Abstract
While it has become common practice for dendrimers to deliver imaging and therapeutic agents there are few reported examples of cellular internalization of dendrimers. Moreover, targeting of dendrimers to the mitochondria in cells has not yet been reported. Previously, we have delivered small molecule imaging agents into glioma and breast cancer cells by targeting the translocator protein (TSPO; formerly known as the peripheral benzodiazepine receptor or PBR) with a family high affinity conjugable ligands. The 18 kiloDalton (kDa.) multimeric TSPO is expressed in steroid-producing cells, primarily on the outer mitochondrial membrane. This protein is a prime candidate for molecular targeting because tumors and other disease related cells contain high densities of TSPO. Here we present the synthesis, characterization and cellular internalization into C6 rat glioma cells of a TSPO targeted dendrimer imaging agent.
INTRODUCTION
Developments of clinically relevant molecular probes to image diseases or biological processes are essential for the advancement of personalized medicine. A variety of biomarkers specific to cancer and other diseases have been identified and used for therapeutic and/or imaging agent specific site delivery(1)(2, 3). For example targeting proteins has been accomplished using antibodies(4), proteins(5), small molecules(6–8), nanoparticles(9–11) and dendrimers(1, 12, 13). Attachment of multiple different species to a single dendrimer have produced a single molecule which targets, images and/or delivers a therapeutic dose to the site of disease(14, 15). The advantages of using dendrimers include a) multiple moieties or multiple copies of the targeting agent on a single particle, b) biocompatibility, c) size solubility and d) tunability. However the challenges of using dendermers include, a) the difficulty in characterization of functionalized molecule, b) nonuniform distribution of the adduct(s), c) complexity in determining the optimum size and d) insuring that the targeting moiety retains activity after attachment to the dendrimer. It is clear that macromolecules will play an important role in personalized medicine; however, the current synthetic scaffolds are particularly ineffective in targeting intra-cellular receptors.
The translocator protein (TSPO, previously known as the peripheral-type benzodiazepine receptor or PBR), spans the mitochondrial membrane and has become an attractive receptor for therapeutics and targeted imaging(16). TSPO is associated with a number of biological processes including cell proliferation, apoptosis, steroidogenesis, and immunomodulation, yet, its exact physiological role is still not fully defined(16–18). Over expression of TSPO has been shown in numerous types of cancer including brain, breast, colorectal, prostate and ovarian cancers, as well as astrocytomas, hepatocellular and endometrial carcinomas(17). Two well characterized cell lines with high TSPO expression are C6 Rat Glioma cells(19) and MDA-MB-231 human breast cancer cells(20). Several small, high binding, selective molecules have been developed for TSPO and made into positron emission tomography(PET), magnetic resonance (MR) and optical imaging agents(21, 22),(6, 23, 24). While attractive, prior to this report macromolecules targeting TSPO have not been reported, possibly due to difficulty in internalizing, synthetic molecules.
Dendrimers have been studied as potential delivery systems to target, image and/or deliver a therapeutic specifically to diseased tissue because of their globular shape, multiple surface attachment sites, biocompatibility, tunable size, monodispersity and low production cost( 25). The multiple surface end groups available on a dendrimer allow for attachment of several moieties and or copies of each drug, ligand or imaging agent. Furthermore, dendrimers can be adapted for advancements in coupling technology allowing them to be used with emerging targets, therapeutics and imaging strategies. Multiple attachment points can provide increased therapeutic efficacy, reduced drug side effects, increased signal to noise for improved imaging detection limits. Methods of targeting include attaching several copies of a binding molecule (small molecules, peptides, proteins or antibodies) to the surface of the dendrimer to capitalize on polyvalency, or the enhanced permeability and retention (EPR) effect were tumor accumulation occurs due to the particle size and the increased vasculature of the tumor(26, 27). Imaging with dendrimers is possible through the attachment of small molecule fluorophores (for optical), metal chelates (for fluorescence, MRI, PET and SPECT) or with a switch activated by a physiological process for enhanced tissue characterization(28, 29). Dendrimers can serve as effective prodrug scaffolds with the drug molecules being attached through a linkage which activates the drug in- vivo once they reach the delivery site. Cleavage from the scaffold can occur through a change in pH, enzyme reaction or slow degradation of the macromolecule( 29). While internalization of large nanoparticles through the cellular membrane has been shown; targeting of specific organelles has scarcely been reported(30, 31). In this paper we present the synthesis, characterization, cellular internalization and mitochondria labeling of a dendrimer nanoparticle. Here we demonstrate that a functionalized forth generation (G(4)-PAMAM) dendrimer particle which targets an internal cellular receptor and binds that target by passing through the cellular membrane under normal physiological conditions and without the external perturbation of the cellular membrane can be prepared, characterized and imaged.
EXPERIMENTAL PROCEDURES
Materials
G(4)-PAMAM™ dendrimer in a 10% w/w solution of methanol was purchased from Fischer Scientific and pipetted into a pre-weighed vial immediately before use. The methanol was evaporated under a stream of Ar(g) and the resulting viscous oil was dissolved in water, frozen and lyophilized to give a fluffy white powder. The mass of the dendrimer was determined by re-weighing the vial and calculating the mass difference. 1-(2-chlorophenyl)isoquinoline-3-carboxylic acid ClPhIQ Acid was synthesized in our laboratory as previously reported(6, 32). All 1,4,7,10-Tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) and DOTA derivatives were purchased form Macrocyclics and used as arrived. All other chemicals were purchased from Fischer Scientific and used as is unless otherwise indicated.
MALDI-TOF MS were obtained on a PerSeptive Biosystems Voyager-DE STR mass spectrometer. Freshly recrystalized trans-indole acrylic acid (IAA) was used as the matrix in a 10 mg/mL solution of DMSO. The plate was spotted with 1 μL of a 10:1 solution of matrix to analyte. NMR spectra were obtained on a 400 MHz Bruker AV-400 instrument with a 5 mm Z-gradient broadband inverse probe. NMR spectra were obtained in d6-dimethyl sulphoxide. UV-vis spectra were obtained on a Shimadzu 1700 UV-vis spectrophotometer. Fluorescence spectra were obtained using a ISS PCI spectrofluorometer at room temperature.
ClPhIQ-G(4)-PAMAM™ (1)
First, ClPhIQ Acid (152.9 mg, 540 μmoles) and benzotriazol-1-yloxytris(dimethylamino)-phosphonium hexafluorophosphate (BOPCl, 288.1 mg, 651 μmoles) were dissolved in 2 mL of DMSO each. Triethyl amine (Et3N, 100 μL) was added to the ClPhIQ Acidsolution. The BOPCl solution was added to the ClPhIQ Acid solution. The mixture was stirred for 5–10 minutes (sometimes a change from clear to bright orange would occur) and was then added to a stirred solution of G(4)-PAMAM™ dendrimer (252.2 mg, 18 nmoles) in DMSO (5 mL). The reaction was stirred at room temperature overnight. Unreacted ClPhIQ Acid was removed by diluting the solution 4 fold with water and placing the solution in an Amicon Centrifugation molecular weight filter (molecular weight cut off = 5 kDa.) and concentrating the solution to approximately 500 μL via centrifuging at 3000 revolutions/minute (1419 × g) for 1 hour. The solution was diluted and re-concentrated three times with water to wash any unreacted materials away. Finally, the solution was lyophilized to give 441.2 mg (100 % yield) of a slightly yellow powder. The same procedure was followed for synthesizing the (ClPhIQ)12-PAMAM using fewer equivalents of ClPhIQ Acid and BOPCl. ClPhIQ30-G(4)-PAMAM: MALDI-TOF MS 19,500 a.m.u. 1H NMR (300 MHz, CDCl3) 8.8 (1H, s), 8.6 (1H, s), 8.2–8.5 (5H, m), 7.5–8.0 (9H, multi), 2.9 (3H, bs) and 3.1–3.5 (28H, m) ppm.
ClPhIQ-G(4)-PAMAM™-Lissamine (2)
G(4)-PAMAM™ (9 mg, 400 nmoles) with or without ClPhIQ Acid was dissolved in 2 mL of dimethylformamide (DMF). Next, 23 μL of a 17.3 mM solution of lissamine sulphonyl chloride™ was added to the stirred dendrimer solution. The reaction was stirred overnight followed by purification in Amicon Centrifugation filters as described above and lyophilized to give 5.6 mg (61% yield) of a pink fluffy solid. The presence of the fluorophor was confirmed with UV-vis (max absorbance at 551 nm) and Fluorescence (ex 551 nm, em. 586 nm) spectroscopies. Specific conditions and yields provided above were for ClPhIQ23-G(4)-PAMAM, but UV-vis and fluorescence spectra were identical for ClPhIQ12-PAMAM and PAMAM-Liss.
Cell internalization experiments with G(4)-PAMAM™-Lissamine derivatives
Either 20,000 C6 rat glioma or 40,000 MDA-MB-231 breast cancer cells were plated into collagen coated glass bottom microscopy (MatTeck™) dishes and allowed to grow for two days in cell media. After two days, the cells had normal morphology and were attached, and approximately 20% confluent. G(4)- PAMAM™-Lissamine compounds (with or without other moieties) were diluted to 1 μM (in ClPhIQ) with media from a 10 mg/mL (12 μM in ClPhIQ) stock solution in DMSO. The media over the cells was replaced with the media containing the fluorophor (either ClPhIQ-PAMAM-Liss or PAMAM-Liss) as diluted noted above and incubated for 6–12 hours. The media was then poured off and the cells were carefully rinsed 3–4 times with Dulbecco’s Phosphate Buffered Saline (DPBS). The cells were imaged live. Both white light and fluorescence pictures were obtained.
For fixed cells, the rinsed cells were incubated at room temperature for 30 minutes with a 4% Para formaldehyde solution. The cells were carefully washed 4 times with DPBS and stored under DPBS at 4 °C until imaged (not longer than 1 week, typically overnight). Immediately before imaging, the fixed cells were incubated with a 25 nM solution of mitotracker green (MTG) for 10 minutes. Excess MTG was removed by 4 washings with DPBS and the cells were imaged. Cellular images were obtained using a A Nikon Eclipse TE2000-U fluorescence microscope (Lewisville, TX) equipped with a Nikon FITC-Texas Red filter sets was employed for thein-vitro imaging. The Texas Red filter (ex: 560–580 nm; polychromatic mirror: 585–665 nm; barrier filter: 600–650 nm) set was used to image lissamine and the FITC filter was used to image MTG (ex: 450–505 nm; polychromatic mirror: 510–555 nm; barrier filter: 515–545 nm). Fluorescence integration times ranged from 100 – 5000 ms, but were typically 100–500 ms. All controls were integrated for at least 5000 ms. Sample homogeneity was maintained by keeping the samples under DPBS the entire time of imaging. For live cells, images were taken within 20 minutes from replacing media with DPBS and were discontinued if the cellular morphology changed in anyway from the media containing morphology.
RESULTS AND DISCUSSION
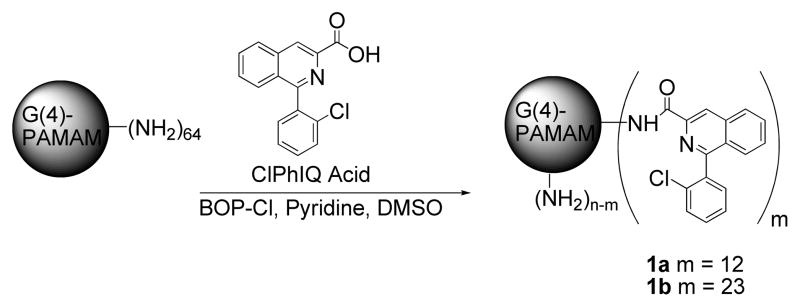
The TSPO targeted dendrimer was synthesized using 1-(2-chlorophenyl)isoquinoline-3-carboxylicacid (ClPhIQ Acid) a precursor of the conjugable TSPO binding molecule previously reported(6). ClPhIQ Acid (Scheme 1) was activated with benzotriazol-1-yloxytris(dimethylamino)-phosphonium hexafluorophosphate (BOPCl), in pyridine and dimethyl sulphoxide (DMSO) and added to a solution of G(4)-PAMAM dendrimer in DMSO. An average of 12 and 23 ClPhIQ Acids were attached by reacting G(4)-PAMAM dendrimer with 15 and 30 molar equivalents of the activated TSPO ligand respectively to obtain two ClPhIQ-PAMAM compounds, 1a and 1b (Scheme 1). Excess ClPhIQ Acid and coupling agent were removed by diafiltration with a molecular weight cut off (MWCO) of 5 kDa. Both ClPhIQ-PAMAM molecules were characterized with nuclear magnetic resonance (NMR) and matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF-MS).
Scheme 1.
Synthesis of ClPhIQ-PAMAM.
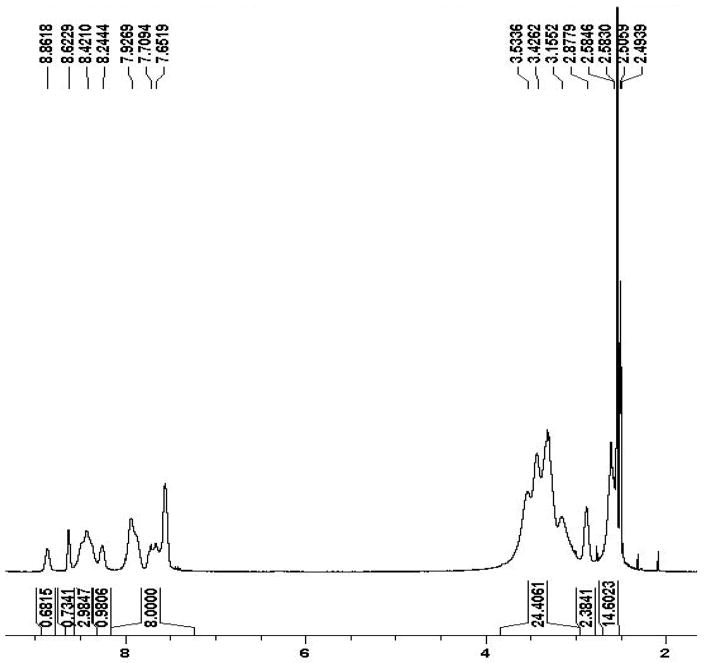
The average number of TSPO ligands per dendrimer was estimated using NMR integrations. It was shown from proton NMR spectra of the PAMAM and ClPhIQ pieces individually that the aromatic proton signals are exclusively from the ClPhIQ Acids attached (7.0–7.9 ppm) to the particle; while the interior PAMAM methylenes are the only signal observed from 2–4 ppm and the amides are found from 8.4–9 ppm (Figure 1). By calculation there are approximately 930 methylene protons in a G(4)-PAMAM dendrimer and 9 protons (all aromatic) per ClPhIQ Acid molecule. To determine the average number of ClPhIQ Acids per dendrimer, the integral from 7.0–8.2 was calibrated to 9 and the resulting integral from 2–4 ppm was divided into 930. As shown in Figure 1, we find 23 ClPhIQ Acids attached to the dendrimer when the dendrimer was reacted with 30 equivalents of ClPhIQ Acid. NMR spectra indicate that 12 ClPhIQ acids are attached when 15 equivalents of ligand are reacted with the dendrimer (data not shown). Interestingly, the amount of ClPhIQ that binds to the dendrimer nearly doubles (from 12 to 23) with doubling the amount of ClPhIQ in the reaction (from 15 to 30).
Figure 1.
NMR of 23 ClPhIQ Acids attached to G(4)-PAMAM (ClPhIQ23-PAMAM).
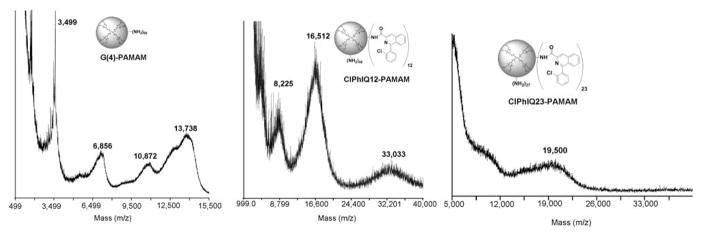
MALDI-TOF MS was also utilized to confirm the average number of ClPhIQ Acids attached to the dendrimer. First a MALDI-TOF MS was acquired of the G(4)-PAMAM with the average molecular weight (MW) found to be 13,244 Daltons, significantly lower than the 14,215 Daltons of a G(4)- PAMAM without any of the widely reported structural defects(33). These defects include incomplete reactions, making one arm of the dendrimer consistently a generation behind, and side reactions that stop the expansion of that branch of dendrimer altogether. The ClPhIQ-PAMAM ( 1a) has an average MW of 16,513; and thus, by calculation has 12 ClPhIQ Acid molecules per PAMAM. This result is consistent with the NMR data; as was the number of ClPhIQ Acids per PAMAM for ClPhIQ-PAMAM (1b) with 23 ClPhIQ ligands attached. Additionally, it was calculated that ClPhIQ-PAMAM, 1b, to have approximately 24 ClPhIQ Acids per PAMAM, consistent with NMR data.
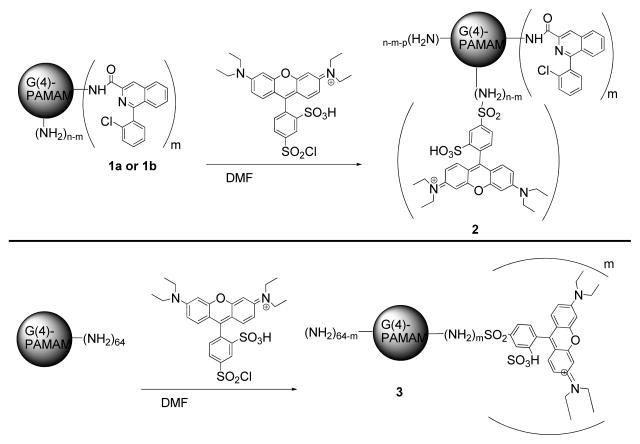
Following characterization of the ClPhIQ-PAMAM molecules with NMR and MALDI-TOF-MS, the dendrimer was further functionalized by reacting it with lissamine rhodamine B sulfonyl chloride™ (Lissamine) in dimethyl formamide (DMF) overnight to give 1 or 2 Lissamine dyes per dendrimer (Scheme 2). The DMF was removed in vacuo and excess dye was removed by diafiltration (MwCO = 5,000) in water. UV-vis and fluorescence were employed to characterize the optical imaging agent. Attaching the dye to the dendrimer produced a compound with an absorbance maximum at 571 nm (versus 563 nm for free dye) and a slightly red shifted emission at 586 nm (versus 583 nm for free dye). A control compound (PAMAM-Liss), the agent absent of the targeting moiety, was synthesized similarly to the targeting agent by reacting G(4)-PAMAM dendrimer with Lissamine™ (Scheme 2). The PAMAM-Liss conjugate was also purified by diafiltration in which the first low molecular weight fraction was slightly pink (from dye) and subsequent washing resulted in lighter color until no color was present. A minimum of three colorless washing in the low molecular weight fraction was completed to provide pure ClPhIQ-PAMAM-Liss with no unreacted lissamine dye. A molecular weight difference for the PAMAM and PAMAM-Liss of 500 – 600 amu was observed in the MALDI-TOF MS spectrum, consistent with an average of 1 dye per dendrimer. The small spectral shift in absorbance and emission was similar to the targeted moiety.
Scheme 2.
Attachment of Lissamine dye to PAMAM.
Finally, the solubility extraction coefficients between octanol and water were determined for both the PAMAM compounds by using the shake method. Briefly, the shake method for finding Log P is determined by mixing an aqueous solution of the PAMAM-dye with a solution PAMAM-dye in octanol and allowing the two solvents to equilibrate. The absorbance of each layer (octanol and water) were then recorded and used to determine the concentration of dye in each layer. The Log P for ClPhIQ- PAMAM-Liss is estimated to be 2.9 while that for PAMAM-Liss (no ClPhIQ) is 1.0. These Log P values indicate that the ClPhIQ-PAMAM-Liss is more lipophilic than the PAMAM-Liss and therefore is more likely to interact favorably with the cellular membrane.
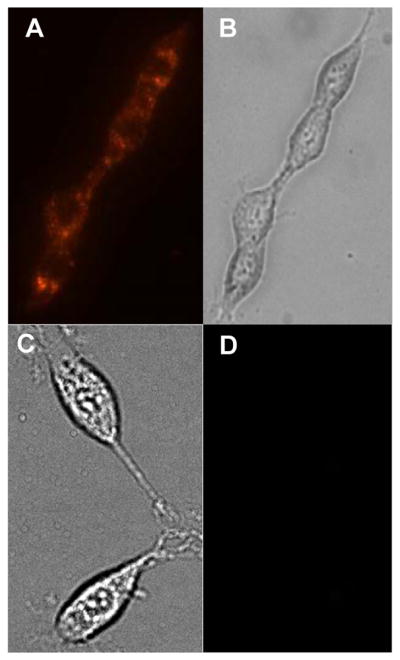
To evaluate the biological activity of the imaging agent, C6 rat glioma cells, shown to express a high concentration of TSPO(34, 35), were incubated with the dendrimer compounds 2b & 3 for 12 hours in collagen coated MatTek™ dishes and visualized by fluorescence microscopy. Live cell imaging was preformed immediately after rinsing the cells with PBS or saline to remove media and unbound or uninternalized imaging agent. A Nikon Eclipse TE2000-U fluorescence microscope (Lewisville, TX) equipped with Texas Red and FITC filter sets was employed for the in-vitro imaging. As shown in Figure 3, the ClPhIQ-PAMAM-Liss labels the cell, while the control (PAMAM-Liss) does not produce fluorescence. As another control, cells were incubated with lissamine rhodamine B sulfonyl chloride™ and again no fluorescence was observed (data not shown). The imaging experiments were performed intriplicate with observations being consistent with respect to absorbance and fluorescence intensity and the absence of fluorescence in the controls. We hypothesize that the dendrimer with the TSPO binding ligand (ClPhIQ-PAMAM-Liss, 2b) produced bright, localized, cellular fluorescence due to its ability to pass through the cellular membrane and bind to the target protein TSPO (Figure 3). Since ClPhIQ Acid conjugated dye is known to bind TSPO, glioma cells overexpress TSPO and it is an organellular protein, we believe that the dendrimer labels the mitochondria by binding to TSPO. In the labeling experiment where the dendrimer without the targeting ligand (compound 3) was used, no fluorescence was found in the cell (either intracellularly or at the membrane) at exposure times as long as 5 seconds. Taken together these results indicate that the ClPhIQ moiety is necessary for cellular uptake of the dendrimerat labeling concentrations of 1 μM and equilibration times of 6–12 hours.
Figure 3.
Live cell images of C6 rat glioma cells. Panal A: Red fluorescence of ClPhIQ-PAMAM-Liss dosed cells; B: DIC image of ClPhIQ-PAMAM-Liss dosed cells; C: DIC image of PAMAM-Liss dosed cells; D: Red Fluorescence image of PAMAM-Liss dosed cells.
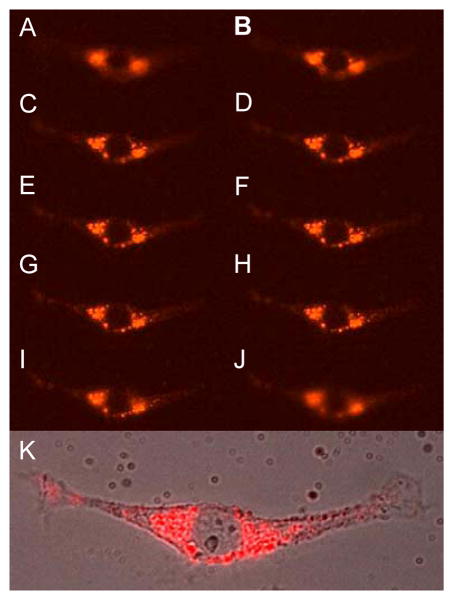
To further confirm that the agent was intracellular, a series of z stacked images (pseudo-confocal) were obtained allowing different regions of the cells to be viewed (Figure 4). These z stacked images were accomplished by changing the lens position incrementally, moving the focal planes of the microscope through cells and collecting the images by viewing fluorescence. In the center focal planes there is a dark hole, consistent with the nucleus as shown in the fluorescence-DIC overlay (Figure 4). This observation suggests that the agent undergoes internalization, passive or active, and does not just label the membrane due to lypophilic nature of the ClPhIQ moiety. Since only a small amount of out-of-focus light is observed in the membrane slices of the z stack (data not shown), labeling of the membrane appears to be negligible. Furthermore, fluorescence signal is observed through the cell to the entire nuclear membrane, but not in the nucleus, indicating uptake of the dendrimer by the cells but not the nucleus. Previously reported small molecule agents have not labeled the nucleus, although low concentrations of TSPO are present there(24). The reported macromolecular agent behaved as expected and targeted the mitochondria and was not uptaken into the nucleus as indicated in Figure 4.
Figure 4.
Selected montage of red fluorescence images of ClPhIQ-PAMAM-Liss dosed cells from the z series. Panel A: slice 10; B: slice 15; C: slice 20; D: slice 21; E slice 22; F: slice 23; G: slice 24, H: slice 325; I: slice 30; J: slice 35; K: DIC Fluorescence overlay.
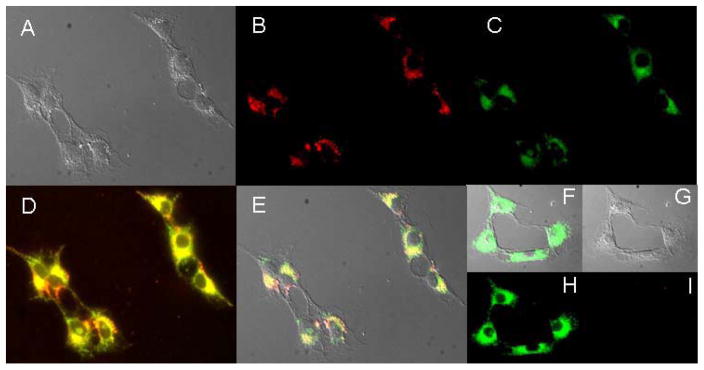
To further explore that ClPhIQ-PAMAM-Liss was targeting the mitochondria, a co-incubation experiment was preformed. This was accomplished by first incubating C6 rat glioma cells with ClPhIQ-PAMAM-Liss as described above for the live cellular imaging, followed by fixation using paraformaldehyde, and then treatment of the cells with Mitotracker Green™ (MTG) according to the manufacturer’s guidelines. The cells were then imaged using a Texas Red filter set to interrogate the Lissamine (red) fluorescence and a FITC filter set to detect the MTG (green) fluorescence. Figure 5 shows C6 rat glioma cells incubated with either ClPhIQ-PAMAM-Liss or the control, PAMAM-Liss. The DIC images in Figure 5A and 5G are present to indicate the location of the cells relative to fluorescence signal of ClPhIQ-PAMAM-Liss (Figure 5A) and PAMAM-Liss dosed cells (Figure 5G). Fluorescence from the imaging agent is displayed in red (Figure 5B) and fluorescence from the MTG as green (Figure 5C). As demonstrated above with live cell imaging, the ClPhIQ-PAMAM-Liss agent labels the fixed cells principally perinuclear (Figure 5B). MTG was used in low concentration (25 nM)to ensure that it primarily labeled mitochondria in the cell. The MTG labels the mitochondria of cells, as shown by numerous investigators(36, 37), particularly when used in low concentrations as those employed here (25nM). More careful interrogation of the fluorescence signals allow an insight about where the species are within the cell. Figure 5 panels D and E display co-registration of ClPhIQ-PAMAM-Liss and MTG with yellow indicating areas of overlap while red and green indicate areas where ClPhIQ-PAMAM-Liss (red) and MTG (green) are not coincident. It is encouraging to see that most of the red and green converge to yellow, indicating that the majority of ClPhIQ-PAMAM-Liss is in the same location of the cell as the MTG. This good co-localization is further evidence that ClPhIQ-PAMAM-Liss is labeling the mitochondria and not other parts of the cell. As demonstrated in Figure 5 panels F – I and in the live cell imaging (Figures 3 & 4), fluorescence from the dendrimer is only detected if ClPhIQ Acid is incorporated into the dendrimer. In other words, images from cells inoculated with the control agent (PAMAM-Liss) exhibit no fluorescence (Figure 5I). MTG fluorescence was not affected (Figure 5C, F&H) by the presence or absence of dendrimer agent and the cellular membrane was not perturbed by non-physiological chemical or physical processes prior to or during the incubation with ClPhIQ-PAMAM-Liss. Of course excess imaging agent was removed prior to fixation in all experiments. Taken together, the fluorescence images in Figure 5 indicate that the ClPhIQ-PAMAM-Liss is targeting the mitochondria and that the ClPhIQ ligand is necessary for this mitochondrial labeling to occur. The z stacked images and co-incubation experimental results indicate that the imaging agent is internalized into the cell through normal cellular functions.
Figure 5.
Fluorescence Images of fixed C6 rat glioma cells. Panal A: DIC of ClPhIQ(23)-PAMAM-Liss and MTG dosed cells; B: Red fluorescence of ClPhIQ(23)-PAMAM-Liss and MTG dosed cells; C: Green fluorescence of ClPhIQ(23)-PAMAM-Lissand MTG dosed cells; D: Red and Green fluorescence overlay of ClPhIQ(23)-PAMAM-Liss and MTG dosed cells; E: DIC, Red and Green fluorescence overlay of ClPhIQ(23)-PAMAM-Liss and MTG dosed cells; F: DIC, Red and Green Fluorescence overlay of PAMAM-Liss and MTG dosed cells; G: DIC of PAMAM-Liss and MTG dosed cells; H: Green fluorescence of PAMAM-Liss and MTG dosed cells; and I: Red fluorescence of PAMAM-Liss and MTG dosed cells.
Although no quantitative binding studies were completed, the agent binds with high enough affinity to remain in the cell after fixation, washing and treatment with MTG. Studies beyond the scope of this report are necessary to determine the exact mechanism of cellular transport and binding, however insight is shown in some initial experiments performed using shorter incubation times with the fluorescent agents (ClPhIQ-PAMAM-Liss and PAMAM-Liss). In these trials, no fluorescence was observed for either compound (with and without ClPhIQ ligand) after incubation for as long as 3 hours (data not shown). It was only after a minimum of 6 hours of incubation that fluorescence was found in the dosed cells. One possible explaination for this observation is that the dendrimeric compound is slowly uptaken into the cell and the ClPhIQ is absolutely necessary for uptake. Another possibility is that the molecule freely enters and exits the cell and eventually binds the target (TSPO), that dye is completely removed (both inter and intracellular) when the cells are rinsed immediately after incubating.
Imaging with IEC- 6 cells was completed as an initial estimate as to whether the ClPhIQ-PAMAM-Liss would be uptaken and bind mitochondria in cells that express low levels of TSPO. Since TSPO is necessary for cellular function, there are no cell lines that do not express TSPO. No fluorescence was seen when imaging the IEC-6 cells with ClPhIQ-PAMAM-Liss at dosing concentrations as high as 1 μM in dye (data not shown). The indication is that the ClPhIQ-PAMAM-Liss imaging agent is TSPO protein specific. Again, the mechanism of cellular uptake into the cell needs further investigation in order to pinpoint the exact mechanism of cellular uptake.
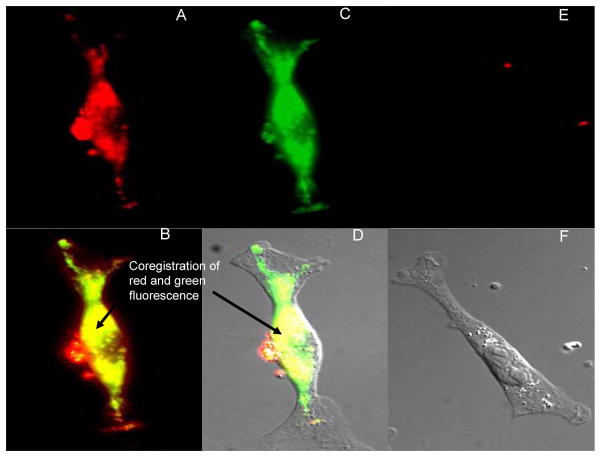
Further evidence of the utility of ClPhIQ-PAMAM-Liss was demonstrated by labeling MDA-MB-231 human breast cancer cells, an aggressive human cancer cell line. Like the C6 rat glioma cells, the MDA-MB-231 human breast cancer cells have been shown to over express TSPO (20). The cells were incubated with ClPhIQ-PAMAM-Liss following the same procedures as with the C6 rat glioma cells, fixed and treated with MTG for co-registration investigations. The individual red and green fluorescence of ClPhIQ-PAMAM-Liss and MTG respectively in fixed MDA-MB-231 cells is demonstrated in panels A and C of Figure 6; while, panels E and F show the lack of red fluorescence in the PAMAM-Liss treated cells. Association between the red and green (shown in yellow) is evident in the overlays of panels B and D indicating co-localization of the ClPhIQ-PAMAM-Liss and MTG. These additional results represent further evidence that ClPhIQ-PAMAM-Liss is able to label TSPO rich mitochondria in cancer cells.
Figure 6.
Microscope images of MDA-MB-231 cells. Panal A: Red fluorescence of ClPhIQ(23)-PAMAM-Liss and MTG dosed cells; B: Red and Green Fluorescence overlay of ClPhIQ(23)-PAMAM-Liss and MTG dosed cells; C: Green fluorescence of ClPhIQ(23)-PAMAM-Liss and MTG dosed cells; D:Red and Green Fluorescence and DIC overlay of ClPhIQ(23)-PAMAM-Liss and MTG dosed cells; E: Red fluorescence of PAMAM-Liss dosed cells; F: DIC image of PAMAM-Liss dosed cells.
The observations made in the cellular imaging experiments show that the ClPhIQ labeled dendrimers enter the cell and bind the mitochondria, but do not appear in the nucleus or cellular membrane. The dendrimers without the ClPhIQ do not label the cell in any significant way. Although the molecules are small enough to enter through the cellular and nuclear membranes, as shown previously the literature suggests that it is not as trivial as size{Najlah, 2006 #1041}. First, dendrimers are structurally different than biological molecules such as proteins and antibodies. Biological molecules are typically well defined, folded structures while dendrimers are floppier, not well defined structures. Second, the end functional groups dictate the interactions with the membranes, which may block or enhance the uptake of the compound, depending on the chemical interactions. In amine terminated dendrimers, the amines can be protonated and do not pass through the lipophilic cellular membrane. Our observations in the cellulare imaging experiments, along with the Log P data indicated that the lipophilicity of the ClPhIQ ligand allows for the dendrimeric molecule to pass into the cell.
In conclusion, a TSPO targeted imaging agent was synthesized using a G(4)-PAMAM backbone, ClPhIQ acid and Lissamine™ (ClPhIQ-PAMAM-Liss). The imaging agent was characterized with MALDI-TOF-MS and NMR and imaged in C6 rat glioma and MDA-MB-231 human breast cancer cells. To our knowledge this is the first example of a TSPO-targeted dendrimer and among the few dendrimeric compounds that are internalized into the cell under normal physiological conditions. Co-localization of the ClPhIQ-PAMAM-Liss and MTG indicates that the compound binds to the mitochondria. The ability for synthetic dendrimers to be internalized into the cell and target a specific protein is a step closer to understanding biological events at the molecular level and ultimately enabling personalized medicine, however further investigation is required to determine exact mechanism of how this agent enters the cell.
Figure 2.
Characterization and determination of the average number of ClPhIQ per Dendrimer by MALDI-TOF MS. Left: MALDI-TOF spectum of G(4)-PAMAM; middle: MALDI-TOF spectrum of ClPhIQ(12)-PAMAM; right: MALDI-TOF spectrum of ClPhIQ(23)-PAMAM.
Acknowledgments
This work was supported by grants from the National Science Foundation (Bes-0323281), Department of Defense (W81XWH-04-1-0432) and the National Institute of Health (5P20 GM072048-4).
LITURATURE CITED
- 1.Tomalia DA, Reyna LA, Svenson S. Dendrimers as multi-purpose nanodevices for oncology drug delivery and diagnostic imaging. Biochemical Society Transactions. 2007;35:61–67. doi: 10.1042/BST0350061. [DOI] [PubMed] [Google Scholar]
- 2.Portney NG, Ozkan M. Nano-oncology: drug delivery, imaging, and sensing. Analytical and Bioanalytical Chemistry. 2006;384:620–630. doi: 10.1007/s00216-005-0247-7. [DOI] [PubMed] [Google Scholar]
- 3.Bremer C, Ntziachristos V, Weissleder R. Optical-based molecular imaging: contrast agents and potential medical applications. European Radiology. 2003;13:231–243. doi: 10.1007/s00330-002-1610-0. [DOI] [PubMed] [Google Scholar]
- 4.Sharkey RM, Cardillo TM, Rossi EA, Chang CH, Karacay H, McBride WJ, Hansen HJ, Horak ID, Goldenberg DM. Signal amplification in molecular imaging by pretargeting a multivalent, bispecific antibody. Nature Medicine. 2005;11:1250–1255. doi: 10.1038/nm1322. [DOI] [PubMed] [Google Scholar]
- 5.Lee LA, Wang Q. Adaptations of nanoscale viruses and other protein cages for medical applications. Nanomedicine. 2006;2:137–49. doi: 10.1016/j.nano.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Manning HC, Goebel T, Marx JN, Bornhop DJ. Facile, efficient conjugation of a trifunctional lanthanide chelate to a peripheral benzodiazepine receptor ligand. Organic Letters. 2002;4:1075–1078. doi: 10.1021/ol017155b. [DOI] [PubMed] [Google Scholar]
- 7.Bai M, Wyatt SK, Han Z, Papadopoulos V, Bornhop DJ. A novel conjugable translocator protein ligand labeled with a fluorescence dye for in vitro imaging. Bioconjug Chem. 2007;18:1118–22. doi: 10.1021/bc060381r. [DOI] [PubMed] [Google Scholar]
- 8.Bai MF, Sexton M, Stella N, Bornhop DJ. MBC94, a conjugable ligand for cannabinoid CB2 receptor imaging. Bioconjugate Chemistry. 2008;19:988–992. doi: 10.1021/bc700419e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulder WJM, Strijkers GJ, van Tilborg GAF, Griffioen AW, Nicolay K. Lipid-based nanoparticles for contrast-enhanced MRI and molecular imaging. Nmr in Biomedicine. 2006;19:142–164. doi: 10.1002/nbm.1011. [DOI] [PubMed] [Google Scholar]
- 10.Morawski AM, Winter PM, Crowder KC, Caruthers SD, Fuhrhop RW, Scott MJ, Robertson JD, Abendschein DR, Lanza GM, Wickline SA. Targeted nanoparticles for quantitative imaging of sparse molecular epitopes with MRI. Magnetic Resonance in Medicine. 2004;51:480–486. doi: 10.1002/mrm.20010. [DOI] [PubMed] [Google Scholar]
- 11.Li KCP, Bednarski MD. Vascular-targeted molecular imaging using functionalized polymerized vesicles. Journal of Magnetic Resonance Imaging. 2002;16:388–393. doi: 10.1002/jmri.10174. [DOI] [PubMed] [Google Scholar]
- 12.Gillies ER, Frechet JMJ. Dendrimers and dendritic polymers in drug delivery. Drug Discovery Today. 2005;10:35–43. doi: 10.1016/S1359-6446(04)03276-3. [DOI] [PubMed] [Google Scholar]
- 13.Lee CC, Yoshida M, Frechet JMJ, Dy EE, Szoka FC. In Vitro and in Vivo Evaluation of Hydrophilic Dendronized Linear Polymers. Bioconjugate Chemistry. 2005;16:535–541. doi: 10.1021/bc0497665. [DOI] [PubMed] [Google Scholar]
- 14.Wickline SA, Neubauer AM, Winter P, Caruthers S, Lanza G. Applications of nanotechnology to atherosclerosis, thrombosis, and vascular biology. Arteriosclerosis Thrombosis and Vascular Biology. 2006;26:435–441. doi: 10.1161/01.ATV.0000201069.47550.8b. [DOI] [PubMed] [Google Scholar]
- 15.Sinha R, Kim GJ, Nie SM, Shin DM. Nanotechnology in cancer therapeutics: bioconjugated nanoparticles for drug delivery. Molecular Cancer Therapeutics. 2006;5:1909–1917. doi: 10.1158/1535-7163.MCT-06-0141. [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacapere JJ, Lindemann P, Norenberg MD, Nutt D, Weizman A, Zhang MR, Gavish M. Translocator protein (18 kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends in Pharmacological Sciences. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Decaudin D, Castedo M, Nemati F, Beurdeley-Thomas A, De Pinieux G, Caron A, Pouillart P, Wijdenes J, Rouillard D, Kroemer G, Poupon MF. Peripheral benzodiazepine receptor ligands reverse apoptosis resistance of cancer cells in vitro and in vivo. Cancer Research. 2002;62:1388–1393. [PubMed] [Google Scholar]
- 18.Casellas P, Galiegue S, Basile AS. Peripheral benzodiazepine receptors and mitochondrial function. Neurochemistry International. 2002;40:475–486. doi: 10.1016/s0197-0186(01)00118-8. [DOI] [PubMed] [Google Scholar]
- 19.Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: Correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Research. 1999;59:831–842. [PubMed] [Google Scholar]
- 20.Olson JM, Mcneel W, Young AB, Mancini WR. Localization of the Peripheral-Type Benzodiazepine Binding-Site to Mitochondria of Human Glioma-Cells. Journal of Neuro-Oncology. 1992;13:35–42. doi: 10.1007/BF00172944. [DOI] [PubMed] [Google Scholar]
- 21.Chen JW, Breckwoldt MO, Aikawa E, Chiang G, Weissleder R. Myeloperoxidase-targeted imaging of active inflammatory lesions in murine experimental autoimmune encephalomyelitis. Brain. 2008;131:1123–33. doi: 10.1093/brain/awn004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nahrendorf M, Sosnovik D, Chen JW, Panizzi P, Figueiredo JL, Aikawa E, Libby P, Swirski FK, Weissleder R. Activatable magnetic resonance imaging agent reports myeloperoxidase activity in healing infarcts and noninvasively detects the antiinflammatory effects of atorvastatin on ischemia-reperfusion injury. Circulation. 2008;117:1153–60. doi: 10.1161/CIRCULATIONAHA.107.756510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammoud DA, Endres CJ, Chander AR, Guilarte TR, Wong DF, Sacktor NC, McArthur JC, Pomper MG. Imaging glial cell activation with [C-11]-R-PK11195 in patients with AIDS. Journal of Neurovirology. 2005;11:346–355. doi: 10.1080/13550280500187351. [DOI] [PubMed] [Google Scholar]
- 24.Manning HC, Goebel T, Thompson RC, Price RR, Lee H, Bornhop DJ. Targeted molecular imaging agents for cellular-scale bimodal imaging. Bioconjugate Chemistry. 2004;15:1488–1495. doi: 10.1021/bc049904q. [DOI] [PubMed] [Google Scholar]
- 25.Svenson S, Tomalia DA. Dendrimers in biomedical applications--reflections on the field. Adv Drug Deliv Rev. 2005;57:2106–29. doi: 10.1016/j.addr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Khandare J, Minko T. Polymer-drug conjugates: Progress in polymeric prodrugs. Progress in Polymer Science. 2006;31:359–397. [Google Scholar]
- 27.Satchi-Fainaro R, Duncan R, Barnes CM. Polymer therapeutics for cancer: Current status and future challenges. Polymer Therapeutics Ii: Polymers as Drugs, Conjugates and Gene Delivery Systems. 2006;193:1–65. doi: 10.1080/10611860600833856. [DOI] [PubMed] [Google Scholar]
- 28.Winter PM, Cai KJ, Chen J, Adair CR, Kiefer GE, Athey PS, Gaffney PJ, Buff CE, Robertson JD, Caruthers SD, Wickline SA, Lanza GM. Targeted PARACEST nanoparticle contrast agent for the detection of fibrin. Magnetic Resonance in Medicine. 2006;56:1384–1388. doi: 10.1002/mrm.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pham W, Choi YD, Weissleder R, Tung CH. Developing a peptide-based near-infrared molecular probe for protease sensing. Bioconjugate Chemistry. 2004;15:1403–1407. doi: 10.1021/bc049924s. [DOI] [PubMed] [Google Scholar]
- 30.Kolonko EM, Kiessling LL. A polymeric domain that promotes cellular internalization. Journal of the American Chemical Society. 2008;130:5626. doi: 10.1021/ja8001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitchens KM, Foraker AB, Kolhatkar RB, Swaan PW, Ghandehari H. Endocytosis and interaction of poly (amidoamine) dendrimers with Caco-2 cells. Pharmaceutical Research. 2007;24:2138–2145. doi: 10.1007/s11095-007-9415-0. [DOI] [PubMed] [Google Scholar]
- 32.Newman AH, Lueddens HWM, Skolnick P, Rice KC. Novel Irreversible Ligands Specific for Peripheral Type Benzodiazepine Receptors - (+/−), (+), and (−)-1-(2-Chlorophenyl)-N-(1-Methylpropyl)-N-(2-Isothiocyanatoethyl)-3-Isoquinolinecarboxamide and 1-(2-Isothiocyanatoethyl)-7-Chloro-1,3-Dihydro-5-(4-Chlorophenyl)-H-2–1,4-Benzodiazepin-2-One. Journal of Medicinal Chemistry. 1987;30:1901–1905. doi: 10.1021/jm00393a036. [DOI] [PubMed] [Google Scholar]
- 33.Hummelen JC, vanDongen JLJ, Meijer EW. Electrospray mass spectrometry of poly(propylene imine) dendrimers - The issue of dendritic purity or polydispersity. Chemistry-a European Journal. 1997;3:1489–1493. [Google Scholar]
- 34.Kozikowski AP, Kotoula M, Ma DW, Boujrad N, Tuckmantel W, Papadopoulos V. Synthesis and biology of a 7-nitro-2,1,3-benzoxadiazol-4-yl derivative of 2-phenylindole-3-acetamide: A fluorescent probe for the peripheral-type benzodiazepine receptor. Journal of Medicinal Chemistry. 1997;40:2435–2439. doi: 10.1021/jm970220w. [DOI] [PubMed] [Google Scholar]
- 35.Starostarubinstein S, Ciliax BJ, Penney JB, Mckeever P, Young AB. Imaging of a Glioma Using Peripheral Benzodiazepine Receptor Ligands. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:891–895. doi: 10.1073/pnas.84.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sehgal I, Sibrian-Vazquez M, Vicente MGH. Photoinduced cytotoxicity and biodistribution of prostate cancer cell-targeted porphyrins. Journal of Medicinal Chemistry. 2008;51:6014–6020. doi: 10.1021/jm800444c. [DOI] [PubMed] [Google Scholar]
- 37.Sacchi S, Bernasconi M, Martineau M, Mothet JP, Ruzzene M, Pilone MS, Pollegioni L, Molla G. pLG72 modulates intracellular D-serine levels through its interaction with D-amino acid oxidase - Effect on schizophrenia susceptibility. Journal of Biological Chemistry. 2008;283:22244–22256. doi: 10.1074/jbc.M709153200. [DOI] [PubMed] [Google Scholar]