Abstract
Eukaryotic translation initiation factor 5A (eIF5A) is the only cellular protein that contains the polyamine-modified lysine, hypusine [Nε-(4-amino-2-hydroxybutyl)lysine]. Hypusine occurs only in eukaryotes and certain archaea, but not in eubacteria. It is formed post-translationally by two consecutive enzymatic reactions catalyzed by deoxyhypusine synthase (DHS) and deoxyhypusine hydroxylase (DOHH). Hypusine modification is essential for the activity of eIF5A and for eukaryotic cell proliferation. eIF5A binds to the ribosome and stimulates translation in a hypusine-dependent manner, but its mode of action in translation is not well understood. Since quantities of highly pure hypusine-modified eIF5A is desired for structural studies as well as for determination of its binding sites on the ribosome, we have used a polycistronic vector, pST39, to express eIF5A alone, or to co-express human eIF5A-1 with DHS or with both DHS and DOHH in Escherichia coli cells, to engineer recombinant proteins, unmodified eIF5A, deoxyhypusine- or hypusine-modified eIF5A. We have accomplished production of three different forms of recombinant eIF5A in high quantity and purity. The recombinant hypusine-modified eIF5A was as active in methionyl-puromycin synthesis as the native, eIF5A (hypusine form) purified from mammalian tissue. The recombinant eIF5A proteins will be useful tools in future structure/function and the mechanism studies in translation.
Keywords: eIF5A, hypusine, polyamine, polycistronic vector, post-translational modification
Introduction
Eukaryotic translation initiation factor 5A (eIF5A) is a small acidic protein and the only cellular protein that contains the unique modified lysine, hypusine [Nε-(4-amino-2-hydroxybutyl)lysine] (Park et al., 1981, 2010). Hypusine is formed by the post-translational modification of one specific lysine residue of the eIF5A precursor, eIF5A(Lys), in two consecutive enzymatic steps (Chen and Liu, 1997; Park, 2006). The first enzyme, deoxyhypusine synthase (DHS) (Joe et al., 1995) catalyzes the transfer of the aminobutyl moiety from the polyamine spermidine to form an intermediate, deoxyhypusine [Nε-(4-aminobutyl)lysine] residue, which is subsequently hydroxylated by deoxyhypusine hydroxylase (DOHH) (Park et al., 2006). Inactivation of the eIF5A genes (both TIF51A and TIF51B) (Schnier et al., 1991; Wöhl et al., 1993) or of the DHS gene causes loss of viability in Saccharomyces cerevisiae (Sasaki et al., 1996; Park et al., 1998). Additional evidence in support of the essential role of hypusine and eIF5A in eukaryotic and mammalian cell proliferation has been reported from several laboratories (Byers et al., 1994; Hanauske-Abel et al., 1994; Park et al., 1994; Chen and Liu, 1997; Chattopadhyay et al., 2008). Hypusine synthesis defines an indispensible function of polyamines in eukaryotic cell proliferation (Byers et al., 1994; Chattopadhyay et al., 2008; Pegg, 2009).
In spite of the essential nature of eIF5A in eukaryotic cell proliferation, the precise cellular function of this putative initiation factor has remained obscure for decades. eIF5A was initially isolated from reticulocyte lysate ribosome as a factor that stimulates methionyl-puromycin synthesis (a model assay for translation initiation; Kemper et al., 1976; Benne and Hershey, 1978). eIF5A appears to bind to the ribosome in a hypusine-dependent manner, since the eIF5A mutant protein (yeast eIF5A K51R), with a Lys to Arg substitution at the hypusine modification site, does not associate with ribosomes (Jao and Chen, 2006; Zanelli et al., 2006). Recent reports of the increase in the polysome/monosome ratios in eIF5A mutant yeast strains (Gregio et al., 2009; Saini et al., 2009) and also in mammalian cells depleted of eIF5A (Landau et al., 2010) suggest a role for eIF5A in the elongation rather than the initiation step of translation. Saccharomyces cerevisiae strains harboring eIF5A temperature-sensitive mutant proteins exhibit diverse cellular changes, suggesting a direct or indirect role for eIF5A in cell wall integrity, mRNA decay, actin polarization, apoptosis and cell cycle progression (Zuk and Jacobson, 1998; Valentini et al., 2002; Chatterjee et al., 2006; Schrader et al., 2006). It is not yet clear how depletion or dysfunction of eIF5A in the mutant strains leads to such pleiotropic phenotypes.
The crystal structures of unmodified eIF5A precursors have been determined for proteins from human (eIF5A-1 with 14 N-terminal and 3 C-terminal amino acids truncated) (Tong et al., 2009) (PDB 3cpf), yeast (PDB 3er0), leishimania (PDB 1xtd, 1x6o) and for the archaeal eIF5A homolog, aIF5A (PDB 1eif, 2eif, 1bkb and 1izb). eIF5A has two domains, a basic N-terminal domain and an acidic C-terminal domain, connected by a hinge. The C-terminal domain resembles an oligonucleotide-binding fold of the Escherichia coli cold shock protein CspA and has been implicated in RNA binding. The lysine that undergoes hypusine modification is located at the tip of an exposed loop (aa46–54 of the human sequence) in the N-terminal domain. The amino acid sequence surrounding this modification site (STSKTGK50HGHAK) is strictly conserved in eukaryotes, and is very basic and hydrophilic. Addition of the 4-amino-2 hydroxybutyl moiety to the ε-amino group of Lys 50 creates a long basic side chain in this loop. The strict conservation of the hypusine loop sequence and the importance of these residues for eIF5A activity (Cano et al., 2008; Dias et al., 2008) suggest that it serves an essential, basic function that has been preserved throughout eukaryotic evolution. However, it is unknown whether hypusine modification causes significant changes in the folding structures of eIF5A and how the hypusine residue and its loop contribute to the binding of eIF5A to the ribosome or other binding partners. Binding of eIF5A to the ribosome appears to be dependent on the hypusine residue as well as on an intact ribosomal complex, since eIF5A binding is disrupted in the presence of EDTA or by RNase A treatment (Jao and Chen, 2006; Zanelli et al., 2006). However, there is no information available as to which parts of the eIF5A molecule or of the ribosome are involved in the interaction. Identification of the exact binding site of the hypusine residue and eIF5A on the ribosome will shed light on its mode of action in translation.
Due to the lack of hypusine modification enzymes in bacteria, expression of the eIF5A gene alone in E.coli leads to overproduction of unmodified eIF5A precursor, eIF5A(Lys), which is devoid of biological activity. Although eIF5A exists mainly as the fully modified hypusine-containing form in mammalian cells and tissues, it is difficult to isolate native eIF5A in quantity and purity suitable for diverse structural and functional studies. This is especially true for the second isoform of eIF5A, eIF5A-2, which exists at a level too low to be detected in normal mammalian cells and tissues (Clement et al., 2006).
Our initial attempt to produce hypusine-containing eIF5A in E.coli using three plasmids with different drug resistance, and individually encoding eIF5A, DHS and DOHH, was not successful, because each plasmid must have a unique but compatible origin of replication.
For this reason, we employed a modular polycistronic system (Tan, 2001) and have cloned human eIF5A-1, human DHS and human DOHH complete coding sequences into one polycistronic vector (Scheme 1). After induction of protein expression by isopropylthiogalactopyranoside (IPTG), all three proteins were produced at high levels and eIF5A(Lys) was effectively modified to the eIF5A(Hpu) in the cellular environment of E.coli. The polycistronic constructs, pST39/eIF5A/DHS and pST39/eIF5A/DHS/DOHH have permitted isolation from E.coli of a large quantity of highly pure, biologically active, recombinant eIF5A containing the deoxyhypusine or hypusine residue.
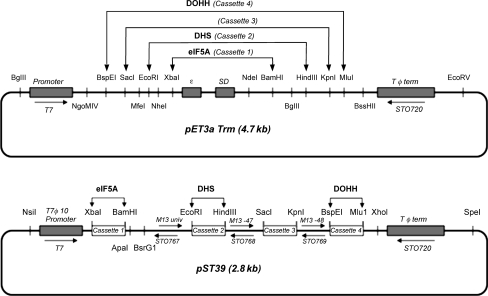
Scheme 1.
Construction of a polycistronic vector encoding eIF5A, DHS and DOHH. Schematic presentations of the cloning of eIF5A, DHS and DOHH in the monocistronic pET3aTrm transfer expression vector (A) and the polycistronic pST39 vector (B). The transcription start and termination sites are indicated. Each ORF of eIF5A, DHS and DOHH was individually subcloned into the pET3aTrm vector at the Nde1/BamH1, Nde1/HinIII and Nde1/Mlu1, to generate three transfer vectors, pET3aTrm/eIF5A, pET3aTrm/DHS, pET3aTrm/DOHH, respectively. The eIF5A cassette 1 was cut from the pET3aTrm/eIF5A and the cassette transferred to pST39 vector at the Xba/BamH1 site. The DHS cassette was inserted in the pST39/eIF5A vector and then the DOHH cassette was subsequently inserted in the pST39/eIF5A/DHS vector at the indicated site (modified from Tan, 2001).
Materials and Methods
Materials
The polycistronic expression vector pST39 and a transfer vector pET3aTr (Tan, 2001) were kindly provided by Dr. Song Tan (Pennsylvania State University). [1,8-3H]Spermidine HCl (15–25 Ci/mmol), was purchased from PerkinElmer/NEN. Precast Tris–glycine and NuPAGE (Bis-Tris) gels, electrophoresis buffers and Simply Blue staining solution were from Invitrogen, ECL Plus Western Blotting Detection system and Q-sepharose from GE Healthcare. The Quick Change Site-Directed Mutagenesis Kit and E.coli competent cells BL21(DE3)pLysS were from Stratagene. The protease inhibitor cocktail was purchased from Roche. DEAE Sephacel and SP-sepharose were from Pharmacia. A monoclonal antibody against recombinant human eIF5A (aa58–154) was purchased from BD Biosciences. Rabbit polyclonal antibodies against human DHS and DOHH were produced by Covance. The oligonucleotide primers were synthesized by Integrated DNA Technologies, Inc. Human eIF5A-1-radiolabeled deoxyhypusine-containing protein was prepared in an in vitro DHS reaction as described (Park et al., 2003). Bovine testis eIF5A was purified following a protocol (S. B. Lee et al., unpublished results) similar to that for recombinant eIF5A(Hpu) described below.
Methods
Construction of polycistronic expression vectors encoding eIF5A-1, DHS and DOHH
The complete coding sequences of human eIF5A-1, human DHS and human DOHH were individually subcloned into the transfer vector pET3aTrm and each cassette was transferred to pST39 in the order eIF5A, DHS and DOHH (Scheme 1). The pET3aTrm vector was generated from pET3aTr by removal of extra EcoRI and HindIII sites outside of the multiple cloning sites. In order to remove the extra HindIII site at 4684 and EcoRI site at 4718, site-directed mutagenesis was performed using the primer sets, 5′-aaactaccgcattagagcttatcgatgataagctg-3′ and 5′-cagcttatcatcgataagctctaatgcggtagttt-3′ for the A4684G substitution and 5′-agctgtcaaacatgagagttcttgaagacgaaaggg-3′ and 5′-ccctttcgtcttcaagaactctcatgtttgacagct-3′ for the A4718G substitution. Human eIF5A-1 ORF was amplified by PCR using the primer sets, 5′-cta cca gta tcg gca cat atg gca gat gac ttg gac ttc g-3′ and 5′-cta cca gta tcg gca gga tcc tta ttt tgc cat ggc ctt gat tgc-3′ and subcloned into NdeI/BamHI sites of pET3aTrm vector. The pET3aTrm/eIF5A was digested with XbaI/BamHI and the resulting cassette 1 (eIF5A) was subcloned into XbaI/BamHI (site 1) of pST39 to generate pST39/eIF5A. Human DHS ORF was amplified using the primer sets 5′-cta cca gta tcg gca cat atg gga ggt ttc ctg gaa cgg g-3′ and 5′-cta cca gta tcg gca aag ctt tca gtc ctc gtt ctt ctc atg cat-3′ and subcloned at the NdeI/HindIII site of pET3aTrm vector. The vector pET3aTrm/DHS was digested with EcoRI/HindIII and the DHS cassette 2 was subcloned at the EcoRI/HindIII site of pST39/eIF5A to generate pST39/eIF5A/DHS. Human DOHH ORF was amplified using the primer sets, 5′-cta cca gta tcg gca cat atg gtg acg gag cag gag gtg g-3′ and 5′-cta cca gta tcg gca acg cgt cta gga ggg ggc ccc gcg cag-3′ and subcloned into NdeI/MluI sites in the pET3aTrm vector. The pET3aTrm/DOHH was digested with BspEI and MluI and the DOHH cassette 4 was subcloned into the BspEI/MluI site of the pST39/eIF5A/DHS vector to generate a polycistronic vector encoding all three proteins. Each recombinant vector was sequenced to confirm the accuracy of the sequence and insertion before progressing to the next step.
Expression and modification of eIF5A in BL21(DE3)pLys cells: detection by labeling with[3H]spermidine
We used BL21(DE3)pLysS competent cells, which allow tight control of background expression under the T7 promoter. BL21(DE3)pLysS cells were transformed with the recombinant pST39 recombinant constructs and grown in the LB medium (2 ml) containing 25 μg/ml of chloramphenicol (pLys vector) and 200 μg/ml of ampicillin (pET3aTrm or pST39 vector). When the cell density reached 0.6 OD600nm, IPTG (1 mM) was added to the culture medium and incubation continued for 1–6 h at 37°C, as indicated, in the presence of 5 μCi/ml of [3H]spermidine. Protein expression was also induced at 18°C and was monitored for 2–18 h by labeling with [3H]spermidine. Cells were harvested by centrifugation and cell pellets were sonicated in 0.2 ml of Buffer A (50 mM Tris acetate buffer pH 7.0, 0.1 mM EDTA, 1 mM DTT) containing protease inhibitor cocktail (Roche). After removal of debris, aliquots of the clarified supernatant were used for SDS/PAGE for detection of proteins by Coomassie-Blue staining or by western blotting and for analysis of hypusine or deoxyhypusine in the acid hydrolysates of TCA-precipitated proteins after thorough washing to remove non-covalently bound radioactive polyamines as described previously (Park et al., 2003).
Purification of recombinant eIF5A(Hpu), eIF5A(Dhp) and eIF5A(Lys) from E.coli BL21(DE3)pLysS cells after IPTG -induction
For protein purification of eIF5A(Hpu), BL21(DE3)pLysS cells transformed with pST39/eIF5A/DHS/DOHH were harvested after 4 h of induction with 1 mM IPTG at 37°C. The cell pellets from 6 l of culture (∼20 g) were resuspended in ice-cold 100 ml Buffer A and were lysed by sonication. The cell lysate was centrifuged at 25 000 × g (in a JA20 rotor) for 30 min and the clarified lysate was separated. To this lysate, the radioactive tracer, human e|IF5A-1([3H]Dhp) (1 000 000 dpm, 50 pmol, ∼0.8 μg) was added to follow purification of eIF5A(Dhp) or eIF5A(Hpu). This mixture was applied to an anion exchange column (70 ml DEAE Sephacel, 2.6 cm × 14 cm) and washed with 100 ml of Buffer A. Proteins were eluted with a stepwise gradient (50 ml at each step) of 0.1, 0.2, 0.3, 0.4 and 0.5 M KCl (in Buffer A) and 25 ml fractions were collected. An aliquot of each fraction was counted and also analyzed by SDS-PAGE. The fractions containing high radioactivity (eluting at 0.3–0.4 M KCl) were pooled. This protein pool was precipitated with ammonium sulfate to obtain the 40–80% fraction and the precipitated proteins were redissolved in Buffer A and dialyzed against Buffer A for 3 h. The dialyzed protein was applied to a SP-sepharose column (20 ml, 1.3 cm × 13 cm). After washing the column with 50 ml of Buffer A, bound proteins were eluted with a stepwise gradient of 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4 and 0.5 M KCl in Buffer A (25 ml each step) and 5 ml fractions were collected. After analysis of each fraction by radioactivity measurement and SDS-PAGE, the early peak containing eIF5A(Lys) and the later radioactive peak containing eIF5A(Hpu) were separately pooled for amino acid analysis and testing of activity. For further purification of eIF5A(Hpu), the later protein pool was dialyzed against buffer A and applied to an anionic exchange resin Q-sepharose (10 ml size). After washing with 25 ml of Buffer A, the protein was eluted with a stepwise gradient of 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4 and 0.5 M KCl in Buffer A (15 ml each step) and 5 ml fractions were collected. The radioactivity and eIF5A(Hpu) protein eluted at 0.3–0.35 M KCl from the Q-sepharose column. eIF5A(Dhp) recombinant protein was isolated from BL21(DE3)pLysS cells transformed with pST39/eIF5A/DHS following the same protocol as described above. eIF5A recombinant proteins eIF5A(Lys), eIF5A(Dhp) and eIF5A(Hpu) induced at 18°C were also purified by this protocol.
In vitro methionyl-puromycin synthesis assay for eIF5A activity
The reaction mixture (in 25 μl) contained 20 mM Tris–HCl pH 7.4, 0.1 M KCl, 1 mM DTT, 3 mM MgCl2, 0.8 mM GTP, 1.5 mM puromycin, 0.15 A260 unit of 60S, 0.07 A260 unit of 40 S, 33 μM AUG, 0.375 μg of eIF1A, 0.75 μg of eIF2, 3.3 μg of eIF3, 0.48 μg of eIF5B and 8 pmol of [3H]Met-tRNA [120 000 dpm, 15 000 dpm/pmol] or [35S]Met-tRNA (64 000 dpm, 8000 dpm/pmol) and the indicated amount of eIF5A(Hpu) or eIF5A(AcHpu). The mixture was incubated at 37°C for 20 min. To each tube, 0.4 ml of 0.2 M Na2HPO4 buffer, pH 8.0, was added. One milliliter of ethyl acetate was added and the tubes shaken for 10 min. The tubes were centrifuged in a microfuge for 10 min. The upper layer (∼0.9 ml) was carefully removed and the radioactivity was counted.
Results
Coexpression of eIF5A, DHS and DOHH and effective modification of eIF5A in BL21(DE3)pLys cells
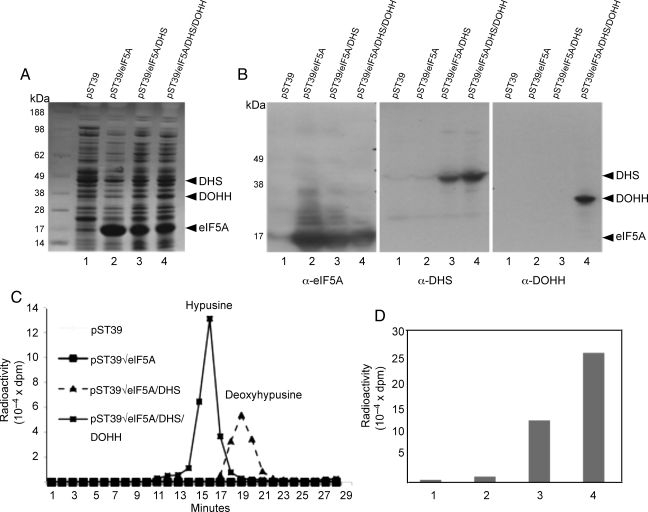
BL21(DE3)pLysS cells were transformed with the empty vector pST39 or vectors encoding eIF5A alone, eIF5A plus DHS or eIF5A plus DHS plus DOHH. eIF5A was expressed at a high level when expressed from the pST39 vector alone or in combination with DHS or DOHH (Fig. 1A, lanes 2–4), although the relative expression level of eIF5A was somewhat reduced in cells coexpressing the enzymes. The expression levels of DHS and DOHH were far less than that of eIF5A, but the enzyme proteins were clearly detectable by staining (Fig. 1A) and also by western blotting with the respective antibodies (Fig. 1B).
Fig. 1.
Expression of eIF5A, DHS and DOHH in BL21(DE3)pLysS cells. E.coli cells transformed with the pST39 empty vector (lane 1), pST39/eIF5A (lane 2), pST39/eIF5A/DHS (lane 3) or pST39/eIF5A/DHS/DOHH (lane 4) were induced with 1 mM IPTG for 4 h in the presence or absence of [3H]spermidine (5 μCi/ml). The protein expression was measured after SDS-PAGE by Coomassie-Blue staining (A) or by western blotting (B). Hypusine modification was measured by ion exchange chromatographic separation of radioactive deoxyhypusine or hypusine in the acid hydrolysate of proteins (C) and the total radioactivity in hypusine (lane 4) or deoxyhypusine peak (lane 3) in each sample is shown (D).
In order to determine whether the eIF5A precursor protein was being modified in E.coli cells coexpressing the enzymes, [3H]spermidine was added to the culture medium at the time of IPTG induction and the radioactivity incorporated into eIF5A after 4 h of incubation was analyzed by ion exchange chromatography after acid hydrolysis of the TCA-precipitated proteins (Fig. 1C). There was virtually no eIF5A protein labeling in E.coli cells transformed with the pST39 empty vector or with that encoding eIF5A alone (Fig. 1C and D). In cells transformed with pST39/eIF5A/DHS vector, only radioactive deoxyhypusine, but no hypusine, was formed in the protein fraction, indicating modification of eIF5A(Lys) by DHS in these cells (Fig. 1C and D). On the other hand, in BL21(DE3)pLysS expressing all three proteins, only radioactive hypusine, but no deoxyhypusine, was detected (Fig. 1C and D) indicating that any eIF5A(Dhp) formed was efficiently hydroxylated by DOHH. The level of radiolabeling of deoxyhypusine was consistently lower (by ∼ 2-fold) than that of hypusine, when the expression level of eIF5A was similar. We previously reported that the DHS reaction is reversible in vitro resulting in back conversion of eIF5A(Dhp) to eIF5A(Lys) (Park et al., 2003). Thus, the low levels of eIF5A(Dhp) labeling, as well as its protein, compared with those of eIF5A(Hpu), may be due to the reversal of the DHS reaction under physiological condition in E.coli cells. Unlike the DHS reaction, the DOHH reaction is irreversible, thereby leading to the accumulation of the product as hypusine-containing eIF5A.
The effects of time and temperature on the expression of recombinant proteins and hypusine modification
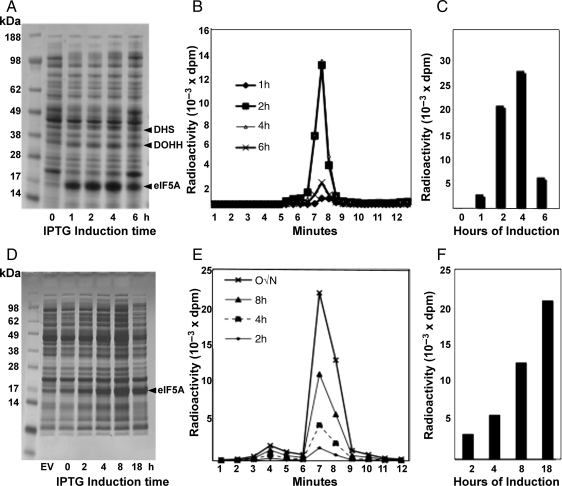
We first examined the time course of protein expression and hypusine modification using the BL21(DE3)pLysS cells transformed with pST39/eIF5A/DHS/DOHH (Fig. 2) upon IPTG induction at 37°C. At this temperature, eIF5A protein was rapidly expressed after addition of IPTG and a prominent band was visible at 1 h of induction. The time course of expression of DHS and DOHH probably followed the same pattern as eIF5A, being produced from the same polycistronic vector. The level of hypusine-modified eIF5A, indicated by the amount of radioactivity incorporation, was quite low at 1 h, but reached a maximum at 4 h of IPTG induction at 37°C (Fig. 2B and C). Since [3H]spermidine was rapidly taken up to reach the maximum level within 1 h (data not shown), the delay in eIF5A modification may reflect the in vivo reaction time required for the enzymatic modification of eIF5A(Lys) in cells. That no deoxyhypusine form was accumulated at any time point indicates that DOHH effectively hydroxylated all the eIF5A(Dhp) formed. The level of eIF5A protein and its hypusine modification was far less at 6 h than at 4 h. This may be due to the degradation of expressed proteins, loss of the enzyme activities and/or loss of viable cells upon prolonged expression of these foreign proteins.
Fig. 2.
Time course of protein expression and hypusine modification upon IPTG induction at 37°C (A–C) and at 18°C (D–F). BL21(DE3)pLysS cells transformed with pST39/heIF5A/hDHS/hDOHH were induced with IPTG at 37°C or 18°C in the absence or presence of [3H]spermidine (5 μCi/ml) for the times indicated. Protein expression was examined by Coomassie-Blue staining after SDS-PAGE (A, D) and the extent of the hypusine modification was measured by measuring radioactivity in the hypusine after ion exchange chromatographic separation (B, E). Total amount of hypusine formed at each induction time point is shown as a bar graph (C, F).
Since eIF5A recombinant protein is rapidly synthesized at 37°C (Fig. 1), eIF5A(Hpu) produced at this temperature may not have proper folding required for its biological activity. IPTG induction at lower temperatures (15–23°C) slows the rate of transcription and translation (Sorensen and Mortensen, 2005; Sahdev et al., 2008) and slow production of recombinant proteins prevents protein aggregation and inclusion body formation. Therefore, we also induced ec-eIF5A(Hpu) at 18°C. Both the rates of synthesis of eIF5A and hypusine modification were markedly reduced at 18°C (Fig. 2D–F). eIF5A protein level and radiolabeling of hypusine continued to increase up to 18 h at 18°C. Only radiolabeled hypusine, but no deoxyhypusine intermediate, was detected in these cells (Fig. 2D–F).
Purification of eIF5A(Hpu), eIF5A(Dhp) and eIF5A(Lys) from BL21(DE3)pLysS cells
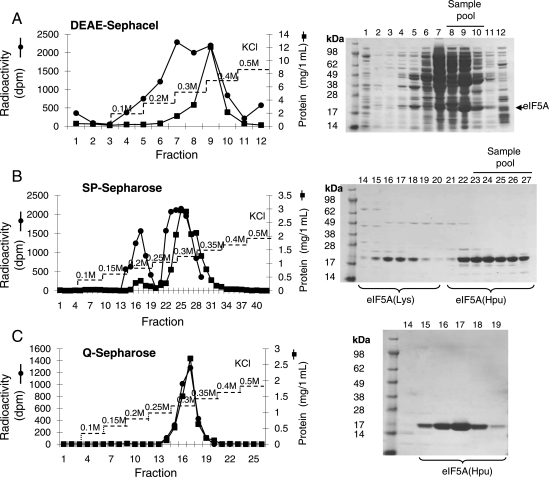
First we purified eIF5A(Hpu) from E.coli cells harboring pST39/eIF5A/DHS/DOHH vector after IPTG induction at 37°C for 4 h. Since eIF5A has a basic N-terminal domain and an acidic C-terminal domain, it binds tightly to both anionic and cationic exchange resins. This bipolar nature enabled us to purify eIF5A in three simple steps, ion exchange chromatography on DEAE Sephacel (anionic exchange resin), ammonium sulfate precipitation (40–80%), followed by ion exchange chromatography on SP-sepharose (cationic exchange resin) (Fig. 3). [3H]Deoxyhypusine-containing eIF5A generated from the in vitro DHS reaction was used as a tracer to follow the purification of eIF5A(Dhp) or eIF5A(Hpu).
Fig. 3.
Purification of eIF5A(Hpu) from BL21(DE3)pLysS/pST39/eIF5A/DHS/DOHH cells. eIF5A(Hpu) was purified from E.coli cells induced with IPTG at 37°C for 4 h as described under the section Materials and methods. Chromatography on (A) DEAE Sephacel, (B) SP-Sepharose and (C) Q-Sepharose. Aliquots from each fraction were used for measurement of radioactivity by liquid scintillation counting, for protein content by the BioRad assay and for purity by SDS-PAGE and Coomassie-Blue staining of the gels. Pooled fractions are indicated by a horizontal line on top of fraction numbers of the SDS gels.
Two peaks of eIF5A proteins were separated in the SP-Sepharose step (Fig. 3B). Conversion of the Lys50 of eIF5A precursor to the hypusine or deoxyhypusine residue adds an extra amino group and increases the pI of the protein from 5.2 to 5.3, leading to a resolution of unmodified and modified eIF5A on SP-Sepharose (Fig. 3B). The early peak of protein in fractions 15–19 contained eIF5A protein (30%) but no radioactivity, indicating that this is the eIF5A precursor, eIF5A(Lys). The later peak (fractions 22–27) contained the 18 kDa protein (70%) and radioactivity indicating that it is the hypusine-containing protein, eIF5A(Hpu). The assignment of the later peak as eIF5A(Hpu) was confirmed by the fluorometric detection of ∼1 mol/mol of hypusine by amino acid analysis of its acid hydrolysate (not shown), whereas no hypusine or deoxyhypusine was detected in the early peak. A refined anionic exchange chromatography on Q-Sepharose resulted in higher purity (Fig. 3C). A total yield of 50 mg of fully modified eIF5A was obtained from a 6 l culture of IPTG-induced cells harboring pST39/eIF5A/DHS/DOHH.
The deoxyhypusine-containing intermediate, eIF5A(Dhp), was purified from BL21(DE3)pLysS cells transformed with pST39/eIF5A/DHS vector following the same protocol as above, and the data obtained (not shown) was very similar to that shown in Fig. 3. In this case, the yield was ∼15 mg from 6 l of culture.
Recombinant eIF5A proteins induced at 18°C showed the same chromatographic properties as those produced at 37°C and were also purified following the same protocol as that shown in Fig. 3, with the yields slightly lower than that induced at 37°C.
Activities of recombinant eIF5A proteins and bovine testis eIF5A
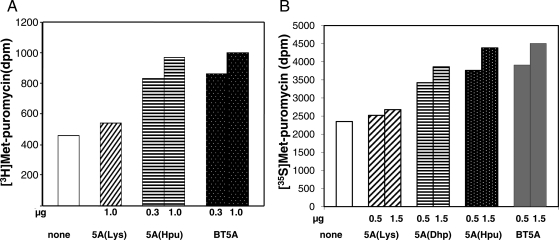
We have compared the activities of recombinant eIF5A(Lys), eIF5A(Dhp), eIF5A(Hpu) with native eIF5A(Hpu) purified from bovine testis in the methionyl-puromycin synthesis assay [Fig. 4, using [3H]Met-tRNA (A) and [35S]Met-tRNA (B)]. The recombinant eIF5A(Hpu) produced at 18°C was nearly as active as bovine testis eIF5A, indicating that this protein has the proper folding as the native eIF5A(Hpu) (A and B). The recombinant deoxyhypusine-containing intermediate, eIF5A(Dhp) produced at 18°C consistently showed partial activity, whereas unmodified eIF5A precursor, eIF5A(Lys) proteins were devoid of activity, underscoring the importance of the hypusine/deoxyhypusine residue in the activity of eIF5A. In contrast to eIF5A(Hpu) induced at 18°C, the recombinant eIF5A(Hpu) induced at 37°C showed fairly low activity (<40%) compared with the mammalian eIF5A (data not shown), probably due to lack of proper folding.
Fig. 4.
Comparison of activities of recombinant eIF5A proteins and bovine testis eIF5A. The methionyl-puromycin synthesis assay was carried out as described under the section ‘Materials and methods’, using either [3H]methionyl-tRNA (A) or [35S]methionyl-tRNA (B) in two independent experiments. The dpm for [3H]methionyl-puromycin formed is twice that of [35S]methionyl-puromycin, since the specific radioactivity of [3H]methionyl-tRNA is approximately twice that of [35S]methionyl-tRNA. Abbreviations are: 5A(Lys), eIF5A(Lys); 5A(Dhp), eIF5A(Dhp); 5A(Hpu), eIF5A(Hpu); BT5A, bovine testis eIF5A.
Discussion
eIF5A is the only protein that undergoes a unique post-translational modification that converts one of its lysine residues to hypusine. The hypusine modification is essential for the activity of eIF5A in vitro and for eukaryotic cell proliferation. Since the hypusine modification creates a long basic side chain on an exposed basic loop (Cano et al., 2008) and since eIF5A binding to ribosome is dependent on the hypusine residue, it is expected to serve as a critical anchoring site on the ribosome. However, there is no experimental evidence on the docking site of eIF5A and its hypusine residue on the ribosome. A highly pure hypusine-containing eIF5A and various mutant proteins would be desirable in probing the binding sites of eIF5A on ribosome. For this purpose and for other structure/function studies, we have undertaken to produce recombinant hypusine-containing protein in E.coli by employing a polycistronic vector and have accomplished a production of a large quantity of highly pure, biologically active eIF5A(Hpu) and eIF5A(Dhp) recombinant human proteins from E.coli. The same polycistronic vector strategy can be used to engineer production of rare, hypusine-containing eIF5A-2 isoforms from mammals and any isoforms from other eukaryotic species including yeast. These recombinant proteins will be useful tools in structure/function studies of a variety of eukaryotic hypusine-containing eIF5A isoforms. Furthermore, the polycistronic coexpression system will also permit isolation of the eIF5A/DHS or eIF5A/DOHH complexes for determination of their crystal structure, which will unveil the secret of the strict specificity of the hypusine modification reactions.
Many cellular processes are regulated by multi-component protein complexes. The efficiency of reconstitution of a multi-protein complex in vitro from each purified component may be low, since it requires optimization of many refolding parameters. A polycistronic vector has been developed to coexpress several recombinant proteins in E.coli and to isolate a multi-protein complex assembled in the cellular in vivo environment (Selleck and Tan, 2008). This approach has been successfully used for coexpression of HisTrxN-VHL-elonginB, HisTrxN-VHL-elonginB-elonginC complexes (Tan, 2001), for the yeast Ada2/Ada3/Gcn5 and Piccolo NuA4 histone acetyltransferase complexes (Barrios et al., 2007) and a variety of other multi-protein complexes (Selleck and Tan, 2008). The two enzymes involved in hypusine biosynthesis, DHS and DOHH, are totally specific for eIF5A and do not modify any other cellular proteins. The strict specificity of the DHS and DOHH reactions is based on the specific protein/protein interaction between eIF5A and the enzymes. The structural basis of this specificity has been characterized by using a variety of site specific or truncated mutants of eIF5A and its modifying enzymes and the reaction has been shown to be dependent on the intact core structure of the N-terminal domain of eIF5A (Park, 2006). In fact, the binding of eIF5A/DHS or eIF5A(Dhp)/DOHH represents the most specific protein–protein interactions known to date. However, the structures of the complexes of eIF5A/DHS or eIF5A(Dhp)/DOHH have not been determined and the key elements in these interactions are not known. The polycistronic coexpression system may permit isolation of the eIF5A/DHS or eIF5A/DOHH complexes for determination of their crystal structure, which will unveil the secret of the strict specificity of the hypusine modification reactions.
Production of biologically active, post-translationally modified eukaryotic protein in E.coli by coexpression of a substrate protein with a modification enzyme using a dual plasmid or dual expression system was accomplished for substrates of protein kinase and N-myristoylation (Duronio et al., 1990; Yue et al., 2000; Sugase et al., 2008). However, for a modification requiring more than two enzymes, coexpression using a polycistronic vector can be successfully applied as demonstrated here for production of eIF5A(Hpu). Such a polycistronic system should be generally applicable for over-production of other mammalian recombinant proteins that require eukaryotic-specific modifications for their activity or even for a bacterial protein requiring multi-step modification, where the endogenous enzyme levels are limiting.
A high-resolution crystal structure of the eukaryotic 80S ribosome is not yet available (Taylor et al., 2009). The mode of ribosomal binding has been determined for only a few eukaryotic initiation factors, including eIF3j (Fraser et al., 2007), eIF1A (Yu et al., 2009) and eIF5B (Unbehaun et al., 2007), by directed hydroxyl radical probing. Recent reports suggest that eIF5A functions at the elongation step rather than the initiation step of translation (Zanelli et al., 2006; Gregio et al., 2009; Saini et al., 2009). Thus, eIF5A may be functionally related to its bacterial ortholog, elongation factor P (EF-P) which has similar sequence and structure as eIF5A, although it lacks the hypusine modification (Glick and Ganoza, 1975; Hanawa-Suetsugu et al., 2004). The crystal structure of EF-P bound on the bacterial 70S ribosome along with the initiator tRNA and a short piece of mRNA reveals its binding between the P-site (peptidyl tRNA site) and E site (exiting tRNA site). Thus, it suggests a role for EF-P in stabilizing initiator tRNA at the P-site and thereby enhancing the first peptide bond formation (Blaha et al., 2009). The long basic side chain of the conserved Arg (R32) or a hypothetical beta-lysyl-lysine derivative (Navarre et al., 2010) at the same position corresponding to that of hypusine in eIF5A binds to the peptidyl transferase center. A homology model with the hypusine residue in place of Arg32 of EF-P shows the long basic side chain of the hypusine reaching into the peptidyl transferase center of the 70S ribosome (Blaha et al., 2009). By analogy, a similar mode of eIF5A(Hpu) binding on the eukaryotic ribosome and of action in eukaryotic translation may be invoked. However, whether the mechanism of eIF5A in translation is similar to that of EF-P awaits experimental determination of the mode of binding of the hypusine-containing eIF5A on the eukaryotic ribosome.
Highly pure hypusine-containing eIF5A protein, or its mutant proteins, produced with the polycistronic vector system will be crucial in future studies designed to determine its binding site on the ribosome and its mode of action in translation. Through the use of the pST39/eIF5A/DHS/DOHH vector, we can generate a variety of eIF5A mutants (with amino acid substitutions outside of the hypusine loop) that still contain the hypusine residue and thereby assess the role of each amino acid in the structure/function of eIF5A independently of the hypusine modification. Such hypusine-containing eIF5A mutant proteins will be useful in probing and identification of the binding sites on the ribosome and also those critical in its activity.
Funding
The research was supported in part by the Intramural Research Program of National Institute of Dental and Craniofacial Research (NIDCR), NIH, NIH extramural Grant R01 GM092927 (to C.S.F.) and by the Human Frontier Science Program fellowship LT00575/2007-L (to M.S.).
Acknowledgments
We thank Dr. Edith C. Wolff (NIDCR, NIH) for critical reading of the manuscript and helpful suggestions.
References
- Barrios A., Selleck W., Hnatkovich B., Kramer R., Sermwittayawong D., Tan S. Methods. 2007;41:271–277. doi: 10.1016/j.ymeth.2006.08.007. doi:10.1016/j.ymeth.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R., Hershey J.W. J. Biol. Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- Blaha G., Stanley R.E., Steitz T.A. Science. 2009;325:966–970. doi: 10.1126/science.1175800. doi:10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers T.L., Lakanen J.R., Coward J.K., Pegg A.E. Biochem. J. 1994;303:363–368. doi: 10.1042/bj3030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano V.S., Jeon G.A., Johansson H.E., Henderson C.A., Park J.H., Valentini S.R., Hershey J.W., Park M.H. FEBS J. 2008;275:44–58. doi: 10.1111/j.1742-4658.2007.06172.x. doi:10.1111/j.1742-4658.2007.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I., Gross S.R., Kinzy T.G., Chen K.Y. Mol. Genet. Genomics. 2006;275:264–276. doi: 10.1007/s00438-005-0086-4. doi:10.1007/s00438-005-0086-4. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M.K., Park M.H., Tabor H. Proc. Natl Acad. Sci. USA. 2008;105:6554–6559. doi: 10.1073/pnas.0710970105. doi:10.1073/pnas.0710970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.Y., Liu A.Y. Biol. Signals. 1997;6:105–109. doi: 10.1159/000109115. doi:10.1159/000109115. [DOI] [PubMed] [Google Scholar]
- Clement P.M., Johansson H.E., Wolff E.C., Park M.H. FEBS J. 2006;273:1102–1114. doi: 10.1111/j.1742-4658.2006.05135.x. doi:10.1111/j.1742-4658.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C.A., Cano V.S., Rangel S.M., et al. FEBS J. 2008;275:1874–1888. doi: 10.1111/j.1742-4658.2008.06345.x. doi:10.1111/j.1742-4658.2008.06345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R.J., Jackson-Machelski E., Heuckeroth R.O., Olins P.O., Devine C.S., Yonemoto W., Slice L.W., Taylor S.S., Gordon J.I. Proc. Natl Acad. Sci. USA. 1990;87:1506–1510. doi: 10.1073/pnas.87.4.1506. doi:10.1073/pnas.87.4.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.S., Berry K.E., Hershey J.W., Doudna J.A. Mol. Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. doi:10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Glick B.R., Ganoza M.C. Proc. Natl Acad. Sci. USA. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. doi:10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregio A.P., Cano V.P., Avaca J.S., Valentini S.R., Zanelli C.F. Biochem. Biophys. Res. Commun. 2009;380:785–790. doi: 10.1016/j.bbrc.2009.01.148. First published on January 29, 2009 doi:10.1016/j.bbrc.2009.01.148 doi:10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- Hanauske-Abel H.M., Park M.H., Hanauske A.R., Popowicz A.M., Lalande M., Folk J.E. Biochim. Biophys. Acta. 1994;1221:115–124. doi: 10.1016/0167-4889(94)90003-5. doi:10.1016/0167-4889(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K., Sekine S., Sakai H., et al. Proc. Natl Acad. Sci. USA. 2004;101:9595–9600. doi: 10.1073/pnas.0308667101. doi:10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao D.L., Chen K.Y. J. Cell. Biochem. 2006;97:583–598. doi: 10.1002/jcb.20658. doi:10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- Joe Y.A., Wolff E.C., Park M.H. J. Biol. Chem. 1995;270:22386–22392. doi: 10.1074/jbc.270.38.22386. doi:10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- Kemper W.M., Berry K.W., Merrick W.C. J. Biol. Chem. 1976;251:5551–5557. [PubMed] [Google Scholar]
- Landau G., Bercovich Z., Park M.H., Kahana C. J. Biol. Chem. 2010;285:12474–12481. doi: 10.1074/jbc.M110.106419. First published on February 24, 2010, doi:10.1074/jbc.M110.106419 doi:10.1074/jbc.M110.106419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W.W., Zou S.B., Roy H., et al. Mol. Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. doi:10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.H. J. Biochem. (Tokyo) 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.H., Cooper H.L., Folk J.E. Proc. Natl Acad. Sci. USA. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. doi:10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.H., Wolff E.C., Lee Y.B., Folk J.E. J. Biol. Chem. 1994;269:27827–27832. [PubMed] [Google Scholar]
- Park M.H., Joe Y.A., Kang K.R. J. Biol. Chem. 1998;273:1677–1683. doi: 10.1074/jbc.273.3.1677. doi:10.1074/jbc.273.3.1677. [DOI] [PubMed] [Google Scholar]
- Park J.-H., Wolff E.C., Folk J.E., Park M.H. J. Biol. Chem. 2003;278:32683–32691. doi: 10.1074/jbc.M304247200. doi:10.1074/jbc.M304247200. [DOI] [PubMed] [Google Scholar]
- Park J.-H., Aravind L., Wolff E.C., Kaevel J., Kim Y.S., Park M.H. Proc. Natl Acad. Sci. USA. 2006;103:51–56. doi: 10.1073/pnas.0509348102. doi:10.1073/pnas.0509348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.H., Nishimura K., Zanelli C.F., Valentini S.R. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. First published on December 8, 2009, doi 10.1007/s00726-009-0408-7 doi:10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A.E. IUBMB Life. 2009;61:880–894. doi: 10.1002/iub.230. doi:10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahdev S., Khattar S.K., Saini K.S. Mol. Cell Biochem. 2008;307:249–264. doi: 10.1007/s11010-007-9603-6. doi:10.1007/s11010-007-9603-6. [DOI] [PubMed] [Google Scholar]
- Saini P., Eyler D.E., Green R., Dever T.E. Nature. 2009;459:118–121. doi: 10.1038/nature08034. First published on March 23, 2009, doi:10.1038/nature08034 doi:10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Abid M.R., Miyazaki M. FEBS Lett. 1996;384:151–154. doi: 10.1016/0014-5793(96)00310-9. doi:10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- Schnier J., Schwelberger H.G., Smit-McBride Z., Kang H.A., Hershey J.W. Mol. Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader R., Young C., Kozian D., Hoffmann R., Lottspeich F. J. Biol. Chem. 2006;281:35336–35346. doi: 10.1074/jbc.M601460200. doi:10.1074/jbc.M601460200. [DOI] [PubMed] [Google Scholar]
- Selleck W., Tan S. Curr. Protoc. Protein Sci., Chapter 5. 2008;52:5.21.1–5.21.21. doi: 10.1002/0471140864.ps0521s52. First published May 1, 2008, doi: 10.1002/0471140864.ps0521s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen H.P., Mortensen K.K. Microb. Cell Fact. 2005;4:1. doi: 10.1186/1475-2859-4-1. doi:10.1186/1475-2859-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugase K., Landes M.A., Wright P.E., Martinez-Yamout M. Protein Expr. Purif. 2008;57:108–115. doi: 10.1016/j.pep.2007.10.018. doi:10.1016/j.pep.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. Protein Expr. Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. doi:10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- Taylor D.J., Devkota B., Huang A.D., Topf M., Narayanan E., Sali A., Harvey S.C., Frank J. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. doi:10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Park I., Hong B.S., Nedyalkova L., Tempel W., Park H.W. Proteins. 2009;75:1040–1045. doi: 10.1002/prot.22378. doi:10.1002/prot.22378. [DOI] [PubMed] [Google Scholar]
- Unbehaun A., Marintchev A., Lomakin I.B., Didenko T., Wagner G., Hellen C.U., Pestova T.V. EMBO J. 2007;26:3109–3123. doi: 10.1038/sj.emboj.7601751. doi:10.1038/sj.emboj.7601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini S.R., Casolari J.M., Oliveira C.C., Silver P.A., McBride A.E. Genetics. 2002;160:393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhl T., Klier H., Ammer H. Mol. Gen. Genet. 1993;241:305–311. doi: 10.1007/BF00284682. [DOI] [PubMed] [Google Scholar]
- Yu Y., Marintchev A., Kolupaeva V.G., et al. Nucleic Acids Res. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. doi:10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue B.G., Ajuh P., Akusjarvi G., Lamond A.I., Kreivi J.P. Nucleic Acids Res. 2000;28:E14. doi: 10.1093/nar/28.5.e14. doi:10.1093/nar/28.5.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli C.F., Maragno A.L., Gregio A.P., Komili S., Pandolfi J.R., Mestriner C.A., Lustri W.R., Valentini S.R. Biochem. Biophys. Res. Commun. 2006;348:1358–1366. doi: 10.1016/j.bbrc.2006.07.195. doi:10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- Zuk D., Jacobson A. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. doi:10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]