SUMMARY
Many animals use their olfactory systems to learn to avoid dangers, but how neural circuits encode naïve and learned olfactory preferences, and switch between those preferences, is poorly understood. Here, we map an olfactory network, from sensory input to motor output, which regulates the learned olfactory aversion of Caenorhabditis elegans for the smell of pathogenic bacteria. Naïve animals prefer smells of pathogens but animals trained with pathogens lose this attraction. We find that two different neural circuits subserve these preferences, with one required for the naïve preference and the other specifically for the learned preference. Calcium imaging and behavioral analysis reveal that the naïve preference reflects the direct transduction of the activity of olfactory sensory neurons into motor response, whereas the learned preference involves modulations to signal transduction to downstream neurons to alter motor response. Thus, two different neural circuits regulate a behavioral switch between naïve and learned olfactory preferences.
INTRODUCTION
In principle, learning can involve alterations to different layers of an animal's nervous system, from sensory neurons to interneurons and motor neurons. To fully understand the neural basis of experience-dependent behavioral plasticity, it is important to map the neuronal pathways that underlie behavioral responses before and after learning, understand how these neuronal pathways interact, and determine what changes occur during learning.
Experience and environmental context can profoundly shape the representations of an odor to an animal. Studies in both vertebrates and invertebrates have identified brain areas, or even specific neurons, that contribute to olfactory learning, such as a few distributed brain areas in the main olfactory system in mammals and mushroom body neurons in flies (Sanchez-Andrade and Kendrick, 2009; Waddell and Quinn, 2001). Specific neurotransmitters can also play regulatory roles in olfactory learning (Menzel and Muller, 1996; Schwaerzel et al., 2003; Zhang et al., 2005). However, a systems-level analysis from sensory input to motor output, showing how both naïve and learned olfactory preferences can be generated by the nervous system, has not yet been possible.
The nematode Caenorhabditis elegans provides an opportunity to study the functional organization of neural networks with comprehensiveness and single-cell resolution. Its entire highly stereotyped nervous system contains just 302 neurons, and all synaptic connections between neurons have been defined by serial reconstruction of electron micrographs (Chen et al., 2006; White et al., 1986). The wiring diagram of the worm nervous system has facilitated the mapping of neural circuits that regulate mechanosensation (Chalfie et al., 1985), olfactory sensation (Bargmann et al., 1993), salt chemotaxis (Bargmann and Horvitz, 1991; Iino and Yoshida, 2009), thermotaxis (Mori and Ohshima, 1995) and navigation (Gray et al., 2005; Tsalik and Hobert, 2003).
In this study, we sought to identify the neural circuits that allow C. elegans to exhibit different olfactory preferences depending on adult-stage experience. The nematode detects hundreds of different odorants, which directs its navigation towards bacterial food sources (Bargmann et al., 1993). C. elegans modulates its behavior in response to food quality, and displays experience-dependent plasticity to avoid ingesting pathogenic bacteria that exist in its habitat, such as Pseudomonas aeruginosa PA14 (Hodgkin et al., 2000; Pujol et al., 2001; Shtonda and Avery, 2006; Tan et al., 1999; Zhang et al., 2005). Animals that are never exposed, and thus naïve, to pathogenic bacteria often prefer the smells of the pathogens. In contrast, animals that have ingested pathogenic bacteria learn to reduce their olfactory preference for the pathogens (Zhang et al., 2005). This form of aversive olfactory learning requires the function of the serotonin biosynthetic enzyme TPH-1 in a pair of serotonergic neurons ADF and the function of a serotonin-gated chloride channel MOD-1 in a few interneurons. Long-term exposure to pathogenic bacteria raises the serotonin content of ADF neurons and increased serotonin promotes learning. Together, these results suggest that ADF serotonin functions as a negative reinforcing signal for aversive olfactory learning on pathogenic bacteria (Zhang et al., 2005).
Here, we asked how an olfactory neural network in C. elegans allows the animal to generate both naive and learned olfactory preferences, and how ADF regulate the switch between those preferences. We combined a systematic laser ablation analysis and an automated behavioral assay that quantifies the olfactory responses of individual animals to measure the contribution of specific neurons to olfactory response and plasticity. These analyses revealed two different groups of neurons that regulate naive and learned olfactory behaviors. One is composed of olfactory sensory neurons AWB and AWC with their downstream interneurons (the AWB-AWC sensorimotor circuit) and is needed for animals to display naïve olfactory preference. Calcium imaging recordings indicate that the naïve preference is determined by the intrinsic properties of AWB and AWC sensory neurons. The other group consists of ADF serotonergic neurons with their downstream interneurons and motor neurons (the ADF modulatory circuit) and is specifically required to display learned olfactory preference. The interplay between the AWB-AWC sensorimotor circuit and the ADF modulatory circuit generates naive and learned olfactory preferences. To the best of our knowledge, this is the first time that a neural network for olfactory learning has been mapped from sensory input to motor output with specific roles assigned to each neuron in the network. Our study has uncovered the functional organization of a neural network that directs olfactory response and learning, demonstrating that C. elegans exhibits naïve and learned olfactory behaviors by regulating the interplay of different neural circuits that transduce olfactory stimuli into motor responses.
RESULTS
An automated olfactory learning assay using individual animals
C. elegans displays olfactory preference on a solid substrate by chemotaxis, using both a “pirouette strategy” and a “weathervane strategy”. In the “pirouette strategy”, locomotion is characterized by periods of forward movements that are interrupted by reorienting maneuvers “pirouettes”, including reversals and Ω turns (sharp turns in which animal’s body shape resembles the Greek letter omega Ω). When an animal experiences improving conditions, such as a positive gradient of attractive chemical cues, it reduces the frequency of reorienting maneuvers; when an animal encounters declining conditions, it increases the frequency of reversals and turns. This behavioral strategy resembles the biased random walk that bacteria exhibit during chemotaxis (Berg and Brown, 1972; Chalasani et al., 2007; Pierce-Shimomura et al., 1999). In the “weathervane strategy”, animals gradually steer themselves during periods of forward movement to move towards an attractant (Iino and Yoshida, 2009). Swimming C. elegans also exhibits chemotaxis and displays olfactory preference by regulating the frequency of omega turns. Attractive odorant molecules suppress turns and their removal evokes turns (Luo et al., 2008; Pierce-Shimomura et al., 2008). Therefore, the frequency of omega turns during swimming is negatively correlated with an animal’s preference for an olfactory stimulus.
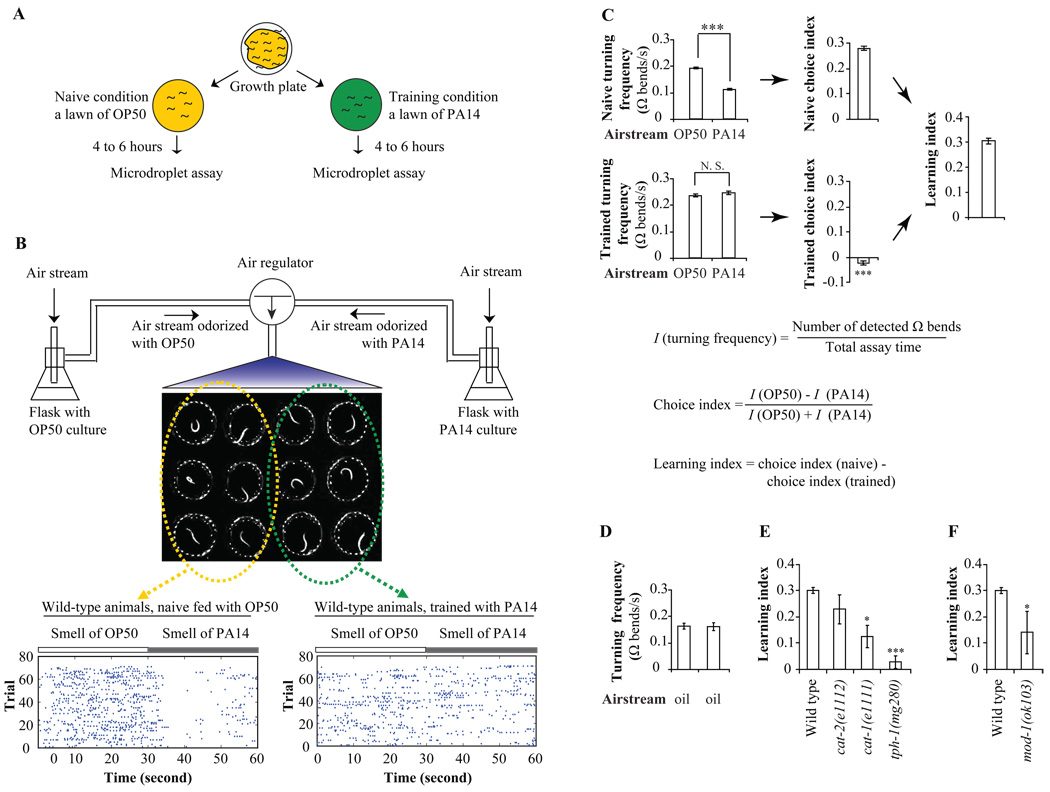
To carry out systematic laser ablation analysis of olfactory learning, we employed a microdroplet assay to automate the analysis of olfactory behaviors using individual animals (Figure 1A and 1B). Previously, it was shown that adult animals exhibit similar levels of olfactory learning whether exposed to pathogenic bacteria only as adults or exposed throughout their lifetimes (Zhang et al., 2005). We raised animals under standard conditions until adulthood and then transferred half of the animals onto a training plate containing a fresh bacterial lawn of a pathogenic bacterium P. aeruginosa PA14 and the other half onto a control plate containing a fresh lawn of a benign bacterium E. coli OP50 (so they would remain naïve to the smell of PA14). After four to six hours, we analyzed trained and naïve animals side by side in the microdroplet assay by subjecting animals, each freely swimming in a microdroplet of buffer, to 12 cycles of alternating airstreams that were odorized with either PA14 or OP50. Switching between airstreams was under computer control (Figure 1A and 1B). We analyzed video records of swimming animals with a machine-vision algorithm that automatically detected omega turns exhibited by each animal. We quantified olfactory preference between the two odorized airstreams with a choice index based on the number of omega turns detected during exposure to each airstream for each assay (Figure 1B and 1C).
Figure 1. C. elegans displays aversive olfactory learning on the pathogenic bacterium PA14 in the microdroplet assay.
A, Training protocol for aversive olfactory learning on adult animals. B, Diagram of the olfactory microdroplet assay. One representative image and data output of 12 animals in one assay are shown. Results are vertically arranged on raster panels and each dot represents one Ω bend. Each row displays the result of one cycle of olfactory assay on one animal and each animal was tested for 12 cycles continuously in every assay. C, The aversive olfactory learning on the smell of PA14 in the microdroplet assays. Turning rates towards OP50 were compared with turning rates towards PA14 in naïve and trained animals with two tailed Student’s t-test. The naïve choice index was compared with the trained choice index with two tailed Student’s t-test. (Data are presented as mean +/− SEM. *** P < 0.001, n > 50 assays, error bars: SEM, N.S.: not significant). D, Wild type animals generate frequent turns when exposed to the clean air that passes mineral oil and carries no odorant (error bars: SEM). E, The aversive olfactory learning ability of wild type animals and mutants defective in neurotransmitter biosynthesis in the microdroplet assay. The learning ability of mutants was compared with that of wild type animals and the P values were calculated by two tailed Student’s t-test. Multiple comparisons were adjusted with Bonferroni correction (Data are presented as mean +/− SEM. * P < 0.05, *** P < 0.001, n ≥ 6 assays, error bars: SEM). F, The mod-1(ok103) mutant is defective in learning in the microdroplet assay. The learning ability of mod-1 was compared with that of wild type animals and the P value was calculated by two tailed Student’s t-test (Data are presented as mean +/− SEM. * P < 0.05, n ≥ 8 assays, error bars: SEM). Please also see Supplemental Figure 1.
Because the frequency of omega turns is negatively correlated with the preference for an olfactory stimulus, a positive choice index indicates an olfactory preference for pathogen PA14 and a negative choice index indicates a preference for OP50. We defined the learning index as the difference in choice indexes exhibited by naïve and trained animals (Figure 1C). We found that naive animals turned much less frequently when exposed to the smell of PA14 and generated a positive choice index, indicating that the naïve olfactory preference is to PA14. However, after training, animals generated similar numbers of turns when exposed either to the smell of PA14 or to the smell of OP50 and produced a choice index close to zero. Therefore, a positive learning index indicates that trained animals learn to decrease their attraction to the smell of PA14 after exposure to PA14 (Figure 1C).
C. elegans exhibits spontaneous omega turns even when subjected to clean air that carries no olfactory stimulus (Figure 1D). Thus, olfactory stimuli do not directly generate omega turns, but instead modulate the frequency of spontaneous turns. When exposed to the alternating airstreams odorized with buffer and bacteria culture of either OP50 or PA14, both naïve and trained animals exhibited a lower turning rate to the smell of either bacterial culture than to buffer, indicating that they are attracted to the smell of food (Supplemental Figure 1A). Thus, learning specifically modulates the olfactory preference between benign and pathogenic bacteria without abolishing the general attraction of food smell.
This form of aversive olfactory learning occurs and reverses rapidly. Adult animals trained on PA14 exhibited a significant amount of learning with two hours of training and became fully trained after 4 hours. When fully-trained adult animals were transferred back to a bacterial lawn of OP50, the learned olfactory preference gradually diminished over 2 hours (Supplemental Figure 1B, 1C), indicating that the training process does not generate permanent alteration to the nervous system. Thus, the fully-developed nervous system of adult C. elegans is capable of experience-dependent modulations to avoid the smell of pathogenic bacteria.
Although the aversive olfactory learning analyzed by the microdroplet assay reverses rapidly, three observations suggest that it is distinct from adaptation. First, no alteration in olfactory preference was detected in this assay when adult animals were trained with a series of non-pathogenic bacterial strains (Supplemental Figure 1D), suggesting that the aversive learning is contingent on the pathogenesis of the training pathogen. Second, two adaptation mutants, egl-4 and adp-1, both displayed normal learning ability in the microdroplet assay (Supplemental Figure 1E). Third, aversive olfactory learning of PA14 requires the negative reinforcing effect that directly results from ingestion of the pathogen. To show this, we put adult animals on a lawn of OP50 while exposing them for 6 hours to the smell of a PA14 lawn, which was grown on the lid of the plate. In this experiment, trained animals were exposed to the smell of PA14, but were fed on OP50. These trained animals exhibited olfactory preference comparable to that of the control animals that fed on OP50 without exposure to the smell of PA14 (Supplemental Figure 1F).
Previously, we used a two-choice assay that quantified the overall movements of populations of crawling worms to elucidate the role of serotonergic neurotransmission in aversive olfactory learning (Zhang et al., 2005). Importantly, the automated microdroplet assay that we utilized in this study recapitulates the phenotypes that were obtained using the two-choice assay and supports the role of serotonin in aversive olfactory learning. The cat-1 mutation, which disrupts both dopamine and serotonin neurotransmission (Duerr et al., 1999), greatly reduced olfactory learning quantified using the microdroplet assay while the cat-2 mutation, which specifically disrupts dopamine production (Lints and Emmons, 1999), had no effect on learning (Figure 1E). The tph-1(mg280) mutant, which is deficient in the only C. elegans tryptophan hydroxylase required for biosynthesis of serotonin (Sze et al., 2000), was completely defective in olfactory learning in the microdroplet assay (Figure 1E). In addition, the mod-1(ok103) mutant, which is defective in a serotonin-gated chloride channel (Ranganathan et al., 2000), also showed greatly reduced learning in the microdroplet assay (Figure 1F). Thus, the microdroplet assay for swimming animals assigns phenotypes that are consistent with the two-choice assay that we previously used. The important advantage of the microdroplet assay is that it allows us to quantify olfactory preference with small numbers of animals.
Aversive olfactory learning requires the AWB and AWC olfactory sensory neurons
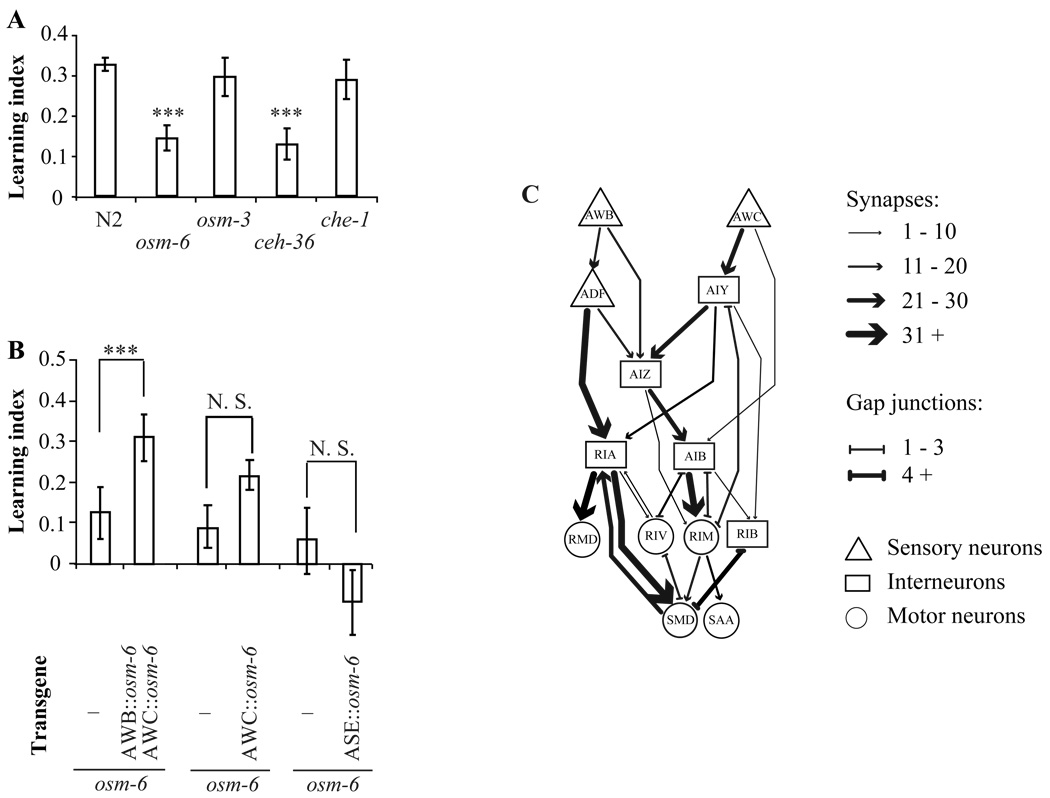
To characterize the neuronal network that regulates the switch of olfactory preference, we began by identifying chemosensory neurons required for olfactory plasticity. We first tested an osm-6 mutant, which is defective in development and sensory function of all ciliated chemosensory neurons (Collet et al., 1998). The osm-6 mutant showed significantly reduced learning to avoid the smell of PA14 (Figure 2A). By comparing choice indexes before and after training, we found that the osm-6 mutant was unable to reduce its olfactory preference for the smell of PA14 after training (Supplemental Figure 2A). These results indicate a requirement for the function of chemosensory neurons in generating a learned preference. The residual learning ability of the osm-6 mutant likely results from its residual olfactory sensory ability in the microdroplet assay (Supplemental Figure 2B). The osm-6 mutant exhibited lower turning frequencies than wild-type animals, especially towards the smell of PA14 (Supplemental Figure 2C), consistent with other studies in which osm-6 mutant exhibited lower omega turn frequencies in crawling and swimming assays (Gray et al., 2005; Srivastava et al., 2009). We also measured the innate immune response of C. elegans to PA14 infection following an established procedure (Tan et al., 1999) and found that the osm-6 mutant displayed the same immune response to PA14 infection as wild type animals (Supplemental Figure 2D), suggesting that its learning defect did not result from an altered immune response.
Figure 2. Aversive olfactory learning requires the AWB and AWC olfactory sensory neurons.
A, The aversive olfactory learning ability of wild type animals and chemosensory mutants. The learning ability of mutants and wild type animals were compared with two tailed Student’s t-test and multiple comparisons were adjusted with Bonferroni correction (Data are presented as mean +/− SEM. *** P < 0.001, n ≥ 6 assays, error bars: SEM). B, Expression of osm-6 cDNA in AWB and AWC olfactory sensory neurons rescues the learning defect of osm-6 mutants, but the expression of osm-6 cDNA in the AWC or in the gustatory neurons ASE does not rescue. The learning ability of transgenic animals was compared with that of non-transgenic siblings with paired two tailed Student’s t-test (Data are presented as mean +/− SEM. *** P < 0.001, n ≥ 4 assays, error bars: SEM, N.S.: not significant). C, A candidate neuronal network downstream of sensory neurons AWB and AWC to regulate aversive olfactory learning. Please also see Supplemental Figure 2 and Supplemental Table 1 and 2.
Since osm-6 is expressed in all ciliated chemosensory neurons, we tested other chemosensory mutants to identify the set of chemosensory neurons that are necessary for learning. An osm-3 mutant, which is defective in function of most chemosensory neurons except olfactory neurons (Tabish et al., 1995), showed learning ability and turning rate comparable to wild type animals. A che-1 mutant, which is impaired in development of the major gustatory neurons ASE (Uchida et al., 2003), also displayed normal learning ability and turning rate (Figure 2A, Supplemental Figure 2A and 2C). However, a mutation in ceh-36, which compromises development of ASE and olfactory sensory neurons AWC (Lanjuin et al., 2003), caused a defective learning and exhibited a much reduced naïve olfactory preference for the smell of bacteria (Figure 2A, Supplemental Figure 2A and 2C). Together, these results suggest that the AWC olfactory neurons are required to detect the smell of bacteria and olfactory sensory inputs are required to generate aversive olfactory learning in the microdroplet assay.
AWA, AWB and AWC represent three pairs of olfactory sensory neurons in the C. elegans nervous system. AWC and AWB sensory neurons mediate attractive and repulsive olfactory responses, respectively, to a variety of odorants (Bargmann et al., 1993; Troemel et al., 1997). It was shown that AWB mediate behavioral response to stay off the lawns of pathogenic bacteria (Pradel et al., 2007). We found that the expression of osm-6 complementary DNA in the olfactory sensory neurons AWB and AWC together fully rescued the learning defect of the osm-6 mutant, while expression of osm-6 cDNA in AWC alone or in the gustatory neurons ASE did not rescue (Figure 2B and Supplemental Figure 2E), suggesting that the combined function of AWB and AWC regulates aversive olfactory learning.
Since different Pseudomonas bacteria strains secrete 2-butanone, iso-amyl alcohol, 2,3-pentanedione and 2,4,5-trimethylthiazole, which are detected by the sensory neurons AWC and AWA as attractive odorants (Bargmann et al., 1993; Zechman and Labows, 1985), one possibility is that training with Pseudomonas aeruginosa PA14 lowers the animal's attraction towards these molecules, or even inverts the animal's response, turning them into repellents. Using the microdroplet assay, we quantified the strength of olfactory responses to this set of chemicals in both naïve animals and animals trained with PA14 and found no significant difference (Supplemental Figure 3). One possibility is that C. elegans recognizes the smell of PA14 as a coherent percept corresponding to a particular mix of different odorant molecules. Another possibility is that the smell of PA14 is recognized through an as yet unidentified odorant molecule that is unique to it.
Two different neural circuits regulate naïve and learned olfactory preferences
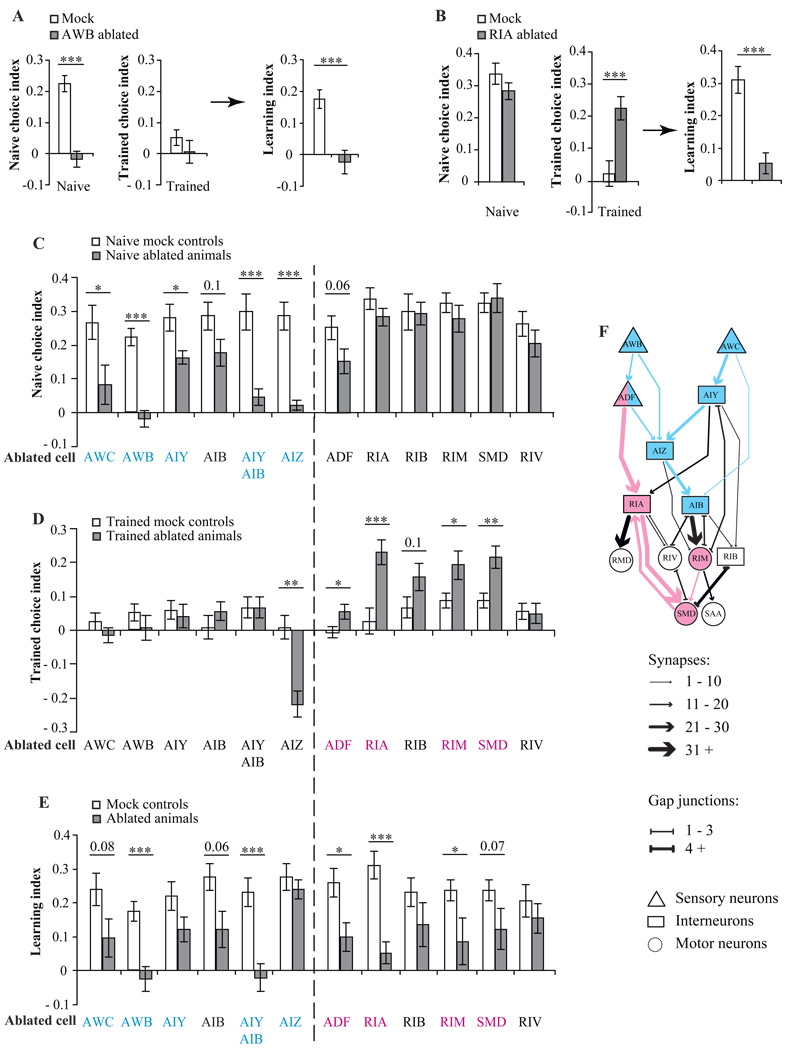
Next, we asked which neurons operate downstream of AWB and AWC to display naïve and learned olfactory preferences. Serial-section electron micrography has revealed the complete wiring diagram of the C. elegans nervous system (Chen et al., 2006; White et al., 1986). Potential functional circuits can be identified as groups of neurons that are heavily interconnected by large numbers of synapses. This straightforward method of counting synapses allowed previous investigators, for example, to map pathways that regulate the ability of crawling worms to generate spontaneous omega turns and reversals (Gray et al., 2005). We applied this method to identify integrated circuits downstream of AWB and AWC based on strong chemical synaptic connections (Supplemental Table 1 and Supplemental Experimental Procedures). This analysis uncovered a multilayered neural network composed of sensory neurons (AWB, AWC, and ADF), interneurons (AIB, AIY, AIZ, RIA, and RIB), and motor neurons (RIM, RIV, RMD, SAA, and SMD) (Figure 2C and Supplemental Table 2). Most neurons in this candidate network represent pairs of bilaterally symmetric neurons except that SMD and SAA are groups of four neurons and RMD are six neurons. This network downstream of the AWB and AWC olfactory sensory neurons overlaps partly with a previously mapped network that regulates the frequency of reversals and omega turns of crawling worms (Gray et al., 2005; Tsalik and Hobert, 2003). For this study, the network downstream of AWB and AWC provides candidate pathways for understanding how olfactory preference and plasticity are generated in the C. elegans nervous system.
To characterize how the neuronal function of the olfactory network in Figure 2C allows animals to display different olfactory preferences before and after learning, we performed a systematic laser ablation analysis of each neuronal type in the network. We conducted laser ablations with a femtosecond laser microbeam (Chung et al., 2006) on L2 larvae and cultivated the operated animals under standard conditions until the adult stage. At this point, we transferred half of the operated animals onto a fresh lawn of OP50 as naïve controls and the other half onto a fresh lawn of PA14 to induce aversive olfactory learning (Figure 1A). After six hours, we measured turning frequency of these naïve and trained animals towards alternating airstreams odorized with OP50 or PA14 (Figure 5G and Figure 6G), which allowed us to analyze choice indexes to quantify olfactory preference (Figure 1B and 1C). We quantified effects of neuronal ablation by comparing the choice indexes of naïve and trained ablated animals with those of matched mock controls. Mock controls went through laser surgery, post-surgery cultivation, and olfactory training and analysis procedures in the same way as the ablated animals but without the neurons being ablated. These comparisons allowed us to define the contribution of each neuronal type in the network to the generation of naïve and learned olfactory preferences.
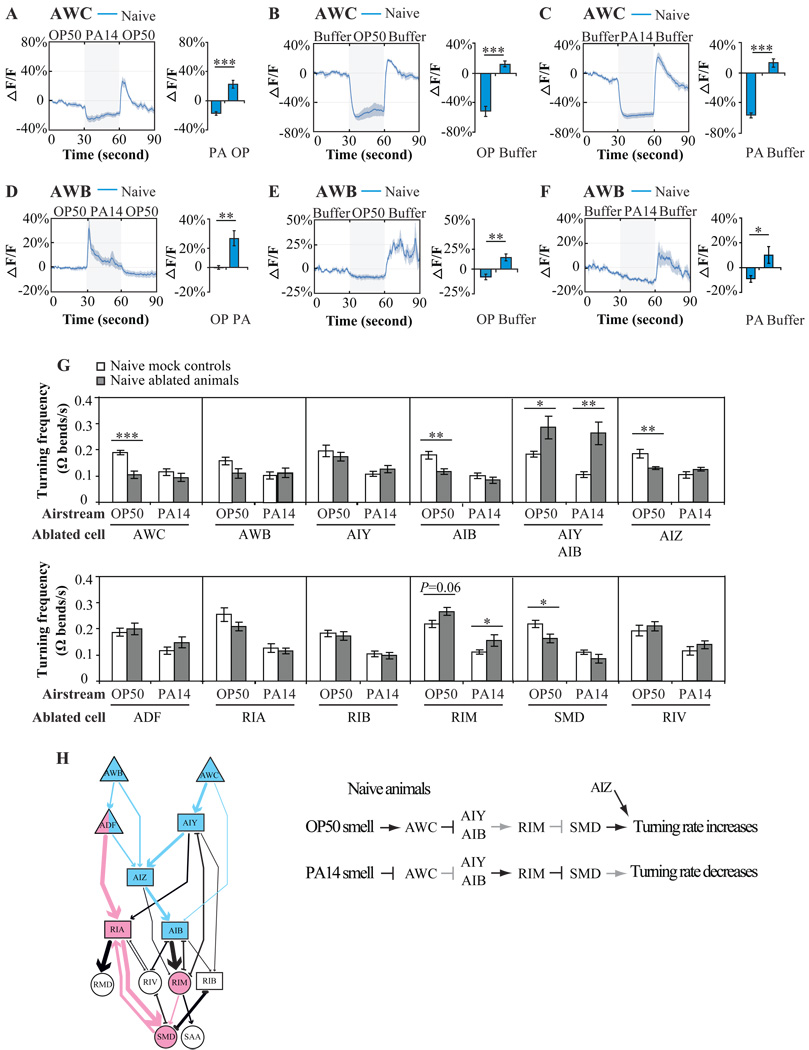
Figure 5. Olfactory sensory neurons in the AWB-AWC sensorimotor circuit mediate naïve olfactory preference.
A–F, G-CaMP calcium response of AWCON (A–C) and AWB (D–F) neurons in naïve animals when stimuli alternated between OP50-condtioined medium and PA14-conditioned medium in the order of OP50-PA14-OP50 (A, D), or when stimuli alternated between buffer and OP50-condtioined medium (B, E), or between buffer and PA14-condtioined medium (C, F). The blue traces indicate the average percentage changes in G-CaMP intensity for multiple recordings and the grey areas around the traces indicate SEM. In A–F, the calcium signals within 3 second window immediately before the switch of the stimuli and the calcium signals within 3 second window that begins 1.5 second after the switch were quantified. Values were compared using a paired two-tailed Student’s t-test (*** P < 0.001, ** P < 0.01, * P < 0.05, n ≥ 5 animals). G, Effects of neuronal ablations on turning rate exhibited by naïve animals when stimuli alternated between the smells of OP50 and PA14. The turning rates of ablated animals were compared with that of matched mock animals with two tailed Student’s t-test (Data are presented as mean +/− SEM. *** P < 0.001, ** P < 0.01, * P < 0.05, n ≥ 6 assays, error bars: SEM). H, In naïve animals, differential neuronal responses of AWC sensory neurons to the smell of OP50 and PA14 propagate through downstream circuit to generate preference. The AWB-AWC sensorimotor circuit is highlighted in blue and the ADF modulatory circuit is highlighted in pink. Please also see Supplemental Figure 4.
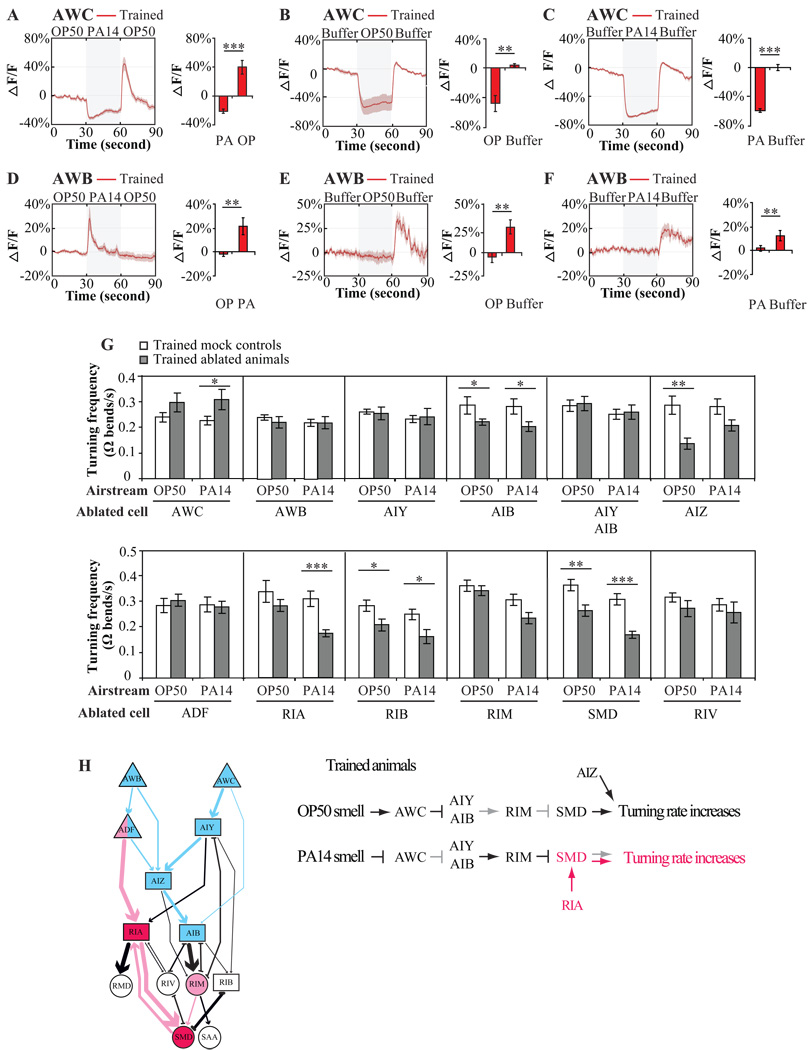
Figure 6. Modulations to signal transduction downstream of sensory neurons generate the learned preference.
A–F, G-CaMP calcium response of AWCON (A–C) and AWB (D–F) neurons in trained animals when stimuli alternated between OP50-condtioined medium and PA14-conditioned medium in the order of OP50-PA14-OP50 (A, D), or when stimuli alternated between buffer and OP50-condtioined medium (B, E), or between buffer and PA14-condtioined medium (C, F). The red traces indicate the average percentage changes in G-CaMP intensity and the grey areas around the traces indicate SEM. In A–F, the calcium signals within 3 second window immediately before the switch of the stimuli and the calcium signals within 3 second window that begins 1.5 second after the switch were quantified. Values were compared using a paired two-tailed Student’s t-test (*** P < 0.001, ** P < 0.01, n ≥ 5 animals). Compared to the calcium response in naïve animals (Figure 5A–F), training had no effect under each condition. G, Effects of neuronal ablations on the turning rates exhibited by trained animals when stimuli alternated between the smell of OP50 and PA14. The turning rates of ablated animals were compared with that of matched mock animals with two tailed Student’s t-test (Data are presented as mean +/− SEM. *** P < 0.001, ** P < 0.01, * P < 0.05, n ≥ 6 assays, error bars: SEM). H, In trained animals, RIA interneurons and SMD motor neurons regulate the turning rate towards the smell of PA14 to generate the trained preference. The AWB-AWC sensorimotor circuit is highlighted in blue and the ADF modulatory circuit is highlighted in pink. Please also see Supplemental Figure 4.
We found that the naïve olfactory preference for PA14 is disrupted by laser ablation of a specific group of neurons. For example, AWB-ablated animals exhibited no naïve olfactory preference for PA14 and trained AWB-ablated animals did not exhibit any olfactory preference either, producing a learning index that was close to zero (Figure 3A). It is important to note that all choice indexes that we measured as being close to zero in this study were due to similar turning rates during the OP50 and the PA14 airstreams, and not due to inability to swim or generate Ω turns. This notion is evidenced by the analyses on turning rates in Figure 5G and Figure 6G for all the ablation results.
Figure 3. Two different neural circuits in an olfactory network regulate the naïve and learned olfactory preferences.
A, Ablation of AWB sensory neurons abolished the naïve olfactory preference and learning. B, Ablation of RIA interneurons abolished the learned olfactory preference and learning without altering the naïve preference. C – E, Effects of ablating different neuronal types on the naïve olfactory preference (C), the learned olfactory preference (D) and learning (E). For A – E, choice indexes and learning indexes of ablated animals were compared with that of matched mock animals with two tailed Student’s t-test. (Data are presented as mean +/− SEM. *** P < 0.001, ** P < 0.01, * P < 0.05, n ≥ 6 assays, P values between 0.05 and 0.1 were indicated above the bar graphs. error bars: SEM). F, Two different neural circuits for the naïve and learned preferences. The AWB-AWC sensorimotor circuit is highlighted in blue and the ADF modulatory circuit is highlighted in pink.
Individually ablating AWC or AIY produced an effect similar to, albeit smaller than, that of ablating AWB. Ablating AIZ or AIY and AIB together generated the same effects on the naïve preference as ablating AWB (Figure 3C). Ablating the ADF serotonergic neurons also moderately reduced the naïve choice index, indicating that ADF might have a small sensory contribution to the naïve olfactory preference for PA14. Ablating any other neurons in the network did not significantly alter naïve olfactory preference (Figure 3C). Thus, AWB, AWC, AIY and AIB, AIZ and possibly ADF play essential roles in generating the naïve olfactory preference between the smells of OP50 and PA14. These neurons are strongly interconnected with chemical synapses. The similar effects caused by ablating these neuronal types suggest that these neurons constitute a functional circuit (an AWB-AWC sensorimotor circuit) that allows C. elegans to encode and display its naïve olfactory preference for PA14 (blue symbols in Figure 3F).
Within the AWB-AWC sensorimotor circuit, the functions of different neurons are diverse. Animals lacking AWB or AWC or AIY and AIB together are not only defective in their naïve preference, but also deficient in generating any clear preference after training and, thus, produce low learning indexes (Figure 3C–3E). The low learning indexes of these animals could be caused either by defects in sensing or distinguishing between the smells of different bacteria, defects in learning, or both. Although the severe defects in the naïve preference caused by ablating AWB, AWC or AIY and AIB together clearly points to their role in producing the naïve preference, their contribution to producing the learned preference cannot be excluded and deserves further examination. In contrast, AIZ-ablated animals exhibited a strong olfactory aversion to the smell of PA14 after training, despite showing no naïve olfactory preference between OP50 and PA14 (Figure 3C and 3D). Both naïve and trained AIZ-ablated animals exhibited lower turning rates when exposed to the smell of OP50 than mock control animals, with a greater decrease in turning rate after training (Figure 5G and Figure 6G), implicating the specific function of AIZ in generating the naïve olfactory preference for PA14. The difference in naïve and trained choice indexes of AIZ-ablated animals yielded a learning index comparable to wild type animals (Figure 3C–3E), indicating that ablating AIZ did not affect olfactory learning ability. The distinct effects of ablating AIZ on olfactory preference and plasticity point to different cellular mechanisms for generating naïve olfactory preference and learning.
We next sought to identify neurons that might regulate olfactory plasticity without affecting naïve olfactory preference. Further laser ablation analysis uncovered such a group of neurons. For example, ablating the RIA interneurons had no effect on the naïve olfactory preference for PA14, but completely abolished the ability to shift olfactory preference away from PA14 after training. Animals without RIA continued to exhibit an olfactory preference for PA14 after training, leading to a low learning index (Figure 3B). Similarly, killing ADF or RIM or SMD significantly changed the learned preference and disrupted learning ability without substantially altering naïve olfactory preference (Figure 3C–3E). Except for the mild effect of killing RIB, ablating any other neuronal types in the network did not generate comparable defects (Figure 3C–3E). The RIA interneurons connect with ADF sensory neurons and SMD motor neurons with large numbers of synapses, and the RIM motor neurons send out a few synapses to SMD. Ablating any neurons in this circuit - RIA, ADF, SMD or RIM - abolished olfactory plasticity without affecting the naïve olfactory preference for PA14. Thus, this circuit (the ADF modulatory circuit) is specifically required to generate experience-dependent plasticity after training with PA14 (pink symbols in Figure 3F). Previously, we found that the serotonergic neurons ADF play an essential role in regulating aversive olfactory learning on pathogenic bacteria (Zhang et al., 2005). Here, by analyzing the function of neurons that are strongly connected to ADF, we identified the pathway downstream of ADF that causes worms to shift their olfactory preference away from PA14 after training.
In summary, two different neural circuits – the AWB-AWC sensorimotor circuit and the ADF modulatory circuit -- allow C. elegans to display the naïve olfactory preference and to change olfactory preference after experience. The ADF neurons contribute to both the naïve olfactory preference and the change in olfactory preference after experience (Figure 3F).
Correspondence between the microdroplet assay for swimming worms and the two-choice assay for crawling worms
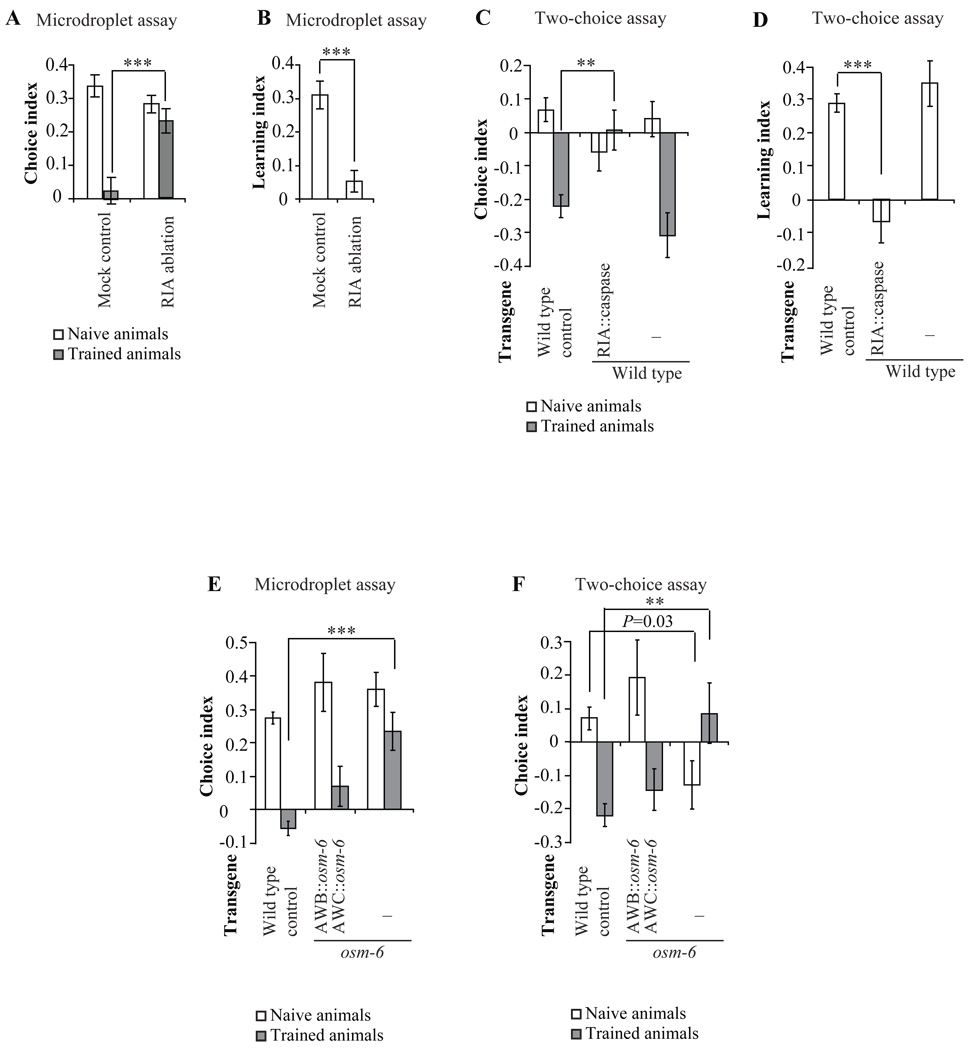
Next, we sought to verify that phenotypes of neuronal ablation that were quantified using individual swimming worms in the microdroplet assay could also be obtained using crawling worms in the two-choice assay that we established earlier (Zhang et al., 2005). Since the two-choice assay requires a large number of animals for each experiment, we were able to do this with the RIA interneurons by ectopically expressing a cell-death molecule caspase with strong cell specific promoter (Brockie et al., 2001; Chelur and Chalfie, 2007). In the microdroplet assay, laser ablation of RIA did not alter the naïve olfactory preference for PA14, but generated a significant deficiency in changing olfactory preference away from PA14 after training (Figure 4A and 4B). Similarly, we found that RIA-genetically-killed animals exhibited a naïve olfactory preference comparable to wild type animals and non-transgenic siblings, but exhibited no ability to shift olfactory preference away from PA14 after training, resulting in a complete loss of learning ability (Figure 4C and 4D). Thus, the results of both assays are consistent in identifying a specific role for RIA in generating olfactory plasticity.
Figure 4. Correspondence between the microdroplet assay and the two-choice assay.
A, B, In the microdroplet assay, laser ablation of RIA generated significant defects in the trained choice index (A) and learning (B) without affecting naïve choice index. The choice indexes or the learning index of ablated animals were compared with that of mock control animals with two tailed Student’s t-test (Data are presented as mean +/− SEM. *** P < 0.001, n ≥ 9 assays, error bars: SEM). C, D, In the two-choice assay, killing RIA generated significant defects in the trained choice index (C) and learning (D) without affecting naïve choice index. The choice indexes or the learning indexes of transgenic animals and non-transgenic siblings were compared with that of wild type animals with two tailed Student’s t-test. Multiple comparisons were adjusted with Bonferroni correction (Data are presented as mean +/− SEM. *** P < 0.001, ** P < 0.01, n ≥ 3 assays, error bars: SEM). E, F, In both the microdroplet assay (E) and the two-choice assay (F), osm-6 mutants exhibited significant defects in the trained choice index and expression of osm-6 cDNA in AWB and AWC neurons rescued the defects. The choice indexes of transgenic animals and non-transgenic siblings were compared with that of wild type animals with two tailed Student’s t-test. Multiple comparisons were adjusted with Bonferroni correction (Data are presented as mean +/− SEM. *** P < 0.001, ** P < 0.01, n ≥ 6 assays, error bars: SEM).
We also compared phenotypes obtained in the microdroplet assay and the two-choice assay using osm-6 mutants and transgenic animals in which function of osm-6 is rescued in olfactory neurons AWB and AWC. We found that in both the microdroplet assay and the two-choice assay the trained choice indexes of osm-6 mutants were significantly different from that of wild type animals and expression of osm-6 cDNA in AWB and AWC neurons fully rescued the learning defect (Figure 4E and 4F). Thus, the microdroplet assay is as reliable as the two-choice assay in defining phenotypes for olfactory preference and learning. Unlike the two-choice assay, however, the microdroplet assay can be combined with systematic laser ablation analysis of any neuron within the circuit.
Olfactory sensory neurons mediate the naïve olfactory preference
As shown above, naïve animals prefer the smell of PA14, evidenced by an increase in their turning rate when airstreams switch from the smell of PA14 to the smell of OP50. In contrast, animals that have been trained by exposure to PA14 display similar turning rates towards the smells of PA14 and OP50, producing a comparatively lower olfactory preference for PA14. We next asked how the neurons for the naïve and learned olfactory preferences regulate turning rate to exhibit olfactory preference.
We first analyzed the AWB-AWC sensorimotor circuit for the naïve olfactory preference (blue symbols in Figure 3F). AWB and AWC mediate repulsive and attractive olfactory responses, respectively. To characterize their function in determining naïve preference, we measured neuronal activity within these sensory neurons upon exposure to the smells of OP50 or PA14 using intracellular calcium imaging. First, we studied transgenic animals expressing the genetically encoded calcium sensitive fluorescent protein G-CaMP in the AWCON cell, one of the two AWC neurons. It was previously shown that the two AWC neurons, AWCON and AWCOFF, generate similar calcium responses to the odorants that they both detect (Chalasani et al., 2007). Removal of attractive odorants stimulates AWC calcium response while exposure to attractants suppresses it (Chalasani et al., 2007). We subjected naïve transgenic animals to alternating streams of PA14-conditioned medium and OP50-conditioned medium in the order of OP50-PA14-OP50, and found that OP50-conditioned medium stimulated calcium transients in AWCON while PA14-conditioned medium suppressed the activation (Figure 5A). We also did control experiments by exposing transgenic animals to the two stimuli in the order of PA14-OP50-PA14 and found similar results (Supplemental Figure 4A). Next, we subjected naïve transgenic animals to alternating streams of clean buffer and streams conditioned with either OP50 or PA14, and found that AWCON calcium transients were suppressed by either type of bacterial conditioned medium (Figure 5B and 5C). Together, these results indicate that the AWC neurons in naïve animals respond to both the smell of PA14 and OP50 as attractants, but respond to the smell of PA14 as a more attractive stimulus than the smell of OP50. Thus, the response properties of the AWC neuron match the olfactory preference of the naïve behaving animal.
We next examined transgenic animals that express G-CaMP in the AWB olfactory sensory neurons, which mediate repulsive olfactory behavioral response to repellants including 2-nonanone (Troemel et al., 1997). We found that removal of 2-nonanone stimulated AWB calcium transients and exposure to 2-nonanone suppressed AWB (Supplemental Figure 4E). This result suggests that the switch from a repellent to the removal of the repellent activates AWB. We subjected these naïve transgenic animals to alternating streams of OP50 and PA14-conditioned mediums in the order of either OP50-PA14-OP50 or PA14-OP50-PA14. We found that the switch from OP50-conditioned medium to PA14-conditioned medium activated AWB calcium transients (Figure 5D and Supplemental Figure 4C). When we alternated streams of clean buffer with streams conditioned by either OP50 or PA14, calcium transients in AWB neurons were activated by switching either type of bacterial conditioned medium to buffer (Figure 5E and 5F). Taken together, these results indicate that both OP50 and PA14-conditioned mediums contain repellents that are detected by AWB and in naïve animals AWB respond to OP50 as a more repulsive stimulus than PA14. Thus, the neuronal response of AWB is consistent with the olfactory preference of naïve animals towards PA14 at the level of behavior.
Next, we asked how the olfactory sensitivities of AWC and AWB are transduced into olfactory behavioral preference by the regulation of turning rate exhibited by swimming worms in response to the smells of OP50 and PA14. To do this, we examined the effects of neuronal ablation on the turning rate of naïve animals. Ablating the AWC sensory neurons, AIB or AIZ interneurons, or SMD motor neurons significantly decreased the turning rate to the smell of OP50, suggesting that the smell of OP50 promotes turns through these neurons. In contrast, ablating both AIY and AIB or ablating the RIM motor neurons increased the turning rate towards the smells of both OP50 and PA14, indicating that these neurons inhibit turns in response to olfactory inputs (Figure 5G). Individually ablating any other neurons did not affect turning rate in naive animals (Figure 5G). The AIB and AIY interneurons are postsynaptic to AWC, and regulate turns during odor sensation in crawling animals (Chalasani et al., 2007). RIM, which receive inputs from both AIY and AIB, synapse onto the four SMD motor neurons. Taken together, these results indicate that these strongly connected interneurons and motor neurons regulate turning rates downstream of AWB and AWC (Figure 5H). When naïve worms are subjected to alternating smells of OP50 and PA14, the smell of OP50 activates AWC neurons and raises turning rate, whereas the smell of PA14 inactivates AWC and lowers turning rates (Figure 5A and 5H). Thus, the differential responses of AWC to the smells of PA14 and OP50 could regulate downstream neurons to generate stimulus-specific turning rates that are displayed as the olfactory preference for PA14 in naïve animals (Figure 5H).
Although the activity of the AWB neurons also reflects the naïve olfactory preference for PA14 (Figure 5D), ablating AWB eliminated naïve olfactory preference without significantly changing turning rates. It is possible that AWB might regulate AIZ directly and/or indirectly through ADF (Figure 5G). Ablating AIZ interneurons specifically lowered the turning rate upon exposure to the smell of OP50 in both naïve and trained animals, eliminating the naïve olfactory preference for PA14 without affecting olfactory learning (Figure 3C–3E, 5G and 6G). Although AIB or RIM or SMD also contribute to the generation of different turning rates upon exposure to the smell of OP50 and PA14 in naïve animals (Figure 5G), the ablation effects were not specific (RIM) or prominent enough (AIB or SMD) to significantly change the naïve olfactory preference for PA14 (Figure 3C). Together, our results indicate that in naïve animals AWB and AWC exhibit stimulus-specific patterns of activity. Differential response of AWC to the smells of OP50 and PA14 regulates downstream circuit to display olfactory preference through the control of turning rate. AIZ contribute to naïve olfactory preference by regulating the response to the smell of OP50 (Figure 5H).
Modulations to signal transduction to the network downstream of olfactory sensory neurons generate the learned preference
Finally, we investigated how this network is changed by training with PA14 to generate learned olfactory preference. First, we studied intracellular calcium responses in the AWB and AWC olfactory neurons upon exposure to the smells of OP50 and PA14 after training. Surprisingly, while AWCON neuronal responses are strongly correlated with the behavioral preference for PA14 over OP50 in naïve animals, AWCON neuronal responses in trained animals did not reflect the shift in olfactory preference away from the smell of PA14. As we did with naïve animals, we subjected trained worms to alternating streams conditioned with either OP50 or PA14. We found that OP50-conditioned medium stimulated AWCON calcium response and PA14-conditioned medium suppressed AWC when animals were exposed to the two stimuli either in the order of OP50-PA14-OP50 or in the order of PA14-OP50-PA14 (Figure 6A and Supplemental Figure 4B). And the switch from buffer to medium conditioned with either OP50 or PA14 also suppressed AWCON calcium transients in trained animals (Figure 6B and 6C). These patterns of AWCON calcium dynamics in trained animals indicate that AWCON continues to respond to PA14 as the more attractive stimulus than OP50, just as in naïve animals. Calcium dynamics in the AWB olfactory sensory neurons were also unchanged by training, continuing to respond to the smell of OP50 as more repulsive than the smell of PA14 (Figure 6D–6F and Supplemental Figure 4D). Thus, the behavioral shift of olfactory preference away from PA14 is not generated by the patterns of AWC or AWB sensory response, pointing to experience-dependent changes to the signal transduction to downstream neurons.
To identify the changes to the network that are caused by training, we examined the turning rates exhibited by trained animals in response to the smells of OP50 and PA14. We found that ablating RIA, the interneurons that are specifically required to shift olfactory preference away from PA14 after training, specifically decreased the PA14-induced turning rate without affecting the OP50-induced turning rate (Figure 6G). Ablating SMD, four motor neurons connecting with RIA, reduced the turning rates towards the smells of both OP50 and PA14, but with a stronger reduction towards the smell of PA14 (Figure 6G). These results suggest that RIA and SMD function downstream of the neural network to increase turning rate towards the smell of PA14 in trained animals. Although ablating AWC or AIB or RIB also altered turning rates of trained animals, the effects on turning rate were similar towards either the smell of OP50 or PA14, resulting in little change on olfactory preference after training (Figure 3D and Figure 6G). These results suggest that olfactory learning to shift olfactory preference away from the smell of PA14 after exposure to PA14 works by modulating the turning rate response upon exposure to the smells of OP50 and PA14, not by modifying the neuronal responses of AWC or AWB sensory neurons to the smell of either bacterium. It was previously shown that the ADF serotonergic neurons, major presynaptic partners of RIA, are essential for aversive olfactory learning in crawling animals. Long term exposure of C. elegans to PA14 increases the serotonin content of ADF, suggesting that serotonin might represent the negative-reinforcing cue (Zhang et al., 2005). Our results suggest that ADF function together with their downstream RIA interneurons and SMD motor neurons to drive aversive olfactory learning. RIA connect with SMD through a large number of reciprocal synapses. Thus, RIA may regulate aversive olfactory learning by integrating the negatively reinforcing serotonergic signal with locomotory response to the olfactory sensory input. This regulation can modulate motor outputs of the olfactory network during training, resulting in an increased turning rate, and thus a decreased olfactory preference towards PA14 in trained animals.
DISCUSSION
Here, by applying systematic single-cell ablation analysis to the C. elegans wiring diagram, we mapped the functional organization of a neural network from sensory input to motor output that regulates the aversive olfactory learning of C. elegans on pathogenic bacteria. This type of learning appears similar to the Garcia’s effect, a common form of learning that animals learn to avoid the taste or smell of a food that makes them ill (Garcia et al., 1955). To our knowledge, our work presents the first systematic analysis on the cellular basis for similar types of learning.
Two different neural circuits drive the naïve and learned olfactory preferences
We have found that two different neural circuits are required for C. elegans to generate its naïve and trained olfactory preferences. The AWB-AWC sensorimotor circuit is required for animals to display their naive olfactory preference; while the ADF modulatory circuit is specifically needed for the animals to modify the naïve olfactory preference after training. Both circuits are connected to downstream motor neurons that control turning rate, suggesting that they regulate motor output during the behavioral display of olfactory preference in either naïve or trained animals (Figure 5H and Figure 6H).
Furthermore, calcium imaging responses of AWB and AWC olfactory sensory neurons in naïve animals are consistent with the behavioral olfactory preference for the smell of PA14 over the smell of OP50. Switching from OP50-conditioned medium to PA14-conditioned medium inhibited intracellular calcium dynamics in AWC and stimulated AWB (Figure 5A, 5D, Supplemental Figure 4A, 4C). The differential effects of OP50 and PA14 stimuli on the activity of these olfactory sensory neurons are likely to be encoded in the intrinsic properties of the neurons, such as expression of a particular group of G-protein coupled receptors. The differential response of these olfactory sensory neurons propagates through downstream neurons to produce different turning rates depending on olfactory inputs (Figure 5H). Our results on olfactory sensory neurons in the learning network suggest that intrinsic neuronal responses of olfactory sensory neurons directly regulate the behavioral olfactory preference of naïve animals.
Interestingly, the olfactory response of AWB and AWC sensory neurons to the smells of benign and pathogenic bacteria were not changed by training (Figure 6A and 6D, Supplemental Figure 4B, 4D). The contrast between the behavioral aversion to PA14 and the neuronal preference of sensory neurons to PA14 in trained animals suggests training-dependent alterations to signal transduction to the downstream of the olfactory learning network. This hypothesis is consistent with our analyses on the turning rate of trained animals, which indicate that aversive experience increases the turning rate towards the training bacterium PA14 through RIA interneurons and SMD motors neurons. Animals lacking RIA or SMD could not learn to reduce their attraction towards PA14 smell after training by increasing their turning rate to PA14 airstream (Figure 3D, 3E and 6G). These findings suggest that the aversive training modulates the function of RIA and SMD neurons, either with an ADF serotonergic negative reinforcing cue or with altered neurotransmission from sensory neurons to downstream circuits or both. These modulations result in the increased turning rate towards PA14 smell in trained animals (Figure 6H).
Roles of neurons in the network for aversive olfactory learning
Our results suggest diverse neuronal functions in the network. Ablating the serotonergic neurons ADF significantly disrupted the learned preference, and also mildly reduced the naïve olfactory preference for PA14, suggesting that ADF play roles in displaying both naïve and learned olfactory preferences (Figure 3C–3F). The overall level of serotonin in C. elegans decreases after food deprivation. The serotonin content of ADF increases after an animal has ingested infectious bacterial food (Chao et al., 2004; Colbert and Bargmann, 1995; Sawin et al., 2000; Sze et al., 2000; Zhang et al., 2005). Thus, different levels of serotonin signaling could regulate different food-related behaviors in naïve and trained worms.
The AIY interneurons regulate several forms of behavioral plasticity and responses to quality of food (Biron et al., 2006; Mori and Ohshima, 1995; Remy and Hobert, 2005; Shtonda and Avery, 2006; Zhang et al., 2005). Here, we found that ablating AIY compromises the naïve preference and ablating AIB generates a mild effect on learning; however, ablating both AIY and AIB together completely abolished the naïve preference and learning (Figure 3C–E). The ablation result clearly indicates the combined function of AIY and AIB in producing naïve olfactory preference, and it is also consistent with the possibility that AIY and AIB interneurons function in a parallel pathway to regulate both naïve preference and learning. The serotonin-gated chloride channel MOD-1 is required for animals to generate aversive olfactory learning (Zhang et al., 2005) (Figure 1F), and expression of wild type MOD-1 activity with ttx-3 promoter in AIY or with odr-2(2b) promoter in AIB, AIZ and several other neurons rescued the learning defect of the mod-1 mutant (Zhang et al., 2005). Taken together, these complementary results from our previous and current studies suggest that AIY and AIB interneurons may contribute to generating both the naïve preference and learning.
It was previously shown that interneurons AIY play important roles in regulating reorienting movements of crawling animals (Gray et al., 2005; Tsalik and Hobert, 2003). In our study, although ablating both AIY and AIB neurons altered the frequency of omega turns, ablating AIY alone did not significantly change the turning rate of swimming animals (Figure 5G and Figure 6G). This difference points to a difference in assay conditions and measurement.
Our ablation analyses indicate that RIA interneurons are specifically required for animals to generate the learned olfactory preference away from pathogenic bacterium PA14 (Figure 3B). RIA are believed to regulate behavioral plasticity in temperature (Mori and Ohshima, 1995) and salt sensation (Stetak et al., 2009). Thus, RIA may play a general role in generating various forms of neural and behavioral plasticity.
Neural networks underlying switch between alternative behavioral outputs
Our systematic laser ablation analysis has identified an inventory of functionally organized neuronal circuits that are needed for experience-dependent switches in olfactory preference in C. elegans. The interplay between neural circuits that are required for C. elegans to display its naive and learned olfactory preferences are reminiscent of those that regulate behavioral switches between swimming and feeding behaviors in the sea slug or fear-extinction and its context-dependent renewal in mice. In the sea slug Pleurobranchaea, activation of the neural network for escape swimming triggered by predatory signals antagonizes the activity of the network for feeding, driving swimming behavior (Jing and Gillette, 2000). In mice, the regulated display of the fear response is mediated by "low fear" and "high fear" neurons in the amygdala. Extinction of fear can be mediated by the inhibition of "high fear" neurons by "low fear" neurons. Renewal of fear can be mediated by inhibition of the "low fear" neurons by hippocampal inputs, allowing the activity of "high fear" neurons to emerge in animal behavior (Herry et al., 2008). Thus, in C. elegans, as in other animals, the switch between alternative behavioral states is generated by the differential usage of different neural circuits under different conditions.
EXPERIMENTAL PROCEDURES
Detailed information on strains and germline transformation is included in Supplemental Information.
Microdroplet assay
In each assay, 12 of 2-µl microdroplets of NGM (nematode growth medium) buffer were placed on a sapphire window (Swiss Jewel Company). One adult animal was placed within each droplet, and the window was placed in a gas-regulated enclosed chamber. Images of swimming animals were recorded by a CCD camera at 10Hz. Olfactory input was provided in the form of two alternating airstreams, one odorized with E. coli OP50 and the other odorized with P. aeruginosa PA14. The airstreams were odorized by passage through liquid cultures of bacterial strains that were prepared overnight at 26°C in NGM medium. The airstreams were automatically switched using solenoid valves controlled by Lab VIEW (National Instruments, Austin, TX). In each experiment, animals were subjected to 12 successive cycles of alternating 30-second exposure to each airstream. The temperature of the sapphire window and the chamber was maintained at 23 °C using a temperature-controlled circulating water bath. The motor responses of individual animals were analyzed using machine-vision software written in MATLAB (Mathworks, Natick, MA). The software calculated a thresholded region corresponding to each worm in each video frame, then determined the eccentricity of an ellipse with the same second moments as the region. The eccentricity of a worm undergoing an Ω bend (which is roughly circular) is smaller than that of a worm exhibiting forward swimming (which is roughly linear). Ω bends were defined as occurring when the eccentricity fell below a threshold value set by measuring the eccentricity in a large number of manually detected Ω bends. The result for turning rate in each assay was the mean value for 3 to 6 animals in the assay.
Aversive olfactory training
Similar number of adult animals were moved onto either a control plate that contained a fresh lawn of the benign bacterium E. coli OP50 or a training plate that contained a fresh lawn of the pathogenic bacterium P. aeruginosa PA14 for aversive training for 6 hours at 20°C. Control and training plates were prepared by inoculating 10cm NGM plates with 0.5 ml overnight NGM culture of OP50 or PA14, respectively, and incubating at 26°C for one and half days.
Calcium imaging
Calcium imaging was performed in a microfluidic device essentially as described (Chalasani et al., 2007; Chronis et al., 2007) with minor modifications. Fluorescence time lapse imaging (100ms exposure) was performed on a Nikon Eclipse Ti-U inverted microscope with a 40x oil immersion objective and a Photometrics CoolSnap EZ camera. The detail of the procedure is included in the Supplemental Information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Caenorhabditis Genetic Center for C. elegans strains, Dr. Cori Bargmann, Dr. Piali Sengupta, Dr. Kyuhyung Kim and Harvard Center for Nanoscale Systems for helps with the microfluidics system, Dr. Linjiao Luo and Dr. Mi Zhang for helps with the femtosecond laser apparatus, Dr. Edward Soucy and Dr. Joel Greenwood at Center for Brain Science Neuroengineering Facility at Harvard University for technical supports and Dr. Junichi Nakai for the recombinant DNA clone pN1-G-CaMP. We thank Dr. Joshua Sanes, Dr. Cori Bargmann, Dr. Kenneth Blum, Dr. Catherine Dulac and Zhang laboratory members for thoughtful comments on the manuscript. This work was supported by the funding from Howard Hughes Medical Institute (K.S.), NIH grant 4R00NS57931 (D.C.-R.), The Esther A. and Joseph Klingenstein Fund, March of Dimes Foundation, The Alfred P. Sloan Foundation, The John Merck Fund, NIH (Y.Z.) and the McKnight Foundation, NSF, and NIH (A.D.T.S.).
Footnotes
AUTHOR CONTRIBUTIONS: Y.Z. conceived of the study; H.H., M.H., A.D.T.S. and Y.Z. designed experiments; H.H., M.H., Y.S., Y.Q. and Y.Z. performed experiments; C.V.G., C.F-Y. and A.D.T.S. provided experimental and analytical tools; D.C-R. and K.S. contributed to genetic reagents; H.H., M.H., A.D.T.S. and Y.Z. analyzed data and wrote the paper.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION accompanies the paper.
REFERENCES
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- Berg HC, Brown DA. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- Biron D, Shibuya M, Gabel C, Wasserman SM, Clark DA, Brown A, Sengupta P, Samuel AD. A diacylglycerol kinase modulates long-term thermotactic behavioral plasticity in C. elegans. Nature neuroscience. 2006;9:1499–1505. doi: 10.1038/nn1796. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MY, Komatsu H, Fukuto HS, Dionne HM, Hart AC. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15512–15517. doi: 10.1073/pnas.0403369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelur DS, Chalfie M. Targeted cell killing by reconstituted caspases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BL, Hall DH, Chklovskii DB. Wiring optimization can relate neuronal structure and function. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4723–4728. doi: 10.1073/pnas.0506806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis N, Zimmer M, Bargmann CI. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nature methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- Chung SH, Clark DA, Gabel CV, Mazur E, Samuel AD. The role of the AFD neuron in C. elegans thermotaxis analyzed using femtosecond laser ablation. BMC neuroscience. 2006;7:30. doi: 10.1186/1471-2202-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron. 1995;14:803–812. doi: 10.1016/0896-6273(95)90224-4. [DOI] [PubMed] [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr JS, Frisby DL, Gaskin J, Duke A, Asermely K, Huddleston D, Eiden LE, Rand JB. The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J Neurosci. 1999;19:72–84. doi: 10.1523/JNEUROSCI.19-01-00072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J, Kimeldorf DJ, Koelling RA. Conditioned aversion to saccharin resulting from exposure to gamma radiation. Science. 1955;122:157–158. [PubMed] [Google Scholar]
- Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herry C, Ciocchi S, Senn V, Demmou L, Muller C, Luthi A. Switching on and off fear by distinct neuronal circuits. Nature. 2008;454:600–606. doi: 10.1038/nature07166. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Kuwabara PE, Corneliussen B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr Biol. 2000;10:1615–1618. doi: 10.1016/s0960-9822(00)00867-8. [DOI] [PubMed] [Google Scholar]
- Iino Y, Yoshida K. Parallel use of two behavioral mechanisms for chemotaxis in Caenorhabditis elegans. J Neurosci. 2009;29:5370–5380. doi: 10.1523/JNEUROSCI.3633-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J, Gillette R. Escape swim network interneurons have diverse roles in behavioral switching and putative arousal in Pleurobranchaea. Journal of neurophysiology. 2000;83:1346–1355. doi: 10.1152/jn.2000.83.3.1346. [DOI] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Developmental cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lints R, Emmons SW. Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFbeta family signaling pathway and a Hox gene. Development. 1999;126:5819–5831. doi: 10.1242/dev.126.24.5819. [DOI] [PubMed] [Google Scholar]
- Luo L, Gabel CV, Ha HI, Zhang Y, Samuel AD. Olfactory behavior of swimming C. elegans analyzed by measuring motile responses to temporal variations of odorants. Journal of neurophysiology. 2008;99:2617–2625. doi: 10.1152/jn.00053.2008. [DOI] [PubMed] [Google Scholar]
- Menzel R, Muller U. Learning and memory in honeybees: from behavior to neural substrates. Annual review of neuroscience. 1996;19:379–404. doi: 10.1146/annurev.ne.19.030196.002115. [DOI] [PubMed] [Google Scholar]
- Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature. 1995;376:344–348. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Chen BL, Mun JJ, Ho R, Sarkis R, McIntire SL. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20982–20987. doi: 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Morse TM, Lockery SR. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 1999;19:9557–9569. doi: 10.1523/JNEUROSCI.19-21-09557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Cannon SC, Horvitz HR. MOD-1 is a serotonin-gated chloride channel that modulates locomotory behaviour in C. elegans. Nature. 2000;408:470–475. doi: 10.1038/35044083. [DOI] [PubMed] [Google Scholar]
- Remy JJ, Hobert O. An interneuronal chemoreceptor required for olfactory imprinting in C. elegans. Science. 2005;309:787–790. doi: 10.1126/science.1114209. [DOI] [PubMed] [Google Scholar]
- Sanchez-Andrade G, Kendrick KM. The main olfactory system and social learning in mammals. Behav Brain Res. 2009;200:323–335. doi: 10.1016/j.bbr.2008.12.021. [DOI] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. The Journal of experimental biology. 2006;209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava N, Clark DA, Samuel AD. Temporal analysis of stochastic turning behavior of swimming C. elegans. Journal of neurophysiology. 2009;102:1172–1179. doi: 10.1152/jn.90952.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetak A, Horndli F, Maricq AV, van den Heuvel S, Hajnal A. Neuron-specific regulation of associative learning and memory by MAGI-1 in C. elegans. PloS one. 2009;4:e6019. doi: 10.1371/journal.pone.0006019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Victor M, Loer C, Shi Y, Ruvkun G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- Tabish M, Siddiqui ZK, Nishikawa K, Siddiqui SS. Exclusive expression of C. elegans osm-3 kinesin gene in chemosensory neurons open to the external environment. Journal of molecular biology. 1995;247:377–389. doi: 10.1006/jmbi.1994.0146. [DOI] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–169. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Tsalik EL, Hobert O. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. Journal of neurobiology. 2003;56:178–197. doi: 10.1002/neu.10245. [DOI] [PubMed] [Google Scholar]
- Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–1224. doi: 10.1242/dev.00341. [DOI] [PubMed] [Google Scholar]
- Waddell S, Quinn WG. Flies, genes, and learning. Annual review of neuroscience. 2001;24:1283–1309. doi: 10.1146/annurev.neuro.24.1.1283. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil. Trans. Roy. Soc. London Ser. B. 1986:314. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Zechman JM, Labows JN., Jr Volatiles of Pseudomonas aeruginosa and related species by automated headspace concentration--gas chromatography. Canadian journal of microbiology. 1985;31:232–237. doi: 10.1139/m85-045. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.