Abstract
Context: Among the genomic loci harboring potential candidate genes for prostatic cancer (PCa) is the 2q31-33 chromosomal region that harbors the gene encoding phosphodiesterase 11A (PDE11A). In addition, the combined cancer genome expression metaanalysis datasets included PDE11A among the top 1% down-regulated genes in PCa.
Objective: In the present study, we screened 50 unrelated PCa patients of Brazilian descent for PDE11A coding defects.
Design: The study consisted of PDE11A sequencing, in vitro functional assays, and immunostaining analysis.
Results: We identified eight different sequence alterations in 15 patients (30%): one stop-codon and seven missense mutations. Three of the variants (R202C, Y658C, and E840K) were novel, and the remaining five (Y727C, R804H, R867G, M878V, and R307X) have been associated with predisposition to adrenal or testicular tumors. The overall prevalence of PDE11A-inactivating sequence variants among PCa patients was significantly higher than in 287 healthy controls (0.16 vs. 0.051, respectively, P < 0.001, odds ratio 3.81, 95% confidence interval 1.86–7.81) and the R202C, Y658C, and E840K substitutions were not found in controls. All missense mutations led to decreased PDE11A activity in human embryonic kidney 293 and PC3M cells and immunostaining of PCa samples with sequence changes showed decreased PDE11A protein expression.
Conclusion: Our data suggest that, like in the adrenal cortex and the testicular germ cells, PDE11A-inactivating genetic alterations may play a role in susceptibility to PCa.
PDE11A, that is polymorphic in the general population and conveys risk to adrenal and testicular tumors, may also be a susceptibility gene for prostatic cancer.
Prostatic cancer (PCa) is the most common noncutaneous cancer among males in the United States, with an estimated incidence in excess of 200,000 new cases per year. In addition, PCa causes approximately 30,000 deaths per year, making PCa the second leading cause of cancer-related death after lung cancer among males in the United States (10% of all cancer deaths in men) (1). Genetic predisposition to PCa is suggested by familial clustering, younger age of diagnosis in families, and increased incidence in certain populations, such as African-American males (2,3). However, identifying specific high-risk susceptibility variants has been challenging due to extreme genetic heterogeneity (4,5). Recently a number of regions harboring potential susceptibility genes for PCa have been located in the genome (2,3,4,5,6,7).
Chromosome 2q31-33, containing the gene encoding phosphodiesterase 11A (PDE11A), is among the most highly linked to PCa genomic regions (2,3). In addition, PDE11A is among the top 1% of mRNAs that are down-regulated in PCa compared with normal prostate, according to expression metaanalyses (8). PDE11A is a dual-specificity phosphodiesterase (PDE) that catalyzes the hydrolysis of both cAMP and cGMP (9). PDE11A protein-truncating mutations were first identified in a series of patients with rare forms of adrenocortical hyperplasias (10); however, other milder PDE11A-inactivating mostly missense mutations were later implicated in the predisposition to a variety of endocrine neoplasms, including adrenocortical tumors, such as adenomas and cancer, and testicular germ-cell tumors (10,11,12,13).
Thus, the PDE11A gene by virtue of its genetic linkage and its involvement in other endocrine tumors appears to be a good candidate for PCa. Additional lines of evidence suggesting PDE11A involvement in PCa included: 1) the 2q chromosomal region has been reported in cytogenetic rearrangements that were identified in PCa (14); 2) PDE11A is highly expressed in normal prostate, and its expression is decreased in PCa as demonstrated by immunostaining (4,15); and 3) cAMP signaling involvement in PCa cell growth and modification of androgen receptor effects (16).
In the present study, we analyzed the coding sequence of PDE11A in patients with PCa. We then assessed the functional effects of the identified genetic variants in vitro and studied PDE11A protein expression in tumor tissues from PCa patients with and without PDE11A sequence defects. Data suggest that germline PDE11A-inactivating variants may confer a certain risk for development of PCa.
Materials and Methods
Subjects and protocol
The PDE11A gene was analyzed in 50 patients of Caucasian descent with PCa (45 sporadic and five familial) (Supplemental Table 1). Written informed consent was obtained from all participants, and the study was approved by the institutional review boards of participating centers. Frozen prostate tumors tissues were collected by the A. C. Camargo Hospital Biobank. For diagnosis and scoring, tumors were fixed in formalin and embedded in paraffin, and 4-μm sections were cut and stained with hematoxylin and eosin. Two control groups were used for this study: 1) 192 previously described individuals (100% Caucasians) with a negative family history of endocrine disorders (designated the endo-negative group) (10,13), and 2) 95 DNA samples from unselected individuals that were negative for the most common adult diseases (designated the Coriell group). Group ii samples were obtained from the commercially available Coriell Institute database. Eighty percent of the Coriell group was of Caucasian descent, and the rest were with mixed or unknown origin. For all individuals, patients and controls, the complete PDE11A-coding and surrounding intronic sequence was analyzed, as previously described (10,11,12).
Functional studies
For the transfection experiments, the PDE11A gene-open reading frame was cloned into pCR3.1, and all missense mutations (including the new ones, R202C, Y658C, E840K) were introduced by overlapping PCR, as previously described (10). Primers used for vector generation are described in Supplemental Table 2. The human embryonic kidney (HEK) 293 and PC3M (derived from prostate adenocarcinoma) cell lines were transiently transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer’s protocol. Cells were transfected with 6 μg of plasmid DNA expressing either the wild-type or the mutated form of PDE11A, harvested 48 h after the transfection, and subjected to PDE activity assay, as previously described (11). The in silico analysis of these single-nucleotide polymorphisms were done using the program PolyPhen (http://coot.embl.de/polyphen).
To assess PDE11A protein expression in affected tumor tissues, we applied immunohistochemistry, as described previously (10).
Results
We identified eight different variants in a total of 15 of our patients (30%): seven nonsynonymous substitutions and one stop-codon mutation. Three of the nonsynonymous substitutions were novel (R202C, Y658C, E840K). We have reported before three nonsynonymous substitutions (R804H, R867G, M878V) that are PDE11A-inactivating, and the stop-codon mutation c.171TdelFS41X, all in patients with adrenocortical and testicular tumors, and, rarely, in the general population (10,11,12). One nonsynonymous substitution (Y727C) was previously known as a polymorphic variant and was also found to significantly impair the ability of the protein to degrade cAMP in vitro (11).
We compared the frequency of PDE11A nonsense and missense variants in the PCa patients with those in 287 healthy control individuals of predominantly Caucasian descent: 192 endo-negative individuals, and 95 Coriell controls (Table 1). Two previously missense variants (p.A349T, p.D609N) reported by our group were identified only among the control individuals (11). Analyzed individually, only the Y727C and E840K variants showed significantly higher frequency in patients with PCa compared with the controls (χ2 = 6.02, P = 0.014, and χ2 = 5.75, P = 0.017, respectively). The cumulative frequency of the coding variants in the patients with PCa was also significantly higher (χ2 = 14.67, P < 0.001, odds ratio 3.81, 95% confidence interval 1.86–7.81). The newly identified substitutions among patients (R202C, Y658C, E840K) were not seen among the controls and have not been seen before in any of our other studies. They were also not present in the extended set of 625 New York Cancer Project control individuals that were genotyped for all identified so far PDE11A variants.
Table 1.
Allele frequency (minor allele) of sequence variations in PDE11A in PCa patients and control individuals
| Sequence change | PCa patients (100 alleles) | Controls
|
PC patients vs. controls
|
|||
|---|---|---|---|---|---|---|
| Endo-negative (384 alleles) | Coriell (190 alleles) | Total (574 alleles) | χ2 | P | ||
| c.171Tdel FS41X | 1 (0.01) | 0 (0.00) | 2 (0.01) | 2 (0.003) | 0.01 | Ns |
| R202Ca | 1 (0.01) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0.98 | Ns |
| A349T | 0 (0.00) | 1 (0.002) | 1 (0.005) | 2 (0.003) | 0.16 | Ns |
| D609N | 0 (0.00) | 0 (0.00) | 1 (0.005) | 1 (0.002) | 0.98 | Ns |
| Y658Ca | 1 (0.01) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0.98 | Ns |
| Y727C | 5 (0.05) | 0 (0.00) | 6 (0.03) | 6 (0.01) | 6.02 | 0.014 |
| R804H | 3 (0.03) | 5 (0.01) | 2 (0.01) | 7 (0.012) | 0.83 | Ns |
| E840Ka | 2 (0.02) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 5.75 | 0.017 |
| R867G | 1 (0.01) | 5 (0.01) | 5 (0.03) | 10 (0.017) | 0.01 | Ns |
| M878V | 2 (0.02) | 0 (0.00) | 1 (0.005) | 1 (0.002) | 2.95 | Ns |
| Total | 16 (0.16) | 11 (0.029) | 18 (0.094) | 29 (0.05) | 14.67 | <0.001 |
χ2 is calculated after Yates correction for continuity (Yates correction was applied for all calculations having a number less than 10 in any cell of the contingency table). Ns, Nonsignificant.
Newly identified in the PCa patients only variations.
All of the mutations were found in the heterozygote state; one patient (PCa no. 23DT) was a carrier of two mutations: the stop codon variant c.171TdelFS41X and the missense change Y727C. So far, the c.171TdelFS41X and Y727C sequence defects have always been observed separately and on different alleles (10,11,12), although lack of tissue prevented us from studying allelic expression in the patient with PCa.
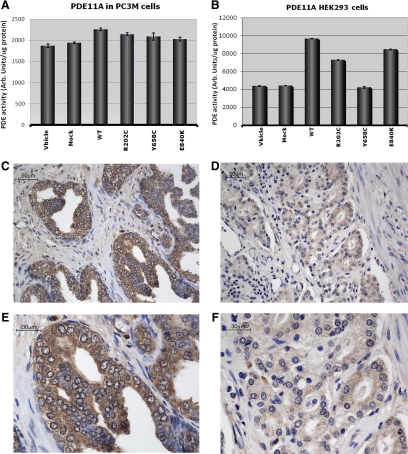
To assess the potential of the newly identified PDE11A mutations (R202C, Y658C, E840K) to impair protein function, we performed transfection experiments in HEK293 cell line and the human prostate cancer cell line PC3M (Fig. 1, A and B). As with the other PDE11A missense variants found in patients with adrenal and testicular tumors, all three substitutions led to decreased PDE activity in both HEK293 and PC3M cells. This effect is likely to be different in various organs and cells. An example of this tissue dependence of PDE11A activity was the observation that the effects of the constructs with mutations were different in the two cell lines, HEK293 and PC3M. We have observed different cell line-dependent effects of PDE11A sequence variants in other settings too: for example, between HEK293 and Hela cells, with regard to cAMP and cGMP handling of this dual-specificity PDE (11). Overall, the most significant inhibitory effect on PDE activity on both cell lines was observed after transfection with the Y658C-harboring construct. This observation is consistent with the in silico analysis that predicted a high potential of this sequence change to impair protein function (Supplemental Table 3).
Figure 1.
PDE activity and immunohistochemistry for PDE11A. PDE activity after transfection of HEK293 cells (A) and PC3M cells (B) with wild-type (WT) and mutant PDE11A expression vectors. C, Expression of PDE11A in surrounding normal prostate cells (magnification, × 20). D, Expression of PDE11A in tumor tissue from the same PCa patient. Magnification of ×40 of PDE11A immunostaining in surrounding normal prostate cells (E) and in tumor tissue (F).
Immunohistochemistry in tumor tissue showed lower PDE11A protein expression compared with the adjacent normal tissue sections for all available for the analysis 16 PC patients: two with no PDE11A mutations and 14 PDE11A mutation carriers; representative immunostaining is shown in Fig. 1, C–F.
Discussion
PCa links to more than 30 distinct genomic regions and is thus considered one of the most genetically heterogeneous malignancies in humans. Fine mapping and sequencing of these regions, however, has not always confirmed the initial association and often produced contradictory results. Failure to identify single high-penetrance disease-causing variants accounting for a reasonable number of cases led the researchers to suggest an interactive network of mild to moderate genetic changes as a likely predisposition model of PCa (8,17). In this study, we observed a significantly higher frequency of PDE11A coding changes in patients with PCa (30%) compared with healthy controls (10%, P < 0.001, odds ratio 3.81, 95% confidence interval 1.86–7.81). When compared with other genes with a potential predisposition to PCa, the prevalence of PDE11A variations in our PCa group among controls was similar or higher.
Other defects that have been recently implicated in predisposition to PCa include macrophage scavenger receptor 1 gene (MSR1), located in the genetic locus 8p22-24, most consistently associated with PCa and considered one of the strongest candidate genes, showed comparable frequencies of deleterious variations in patients and controls and was not identified as a major PCa susceptibility gene (18). ELAC2, extensively studied for association with PCa, was also found to eventually have similar genetic variation in disease cases and controls and to play an apparently weak role in susceptibility to PCa (19).
Considering the possible additive effect of different PDE11A genetic variants in PCa, it is noteworthy that our patient PCa no. 23 carrier of two different PDE11A coding changes, Y727C and the stop codon mutation c.171TdelFS41X and was most likely a compound heterozygote; he was one of five patients with a familial history of PCa (affected brother), relatively early disease onset (58 yr) and Gleason score 7 (see Supplemental Table 1). Despite the segregation model, however, the association of a more severe phenotype with a stop codon PDE11A mutation favors its involvement in the PCa pathogenesis, among the PCa patients, only PCa no. 23 carried a PDE11A protein-truncating variant.
Incomplete penetrance of PDE11A variations is not clearly understood but likely relates to the versatility and redundancy of the PDE family: a PDE11A defect can easily be compensated by other cAMP- or cGMP-specific PDEs. The presence of a defect almost certainly predisposes to mild effects at least in endocrine tissues, as our studies demonstrate. This suggests that the endo-negative group has a rate of PDE11A sequencing variants lower than the general unscreened population (13). Our previous experiments with inactivating PDE11A mutations in the adrenal cortex are suggestive of partial loss of heterozygosity in tumor tissues; a similar mechanism may take place in prostate cancer tumors (10). However, other mechanisms responsible for the reduction in PDE11A level expression in tumor tissue may be present.
In conclusion, after the adrenal cortex and the testes, prostate is the third gland in which tumors develop in patients with PDE11A-inactivating sequence variants. These tissues are extremely sensitive to cAMP levels and have the highest expression of PDE11A (10). In fact, compared with patients with adrenocortical and testicular tumors, patients with PCa display a higher frequency of PDE11A defects (with an allelic frequency of 0.16 vs. 0.10 and 0.13 in the adrenocortical and testicular, respectively, tumor groups, P < 0.05) (Supplemental Table 4). Whether PDE11A sequence defects predispose to other endocrine tumors remains unclear. It should also be noted that PDE11A is inhibited by some of the PDE inhibitors used for erectile dysfunction in patients with a similar age to those at risk for PCa. One of the several related trials assessed the relation between daily use of tadalafil and benign prostatic hyperplasia (20). Future studies should address the frequency of prostatic side effects taking into consideration the PDE11A genotypes.
Supplementary Material
Footnotes
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development intramural National Institutes of Health Project Z01-HD-000642-04 (to C.A.S.); and, in part, by a grant from the Secretaria de Estado, da Ciência, Tecnologia e Ensino Superior/Unidade Gestora do Fundo Paraná (SETI/UGF), Project 098/07 (to F.R.F.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online September 29, 2010
Abbreviations: HEK, Human embryonic kidney; PCa, prostatic cancer; PDE, phosphodiesterase; PDE11A, PDE 11A.
References
- http://www.cancer.gov/cancertopics/types/prostate
- Suarez BK, Lin J, Burmester JK, Broman KW, Weber JL, Banerjee TK, Goddard KA, Witte JS, Elston RC, Catalona WJ 2000 A genome screen of multiplex sibships with prostate cancer. Am J Hum Genet 66:933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, Schleutker J, Hamdy FC, Neal DE, Donovan JL, Stanford JL, Ostrander EA, Ingles SA, John EM, Thibodeau SN, Schaid D, Park JY, Spurdle A, Clements J, Dickinson JL, Maier C, Vogel W, Dörk T, Rebbeck TR, Cooney KA, Cannon-Albright L, Chappuis PO, Hutter P, Zeegers M, Kaneva R, Zhang HW, Lu YJ, Foulkes WD, English DR, Leongamornlert DA, Tymrakiewicz M, Morrison J, Ardern-Jones AT, Hall AL, O'Brien LT, Wilkinson RA, Saunders EJ, Page EC, Sawyer EJ, Edwards SM, Dearnaley DP, United Kingdom Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; United Kingdom ProtecT Study Collaborators; PRACTICAL Consortium, et al. 2009 Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet 2009 41:1116–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Stanford JL 2000 Genetics of prostate cancer: too many loci, too few genes. Am J Hum Genet 67:1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ 2004 The complex genetic epidemiology of prostate cancer. Hum Mol Genet 13:R103–R121 [DOI] [PubMed] [Google Scholar]
- Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, Yu K, Hutchinson A, Wang Z, Amundadottir L, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson B, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Key TJ, Kaaks R, Isaacs W, Isaacs S, Wiley KE, Gronberg H, Wiklund F, Stattin P, Xu J, Zheng SL, Sun J, Vatten LJ, Hveem K, Kumle M, Tucker M, Gerhard DS, Hoover RN, Fraumeni Jr JF, Hunter DJ, Thomas G, Chanock SJ 2009 Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet 41:1055–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M, Stacey SN, Sigurdsson A, Wahlfors T, Tammela T, Breyer JP, McReynolds KM, Bradley KM, Saez B, Godino J, Navarrete S, Fuertes F, Murillo L, Polo E, Aben KK, van Oort IM, Suarez BK, Helfand BT, Kan D, Zanon C, Frigge ML, Kristjansson K, Gulcher JR, Einarsson GV, Jonsson E, Catalona WJ, Mayordomo JI, Kiemeney LA, Smith JR, Schleutker J, Barkardottir RB, Kong A, Thorsteinsdottir U, Rafnar T, Stefansson K 2009 Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet 41:1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlov IP, Gallick GE, Gorlova OY, Amos C, Logothetis CJ 2009 GWAS meets microarray: are the results of genome-wide association studies and gene-expression profiling consistent? Prostate cancer as an example. PLoS One 4:e6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett L, Baxendale R, Stacey P, McGrouther C, Harrow I, Soderling S, Hetman J, Beavo JA, Phillips SC 2000 Molecular cloning and characterization of a distinct human phosphodiesterase gene family. Proc Natl Acad Sci USA 97:3702–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Boikos S, Giatzakis C, Robinson-White A, Groussin L, Griffin KJ, Stein E, Levine E, Delimpasi G, Hsiao HP, Keil M, Heyerdahl S, Matyakhina L, Libè R, Fratticci A, Kirschner LS, Cramer K, Gaillard RC, Bertagna X, Carney JA, Bertherat J, Bossis I, Stratakis CA 2006 A genome-wide scan identifies mutations in the gene encoding phosphodiesterase 11A4 (PDE11A) in individuals with adrenocortical hyperplasia. Nat Genet 38:794–800 [DOI] [PubMed] [Google Scholar]
- Horvath A, Korde L, Greene MH, Libe R, Osorio P, Faucz FR, Raffin-Sanson ML, Tsang KM, Drori-Herishanu L, Patronas Y, Remmers EF, Nikita ME, Moran J, Greene J, Nesterova M, Merino M, Bertherat J, Stratakis CA 2009 Functional phosphodiesterase 11A mutations may modify the risk of familial and bilateral testicular germ cell tumors. Cancer Res 69:5301–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Giatzakis C, Robinson-White A, Boikos S, Levine E, Griffin K, Stein E, Kamvissi V, Soni P, Bossis I, de Herder W, Carney JA, Bertherat J, Gregersen PK, Remmers EF, Stratakis CA 2006 Adrenal hyperplasia and adenomas are associated with inhibition of phosphodiesterase 11A in carriers of PDE11A sequence variants that are frequent in the population. Cancer Res 66:11571–11575 [DOI] [PubMed] [Google Scholar]
- Libé R, Fratticci A, Coste J, Tissier F, Horvath A, Ragazzon B, Rene-Corail F, Groussin L, Bertagna X, Raffin-Sanson ML, Stratakis CA, Bertherat J 2008 Phosphodiesterase 11A (PDE11A) and genetic predisposition to adrenocortical tumors. Clin Cancer Res 14:4016–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothman AR 1997 Cytogenetic studies in prostate cancer: are we making progress? Cancer Genet Cytogenet 95:116–121 [DOI] [PubMed] [Google Scholar]
- D'Andrea MR, Qiu Y, Haynes-Johnson D, Bhattacharjee S, Kraft P, Lundeen S 2005 Expression of PDE11A in normal and malignant human tissues. J Histochem Cytochem 53:895–903 [DOI] [PubMed] [Google Scholar]
- Bagchi G, Wu J, French J, Kim J, Moniri NH, Daaka Y 2008 Androgens transduce the Gαs-mediated activation of protein kinase A in prostate cells. Cancer Res 68:3225–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CC, Magalhaes WC, Gonzalez-Bosquet J, Chanock SJ 2010 Genome-wide association studies in cancer—current and future directions. Carcinogenesis 31:111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier C, Vesovic Z, Bachmann N, Herkommer K, Braun AK, Surowy HM, Assum G, Paiss T, Vogel W 2006 Germline mutations of the MSR1 gene in prostate cancer families from Germany. Hum Mutat 27:98–102 [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, Dayananth P, Desrochers M, Dumont M, Farnham JM, Frank D, Frye C, Ghaffari S, Gupte JS, Hu R, Iliev D, Janecki T, Kort EN, Laity KE, Leavitt A, Leblanc G, McArthur-Morrison J, Pederson A, Penn B, Peterson KT, Reid JE, Richards S, Schroeder M, Smith R, Snyder SC, Swedlund B, Swensen J, Thomas A, Tranchant M, Woodland AM, Labrie F, Skolnick MH, Neuhausen S, Rommens J, Cannon-Albright LA 2001 A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet 27:172–180 [DOI] [PubMed] [Google Scholar]
- Porst H, McVary KT, Montorsi F, Sutherland P, Elion-Mboussa A, Wolka AM, Viktrup L 2009 Effects of once-daily tadalafil on erectile function in men with erectile dysfunction and signs and symptoms of benign prostatic hyperplasia. Eur Urol 56:727–735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.