher organisms is thought to have an important regulatory role [53]; and (iv) a critical importance of post-transcriptional and post-translational regulation in the control of the function of gene products, which is both spatially and temporally regulated [54]. As a result, in the past five years, the concept of the gene has been subject of substantial revisions [44–48].
Only temporarily overshadowed with the excitement about generating genome sequences, the true complexity of the relationship between an organism's genome and phenotype was recognized early. Even in the initial development of genomics we can recognize the elements of “systems” thinking. In 1997, Hieter and Boguski wrote “Functional genomics will … supplement the detailed understanding of gene function provided by traditional approaches with a powerful new perspective on the holistic operation of biological systems” [40]. In the next few years, the idea of a “holistic understanding” was further articulated in terms of mathematical models, whole-genome data sets, and the experience accumulated in studies of complex systems [1–3]. Almost simultaneously, the need for an engineering mindset in molecular biology was suggested in an influential (and humorous) article written by a prominent biologist [55]. An aspect of systems thinking is the recognition that biological systems are “complex” in the mathematical sense [56–58]. Such complex systems have long been of interest in physics and mathematics, and the direct relevance of the knowledge accumulated in these disciplines to biology was realized [58, 59]. It is now widely recognized that the availability of fully sequenced genomes and high-throughput (‘omics) data sets makes the aspirations of “systems” thinking in biology an achievable goal [60–64].
3. System-Level Description, System-Level Understanding, and the System Itself
There are two frequently quoted approaches to systems biology, namely, “top-down” and “bottom-up” [5, 65, 66]. Furthermore, systems biology practitioners can be arbitrarily divided into two (not mutually exclusive) camps: “pragmatic” and “systems oriented” [67, 68]. O'Malley and Dupre suggested that both camps of systems biologists lack a clear definition of what constitutes a “system” [68]. Indeed, the literature abounds with different definitions and calls for “system-level description” and “system-level understanding”. This only confounds the matter since the universally accepted definition of “system” is lacking. While confusing at first sight, the meaning of “system” in systems biology depends on the problem at hand, the objectives of the study, and the choices made in the art of mathematical modeling.
Mathematical modeling is often used in genomics and molecular biology, but in systems biology, it takes center stage, as “no more, but no less, than a way of thinking clearly” [69]. Biological systems consist of a large number of functionally diverse components, which interact highly selectively and often nonlinearly to produce coherent behaviors [2]. These components may be individual molecules (such as in signaling or metabolic networks), assemblies of interacting complexes, sets of physical factors that guide the development of an organism (genes, mRNA, associated proteins and protein complexes), cells in tissues or organs, and even entire organisms in ecological communities. What is common to all these examples is the sheer number of components, and their selective, non-linear interactions that render the behaviors of these systems beyond the intuitive grasp. Take, for example, the cell cycle in the yeast Schizosaccharomyces pombe: the model of its cell-cycle regulatory network involves about twenty components, whose interactions can be approximately described with a dozen differential equations and about 30 kinetic parameters [70]. The dynamic behavior of this network of interactions is possible to grasp only with the help of computer simulations and dynamical systems theory [70, 71]. Another example is the cellular response of yeast to hyperosmotic shock: it is only with mathematical modeling that a coherent picture emerges, connecting various known components of the system with the observed properties [72].
There are other reasons why the concept of “system” is so elusive. The role of mathematical models, particularly in generating experimentally testable hypotheses, has been discussed extensively [2, 5, 19]. Perhaps less widely appreciated is that mathematical models of biological systems are increasingly being used to represent our knowledge about these systems. For example, the iAF1260 model of Escherichia coli's metabolic network not only predicts experimentally observed behavior of E. coli under genetic perturbations [73], but also in itself is a representation of the E. coli metabolic network. Similarly, the kinetic model of glycolysis in the bloodstream form of Trypanosoma brucei [74] is the state-of-the-art representation of glycolysis in this organism. There is no alternative way of quantitative thinking about these complex systems but through models that rely on precise mathematical descriptions. These mathematical or computational models are essentially beyond a simple intuitive grasp, and represent concise summaries of our current knowledge of respective systems.
There may be significant differences in scope and scale between different models used in systems biology. Consider, for example, the model of the yeast genome-scale metabolic network [75] and the model of glycolysis in yeast [76]. It is not that one model is better than the other, rather the two models have different motivations, objectives, scales, and capabilities: the first is the genome-scale model of the entire metabolic network, while the second is a model of a single metabolic pathway which includes detailed descriptions of kinetics of individual enzyme catalyzed reactions. This illustrates an important general principle of mathematical modeling, highly relevant to systems biology: every mathematical model aims for a certain level of description, which depends on the objective of the study, limitations in our knowledge about the system of interest, and our ability to experimentally observe the system/phenomena of interest (necessary for testing the model's predictions).
Genome-scale metabolic models typically ignore kinetic parameters of individual reactions because such models aim to be comprehensive, and the kinetic parameters for most reactions are unknown (but see recent theoretical advances [77]). In contrast, kinetic models are much more detailed but less comprehensive; however, they can provide not only the information about the steady state but also the time course given some initial conditions. Choosing the correct “level of description” is one of the more difficult aspects in mathematical modeling and is a pervasive challenge in systems biology. Another challenge is choosing the boundaries of the model (note: this amounts to defining the “system”). This usually requires exquisite familiarity with the phenomena of interest, and a considerable experience in mathematical modeling. Complex dynamical systems form structures [59], and nature often provides modular designs [78]. This modularity must be both understood and exploited correctly for optimal modeling. In genome-scale studies of microbial organisms, a convenient system boundary is the cell boundary; in most other cases, the question of the appropriate systems boundary is more opaque and must be addressed based on the prior knowledge of components and the coupling between these components. Trivial examples of this include tissue structure in a multicellular organism or subcellular compartmentalization of metabolites. Hence all the difficulties in defining the “system” in systems biology.
Modern systems biology is a rapidly evolving discipline. In the past, systems thinking was invoked in the context of a variety of systems and processes: from humans [79, 80] to microorganisms [5, 60, 81], animals [82, 83], and plants [84], and in regards to different levels of biological organization, from molecular subnetworks [70, 74, 76, 85, 86], to cellular interaction networks [87], cells, entire organs [88], organisms [80, 83], and even communities of organisms [25, 30]. Areas that have proven particularly fruitful for systems biology include studies of biochemical networks and applications to microorganisms [60, 61, 73, 89–93].
4. Biochemical Networks in Microorganisms
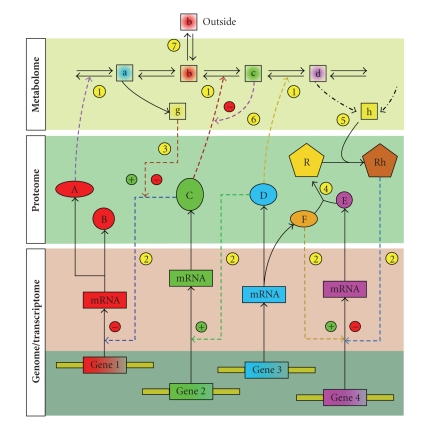
We are only beginning to appreciate the full complexity and the multidimensional nature of biochemical networks operating in all living organisms (Figure 2). Studies of metabolic networks, gene regulatory networks, and protein-protein interaction networks in microbial organisms have significantly contributed to this, and indeed to the identity of systems biology. Microorganisms are convenient models for systems studies for several reasons: (i) decades of genetic and biochemical work have generated deep biological insights, and resulted in sophisticated molecular biology techniques for experimental manipulation, (ii) they can be readily and rapidly cultured in inexpensive media, providing ample material for controlled experiments, (iii) many are pathogens of humans, plants, and domestic animals, and therefore are of medical or environmental interest, and (iv) many are important in industrial processes and therefore are relevant for biotechnology. Furthermore, microbial organisms are unicellular, and the cell membrane provides a convenient boundary that delineates the “system” for genome-wide studies. In the past, microbes have been used in numerous systems studies, including that of genetic networks [90, 94], protein-protein interactions [89, 95, 96], metabolic networks [97–100], cell cycle regulation [70, 71], and signal transduction networks [101].
Figure 2.
A conceptualization of biochemical networks showing genome, transcriptome, proteome, and metabolome-level networks, highlighting their complexity and mutual interdependence. In biological systems a large number of structurally and functionally diverse components (genes, proteins, metabolites) are involved in dynamic, non-linear interactions, which in turn involve a range of time scales and interaction strengths. Direct conversions of species shown in solid lines, while some possible interactions (not necessarily one-step) are designated in dashed lines. Several types of interactions are shown: (1) enzyme catalysis, (2) posttranscriptional control of gene expression by proteins/protein complexes, including mechanisms that act on mRNAs (deadenylation, storage granulation) and mechanisms that act either directly or indirectly on DNA (histone modification, methylation), (3) effect of metabolite on gene transcription mediated by a protein, (4) protein-protein interaction, (5) effect of a downstream (“reporter”) metabolite on transcription through binding to a protein, (6) feedback inhibition/activation of an enzyme by a downstream metabolite, and (7) exchange of a metabolite with outside of the system (cell, organism).
Three types of biochemical networks, roughly corresponding to three different levels or “omes”, have been mostly studied in the past: gene regulatory networks [90, 94, 102], protein interaction networks [89, 95, 96, 103] and metabolic networks [97–100, 104]. When prior knowledge of modularity allowed the assumption of decoupling, systems studies on biochemical subnetworks or cross-networks were possible. Examples of this include modeling of the cell cycle in yeast [70, 71], specific metabolic pathways [74, 76, 85], and signal transduction pathways [72, 101, 105]. Several pioneering studies integrated responses across individual “omes”. Examples of this include studies of transcriptome and proteome responses to perturbations in metabolic pathways [61, 106], the effects of a transcriptional regulator on central carbon metabolism in Bacillus subtilis [107], and coordinated analysis of the minimal bacterium Mycoplasma pneumoniae, including analysis of its mRNA [91], protein complexes [92], and the metabolic network [93].
What are these studies telling us? Integrating the information from different biological levels reveals complex and unanticipated global behaviors in what were thought to be “simple” organisms and biochemical systems. For example, the metabolic network in E. coli appears remarkably stable with respect to various types of perturbations, but the mechanism for how this stability is achieved appears profoundly different for environmental and genetic perturbations [106]. Surprisingly, the flux through the E. coli pentose phosphate pathway is reversed in response to a blocking mutation, and yet, this is achieved with only subtle changes in the enzyme levels [106]. Another telling example is the smallest self-replicating organism, the bacterium M. pneumoniae whose genome encodes merely 689 proteins [93]. Compared to more complex bacteria (E. coli encodes ~4,200 proteins), M. pneumoniae lacks most transcription factors and other regulators; yet this organism shows a highly complex, intrinsically structured transcriptional response, with many alternative transcripts and multiple regulators per gene [91]. In spite of its minimal genome, the proteome of M. pneumoniae exhibits modularity and extensive reuse of functional components, with a substantial crosstalk between different cellular processes [92]. Furthermore, M. pneumoniae shows highly coordinated changes in gene expression, specific responses to metabolic perturbations, and adaptability to carbon sources similar to that observed in E. coli [93]. It is unlikely that M. pneumoniae is a fundamentally unusual organism; rather, these observations suggest a host of unknown regulatory mechanisms that operate across the levels of transcriptome, proteome, and metabolome [108].
As a result of decades of detailed biochemical work, metabolic networks are the best understood of all biochemical networks [109, 110]. We have near-complete collections of components and topologies of metabolic networks in model microorganisms such as E. coli [73] and Saccharomyces cerevisiae [75]. For the model organism E. coli, the majority of metabolic reactions, enzymes, cofactors, substrates, and products are known [73]. This, however, represents only the first step towards understanding how these components function in spatial and temporal integration, and precisely what are the controls exerted on them. While the topologies of metabolic networks are well understood, we are only beginning to understand interactions that control metabolism [111, 112]. Metabolite equilibrium concentrations are accessible experimentally through quantitative metabolomic approaches [11, 12, 113], which is directly comparable to the measurement of mRNA and protein levels in transcriptomics and proteomics, respectively. In contrast to all other types of biochemical networks, experimental approaches for assessing in vivo reaction rates (fluxes) are also well developed for metabolic networks [109, 114–116]. This is of great importance, as metabolic fluxes are the key determinants of cellular physiology and cannot be predicted from mRNA, protein, or even metabolite levels [114]. Thus, measurement of metabolic flux is equivalent to the measurement of information flow through a signaling pathway, or the information flow between genes residing on the same control circuit. New theoretical frameworks for more efficient extraction of information from experimental data continue to be proposed [117], and a considerable progress has been made in the analysis of metabolic fluxes under isotopic nonstationary conditions [115, 118]. Since nonstationary flux analysis relies on shorter, transient experiments, this opens an array of new possibilities for flux analysis in higher organisms, improving the scope of systems biology studies of metabolic networks [118].
5. Bioinformatic Tools for Systems Biology
While many systems biology approaches involve mathematical and computational modeling, the development, maintenance, and dissemination of tools for systems biology is in itself a significant challenge. Examples of this include development of data repositories, data standards and software tools for simulation, analysis and visualization of system components such as biochemical networks. Another example are applications of high-throughput molecular profiling technologies which often require sophisticated data processing and analysis, and typically involve elements of signal processing and statistical analysis. As the resulting quantitative measurements are transferred to formal mathematical models for the purpose of modeling, the endeavor becomes perhaps more systems biology and less bioinformatics. However, that is only a matter of a degree, with often no clear boundary between bioinformatics and systems biology.
The need for effective exchange of formal, quantitative systems biology models has driven the development of the Systems Biology Markup Language (SBML) [119]. The SBML project aims for the development of the computer-readable format for the representation of biological processes. SBML provides a well-defined format which different software tools can use for the exchange of biological models with high fidelity. A testimony to the importance of SBML is its adoption by software tools concerned with biological modeling (at the time of this writing, over 180 software tools support SBML). The graphical notation for the representation of biological processes has been proposed recently (Systems Biology Graphical Notation, SBGN) [120]. The current SBGN specification consists of three complementary languages which aim to describe biological processes and relationships between biological entities [120].
Since studies of biochemical networks are particularly successful aspect of systems biology, it is not surprising that a plethora of computational tools that address different needs in the analysis of biochemical networks have been reported, and in many cases, these tools are freely accessible. Without attempting to be comprehensive, we highlight some of the widely used research and training tools. Systems Biology Workbench (SWB) is a framework that allows different components for systems biology to communicate, exchange models via SBML, and reuse capabilities without understanding all the details of the each component implementation [121]. From the user's perspective, SWB is a collection of tools for systems biology that includes programs for building, viewing, and editing of biochemical networks, tools for simulation, and tools for import and translation of models. Another highly useful tool is CellDesigner, a Java-based program for constructing and editing of biochemical networks [122]. Recent versions of CellDesigner are able to import models in SBML and support display of biochemical networks based on process diagram language specified by SBGN. In CellDesigner models can be simulated either with a built-in simulator, or alternatively CellDesigner can connect to external simulators, such as those provided by SWB [121]. An independent simulator of models encoded in SBML is COPASI [123]. COPASI can simulate models based on ordinary differential equations (ODEs) as well as stochastic models by using the Gillespie's algorithm. COPASI provides tools for visual analysis of simulation results, and can also perform steady-state and metabolic control analyses [123].
As biological research accelerates through the development of new technologies and instrumentation, biological databases have become an indispensable partner in such research. Building and maintaining of primary databases such as GenBank [124] or Protein Data Bank [125] have long been recognized as important bioinformatics work. Primary biological databases serve both as repositories of experimentally derived information and are the basis for the development of secondary databases that capture higher-level knowledge. An example of such secondary database is Pfam database of proteins families and domains [126]. Concomitantly with the development of the biochemical systems biology, an important niche of secondary biological databases has emerged: the databases that capture the properties and processes in biochemical networks. The ecosystem of such databases and associated tools is rapidly growing and includes metabolic pathways databases organized around the BioCyc project [127], database of human biological pathways [128], database of interactions between small molecules and proteins [129], and databases of protein-protein interactions [130]. As these databases attempt to reconstruct and organize information about interactions between cellular components, they also attempt to build higher-level knowledge and theories about the biological processes they are concerned with. Such in silico knowledge is much needed, as the integral complexity of most biological processes is beyond what is comprehensible to the human mind. Therefore, these “systems biology databases” often represent important foundations for quantitative modeling of biological systems. In some cases, these databases allow a direct export of mathematical models. Also, the first collections of mathematical models of biological processes have been developed (databases of models), concerned solely with archiving and curating the models in SBML for future reuse and refinement [131]. Much needed bioinformatics tools for systems biology research are the tools for visualization of network structures and network overlay of simulated and experimental data. These tools include yEd graph editor for editing networks, and tools for visualization of ‘omics data in the context of biochemical networks, such as Cytoscape [132] and Pathway Tools Omics Viewer [133].
6. Future Perspectives
Systems biology is rapidly gaining momentum, as evidenced by the number of publications referencing the term (Figure 1). To understand the relevance of “systems” thinking for future biochemical research, one needs only to remember that we know most of the components in many biochemical systems, often in exquisite detail, yet understand very little about how these components interact to produce coherent temporal and spatial behaviors that are the hallmark of biological systems [2]. On the other hand, bioinformatics has originally grown from the need to provide tools and handle increasingly large amounts of biological data. As a discipline bioinformatics continues to grow in this important role, but is also increasingly merging and contributing to systems approaches to provide tools necessary for perhaps the most exciting phase in the development of biological sciences.
One of the defining features of systems biology is the use of mathematical and computational models, which are essential to rigorously account for the inherent complexity of biological systems. This complexity arises from the diversity of components (genes, proteins, and metabolites), the high selectivity of their interactions, and a non-linear nature of these interactions. These properties together render the behavior of biological systems intractable to pure intuition. The computational models used in biochemical systems biology typically require iterative building and stepwise improvements based on the comparison with experiments [134]. Once sufficiently refined, such models have the ability to predict the behavior of the biochemical system under different perturbations, or hypothetical conditions that may be of interest but are not feasible in experimental settings (e.g., when they are too expensive for practical implementation, or when the analysis of many different conditions is desirable [135]). However, in the new era of systems biology, mathematical models are more than just tools for integrating observations, making testable predictions, or for high throughput in silico experimentation. Highly refined mathematical models also serve as the embodiments of our current knowledge about specific biochemical systems.
Mathematical and computational models that underpin biochemical studies may involve different levels of detail and scale, depending on the objectives of the study, what is known a priori, and what additional information is accessible experimentally. For example, protein complexes may be studied comprehensively [92], or the focus may be on a subset of proteins responsible for a specific function, such as protein import into mitochondria [136]. Most of the so-called bottom-up approaches, which start from the descriptions of interactions, focus on a part of the biological system because we lack a comprehensive information about the system of interest [137]. Nevertheless, bottom-up approaches provide highly useful frameworks for the integration of diverse knowledge, for example, the principles established from decades of biochemical work with the information accessible only with the latest experiments. In contrast, top-down approaches are largely data driven, with the caveat that their comprehensiveness is limited by the limitations in experimental approaches. For example, in one of the most comprehensive metabolomic studies to date, 198 out of an expected 453 primary metabolites were quantified simultaneously in cells grown in minimal medium [138]. Therefore, in such applications advances in technology drive the level of “comprehensiveness” that can be achieved.
Many biochemical processes can be conceptualized as complex dynamic networks on the molecular level (Figure 2), and studies of biochemical networks are assuming centre stage in systems biology [65–67, 139, 140]. Measurements on different ‘omics levels provide different, often complementary views of the functions of molecular networks. Increasingly, we are interested in the crosstalk between the genes, transcripts, proteins, and metabolites that the gene's expression impacts upon [112, 141]. Increasingly sophisticated models will be required to account for increasingly accurate and comprehensive experimental measurements. Systems approaches have already provided a deeper understanding of diverse biochemical processes, from individual metabolic pathways [74, 76], to signaling networks [70–72], to genome-scale metabolic networks [73, 75]. Therefore, we can safely predict that systems thinking will become even more pervasive in future. The role of formal mathematical and computational models in systems approaches renders the role of bioinformatics increasingly important for systems biology research.
Acknowledgments
The authors acknowledge support from Metabolomics Australia (V. A. Likić, M. J. McConville, A. Bacic) and Australian Centre for Plant Functional Genomics (A. Bacic). T. Lithgow is a Federation Fellow of the Australian Research Council. M. J. McConville is a Principal Research Fellow of the Australian National Health and Medical Research Council.
References
- 1.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annual Review of Genomics and Human Genetics. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 2.Kitano H. Computational systems biology. Nature. 2002;420(6912):206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 3.Kitano H. Systems biology: a brief overview. Science. 2002;295(5560):1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 4.Westerhoff HV, Palsson BO. The evolution of molecular biology into systems biology. Nature Biotechnology. 2004;22(10):1249–1252. doi: 10.1038/nbt1020. [DOI] [PubMed] [Google Scholar]
- 5.Stelling J. Mathematical models in microbial systems biology. Current Opinion in Microbiology. 2004;7(5):513–518. doi: 10.1016/j.mib.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Craig Venter J, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 8.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 9.Lashkari DA, DeRisi JL, Mccusker JH, et al. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(24):13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patterson SD, Aebersold RH. Proteomics: the first decade and beyond. Nature Genetics. 2003;33:311–323. doi: 10.1038/ng1106. [DOI] [PubMed] [Google Scholar]
- 11.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends in Biotechnology. 1998;16(9):373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 12.Fiehn O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comparative and Functional Genomics. 2001;2(3):155–168. doi: 10.1002/cfg.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiener N. Cybernetics: Or Control and Communication in the Animan and the Machine. Cambridge, Mass, USA: MIT Press; 1946. [Google Scholar]
- 14.Mesarovic MD. Systems theory and biology—view of a theoretician. In: Mesarovic MD, editor. Systems Theory and Biology. New York, NY, USA: Springer; 1968. pp. 59–87. [Google Scholar]
- 15.von Bertalanffy L. General System Theory. New York, NY, USA: George Braziller; 1969. [Google Scholar]
- 16.Kacser H, Burns JA. The control of flux. Symposia of the Society for Experimental Biology. 1973;27:65–104. [PubMed] [Google Scholar]
- 17.Heinrich R, Rapoport TA. A linear steady state treatment of enzymatic chains: general properties, control and effector strength. European Journal of Biochemistry. 1974;42(1):89–95. doi: 10.1111/j.1432-1033.1974.tb03318.x. [DOI] [PubMed] [Google Scholar]
- 18.Kacser H, Burns JA. The molecular basis of dominance. Genetics. 1981;97(3-4):639–666. doi: 10.1093/genetics/97.3-4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolkenhauer O. Systems biology: the reincarnation of systems theory applied in biology? Briefings in Bioinformatics. 2001;2(3):258–270. doi: 10.1093/bib/2.3.258. [DOI] [PubMed] [Google Scholar]
- 20.Benner SA, Sismour AM. Synthetic biology. Nature Reviews Genetics. 2005;6(7):533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDaniel R, Weiss R. Advances in synthetic biology: on the path from prototypes to applications. Current Opinion in Biotechnology. 2005;16(4):476–483. doi: 10.1016/j.copbio.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Heinemann M, Panke S. Synthetic biology—putting engineering into biology. Bioinformatics. 2006;22(22):2790–2799. doi: 10.1093/bioinformatics/btl469. [DOI] [PubMed] [Google Scholar]
- 23.Barrett CL, Kim TY, Kim HU, Palsson BØ, Lee SY. Systems biology as a foundation for genome-scale synthetic biology. Current Opinion in Biotechnology. 2006;17(5):488–492. doi: 10.1016/j.copbio.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Mukherji S, van Oudenaarden A. Synthetic biology: understanding biological design from synthetic circuits. Nature Reviews Genetics. 2009;10(12):859–871. doi: 10.1038/nrg2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vieites JM, Guazzaroni M-E, Beloqui A, Golyshin PN, Ferrer M. Metagenomics approaches in systems microbiology. FEMS Microbiology Reviews. 2009;33(1):236–255. doi: 10.1111/j.1574-6976.2008.00152.x. [DOI] [PubMed] [Google Scholar]
- 26.Liu ET. Systems biology, integrative biology, predictive biology. Cell. 2005;121(4):505–506. doi: 10.1016/j.cell.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 27.O’Callaghan PM, James DC. Systems biotechnology of mammalian cell factories. Briefings in Functional Genomics and Proteomics. 2008;7(2):95–110. doi: 10.1093/bfgp/eln012. [DOI] [PubMed] [Google Scholar]
- 28.Tomlins SA, Rubin MA, Chinnaiyan AM. Integrative biology of prostate cancer progression. Annual Review of Pathology. 2006;1:243–271. doi: 10.1146/annurev.pathol.1.110304.100047. [DOI] [PubMed] [Google Scholar]
- 29.Liu ET. Integrative biology—a strategy for systems biomedicine. Nature Reviews Genetics. 2009;10(1):64–68. doi: 10.1038/nrg2488. [DOI] [PubMed] [Google Scholar]
- 30.Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annual Review of Genetics. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 31.Kirschner MW. The meaning of systems biology. Cell. 2005;121(4):503–504. doi: 10.1016/j.cell.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Cowley AW., Jr. The elusive field of systems biology. Physiological Genomics. 2004;16:285–286. doi: 10.1152/physiolgenomics.00007.2004. [DOI] [PubMed] [Google Scholar]
- 33.Kohl P, Crampin EJ, Quinn TA, Noble D. Systems biology: an approach. Clinical Pharmacology and Therapeutics. 2010;88(1):25–33. doi: 10.1038/clpt.2010.92. [DOI] [PubMed] [Google Scholar]
- 34.McKusick VA, Ruddle FH. A new discipline, a new name, a new journal. Genomics. 1987;1(1):1–2. [Google Scholar]
- 35.Maxam AM, Gilbert W. A new method for sequencing DNA. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. Journal of Molecular Biology. 1975;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 37.Smith LM, Sanders JZ, Kaiser RJ, et al. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321(6071):674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 38.Hood LE, Hunkapiller MW, Smith LM. Automated DNA sequencing and analysis of the human genome. Genomics. 1987;1(3):201–212. doi: 10.1016/0888-7543(87)90046-2. [DOI] [PubMed] [Google Scholar]
- 39.Collins FS, Morgan M, Patrinos A. The human genome project: lessons from large-scale biology. Science. 2003;300(5617):286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 40.Hieter P, Boguski M. Functional genomics: it’s all how you read it. Science. 1997;278(5338):601–602. doi: 10.1126/science.278.5338.601. [DOI] [PubMed] [Google Scholar]
- 41.Rastan S, Beeley LJ. Functional genomics: going forwards from the databases. Current Opinion in Genetics and Development. 1997;7(6):777–783. doi: 10.1016/s0959-437x(97)80040-8. [DOI] [PubMed] [Google Scholar]
- 42.Somerville C, Somerville S. Plant functional genomics. Science. 1999;285(5426):380–383. doi: 10.1126/science.285.5426.380. [DOI] [PubMed] [Google Scholar]
- 43.Brent R. Genomic biology. Cell. 2000;100(1):169–183. doi: 10.1016/s0092-8674(00)81693-1. [DOI] [PubMed] [Google Scholar]
- 44.Griffiths PE, Stotz K. Genes in the postgenomic era. Theoretical Medicine and Bioethics. 2006;27(6):499–521. doi: 10.1007/s11017-006-9020-y. [DOI] [PubMed] [Google Scholar]
- 45.Rédei GP, Koncz C, Phillips JD. Changing images of the gene. Advances in Genetics. 2006;56:53–100. doi: 10.1016/S0065-2660(06)56002-X. [DOI] [PubMed] [Google Scholar]
- 46.Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Research. 2007;17(6):682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 47.Gerstein MB, Bruce C, Rozowsky JS, et al. What is a gene, post-ENCODE? History and updated definition. Genome Research. 2007;17(6):669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 48.Portin P. The elusive concept of the gene. Hereditas. 2009;146(3):112–117. doi: 10.1111/j.1601-5223.2009.02128.x. [DOI] [PubMed] [Google Scholar]
- 49.Carninci P. Tagging mammalian transcription complexity. Trends in Genetics. 2006;22(9):501–510. doi: 10.1016/j.tig.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 50.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nature Reviews Genetics. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 51.Avise JC. Evolving genomic metaphors: a new look at the language of DNA. Science. 2001;294(5540):86–87. doi: 10.1126/science.294.5540.86. [DOI] [PubMed] [Google Scholar]
- 52.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319(5871):1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 53.Mattick JS. The functional genomics of noncoding RNA. Science. 2005;309(5740):1527–1528. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- 54.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136(4):719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lazebnik Y. Can a biologist fix a radio?—Or, what I learned while studying apoptosis. Cancer Cell. 2002;2(3):179–182. doi: 10.1016/s1535-6108(02)00133-2. [DOI] [PubMed] [Google Scholar]
- 56.Weng G, Bhalla US, Iyengar R. Complexity in biological signaling systems. Science. 1999;284(5411):92–96. doi: 10.1126/science.284.5411.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitesides GM, Ismagilov RF. Complexity in chemistry. Science. 1999;284(5411):89–92. doi: 10.1126/science.284.5411.89. [DOI] [PubMed] [Google Scholar]
- 58.Ross J, Arkin AP. Complex systems: from chemistry to systems biology. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(16):6433–6434. doi: 10.1073/pnas.0903406106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldenfeld N, Kadanoff LP. Simple lessons from complexity. Science. 1999;284(5411):87–89. doi: 10.1126/science.284.5411.87. [DOI] [PubMed] [Google Scholar]
- 60.Castrillo JI, Oliver SG. Metabolomics and systems biology in Saccharomyces cerevisiae . In: Brown AJP, editor. The Mycota: A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research. Heidelberg, Germany: Springer; 2006. pp. 3–18. [Google Scholar]
- 61.Ideker T, Thorsson V, Ranish JA, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292(5518):929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 62.Ge H, Walhout AJM, Vidal M. Integrating ’omic’ information: a bridge between genomics and systems biology. Trends in Genetics. 2003;19(10):551–560. doi: 10.1016/j.tig.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 63.de Hoog CL, Mann M. Proteomics. Annual Review of Genomics and Human Genetics. 2004;5:267–293. doi: 10.1146/annurev.genom.4.070802.110305. [DOI] [PubMed] [Google Scholar]
- 64.Bruggeman FJ, Westerhoff HV. The nature of systems biology. Trends in Microbiology. 2007;15(1):45–50. doi: 10.1016/j.tim.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 65.Ideker T, Lauffenburger D. Building with a scaffold: emerging strategies for high- to low-level cellular modeling. Trends in Biotechnology. 2003;21(6):255–262. doi: 10.1016/S0167-7799(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 66.Laubenbacher R, Jarrah AS. Algebraic models of biochemical networks. Methods in Enzymology. 2009;467(C):163–196. doi: 10.1016/S0076-6879(09)67007-5. [DOI] [PubMed] [Google Scholar]
- 67.Huang S. Back to the biology in systems biology: what can we learn from biomolecular networks? Brief Funct Genomic Proteomic. 2004;2(4):279–297. doi: 10.1093/bfgp/2.4.279. [DOI] [PubMed] [Google Scholar]
- 68.O’Malley MA, Dupré J. Fundamental issues in systems biology. BioEssays. 2005;27(12):1270–1276. doi: 10.1002/bies.20323. [DOI] [PubMed] [Google Scholar]
- 69.May RM. Uses and abuses of mathematics in biology. Science. 2004;303(5659):790–793. doi: 10.1126/science.1094442. [DOI] [PubMed] [Google Scholar]
- 70.Tyson JJ, Chen K, Novak B. Network dynamics and cell physiology. Nature Reviews Molecular Cell Biology. 2001;2(12):908–916. doi: 10.1038/35103078. [DOI] [PubMed] [Google Scholar]
- 71.Chen KC, Calzone L, Csikasz-Nagy A, Cross FR, Novak B, Tyson JJ. Integrative analysis of cell cycle control in budding yeast. Molecular Biology of the Cell. 2004;15(8):3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klipp E, Nordlander B, Krüger R, Gennemark P, Hohmann S. Integrative model of the response of yeast to osmotic shock. Nature Biotechnology. 2005;23(8):975–982. doi: 10.1038/nbt1114. [DOI] [PubMed] [Google Scholar]
- 73.Feist AM, Henry CS, Reed JL, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Molecular Systems Biology. 2007;3, article 121 doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bakker BM, Michels PAM, Opperdoes FR, Westerhoff HV. What controls glycolysis in bloodstream form Trypanosoma brucei? Journal of Biological Chemistry. 1999;274(21):14551–14559. doi: 10.1074/jbc.274.21.14551. [DOI] [PubMed] [Google Scholar]
- 75.Duarte NC, Herrgård MJ, Palsson BØ. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Research. 2004;14(7):1298–1309. doi: 10.1101/gr.2250904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teusink B, Passarge J, Reijenga CA, et al. Can yeast glycolysis be understood terms of vitro kinetics of the constituent enzymes? Testing biochemistry. European Journal of Biochemistry. 2000;267(17):5313–5329. doi: 10.1046/j.1432-1327.2000.01527.x. [DOI] [PubMed] [Google Scholar]
- 77.Smallbone K, Simeonidis E, Swainston N, Mendes P. Towards a genome-scale kinetic model of cellular metabolism. BMC Systems Biology. 2010;4, article 6 doi: 10.1186/1752-0509-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 79.Hornberg JJ, Bruggeman FJ, Westerhoff HV, Lankelma J. Cancer: a systems biology disease. BioSystems. 2006;83(2-3):81–90. doi: 10.1016/j.biosystems.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 80.Kohl P, Noble D. Systems biology and the virtual physiological human. Molecular Systems Biology. 2009;5, article 292 doi: 10.1038/msb.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sang YL, Lee D-Y, Tae YK. Systems biotechnology for strain improvement. Trends in Biotechnology. 2005;23(7):349–358. doi: 10.1016/j.tibtech.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 82.Argmann CA, Chambon P, Auwerx J. Mouse phenogenomics: the fast track to "systems metabolism". Cell Metabolism. 2005;2(6):349–360. doi: 10.1016/j.cmet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 83.Zahn JM, Kim SK. Systems biology of aging in four species. Current Opinion in Biotechnology. 2007;18(4):355–359. doi: 10.1016/j.copbio.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yuan JS, Galbraith DW, Dai SY, Griffin P, Stewart CN., Jr. Plant systems biology comes of age. Trends in Plant Science. 2008;13(4):165–171. doi: 10.1016/j.tplants.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 85.Hynne F, Danø S, Sørensen PG. Full-scale model of glycolysis in Saccharomyces cerevisiae. Biophysical Chemistry. 2001;94(1-2):121–163. doi: 10.1016/s0301-4622(01)00229-0. [DOI] [PubMed] [Google Scholar]
- 86.Albert M-A, Haanstra JR, Hannaert V, et al. Experimental and in silico analyses of glycolytic flux control in bloodstream form Trypanosoma brucei. Journal of Biological Chemistry. 2005;280(31):28306–28315. doi: 10.1074/jbc.M502403200. [DOI] [PubMed] [Google Scholar]
- 87.Kim PS, Levy D, Lee PP. Modeling and simulation of the immune system as a self-regulating network. Methods in Enzymology. 2009;467(C):79–109. doi: 10.1016/S0076-6879(09)67004-X. [DOI] [PubMed] [Google Scholar]
- 88.Noble D. Modeling the heart—from genes to cells to the whole organ. Science. 2002;295(5560):1678–1682. doi: 10.1126/science.1069881. [DOI] [PubMed] [Google Scholar]
- 89.Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nature Biotechnology. 2000;18(12):1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 90.Lee TI, Rinaldi NJ, Robert F, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298(5594):799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 91.Güell M, van Noort V, Yus E, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326(5957):1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 92.Kühner S, van Noort V, Betts MJ, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326(5957):1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 93.Yus E, Maier T, Michalodimitrakis K, et al. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326(5957):1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- 94.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genetics. 2002;31(1):64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 95.Krogan NJ, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 96.Tarassov K, Messier V, Landry CR, et al. An in vivo map of the yeast protein interactome. Science. 2008;320(5882):1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 97.Tang Y, Pingitore F, Mukhopadhyay A, Phan R, Hazen TC, Keasling JD. Pathway confirmation and flux analysis of central metabolic pathways in Desulfovibrio vulgaris Hildenborough using gas chromatography-mass spectrometry and Fourier transform-ion cyclotron resonance mass spectrometry. Journal of Bacteriology. 2007;189(3):940–949. doi: 10.1128/JB.00948-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.del Castillo T, Ramos JL, Rodríguez-Herva JJ, Fuhrer T, Sauer U, Duque E. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. Journal of Bacteriology. 2007;189(14):5142–5152. doi: 10.1128/JB.00203-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schilling O, Frick O, Herzberg C, et al. Transcriptional and metabolic responses of Bacillus subtilis to the availability of organic acids: transcription regulation is important but not sufficient to account for metabolic adaptation. Applied and Environmental Microbiology. 2007;73(2):499–507. doi: 10.1128/AEM.02084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saito N, Robert M, Kochi H, et al. Metabolite profiling reveals YihU as a novel hydroxybutyrate dehydrogenase for alternative succinic semialdehyde metabolism in Escherichia coli. Journal of Biological Chemistry. 2009;284(24):16442–16451. doi: 10.1074/jbc.M109.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krantz M, Ahmadpour D, Ottosson L-G, et al. Robustness and fragility in the yeast high osmolarity glycerol (HOG) signal-transduction pathway. Molecular Systems Biology. 2009;5, article 281 doi: 10.1038/msb.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koide T, Hayata T, Cho KWY. Xenopus as a model system to study transcriptional regulatory networks. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(14):4943–4948. doi: 10.1073/pnas.0408125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Giot L, Bader JS, Brouwer C, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302(5651):1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 104.Tang YJ, Chakraborty R, Martín HG, Chu J, Hazen TC, Keasling JD. Flux analysis of central metabolic pathways in Geobacter metallireducens during reduction of soluble Fe(III)-nitrilotriacetic acid. Applied and Environmental Microbiology. 2007;73(12):3859–3864. doi: 10.1128/AEM.02986-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nature Cell Biology. 2006;8(11):1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 106.Ishii N, Nakahigashi K, Baba T, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316(5824):593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 107.Tännler S, Fischer E, Le Coq D, et al. CcpN controls central carbon fluxes in Bacillus subtilis. Journal of Bacteriology. 2008;190(18):6178–6187. doi: 10.1128/JB.00552-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ochman H, Raghavan R. Systems biology. Excavating the functional landscape of bacterial cells. Science. 2009;326(5957):1200–1201. doi: 10.1126/science.1183757. [DOI] [PubMed] [Google Scholar]
- 109.Stephanopoulos G. Metabolic fluxes and metabolic engineering. Metabolic Engineering. 1999;1(1):1–11. doi: 10.1006/mben.1998.0101. [DOI] [PubMed] [Google Scholar]
- 110.Sauer U. Metabolic networks in motion: 13C-based flux analysis. Molecular Systems Biology. 2006;2, article 62 doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gianchandani EP, Brautigan DL, Papin JA. Systems analyses characterize integrated functions of biochemical networks. Trends in Biochemical Sciences. 2006;31(5):284–291. doi: 10.1016/j.tibs.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 112.Grüning N-M, Lehrach H, Ralser M. Regulatory crosstalk of the metabolic network. Trends in Biochemical Sciences. 2010;35(4):220–227. doi: 10.1016/j.tibs.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 113.Nielsen J, Oliver S. The next wave in metabolome analysis. Trends in Biotechnology. 2005;23(11):544–546. doi: 10.1016/j.tibtech.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 114.Zamboni N, Sauer U. Novel biological insights through metabolomics and 13C-flux analysis. Current Opinion in Microbiology. 2009;12(5):553–558. doi: 10.1016/j.mib.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 115.Wiechert W, Nöh K. From stationary to instationary metabolic flux analysis. Advances in Biochemical Engineering/Biotechnology. 2005;92:145–172. doi: 10.1007/b98921. [DOI] [PubMed] [Google Scholar]
- 116.Tang YJ, Martin HG, Myers S, Rodriguez S, Baidoo EEK, Keasling JD. Advances in analysis of microbial metabolic fluxes via 13C isotopic labeling. Mass Spectrometry Reviews. 2009;28(2):362–375. doi: 10.1002/mas.20191. [DOI] [PubMed] [Google Scholar]
- 117.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metabolic Engineering. 2007;9(1):68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnology and Bioengineering. 2008;99(3):686–699. doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
- 119.Hucka M, Finney A, Bornstein BJ, et al. Evolving a lingua franca and associated software infrastructure for computational systems biology: the Systems Biology Markup Language (SBML) project. Systems Biology. 2004;1(1):41–53. doi: 10.1049/sb:20045008. [DOI] [PubMed] [Google Scholar]
- 120.Novère NL, Hucka M, Mi H, et al. The systems biology graphical notation. Nature Biotechnology. 2009;27(8):735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- 121.Sauro HM, Hucka M, Finney A, et al. Next generation simulation tools: the systems biology workbench and BioSPICE integration. OMICS. 2003;7(4):355–372. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- 122.Funahashi A, Morohashi M, Kitano H, Tanimura N. CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. BIOSILICO. 2003;1:159–162. [Google Scholar]
- 123.Mendes P, Hoops S, Sahle S, Gauges R, Dada J, Kummer U. Computational modeling of biochemical networks using COPASI. Methods in Molecular Biology. 2009;500:17–59. doi: 10.1007/978-1-59745-525-1_2. [DOI] [PubMed] [Google Scholar]
- 124.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Research. 2010;37(1):D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kouranov A, Xie L, de la Cruz J, et al. The RCSB PDB information portal for structural genomics. Nucleic Acids Research. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Research. 2010;38(supplement 1):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Caspi R, Altman T, Dale JM, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Research. 2010;38(supplement 1):D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Matthews L, Gopinath G, Gillespie M, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Research. 2009;37(1):D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kuhn M, Szklarczyk D, Franceschini A, et al. STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Research. 2010;38(supplement 1):D552–D556. doi: 10.1093/nar/gkp937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lehne B, Schlitt T. Protein-protein interaction databases: keeping up with growing interactomes. Human Genomics. 2009;3(3):291–297. doi: 10.1186/1479-7364-3-3-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Le Novère N, Bornstein B, Broicher A, et al. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Research. 2006;34:D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software Environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Karp PD, Paley SM, Krummenacker M, et al. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Briefings in Bioinformatics. 2009;11(1):40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Reed JL, Palsson BØ. Thirteen years of building constraint-based in silico models of Escherichia coli. Journal of Bacteriology. 2003;185(9):2692–2699. doi: 10.1128/JB.185.9.2692-2699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schilling CH, Edwards JS, Palsson BO. Toward metabolic phenomics: analysis of genomic data using flux balances. Biotechnology Progress. 1999;15(3):288–295. doi: 10.1021/bp9900357. [DOI] [PubMed] [Google Scholar]
- 136.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313(5785):314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 137.Nielsen J. Systems biology of lipid metabolism: from yeast to human. FEBS Letters. 2009;583(24):3905–3913. doi: 10.1016/j.febslet.2009.10.054. [DOI] [PubMed] [Google Scholar]
- 138.Ohashi Y, Hirayama A, Ishikawa T, et al. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Molecular BioSystems. 2008;4(2):135–147. doi: 10.1039/b714176a. [DOI] [PubMed] [Google Scholar]
- 139.Brazhnik P, de la Fuente A, Mendes P. Gene networks: how to put the function in genomics. Trends in Biotechnology. 2002;20(11):467–472. doi: 10.1016/s0167-7799(02)02053-x. [DOI] [PubMed] [Google Scholar]
- 140.Alm E, Arkin AP. Biological networks. Current Opinion in Structural Biology. 2003;13(2):193–202. doi: 10.1016/s0959-440x(03)00031-9. [DOI] [PubMed] [Google Scholar]
- 141.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]